Abstract
The weighted ensemble (WE) methodology orchestrates quasi-independent parallel simulations run with intermittent communication that can enhance sampling of rare events such as protein conformational changes, folding, and binding. The WE strategy can achieve superlinear scaling—the unbiased estimation of key observables such as rate constants and equilibrium state populations to greater precision than would be possible with ordinary parallel simulation. WE software can be used to control any dynamics engine, such as standard molecular dynamics and cell-modeling packages. This article reviews the theoretical basis of WE and goes on to describe successful applications to a number of complex biological processes—protein conformational transitions, (un)binding, and assembly processes, as well as cell-scale processes in systems biology. We furthermore discuss the challenges that need to be overcome in the next phase of WE methodological development. Overall, the combined advances in WE methodology and software have enabled the simulation of long-timescale processes that would otherwise not be practical on typical computing resources using standard simulation.
Keywords: cell modeling, kinetics, molecular dynamics, path sampling, rare events, weighted ensemble
INTRODUCTION
Dramatic improvements in achievable computing speed based on hardware advances (44, 56, 60) have transformed the field of biomolecular simulation. In addition to record-setting molecular dynamics (MD) simulations in terms of both length (57) and system size (71), it has become routine to carry out simulations on the microsecond timescale, a critical regime for biological processes such as protein conformational changes and binding processes. Nonetheless, the computational cost of standard MD simulations remains prohibitive for many biological processes, particularly when the intention is to generate a sufficiently large number of corresponding events to characterize the kinetics. Furthermore, the development of more accurate simulation models [e.g., polarizable force fields (58) and quantum mechanics/molecular mechanics models (42)] requires benchmark simulations with significantly higher computational efficiency.
Path sampling approaches can greatly enhance the efficiency of simulating rare events such as protein folding, protein binding, and cellular signaling processes by focusing the computing effort on functional transitions rather than stable states [e.g., milestoning (27), transition interface sampling (65), and forward flux sampling (5)]. Such approaches typically exploit the fact that rare events are infrequent, but relatively fast, once the actual transition occurs; in particular, the duration of the transition event itself (tb) is typically orders of magnitude less than the dwell time (tdwell) in the preceding stable or metastable region (tb ≪ tdwell). Importantly, no bias is introduced into the dynamics by rigorous path sampling methods. Thus, in contrast to other approaches for accessing long-timescale motions such as metadynamics (43), accelerated MD (34), and replica exchange (35, 47), path sampling approaches enable rigorous, straightforward calculations of rate constants without additional assumptions.
The ability to efficiently provide kinetics observables and ensembles of unbiased pathways is highly complementary to state-of-the-art kinetics experiments. Although such experiments can measure only the overall rate constants of biological processes (40), path sampling approaches can be used to directly calculate the rate constants of each individual step. In addition, the pathways that are generated provide insights into the degree of diversity of pathways as well as yield ensembles of atomically detailed structures of transient states such as transition states and metastable intermediate states. These structures are not attainable by experiment and are particularly valuable for identifying residues of possible kinetic significance that could be mutated to alter the overall rate of the biological process.
The weighted ensemble (WE) approach was one of the first rare-events methods introduced for molecular systems that was capable of yielding unbiased estimates of nonequilibrium observables, but WE can also be seen as a rediscovery of the splitting strategy. The splitting (replication) of trajectories was described by Kahn in 1951 as an idea of von Neumann’s: “[W]hen the sampled particle goes from a less important to a more important region, it is split into two independent particles, each one-half the weight of the original” (38, p. 27). The context for Kahn and von Neumann was calculating neutron transmission through shielding materials, but the analogy to biomolecular and systems biology problems is essentially exact. Nevertheless, the original WE approach required further modifications, described below, for calculating kinetics generally and describing steady states (10, 62).
Indeed, the basic splitting idea has been rediscovered numerous times, both before and after the 1996 WE paper by Huber & Kim (37). In 1959, prior to WE, Wall & Erpenbeck (68) introduced a splitting approach for calculating ensemble properties of polymers and self-avoiding walks, and the RESTART approach of 1994 was introduced to aid assessment of physical network reliability (67; see also works described in 54). A number of splitting methods have been introduced since WE, including subset simulation (6), adaptive multilevel splitting (13), diffusion map–directed MD(50), and FAST (72). Also, recent versions of nonequilibrium umbrella sampling and forward flux sampling use WE/splitting ideas (8, 18).
Here, we review recent advances and applications of the WE path sampling approach, which has enabled a number of otherwise unfeasible applications (2, 17, 19, 22, 55, 59, 76). We present the theoretical basis of WE and also discuss ongoing challenges and future directions for this promising approach.
