Abstract
Objective
There is growing evidence that circulating microRNAs (miRNAs) play an important role in obesity. However, whether they can contribute to adult weight gain is still unclear.
Methods
In the training set with 40 nonsmoking, healthy women identified from the Mano-A-Mano Mexican American Cohort study, global circulating miRNA profiles in plasma samples were assessed. Cox proportional hazard regression was used to assess the effects of plasma miRNAs on significant weight gain during a 5-year follow-up. Plasma miRNAs associated with significant weight gain were further validated in two testing sets (N=160 and 100, respectively).
Results
A total of 23 significant plasma miRNAs were identified in the training set. Among them, eight were validated in two testing sets. They were miR-142, miR-122, miR-125b, miR-15b, miR-130b, miR222, miR-519d, and miR-31. Using those eight miRNAs, a risk score for significant weight gain was created. Study participants with a high risk score had 3.01-fold increased risk of having significant weight gain in the whole study population (hazard ratio: 3.01, 95% confidence interval: 1.70–5.47).
Conclusions
The findings provide evidence that circulating miRNAs play important roles in obesity and weight gain and suggest new targets for understanding the mechanisms of weight gain and developing weight loss intervention strategies.
Introduction
Adult weight gain, irrespective of weight status, has emerged as a significant risk factor for various types of obesity-related chronic diseases, including diabetes, cardiovascular disease, and cancers (1–5). Although weight gain happens gradually and insidiously throughout adulthood, the rate, amount, and timing vary significantly among individuals. Such variation may provide an opportunity for targeted weight loss intervention. A few determinants have been linked with adult weight gain, including physical activity, education, smoking cessation, various dietary components, and body fat mass and obesity-associated genotype (6–12). However, even with those identified determinants taken together, the predictive power is still very low, and a significant amount of variation in weight gain is still unexplained (6).
Identifying high-risk Mexican Americans is particularly relevant because not only are they the fastest-growing minority group in the United States, but they also have an elevated prevalence of obesity/overweight. According to the National Health and Nutrition Examination Survey conducted from 2011 to 2014, 43.5% of adult men and 48.6% of adult women of Mexican origin have obesity (13). More alarmingly, the rate of obesity has been steadily increasing. Clearly, excessive weight gain and obesity-related chronic diseases have posed enormous threats to the health of Mexican Americans.
Extensive efforts are being made to identify obesity- and weight gain-related biomarkers to better understand pathogenesis, find new targets for clinical therapy, and allow early prediction of metabolic complications. A potentially promising set of biomarkers is circulating micro-RNAs (miRNAs), which are small, noncoding, highly conserved RNAs. Since the discovery of miRNAs in 1993, their expression profiles and functions have been extensively studied (14–16). Through modifying messenger RNA (mRNA) availability and protein synthesis, miRNAs regulate many cellular processes such as cell growth, proliferation, differentiation, and apoptosis (15). Moreover, because miRNA expression is closely related to cellular behavior and, eventually, the normal development and function of body tissues, changes in miRNA profiles are being increasingly analyzed in cancer, osteoporosis, ischemic heart disease, and heart failure (17–20).
Their recent discovery in the circulation has promoted further exploration of their potential use as novel minimally invasive biomarkers of diseases. Indeed, circulating miRNAs show a high degree of reproducibility within individuals. A few studies have reported an altered expression of circulating miRNAs in human subjects with obesity (21–23). For example, Ortega et al. reported that circulating miRNAs were deregulated in severe obesity (21). Surgery-induced weight loss could induce changes in circulating miRNA profiles, suggesting a potential mechanistic relevance. Furthermore, Prat-Puig et al. identified a panel of miRNAs in plasma samples associated with 3-year weight change in children (24). However, to date, there has been no study to investigate whether circulating miRNAs might predict future weight gain prospectively in adult Mexican-American participants.
In the current study, we used TaqMan® low-density arrays and reverse transcription-quantitative PCR to define the pattern of circulating miRNAs in plasma samples from three sets of healthy Mexican-American women from the Mano-A-Mano Mexican American Cohort study (25). The objectives were twofold. First, we investigated prospectively whether baseline plasma miRNA levels predicted significant weight gain during a 5-year follow-up; second, we examined cross-sectional associations between plasma miRNA levels with BMI and obesity at baseline.
Methods
Study population
The samples for the current study were drawn from participants in a large population-based cohort of Mexican-origin households recruited from the Houston area in Texas. This ongoing prospective cohort of first- and second-generation Mexican-origin immigrant households was initiated in July 2001 and maintained by the Department of Epidemiology at the University of Texas MD Anderson Cancer Center. A detailed description of the sampling and recruitment strategy has been published previously (25). Briefly, participants have been recruited through block walking in predominantly Mexican-American neighborhoods, through community centers and local health clinics, and by networking through currently enrolled participants. Of the identified eligible households, ~88% agreed to participate in the study. After written informed consent was obtained, trained bilingual research interviewers conducted a structured face-to-face interview lasting ~45 minutes, using a standardized and validated questionnaire in the participant’s preferred language, either Spanish or English. The questionnaire elicited information on birthplace and residential history, social-demographic characteristics, lifestyle behaviors, levels of physical activity, personal medical history, family history of chronic disease, acculturation, and occupational exposure. Participants have been actively followed up via annual telephone recontact to update body weight, selected exposures, and new diagnosis of selected chronic diseases, including cancer, diabetes, and hypertension.