THEORY
The Original Huber–Kim Algorithm
To understand the WE algorithm of Huber & Kim (37), we require an elementary understanding of nonequilibrium trajectory physics. The essential point is that given some kind of stochastic dynamics, such as MD with a stochastic thermostat, an arbitrary initial distribution ρ0(x0) of phase-space points x0 leads to a well-defined distribution of trajectories. For simplicity, a continuous trajectory ζ is described here at a set of discrete time points as ζ = (xo, x1, x2, …, xN), where xi = x(ti), and we refer to phase-space points as configurations because velocity information does not enter our discussion. If we examine the distribution of trajectories at any later time ti, we can infer the time-evolved distribution ρi(xi), which may be familiar from Fokker–Planck descriptions (32, 51) (Figure 1). For very large t, the distribution will relax to a nonequilibrium steady-state ρss if there is a steady addition and removal of particles, probability, or energy; the distribution will relax to the equilibrium distribution ρeq if there is no addition or removal. Importantly, the trajectory distribution ρζ[ζ] has more information than the set of configurational distributions ρi(xi) at all times ti because a trajectory ζ gives the sequence of configurations: This is detailed mechanistic information.
Figure 1.
From trajectories to configurational distributions. (a) Example trajectories (red, blue, brown, green) from a schematic trajectory ensemble, generated in the potential U. (b) Configurational distributions can be obtained from trajectory ensembles by histogramming the configurations at fixed time points t1, t2, and t3. The configurational distributions will relax to the equilibrium distribution at sufficiently long times if there is no injection and removal of probability.
Any of the phase-space or trajectory distributions can be studied numerically by a finite representative (or statistical) sample, and importantly for WE, there are different types of valid samples. Converting from one type of valid sample to another for the same distribution is called resampling (70). Consider starting from a uniform random distribution of 200 points in the interval −1 < x < 1, with each point assigned an equal weight in the sample. One can resample only the negative x values by randomly selecting half of them, as long as the remaining negative values are assigned twice the weight of the positives. Similarly, adding another 100 randomly and uniformly chosen positive values with all positives now halved in weight also yields a valid sample of the uniform distribution.
With those preliminaries, the basic procedure of WE simulation as originally described by Huber & Kim (37) can be framed simply as two alternating steps, once a sample of initial configurations has been chosen that defines a set of trajectory walkers (Figure 2). The steps are (a) simulate (or continue simulating) all walkers for an arbitrary, brief interval—the same interval for all walkers—using any stochastic dynamics and (b) resample trajectories statistically, maintaining the distribution of trajectories at the given time point.
Figure 2.
Basic weighted ensemble (WE) algorithm illustrated for a configurational space divided into bins. Multiple trajectory walkers are run using any dynamics and evaluated for resampling after every interval τ. Walkers are assigned statistical weights (filled circles) that sum to a total probability of 1 and are split/combined in a rigorous manner such that no bias is introduced in the dynamics. Figure adapted with permission from Reference 21.
The WE algorithm explicitly relies on stochastic dynamics, but we do not consider this a limitation. Although deterministic thermostats are often used (7, 31), we note that their physical basis can be questioned: The finite systems of interest should be coupled to a thermal bath, which is intended to represent a thermodynamically large system with known statistical properties but unknown dynamics. Representing that bath stochastically is thus a physically sound choice (73).
Typically, resampling in WE is performed using bins, which are simply regions into which configuration space has been subdivided. Usually, WE targets maintaining a fixed number of walkers per bin, M (37). Resampling is then performed in two basic ways. If there are fewer than M walkers in a bin at a resampling time, each walker is replicated, or split, to create multiple identical copies of the trajectory (including its full history). The weights of all the child trajectories must sum to the weight of the parent; usually child weights are made equal. In contrast, in a bin with more than M walkers, trajectories must be pruned and typically a pairwise procedure is used: An arbitrary pair of walkers is selected and one is chosen to survive, in proportion to the relative weights, and the survivor inherits the additional weight of the pruned walker; this process is also called merging. Both procedures rigorously resample the trajectory ensemble and do not change its statistical properties (70).
We note that the split/merge steps introduce correlations in the trajectory ensemble produced by WE. For example, if one considers two child trajectories of the same parent, these were the very same trajectory until the split point and are hence correlated in history. Accounting for such correlations is critical in error analysis, and efforts to improve WE efficiency amount to reducing correlations. These points are addressed below in the section titled Challenges and Future Directions.
There is significant flexibility when using bins to perform unbiased resampling (70). The bins can be of arbitrary sizes, and there is no volume correction required. Each bin can have a different targeted number of walkers. The bins can be changed on the fly (3, 16, 70).
We note that it is possible to resample even without bins. For example, one could replicate only the single walker (from the ensemble) making the most progress at each resampling step. It seems that any binless procedure can be recast formally in terms of bins: In the example just given, the lead walker can be said to occupy a single bin that is updated adaptively, with all other walkers in another bin.
The preceding discussion of bin-based WE makes clear that there are a number of parameters that must be chosen in a typical setup. Bins must be constructed, the number of walkers selected, and the resampling interval chosen, but ideal choices for these parameters are system specific. Some guidance on parameter selection can be found in Reference 74 and at the WESTPA (Weighted Ensemble Simulation Toolkit with Parallelization and Analysis) website, https://westpa.github.io/westpa.
Equilibrium, Steady States, and Kinetics
Although the original WE algorithm yields the unbiased time evolution of the trajectory and configurational distributions, it is not a practical means to infer steady-state information in complex systems. In particular, we expect that an extended amount of time is required for relaxation to steady-state behavior—longer than is available by straightforward sampling, because otherwise WE would not be of great value in the first place. Further, because multiple trajectories are run in WE, the amount of “physical” time available for relaxation is reduced accordingly; for example, an overall investment of 10 μs spread over 100 trajectories yields only 100 ns of relaxation in the Huber–Kim algorithm.
Special procedures have therefore been developed for inferring steady-state information, including rate constants and equilibrium populations (10, 62). These approaches are based on the simple observation that conditional probabilities for transitions among bins can typically be estimated with much less computing than would be required by the full relaxation process for the whole system. These bin-to-bin transition probabilities can be estimated during “ordinary” WE simulation and then used to infer steady-state behavior.
To calculate macroscopic rate constants of transitions without bias between states designated A and B, one can use the Hill relation between the flux and mean first-passage time (MFPT):
| 1 |
where Flux(A → B; SS) is the probability per unit time entering state B in an A-to-B steady state. The overall flux into B can be estimated using bins as long as states A and B each consist precisely of one or more bins, which generally is not a limitation, because bin boundaries can be adjusted during a post-simulation analysis.
More specifically, the flux from A to B can be computed based on the matrix of “color”-labeled bin-to-bin transition probabilities for an interval τ among bins i and j (62; see also 15). The superscripts μ and ν refer to the “color” state before and after the interval, which simply tracks whether state A (α color) or B (β color) was more recently occupied (see 62 for full details of calculating fluxes in nonequilibrium steady states). We note that tracking the most-recent-state history avoids a Markov assumption and is necessary for estimating an unbiased MFPT because first-passage processes refer explicitly to events occurring in a given direction: A to B, or B to A (20, 28, 65, 66). Mixing the two types of trajectories in a history-agnostic Markov analysis can lead to significant bias in the MFPT and its reciprocal, the rate constant (62), and this also holds for analyzing standard MD trajectories (61, 63). Equilibrium probabilities can also be calculated via the transition rates, although the color information is not strictly necessary (10, 62).
Estimating kinetic and equilibrium information in a post-analysis procedure offers several advantages. First, different macroscopic states (A, B) of interest can be investigated as they need not be defined in advance (62). Thus, for example, state definitions could be refined based on kinetic behavior, such as minimum sensitivity to state boundaries. Bin definitions different from those used during dynamics propagation of the WE simulation can also be used in a post-analysis. This could be important, for example, in determining whether coordinates originally orthogonal to the binning directions (i.e., coordinates that were not subdivided in the original bins) affect estimates of rate constants and state populations. The ability to further subdivide slow coordinates after a simulation could help to “rescue” a suboptimal original choice of coordinates, though in principle some exploration of every slow coordinate is necessary for quantitative accuracy.
SOFTWARE
The power of an enhanced sampling approach relies on its algorithm as well as its software implementation, which must ultimately be optimized for running large-scale simulations of long-timescale processes. The WE strategy is particularly well suited for highly scalable software implementations due to its inherently parallel algorithm. In addition, the software can be designed to be interoperable, that is, interfacing with any dynamics engine, because the algorithm does not require under-the-hood modifications of the dynamics engine. To our knowledge, the first freely available, highly scalable, and interoperable WE software package was WESTPA (74), which has been widely applied to a variety of systems ranging in scale from atomistic to cellular (2–4, 21, 22, 59, 62, 63, 75, 76) and interfaced with a diversity of dynamics engines, for example, with GROMACS(36), NAMD(49), OpenMM (23), and AMBER (12) at the atomistic/molecular scale, including GPU versions, and with UIOWA-BD (25, 30), BioNetGen (11, 26), and MCell (39) at the cellular scale. The WESTPA package embodies a wide range of WE capabilities, including the calculation of both equilibrium and kinetic observables using on-the-fly reweighting (10) or a post-analysis procedure (62) as well as plug-ins for using the WE-based string method (3) and WExplore, a recently developed WE strategy that defines sampling regions in a hierarchical fashion (16). Other WE packages include in-house codes (9, 16) and a publicly available distributed computing implementation called AWE-WQ, which is interfaced with GROMACS (1).