The current study included 300 healthy, female nonsmokers with no major chronic diseases reported at baseline (e.g., hypertension, cardiovascular disease, diabetes, cancer). Height and weight were measured during the in-person interview according to a standard protocol by trained interviewers at baseline. Self-reported body weight was also collected during the follow-up. the study protocol was approved by the Institutional Review Board of the University of Texas MD Anderson Cancer Center. The entire study population was divided into three sets: training set (n = 40), testing set 1 (n =160), and testing set 2 (n =100).
Total RNA isolation
Total RNA was isolated from plasma samples using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. For each study subject, 400 μL plasma was used for RNA isolation. Synthetic cel-miR-39 and cel-miR-54 were added to each sample as internal controls for evaluation of successful extraction. RNAs were eluted with 20 μL of water. The initial quality check of small RNA molecules was performed using NanoDrop ND-1000 spectrometer (Thermo Scientific, Waltham, Massachusetts). We carefully kept consistent the amount of plasma samples and spike-in miRNAs prepared from the same batch for isolation throughout all experiments. In addition, the raw Ct value for each miRNA was standardized to the raw Ct value for spike-in miRNAs obtained from each individual sample to eliminate potential variations introduced from the isolation process and RNA quantification. As described in our previous study (26), we selected miR-93 as the endogenous control miRNA for normalization. In addition, to assess the reproducibility, we used a global mean of the top 100 expressed miRNAs as an alternative mean for normalization. We found that the results were similar between the two methods. To better present our findings here, we only presented the results normalized by miR-93.
Whole-genome plasma miRNA profiling
Whole-genome plasma miRNA profiling was performed on the 40 plasma samples (training set) using the TaqMan Array Human Microarray Card Set v3.0 (Applied Biosystems, Foster City, California). In brief, purified plasma RNA samples were reverse-transcribed using Megaplex™ RT Primers followed by a preamplification step using Megaplex PreAmp Primers for maximum sensitivity of detection. For final quantification, the TaqMan Universal Master Mix II was added, and each sample was loaded on the miRNA microarray cards. Quantitative PCR was performed on the 7900HT Fast Real-Time PCR System (Applied Biosystems). The expression threshold for each miRNA detector was automatically determined. A miRNA candidate was considered highly expressed when more than 75% of samples in both study groups generated detectable Ct values of less than 35. A list of miRNA candidates is provided in Supporting Information Table S1.
Quantitative real-time PCR assay
Significant miRNAs from the training set were further validated in two testing sets, using individual TaqMan miRNA assays (Applied Biosystems) to quantitatively measure the difference in their expression levels between the study participants with significant weight gain and the group without significant weight gain with varied follow-up times. Each assay was tested in duplicates. Data points that generated duplicated Ct values with over one cycle variance were excluded from analysis. The mean Ct value obtained from each sample was normalized to the averaged expression of spike-in miRNAs and then subjected to analysis with the 2−ΔΔCt method.
Statistical analysis
All analyses were performed with Stata® software version 13.1 (StataCorp, College Station, Texas) and the R statistical language. First, we examined the association between plasma miRNAs and 5-year significant weight gain. During the 5-year follow-up, each study participant had at least three self-reported body weights. Because the focus of this study was on weight gain, 15 study participants who lost weight during follow-up were excluded from further analysis. Among them, 10 were from the testing set 1 and 5 were from the testing set 2. To assess the levels of weight gain during the follow-up, we created a variable, termed “significant weight gain,” which was defined as increase in BMI by at least one category between the baseline and the follow-up. The creation of this variable was based on several reasons. First, a one-category change was considered major weight gain in relation to the time horizon over a 5-year period of prediction. Second, the change was considered high enough to exclude random variation in body weight while simultaneously allowing for some weight gain as a natural part of the aging process. Last, self-reported BMI was used during the follow-up. Self-reported BMI correlates reasonably well with measured BMI values in adults (27), but misclassification is more common among Mexican-American women than men (28).
Each individual plasma miRNA was dichotomized into two categories (high vs. low levels), using the median levels of the miRNA in the study participants as the cutoff point. Cox regression analysis was performed to assess the association of high level of individual plasma miRNA with significant weight gain, adjusting for covariates, including age, HbA1c, acculturation score, nativity, years of living in the United States, sedentary lifestyle, sitting time, and biospecimen storage time. Study participants with follow-up times over 5 years and without significant weight gain were censored at 60 months. In all statistical analyses, P ≤ 0.05 was considered significant. A combined eight-miRNA risk score for each study participant was derived by linear combination of the product of reference-normalized expression level of each miRNA by its Cox regression corresponding coefficient. All study participants were further dichotomized by the median risk score, and individuals with a risk score higher or lower than the median were classified as high- or low-risk groups, respectively. Then we examined the effect of baseline BMI as continuous variables in relation to plasma miRNA levels using linear aggression, adjusting for covariates. The study participants were further grouped into five categories based on their BMI, including normal weight (<25), overweight (25–29.90), class I obesity (30–34.90), class II obesity (35–39.90), and class III obesity (40 and above). Analysis of variance (ANOVA) was performed to determine the relationship between baseline BMI category and plasma miRNA levels.
Results
At baseline, 47.5%, 47.5%, and 57.0% of study participants in the training set and testing sets 1 and 2 had obesity (Table 1). The obesity (BMI ≥ 30) prevalence was further increased to 72.5%, 54.4% and 63.0% during the follow-up. In the training set, 20 (50%) study participants experienced significant weight gain during the follow-up. The percentages of significant weight gain were 21.9% and 22.0% in testing sets 1 and 2, respectively. Fifteen study participants lost weight during the follow-up, ten in testing set 1 and five in testing set 2. The follow-up time was longer in testing set 2 than the other two sets. The three sets were comparable with regard to other baseline variables, including baseline age, HbA1c levels, nativity, years of living in the United States, acculturation score, sedentary lifestyle, and sitting time.
TABLE 1.
Basic characteristics of study participants in the training and testing cohorts
| Training (n =40) | Testing 1 (n =160) | Testing 2 (n =100) | |
|---|---|---|---|
| BMI at baseline, mean (SD) | 30.2 (5.4) | 30.7 (5.9) | 31.5 (6.0) |
| BMI category at baseline (kg/m2) | |||
| <25.0 | 6 (15.0%) | 26 (16.3%) | 8 (8.0%) |
| 25.0–29.9 | 15 (37.5%) | 58 (36.3%) | 35 (35.0%) |
| 30.0–34.9 | 10 (25.0%) | 44 (27.5%) | 33 (33.0%) |
| 35.0–39.9 | 5 (12.5%) | 19 (11.9%) | 14 (14.0%) |
| 40.0 and above | 4 (10.0%) | 13 (8.1%) | 10 (10.0%) |
| BMI at follow-up, mean (SD) | 31.9 (5.1) | 31.8 (6.0) | 32.6 (5.6) |
| BMI category at last follow-up (kg/m2) | |||
| <25.0 | 2 (5.0%) | 14 (8.8%) | 5 (5.0%) |
| 25.0–29.9 | 9 (22.5%) | 59 (36.8%) | 32 (32.0%) |
| 30.0–34.9 | 16 (40.0%) | 51 (31.8%) | 33 (33.0%) |
| 35.0–39.9 | 8 (20.0%) | 22 (13.8%) | 22 (22.0%) |
| 40.0 and above | 5 (12.5%) | 14 (8.8%) | 8 (8.0%) |
| Weight change | |||
| Significant weight gain | 20 (50.0%) | 35 (21.9%) | 22 (22.0%) |
| No significant weight gain | 20 (50.0%) | 115 (71.9%) | 73 (73.0%) |
| Weight loss | 0 (0.0%) | 10 (6.2%) | 5 (5.0%) |
| HbA1C, mean (SD) | 4.9 (0.53) | 4.9 (0.83) | 4.9 (0.44) |
| Follow-up time (d) | |||
| Mean (SD) | 953 (326) | 997 (436) | 1459 (326) |
| Median (range) | 942 (265–1,643) | 964 (229–1,825) | 1481 (932–1,825) |
| Age at baseline (y) | |||
| Mean (SD) | 40 (8.1) | 40 (8.5) | 40 (10.7) |
| Median (range) | 37 (21–70) | 39 (20–72) | 38 (21–69) |
| Nativity | |||
| United States | 6 (15.0%) | 29 (18.1%) | 13 (13.0%) |
| Mexico | 34 (85.0%) | 131 (81.9%) | 87 (87.0%) |
| Years of living in United States, mean (SD) | 18.2 (9.6) | 19.9 (13.4) | 19.9 (14.0) |
| Acculturation score, mean (SD) | 2.22 (0.72) | 2.27 (0.86) | 2.10 (0.90) |
| Sedentary lifestyle | |||
| No | 8 (20.0%) | 32 (20.0%) | 12 (17.0%) |
| Yes | 32 (80.0%) | 128 (80.0%) | 88 (83.0%) |
| Sitting h/d, mean (SD) | 2.1 (0.6) | 2.1 (0.8) | 2.0 (1.0) |
First, we explored whether plasma miRNA levels at baseline might prospectively predict significant weight gain during 5-year follow-up in the training set. Using the median levels of plasma miRNAs in study participants as the cutoff points, a high level of 23 plasma miRNAs was significantly associated with significant weight gain during the follow-up, after adjusting for age, acculturation, nativity, years living in the United States, baseline BMI, HbA1c, sedentary lifestyle, sitting time, and biospecimen storage time (Table 2). The top three significant plasma miRNAs were miR-142, miR-122, and miR-125b (P =0.011, 0.014, and 0.021, respectively). High levels of miR-142 were associated with 1.96-fold increased risk of having significant weight gain during the follow-up (hazard ratio [HR] =1.96, 95% confidence interval [CI]: 1.11–4.36). Those 23 significant plasma miRNAs were further evaluated in two testing sets. In testing set 1, testing set 2, and both sets combined, nine, eight, and eight plasma miRNAs, respectively, remained significant. The plasma miRNAs significant in both testing sets included miR-142, miR-122, miR-125b, miR-15b, miR-130b, miR-222, miR-519d, and miR-31. An elevated risk of having significant weight gain during the follow-up was associated with high levels of miR-142, miR-122, miR-15b, miR-222, miR-130b, and miR-519d, but with reduced levels of miR-125b and miR-31.