APPLICATIONS
Early applications of the WE strategy were carried out in the late 1990s and involved Brownian dynamics (BD) simulations of (a) protein–protein association using hard-sphere models with specific binding contacts (37) and (b) protein folding of a four-helix bundle in which the helices were represented by cylinders connected by frictionless strings (52). Several years later, the WE strategy was proven to yield continuous pathways and rate constants in a rigorous manner for any type of stochastic dynamics, including Monte Carlo, BD, and MD, thereby highlighting the generality of the strategy (70). Regardless of the type of dynamics or scale of the system, numerous studies have demonstrated that the WE strategy exhibits significant super-linear scaling in estimating observables; that is, quantities such as rate constants can be estimated with orders of magnitude fewer overall computing resources compared to standard parallelized simulations because WE-spawned trajectories can be focused on sampling bottlenecks (3, 17, 21, 22, 53, 55, 69, 75). Given this efficiency, there has been a resurgence of interest in the WE strategy over the past decade, resulting in the successful generation of pathways and calculation of rate constants for long-timescale processes that would otherwise be infeasible to simulate. These processes include phenomena at atomistic/molecular scales as well as those at network/cellular scales (Figure 3).
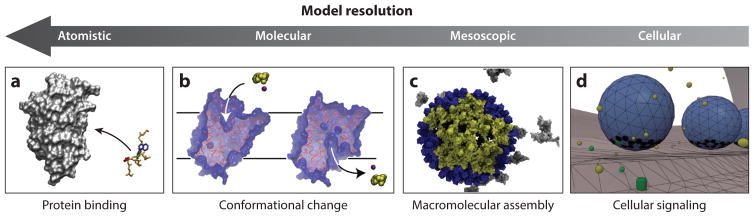
Figure 3.
Weighted ensemble (WE) applications from atomistic to cellular scales. (a) Atomistic scale: protein–peptide binding. Panel a adapted with permission from Reference 76. Copyright 2016 American Chemical Society. (b)Molecular scale: large conformational changes in a membrane protein. Panel b adapted with permission from Reference 2. Copyright 2011 Elsevier. (c) Mesoscopic scale: virus capsid assembly (59). (d) Cellular scale: cellular signaling (22).
Molecular-Scale Processes
Successful applications of the WE strategy at the atomistic level include simulations of the following:
Conformational sampling. At only a fraction of the cost of a conventional simulation carried out on the Anton special-purpose supercomputer, a WE simulation has yielded an unprecedented amount of conformational sampling in explicit solvent for the HIV-1 trans-activating response (TAR) RNA hairpin (19). These encouraging results indicate that WE strategies may become an attractive alternative to metadynamics (43), adaptive biasing force (14), and replica exchange MD (35, 47) for the enhanced sampling of biomolecular conformations, particularly when the calculation of kinetics observables is also of interest.
Protein folding. Folding pathways have been generated for the Fip35 WW domain in implicit solvent at its in silico melting temperature of 395 K and low solvent viscosity (1). Although these conditions are far from those of experiment, the computed rate constants for folding and unfolding are consistent with those from standard simulations under the same conditions, providing internal validation of the WE strategy for a complex biological process.
Protein–peptide binding. Thus far, the largest-scale application of the WE strategy to a complex biological process at the experimental temperature is the simulation of a protein–peptide binding process involving an N-terminal intrinsically disordered p53 peptide and the MDM2 oncoprotein (76) (Figure 3). Using 3,500-CPU cores on XSEDE’s Stampede, the simulation was carried out with implicit solvent at water-like viscosity and generated >180 pathways for the binding process within 15 days, yielding a kon that is in good agreement with experiment and identifying a residue that may be kinetically important. To our knowledge, this simulation is the first to generate atomistic pathways and rigorous rate constants for a protein–peptide binding process.