TABLE 2.
Individual plasma miRNAs predicting significant weight gain during follow-up
| miRNA | Training | Testing 1 | Testing 2 | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| HR (95% CI)a | P value | HR (95% CI)a | P value | HR (95% CI)a | P value | |
| miR-142b | 1.96 (1.11–4.36) | 0.011 | 1.70 (1.17–3.52) | 0.004 | 1.66 (1.07–4.35) | 0.009 |
| miR-122b | 2.16 (1.10–4.47) | 0.014 | 1.63 (1.15–3.69) | 0.007 | 1.73 (1.05–4.69) | 0.011 |
| miR-30a | 2.23 (1.11–3.89) | 0.017 | 1.38 (0.63–3.64) | 0.369 | 1.54 (0.79–5.23) | 0.214 |
| miR-125bb | 0.61 (0.42–0.90) | 0.021 | 0.53 (0.38–0.88) | 0.005 | 0.63 (0.32–0.92) | 0.008 |
| miR-21a | 0.60 (0.41–0.89) | 0.022 | 1.32 (0.56–1.86) | 0.548 | 1.13 (0.43–3.93) | 0.624 |
| miR-15a | 1.68 (1.17–4.72) | 0.023 | 1.24 (0.73–2.57) | 0.425 | 1.24 (0.73–2.57) | 0.436 |
| miR-15bb | 1.75 (1.09–4.57) | 0.021 | 1.69 (1.14–3.43) | 0.011 | 1.71 (1.04–4.06) | 0.025 |
| miR-130a | 0.52 (0.41–0.98) | 0.039 | 0.54 (0.33–1.10) | 0.114 | 0.59 (0.33–1.09) | 0.147 |
| miR-130bb | 1.80 (1.09–4.37) | 0.024 | 1.54 (1.12–3.95) | 0.018 | 1.77 (1.02–3.95) | 0.037 |
| miR-222b | 1.90 (1.08–5.15) | 0.035 | 1.85 (1.20–3.06) | <0.001 | 1.72 (1.01–3.83) | 0.048 |
| miR-138 | 1.79 (1.04–4.97) | 0.047 | 1.65 (0.73–3.89) | 0.436 | 1.48 (0.69–4.05) | 0.623 |
| miR-335 | 2.04 (1.03–4.63) | 0.048 | 1.66 (0.71–3.78) | 0.513 | 1.25 (0.71–5.02) | 0.735 |
| miR-758 | 0.54 (0.38–0.97) | 0.037 | 1.53 (0.69–3.07) | 0.425 | 1.03 (0.81–3.87) | 0.913 |
| miR-27 | 1.75 (1.05–5.18) | 0.041 | 1.23 (0.73–3.79) | 0.658 | 1.36 (0.79–3.91) | 0.814 |
| miR-103 | 1.99 (1.07–4.92) | 0.038 | 1.09 (0.64–3.52) | 0.821 | 1.48 (0.79–4.15) | 0.279 |
| miR-423 | 0.50 (0.38–0.96) | 0.041 | 0.72 (0.48–2.54) | 0.569 | 1.31 (0.88–3.54) | 0.292 |
| miR-519db | 1.71 (1.06–5.02) | 0.032 | 1.89 (1.04–4.02) | 0.024 | 1.74 (1.04–3.99) | 0.037 |
| miR-448 | 0.60 (0.38–0.92) | 0.042 | 0.69 (0.35–1.04) | 0.077 | 1.02 (0.52–4.02) | 0.926 |
| miR-210 | 1.95 (1.05–4.89) | 0.044 | 1.33 (0.56–3.46) | 0.872 | 1.73 (0.65–4.14) | 0.642 |
| miR-107 | 2.03 (1.03–5.01) | 0.046 | 1.89 (0.81–4.09) | 0.264 | 1.54 (0.57–3.87) | 0.764 |
| miR-31b | 0.62 (0.39–0.99) | 0.045 | 0.55 (0.34–0.97) | 0.031 | 0.57 (0.33–0.99) | 0.049 |
| miR-150 | 2.04 (1.04–5.53) | 0.048 | 1.05 (0.62–1.95) | 0.762 | 1.63 (0.80–4.21) | 0.762 |
| miR-200 | 1.89 (1.01–5.47) | 0.049 | 1.67 (1.01–4.33) | 0.048 | 1.54 (0.48–3.86) | 0.638 |
Cox hazard regression adjusted for age-acculturation score-nativity-years living in United States-baseline BMI-HbA1C-sedentary lifestyle-sitting time-and biospecimen storage time.
Significant in both training and two testing sets.
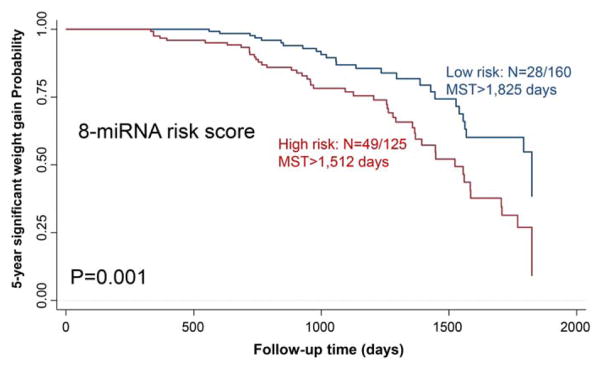
We further assessed the combined effects of these eight plasma miR-NAs on significant weight gain. We generated a risk score using the linear combination of expression levels of these eight miRNAs for each individual in the training data set, based on the median levels of each miRNA (Table 3). Study participants with high risk scores were more likely to have significant weight gain compared to those with low risk scores, although the strength of association was not statistically significant (HR =2.75, 95% CI: 0.64–12.26). When the same cutoff was applied to the two testing sets, stronger associations than the training set were observed (HR =2.91, 95% CI: 1.23–7.06 for testing set 1; and HR =3.13, 95% CI: 1.03–9.86 for testing set 2). When all three sets were combined, the corresponding risk of having significant weight gain was 3.01 (95% CI: 1.70–5.47), P = 3.58E-06. Kaplan-Meier 5-year survival curve shows that study participants with high risk scores had a median survival time of 4.14 years compared to over 5 years in the low-risk group (P =0.001; Figure 1).
TABLE 3.
Plasma miRNA 8-marker risk score associated with significant weight gain
| Risk score | Significant weight gain, n (%) | Nonsignificant weight gain, n (%) | Adjusted HR (95% CI)a |
P value | |
|---|---|---|---|---|---|
| Training | Low | 7 (35.0) | 12 (60.0) | 1 (Reference) | |
| High | 13 (65.0) | 8 (40.0) | 2.75 (0.64–12.26) | 0.127 | |
| Testing 1 | Low | 13 (37.1) | 73 (63.5) | ||
| High | 22 (62.9) | 42 (36.5) | 2.91 (1.23–7.06) | 0.006 | |
| Testing 2 | Low | 8 (36.4) | 47 (64.4) | ||
| High | 14 (63.6) | 26 (35.6) | 3.13 (1.03–9.86) | 0.023 | |
| Combined | Low | 28 (36.4) | 132 (63.5) | ||
| High | 49 (63.6) | 76 (36.5) | 3.01 (1.70–5.47) | 3.58E-06 |
Cox hazard regression adjusted for age, acculturation score, nativity, years living in United States, baseline BMI, HbA1C, sedentary lifestyle, sitting time, and biospecimen storage time.
Figure 1.
Kaplan-Meier 5-year significant weight gain curves for study subjects grouped by low (blue line) and high (red line) risk score. N =number of study subjects with an event (significant weight gain) at 5 years/total number of study subjects in the data set. MST =median survival time. [Color figure can be viewed at wileyonlinelibrary.com]
Finally, using plasma miRNA data generated above, we examined the associations between plasma miRNAs and BMI at baseline. In the training set, a total of 29 plasma miRNAs were significantly associated with BMI at baseline after adjusting for potential confounders. Among them, 10 were also associated with significant weight gain (Table 4). Their relationships with BMI were further confirmed in the two testing sets. Five out of ten were significantly associated with BMI in both testing sets. Levels of plasma miR-140, miR-122, miR-142, and miR-519d were significantly positively associated with BMI, and levels of plasma miR-125b were significantly inversely associated with BMI. When BMI was further grouped into five categories, namely, normal weight, overweight, and class I, II, and III obesity, the relationships between those five plasma miRNAs and BMI category remained significant in the training and both testing sets.
TABLE 4.