Protein–ligand unbinding. Protein–ligand unbinding pathways have been generated using explicit-solvent simulations for the FK506 binding protein and several low-affinity, small-molecular inhibitors that rapidly dissociate on timescales up to tens of nanoseconds (17). In addition to yielding koff values that are consistent with those from standard simulations, this study apparently conducted the first analysis of ligand-exit distributions.
Even coarse-grained (CG) simulations with molecular-level models have benefited from WE simulations. CG models in themselves do not guarantee the accessibility of timescales of interest by straight-ahead simulation. CG applications have included simulation of the following:
Large-scale protein conformational transitions. The WE strategy has efficiently yielded pathways and rate constants for transitions between the apo and holo forms of calmodulin (69) and the outward-to-inward-facing transitions in a membrane protein, the sodium symporter Mhp1 (2) (Figure 3).
Ion permeation. A proof-of-principle study involving the use of a simplified model of a narrow ion channel has demonstrated that the WE strategy can efficiently reproduce the current-voltage dependence of an ion channel from atomistic simulations, especially when permeation events through the channel are rare (4).
Protein–protein binding. Using flexible molecular models that reproduce the molecular shapes, electrostatic potentials, and diffusion properties of all-atom models, the WE strategy in combination with the Northrup-Allison-McCammon (NAM) approach (48) has reproduced the experimental kon for wild-type and mutant pairs of the barnase and barstar proteins (55). Notably, this study reported highly efficient simulation of the slow association between the exact hydrophobic, uncharged isosteres of the proteins, yielding the basal kon—a quantity of fundamental interest for determining the extent to which electrostatic interactions enhance the kon of the wild-type proteins.
Virus capsid assembly. The WE strategy has enabled simulation of the final stages of hepatitis B capsid assembly using a mesoscopic model between molecular and cellular scales that incorporates dramatically more structural detail than was previously feasible (59) (Figure 3).
Network/Cellular-Scale Processes
The WE strategy has proved highly valuable for systems biology simulations at the cellular scale for both well-mixed and spatially resolved models:
Well-mixed systems biology models. Because of the recognized importance of stochasticity in cellular-scale biology, stochastic simulations are an important complement to traditional deterministic ordinary differential equation modeling (41). However, the substantially greater cost of stochastic simulation often leads to challenges analogous to those of molecular simulation: Complex systems can exhibit long timescales relative to accessible simulation lengths. WE simulation has led to significant efficiency gains in characterizing models of cell-signaling and genetic switching (21, 64).
Spatially resolved cellular-scale models. When spatial degrees of freedom are introduced into cellular-scale models, the simulation challenges are still greater. WE was able to sample previously inaccessible low calcium concentrations in a neuromuscular junction model and reproduce experimentally reported power-law behavior (Figure 3), as well as accessing rare events in a complex signaling model embedded in realistic cell and organelle geometry (22).
CHALLENGES AND FUTURE DIRECTIONS
A fundamental limitation of all rare-events approaches, including WE, is that the most probable pathways may be missed due to inadequate sampling of the slowest relevant motions. As emphasized in the section Theory, the WE approach is extremely flexible within the boundaries of its rigorous underpinnings in statistical mechanics. Not only can bin boundaries be updated on the fly, for example, but alternative bins can be used for analysis and bins themselves are not strictly necessary. This flexibility implies that considerable variations in implementations are possible, and it is fair to say that the WE community has not fully optimized the approach. Put a different way, because WE intrinsically generates correlated trajectories, the basic ongoing challenge is to explore WE variations that reduce the correlations and hence improve sampling.
Binning
Bins are at the heart of traditional WE simulations, and their construction can dramatically affect efficiency. The criteria for setting up sufficient bins for a given system are not completely straightforward, however.
Most fundamentally, bins should divide slow coordinates into regions that are traversable with short trajectory segments. This requirement may make it seem impossible to construct effective bins for even modestly complex systems, but some examples can demonstrate that this perspective is not fully correct. Consider, for example, the association of simple solutes in explicit solvent, which has been extensively investigated using WE (62, 63, 75): As two solutes come together, the pathways of the water behavior are extremely difficult to describe, and predicting good progress coordinates is more challenging; however, because the water motion is highly correlated with the solute motion (the solutes cannot approach one another without water rearranging), it is sufficient to use bins that divide only the distance between simple solutes. In short, because WE uses only unbiased trajectory segments, only uncorrelated slow coordinates need to be binned. This point can also be appreciated within the canonical view that evolutionarily tuned biomolecular transitions tend to occur along “tubes” in conformation space representing a dominant pathway and fluctuations around it (46); in this perspective, the important subspace can be described in principle by a small number of coordinates.