Association of plasma miRNAs with BMI at baseline in the training and testing cohorts
| miRNA | Training | Testing 1 | Testing 2 | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| ρ | P valuea | ρ | P valuea | ρ | P valuea | |
| Also associated with significant weight gain | ||||||
| miR-142b | 0.374 | 0.002 | 0.342 | <0.001 | 0.307 | <0.001 |
| miR-122b | 0.405 | 0.001 | 0.287 | <0.001 | 0.292 | <0.001 |
| miR-30ab | 0.411 | <0.001 | 0.305 | <0.001 | 0.316 | <0.001 |
| miR-125bb | −0.316 | 0.009 | −0.217 | 0.012 | −0.264 | 0.002 |
| miR-222 | 0.279 | 0.038 | 0.168 | 0.435 | 0.192 | 0.378 |
| miR-27 | −0.298 | 0.024 | −0.161 | 0.426 | −0.132 | 0.325 |
| miR-519db | 0.313 | 0.011 | 0.259 | 0.001 | 0.247 | 0.006 |
| miR-103 | 0.362 | 0.003 | 0.184 | 0.207 | 0.193 | 0.332 |
| miR-210 | 0.346 | 0.007 | 0.153 | 0.572 | 0.107 | 0.743 |
| miR-150 | 0.358 | 0.006 | 0.163 | 0.538 | 0.124 | 0.652 |
Adjusted by age, acculturation score, nativity, years living in United States, baseline BMI, HbA1C, sedentary lifestyle, sitting time, and biospecimen storage time.
Significant in both testing and two training sets.
Discussion
Identifying individuals who are at risk of gaining excessive weight in the near future has significant implication in reducing the burden of obesity and its related chronic diseases. However, how to identify such individuals remains challenging. In the current study, we provide the first evidence, to our knowledge, of a plasma miRNA signature in predicting 5-year weight gain in Mexican-American women. A panel of eight plasma miRNAs, including miR-142, miR-122, miR-125b, miR-15b, miR-130b, miR222, miR-519d, and miR-31, was identified with levels significantly associated with significant weight gain, defined as weight gain of one category or more, during 5-year follow-up. In addition, we observed several plasma miRNAs associated with BMI at baseline.
Although none of the eight plasma miRNAs identified in our study has been linked to adult weight gain, most of them have been associated with obesity and/or other metabolic phenotypes previously. For example, Ortega et al. reported that levels of plasma miR-142 were significantly higher and levels of plasma miR-125b were significantly lower in study participants with severe obesity than ones without severe obesity (21). The expression of miR-142 was considered as a marker of acute and chronic inflammation. Increased expression measures of miR-142 have been reported in serum from patients with chronic inflammation, autoimmune attack, and vascular damage (29). miR-125b was found to be downregulated both in mature adipocytes and in subcutaneous fat from subjects with obesity with or without type 2 diabetes (30). Giroud et al. found that miR-125b plays an important role in the repression of brown-in-white or beige adipocyte function by modulating oxygen consumption and mitochondrial gene expression (31). In the current study, we not only confirmed the relationships of miR-142 and miR-125b with obesity/BMI, but also demonstrated that both miRNAs were associated with significant weight gain.
The previous reports on the relationship between miR-130b and obesity were inconsistent. Ortega et al. reported that levels of miR-130b were downregulated in both mature adipocytes and subcutaneous fat from subjects with obesity (30). However, Wang et al. found that the circulating level of miR-130b was elevated in Chinese individuals with obesity and correlated positively with BMI (32). They further suggested that TGF-β, which was proportionately increased with obesity, stimulated miR-130b secretion from adipocytes during adipogenesis. In another study by Ortega et al., levels of plasma miR-130b were higher in study participants with obesity but significantly lower in those with severe obesity than with normal weight (21). In our study, plasma levels of miR-130b were not correlated with BMI at baseline. But we found that high plasma levels of miR-130b were associated with significant weight gain during the follow-up.
Another interesting finding in the current study is the positive relationships of plasma levels of miR-122, significant weight gain, and BMI at baseline. Our results are in line with the report from Wang et al. (22), which showed that elevated circulating miR-122 was associated with obesity and insulin resistance in young adults. miR-122 is primarily expressed in the liver and has been shown to affect hepatic cholesterol and fatty acid metabolism (33,34). In humans, miR-122 expression was significantly associated with hepatic enzymes and was an independent association of miR-122 with ALT (22). In our study, we found that elevated plasma levels of miR-222 were significantly associated with weight gain of one category or more during the follow-up. Functionally, miR-222 is a negative regulator of adipocyte insulin sensitivity, seemingly via repression of ERα and GLUT4 (35). Human data from Shi et al. also reported higher miR-222 expression in the omental adipose tissue of women with gestational diabetes at the time of cesarean delivery compared with pregnant women with nongestational diabetes (35). miR-222 was found to be significantly higher in the whole plasma of two distinct cohorts of human patients with obesity (21,36). Thus, our current findings are consistent with a role of miR-222 in the development of obesity.
We also found that high levels of plasma miR-519d and low levels of miR-31 were significantly associated with significant weight gain. The relationship among circulating miR-519d, miR-31, and obesity/weight gain has not been reported previously. However, studies using in vitro cell lines and adipose tissues have shown that both miRNAs are involved in adipogenesis. For example, Martinelli et al. reported that miR-519d overexpression was associated with human obesity (37). miR-519d was shown to specifically and dose-dependently suppress translation of the PPARα protein (a predicted target for miR-519d) and increase lipid accumulation during preadipocyte differentiation. miR-31 directly targeted C/EBPα, and levels of this miRNA were downregulated in adipogenesis in human adipose stem cells (38).
The Mexican-American population is plagued with a high prevalence of overweight and obesity. More alarmingly, they are continuing to gain weight. In our cohort, 20% of study subjects have gained at least 10% of body weight from baseline in just 5 years. Such rapid weight gain has significantly increased their risk for obesity-related chronic diseases. Thus, the identification of circulating miRNAs that can be used as predictors for weight gain can potentially help identify individual Mexican Americans who are more likely to gain excess body weight in the near future. Those individuals can be targeted for aggressive weight control programs to reduce their likelihood of gaining excessive weight and thereby decrease their risk of obesity-related chronic diseases in later life. In addition, because most of the identified circulating miRNAs are biologically associated with obesity and/or other metabolic phenotypes, we expect the results from this study can be applied to other populations. Certainly, population differences in genotypic and phenotypic characteristics have to be considered.
Several potential limitations should be considered. The study sample size of 300 participants is relatively small. Considering that the effect size of certain plasma miRNAs may be small, some of the associations may not be detected in the current study. The study subject characteristics among the three sets were not identically balanced. For example, the mean and median follow-up time was significantly longer in testing set 2 than the other two sets. The plasma miRNA levels in our study were based on blood samples from a single point in time that may not reflect the true associations over time. Self-reported BMI was used during the follow-up. Self-reported BMI correlates reasonably well with measured BMI values in adults (27), but misclassification is more common among Mexican-American women than men (28). Nevertheless, this is the first global miRNA profiling study to identify plasma miRNAs prospectively predicting weight gain in Mexican-American women. The results from this study, if confirmed in additional large studies, can serve as a basis to develop circulating miRNA-based tools to identify individuals most at risk of gaining excessive amounts of weight in the near future.
Supplementary Material
Acknowledgments
Funding agencies: The Mexican American Cohort receives funds collected pursuant to the Comprehensive Tobacco Settlement of 1998 and appropriated by the 76th legislature to the University of Texas MD Anderson Cancer Center and from the Caroline W. Law Fund for Cancer Prevention and the Duncan Family Institute for Risk Assessment and Cancer Prevention.
We thank the field staff for their ongoing work with participant recruitment and follow-up. Most importantly, we thank our study participants and their parents for their cooperation and participation, without which this research would not be possible.
Footnotes
Disclosure: The authors declared no conflict of interest.
Additional Supporting Information may be found in the online version of this article
References
- 1.Thow AM, Jan S, Leeder S, Swinburn B. The effect of fiscal policy on diet, obesity and chronic disease: a systematic review. Bull World Health Organ. 2010;88:609–614. doi: 10.2471/BLT.09.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renzaho AM, Halliday JA, Nowson C. Vitamin D, obesity, and obesity-related chronic disease among ethnic minorities: a systematic review. Nutrition. 2011;27:868–879. doi: 10.1016/j.nut.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Kushner RF. Weight loss strategies for treatment of obesity. Prog Cardiovasc Dis. 2014;56:465–472. doi: 10.1016/j.pcad.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Egger G, Dixon J. Beyond obesity and lifestyle: a review of 21st century chronic disease determinants. Biomed Res Int. 2014;2014:731685. doi: 10.1155/2014/731685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaffee BW, Weston SJ. Association between chronic periodontal disease and obesity: a systematic review and meta-analysis. J Periodontol. 2010;81:1708–1724. doi: 10.1902/jop.2010.100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffen A, Sorensen TI, Knuppel S, et al. Development and validation of a risk score predicting substantial weight gain over 5 years in middle-aged European men and women. PLoS One. 2013;8:e67429. doi: 10.1371/journal.pone.0067429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.May AM, Romaguera D, Travier N, et al. Combined impact of lifestyle factors on prospective change in body weight and waist circumference in participants of the EPIC-PANACEA study. PLoS One. 2012;7:e50712. doi: 10.1371/journal.pone.0050712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohrmann S, Steinbrecher A, Linseisen J, et al. The association of education with long-term weight change in the EPIC-PANACEA cohort. Eur J Clin Nutr. 2012;66:957–963. doi: 10.1038/ejcn.2012.55. [DOI] [PubMed] [Google Scholar]
- 9.Vimaleswaran KS, Angquist L, Hansen RD, et al. Association between FTO variant and change in body weight and its interaction with dietary factors: the DiOGenes study. Obesity (Silver Spring) 2012;20:1669–1674. doi: 10.1038/oby.2012.49. [DOI] [PubMed] [Google Scholar]