Although a dominant quasi-one-dimensional pathway may characterize many systems, we can expect that some systems will exhibit multiple uncorrelated slow coordinates, some of which may not be easy to guess in advance. Two overall strategies for this problem have been developed. First, the use of adaptive binning based on Voronoi cells has been proposed as a means for spanning complex spaces without a combinatorial expansion of the number of bins (16, 70). Second, post-analysis of alternative bin structures can be used as a means of accounting for slow coordinates orthogonal to the original bins that may not have fully relaxed, because matrix calculations based on transitions rates among a larger set of bins (see the section titled Theory) are inexpensive in comparison to running additional trajectories. Nevertheless, such alternative post-analyses will be insufficient if no trajectories have explored the orthogonal (originally unbinned) slow coordinates.
Alternative Pruning and Merging Strategies
As noted in the section Theory, in the merging step of the WE protocol, any two trajectories can be chosen, which suggests that improved merge procedures may reduce trajectory correlations and hence improve WE efficiency. The diversity of trajectories should be increased to reduce correlations. In current practice, however, regardless of correlations, a low-weight trajectory is typically chosen to be merged with a high-weight trajectory, a process that tends to lead to pruning of the lower-weight trajectory. As in sequential importance sampling (45), such a procedure will suppress diversity because one can expect that low- and high-weight trajectories likely diverged from one another multiple iterations prior (compared to trajectories of similar weights). It makes sense, therefore, to pursue merge-pair selection to maximize similarity. A simple approach would be to choose merge-pair candidates based on RMSD (root-mean-square deviation) similarity of the current configurations, and we have begun to explore this and related strategies.
Error and Efficiency Analysis
Because of the complex correlation structure among WE trajectories, discussed in several sections above, error analysis is not straightforward. Consider calculation of the rate constant for a transition from state A to B using the Hill relation (Equation 1), where the probability (weight) per unit time arriving to B will be averaged. This flux will fluctuate in time, of course, but there will be sequential/temporal correlations of an unknown nature. It is therefore necessary to use error analysis that accounts for such correlations based on established ideas of time correlations and block analysis (24, 29, 33). However, different observables may have somewhat different time correlations. Instead of the rate constant, for example, one may be interested in pathways and hence attempt to estimate either the diversity of pathways [e.g., distribution of duration times (69, 75) or first-passage times (63)] or the number of independent trajectories for that purpose.
The efficiency of the WE approach is best assessed relative to standard parallelized simulations as the gold standard. We note that WE simulations can be only as accurate as the corresponding, fully converged standard simulations using the same underlying models and dynamics engine/software. In the case of benchmark systems for which standard simulations have been feasible, the efficiency of WE has been determined by considering the aggregate simulation time that has been required to yield the same rate constant and corresponding statistical precision as the standard simulations for the biological process of interest (3, 37, 53, 69, 75). However, as the motivation for using the WE approach is to enable otherwise unfeasible applications, converged standard simulations are typically not available for determining the efficiency of WE. In these cases, the efficiency of WE has been calculated by estimating the aggregate simulations time that would be required by standard simulations to yield the same observable of interest, for example, the same number of distinct pathways (55, 76) or the same rate constant and statistical precision as predicted by single-exponential kinetics, which is a reasonable model for the kinetics of two-state processes (21).
SUMMARY POINTS.
Path sampling strategies focus computational effort on transition events of interest rather than the stable states and hence can offer substantial efficiencies compared to ordinary MD simulation.
The WE path sampling strategy is based on rigorous nonequilibrium statistical mechanics and is capable of yielding unbiased estimates of kinetics and equilibrium quantities.
A non-Markovian formulation is required to obtain unbiased kinetics estimates, and the analysis carries over directly to straight-ahead MD simulations.
A strength of WE is that it requires examination of trajectories only at fixed time points and hence does not require under-the-hood modifications of the dynamics engine. This in turn has enabled facile interoperability with applications across a variety of dynamics engines and scales ranging from molecular to cellular.
WE has been successfully applied to molecular simulations of complex biological processes such as large protein conformational transitions, protein folding, and protein binding as well as cell modeling in systems biology.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant RO1GM115805 to L.T.C. and D.M.Z., NIH grant P41GM103712 and National Science Foundation (NSF) grant MCB-1119091 to D.M.Z., and NSF Faculty Early Career Development (CAREER) award MCB-0845216 to L.T.C.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abdul-Wahid B, Feng H, Rajan D, Costaouec R, Darve E, et al. AWE-WQ: fast-forwarding molecular dynamics using the accelerated weighted ensemble. J Chem Inf Model. 2014;54:3033–43. doi: 10.1021/ci500321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adelman JL, Dale AL, Zwier MC, Bhatt D, Chong LT, et al. Simulations of the alternating access mechanism of the sodium symporter Mhp1. Biophys J. 2011;101:2399–407. doi: 10.1016/j.bpj.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adelman JL, Grabe M. Simulating rare events using a weighted ensemble-based string method. J Chem Phys. 2013;138:044105. doi: 10.1063/1.4773892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adelman JL, Grabe M. Simulating current-voltage relationships for a narrow ion channel using the weighted ensemble method. J Chem Theory Comput. 2015;11:1907–18. doi: 10.1021/ct501134s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen RJ, Warren PB, ten Wolde PR. Sampling rare switching events in biochemical networks. Phys Rev Lett. 2005;94:4. doi: 10.1103/PhysRevLett.94.018104. [DOI] [PubMed] [Google Scholar]
- 6.Au S-K, Beck JL. Estimation of small failure probabilities in high dimensions by subset simulation. Probab Eng Mech. 2001;16:263–77. [Google Scholar]
- 7.Basconi JE, Shirts MR. Effects of temperature control algorithms on transport properties and kinetics in molecular dynamics simulations. J Chem Theory Comput. 2013;9:2887–99. doi: 10.1021/ct400109a. [DOI] [PubMed] [Google Scholar]
- 8.Becker NB, Allen RJ, ten Wolde PR. Non-stationary forward flux sampling. J Chem Phys. 2012;136:18. doi: 10.1063/1.4704810. [DOI] [PubMed] [Google Scholar]
- 9.Bhatt D, Bahar I. An adaptive weighted ensemble procedure for efficient computation of free energies and first passage rates. J Chem Phys. 2012;137:104101. doi: 10.1063/1.4748278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatt D, Zhang BW, Zuckerman DM. Steady-state simulations using weighted ensemble path sampling. J Chem Phys. 2010;133:014110. doi: 10.1063/1.3456985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. BioNetGen: software for rule-based modeling of signaling transduction based on the interactions of molecular domains. Bioinformatics. 2004;20:3289–91. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 12.Case DA, Cheatham I, Darden TETA, Gohlke H, Luo R, et al. The Amber biomolecular simulation programs. J Comp Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerou F. Adaptive multilevel splitting for rare event analysis. Stochastic Anal Appl. 2007;25:417–43. [Google Scholar]
- 14.Darve E, Rodriguez-Gomez D, Pohorille A. Adaptive biasing force method for scalar and vector free energy calculations. J Chem Phys. 2008;128:144120. doi: 10.1063/1.2829861. [DOI] [PubMed] [Google Scholar]
- 15.Darve E, Ryu E. Computing reaction rates in bio-molecular systems using discrete macro-states. In: Schlick T, editor. Innovations in Biomolecular Modeling and Simulations. Vol. 1. London: R. Soc. Chem; 2012. pp. 138–206. [Google Scholar]
- 16.Dickson A, Brooks CL. WExplore: hierarchical exploration of high-dimensional spaces using the weighted ensemble algorithm. J Phys Chem B. 2014;118:3532–42. doi: 10.1021/jp411479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickson A, Lotz SD. Ligand release pathways obtained with WExplore: residence times and mechanisms. J Phys Chem B. 2016;120:5377–85. doi: 10.1021/acs.jpcb.6b04012. [DOI] [PubMed] [Google Scholar]
- 18.Dickson A, Maienschein-Cline M, Tovo-Dwyer A, Hammond JR, Dinner AR. Flow-dependent unfolding and refolding of an RNA by nonequilibrium umbrella sampling. J Chem Theory Comput. 2011;7:2710–20. doi: 10.1021/ct200371n. [DOI] [PubMed] [Google Scholar]
- 19.Dickson A, Mustoe AM, Salmon L, Brooks CL. Efficient in silico exploration of RNA interhelical conformations using Euler angles and WExplore. Nucleic Acids Res. 2014;42:12126–37. doi: 10.1093/nar/gku799. Demonstrates the power of WE for conformational sampling of RNA molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson A, Warmflash A, Dinner AR. Separating forward and backward pathways in nonequilibrium umbrella sampling. J Chem Phys. 2009;131:10. doi: 10.1063/1.3244561. [DOI] [PubMed] [Google Scholar]