- 10.Travier N, Agudo A, May AM, et al. Longitudinal changes in weight in relation to smoking cessation in participants of the EPIC-PANACEA study. Prev Med. 2012;54:183–192. doi: 10.1016/j.ypmed.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Ekelund U, Besson H, Luan J, et al. Physical activity and gain in abdominal adiposity and body weight: prospective cohort study in 288,498 men and women. Am J Clin Nutr. 2011;93:826–835. doi: 10.3945/ajcn.110.006593. [DOI] [PubMed] [Google Scholar]
- 12.Forouhi NG, Sharp SJ, Du H, et al. Dietary fat intake and subsequent weight change in adults: results from the European Prospective Investigation into Cancer and Nutrition cohorts. Am J Clin Nutr. 2009;90:1632–1641. doi: 10.3945/ajcn.2009.27828. [DOI] [PubMed] [Google Scholar]
- 13.National Center for Health Statistics. Health, United States. 2015: With Special Feature on Racial and Ethnic Health Disparities. Hyattsville, MD: 2016. DHHS Publication No. 2016–1232. [PubMed] [Google Scholar]
- 14.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA--novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201–211. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 15.Williams MD, Mitchell GM. MicroRNAs in insulin resistance and obesity. Exp Diabetes Res. 2012;2012:484696. doi: 10.1155/2012/484696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomaru Y, Hayashizaki Y. Cancer research with non-coding RNA. Cancer Sci. 2006;97:1285–1290. doi: 10.1111/j.1349-7006.2006.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Wijnen AJ, van de Peppel J, van Leeuwen JP, et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr Osteoporos Rep. 2013;11:72–82. doi: 10.1007/s11914-013-0143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Huang M, Li Z, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–S131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong LL, Wang J, Liew OW, Richards AM, Chen YT. MicroRNA and Heart Failure. Int J Mol Sci. 2016;17:502. doi: 10.3390/ijms17040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortega FJ, Mercader JM, Catalan V, et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013;59:781–792. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 22.Wang R, Hong J, Cao Y, et al. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur J Endocrinol. 2015;172:291–300. doi: 10.1530/EJE-14-0867. [DOI] [PubMed] [Google Scholar]
- 23.Carreras-Badosa G, Bonmati A, Ortega FJ, et al. Altered circulating miRNA expression profile in pregestational and gestational obesity. J Clin Endocrinol Metab. 2015;100:E1446–E1456. doi: 10.1210/jc.2015-2872. [DOI] [PubMed] [Google Scholar]
- 24.Prats-Puig A, Ortega FJ, Mercader JM, et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab. 2013;98:E1655–E1660. doi: 10.1210/jc.2013-1496. [DOI] [PubMed] [Google Scholar]
- 25.Chow WH, Chrisman M, Daniel RC, et al. Cohort profile: the Mexican American mano a mano cohort [published online March 8, 2015] Int J Epidemiol. doi: 10.1093/ije/dyv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Hu Q, Schrauder M, et al. Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget. 2014;5:5284–5294. doi: 10.18632/oncotarget.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuczmarski MF, Kuczmarski RJ, Najjar M. Effects of age on validity of self-reported height, weight, and body mass index: findings from the Third National Health and Nutrition Examination Survey, 1988–1994. J Am Diet Assoc. 2001;101:28–34. doi: 10.1016/S0002-8223(01)00008-6. [DOI] [PubMed] [Google Scholar]
- 28.Villanueva EV. The validity of self-reported weight in US adults: a population based cross-sectional study. BMC Public Health. 2001;1:11. doi: 10.1186/1471-2458-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino K, Jinnin M, Kajihara I, et al. Circulating miR-142-3p levels in patients with systemic sclerosis. Clin Exp Dermatol. 2012;37:34–39. doi: 10.1111/j.1365-2230.2011.04158.x. [DOI] [PubMed] [Google Scholar]
- 30.Ortega FJ, Moreno-Navarrete JM, Pardo G, et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One. 2010;5:e9022. doi: 10.1371/journal.pone.0009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giroud M, Pisani DF, Karbiener M, et al. miR-125b affects mitochondrial biogenesis and impairs brite adipocyte formation and function. Mol Metab. 2016;5:615–625. doi: 10.1016/j.molmet.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang YC, Li YY, Wang XY, et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia. 2013;56:2275–2285. doi: 10.1007/s00125-013-2996-8. [DOI] [PubMed] [Google Scholar]
- 33.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 35.Shi Z, Zhao C, Guo X, et al. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERalpha expression in estrogen-induced insulin resistance. Endocrinology. 2014;155:1982–1990. doi: 10.1210/en.2013-2046. [DOI] [PubMed] [Google Scholar]
- 36.Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 37.Martinelli R, Nardelli C, Pilone V, et al. miR-519d overexpression is associated with human obesity. Obesity (Silver Spring) 2010;18:2170–2176. doi: 10.1038/oby.2009.474. [DOI] [PubMed] [Google Scholar]
- 38.Tang YF, Zhang Y, Li XY, Li C, Tian W, Liu L. Expression of miR-31, miR-125b-5p, and miR-326 in the adipogenic differentiation process of adipose-derived stem cells. OMICS. 2009;13:331–336. doi: 10.1089/omi.2009.0017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.