- 21.Donovan RM, Sedgewick AJ, Faeder JR, Zuckerman DM. Efficient stochastic simulation of chemical kinetics networks using a weighted ensemble of trajectories. J Chem Phys. 2013;139:115105. doi: 10.1063/1.4821167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donovan RM, Tapia J-J, Sullivan DP, Faeder JR, Murphy RF, et al. Unbiased rare event sampling in spatial stochastic systems biology models using a weighted ensemble of trajectories. PLOS Comput Biol. 2016;12:e1004611. doi: 10.1371/journal.pcbi.1004611. Application of WE to cellular-scale systems, including complex biochemical networks embedded in inhomogeneous spatial environments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eastman P, Friedrichs MS, Chodera JD, Radmer RJ, Bruns CM, et al. OpenMM 4: a reusable, extensible, hardware independent library for high performance molecular simulation. J Chem Theory Comput. 2013;9:461–69. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efron BYB, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat Sci. 1986;1:54–75. [Google Scholar]
- 25.Elcock AH. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. PLOS Comput Biol. 2006;2:e98. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faeder JR, Blinov ML, Hlavacek WS. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol Biol. 2009;500:113–67. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- 27.Faradjian AK, Elber R. Computing time scales from reaction coordinates by milestoning. J Chem Phys. 2004;120:10880–89. doi: 10.1063/1.1738640. [DOI] [PubMed] [Google Scholar]
- 28.Feng HY, Costaouec R, Darve E, Izaguirre JA. A comparison of weighted ensemble and Markov state model methodologies. J Chem Phys. 2015;142:214113. doi: 10.1063/1.4921890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flyvbjerg H, Peterson HG. Error estimates on averages of correlated data. J Chem Phys. 1989;91:461. [Google Scholar]
- 30.Frembgen-Kesner T, Elcock AH. Striking effects of hydrodynamic interactions on the simulated diffusion and folding of proteins. J Chem Theory Comput. 2009;5:242–56. doi: 10.1021/ct800499p. [DOI] [PubMed] [Google Scholar]
- 31.Frenkel D, Smit B. Understanding Molecular Simulation. Orlando, FL: Academic; 2001. [Google Scholar]
- 32.Gardiner C. Stochastic Methods. Berlin: Springer-Verlag; 2010. [Google Scholar]
- 33.Grossfield A, Zuckerman DM. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu Rep Comput Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–29. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 35.Hansmann U. Parallel tempering algorithm for conformational studies of biological macromolecules. Chem Phys Lett. 1997;281:140–50. [Google Scholar]
- 36.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–47. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 37.Huber GA, Kim S. Weighted-ensemble Brownian dynamics simulations of protein association reactions. Biophys J. 1996;70:97–110. doi: 10.1016/S0006-3495(96)79552-8. The original WE algorithm paper, which clearly describes and applies the simple, powerful strategy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahn H, Harris TE. Estimation of particle transmission by random sampling. Natl Bur Stand Appl Math Ser. 1951;12:27–30. The earliest known description of the splitting idea, which is the essence of WE. [Google Scholar]
- 39.Kerr R, Bartol TM, Kaminsky B, Dittrich M, Chang JCJ, et al. Fast Monte Carlo simulation methods for biological reaction-diffusion systems in solution and on surfaces. SIAM J Sci Comput. 2008;30:3126–49. doi: 10.1137/070692017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiefhaber T, Bachmann A, Jensen KS. Dynamics and mechanisms of coupled protein folding and binding reactions. Curr Opin Struct Biol. 2012;22:21–29. doi: 10.1016/j.sbi.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Kitano H. Computational systems biology. Nature. 2002;420:206–10. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 42.Kratz EG, Duke RE, Cisneros GA. Long-range electrostatic corrections in multipolar/polarizable QM/MM simulations. Theor Chem Acc. 2016;135:166. doi: 10.1007/s00214-016-1923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laio A, Parrinello M. Escaping free-energy minima. PNAS. 2002;99:12562–66. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Grand S, Gotz AW, Walker RC. SPFP: speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput Phys Commun. 2013;184:374–80. [Google Scholar]
- 45.Liu JS. Monte Carlo Strategies in Scientific Computing. New York: Springer-Verlag; [Google Scholar]
- 46.Metzner P, Schutte C, Vanden-Eijnden E. Illustration of transition path theory on a collection of simple examples. J Chem Phys. 2006;125:084110. doi: 10.1063/1.2335447. [DOI] [PubMed] [Google Scholar]
- 47.Mitsutake A, Sugita Y, Okamoto Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers. 2001;60:96–123. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 48.Northrup SH, Allison SA, McCammon JA. Brownian dynamics simulation of diffusion-influenced bimolecular reactions. J Chem Phys. 1984;80:1517–24. [Google Scholar]
- 49.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Preto J, Clementi C. Fast recovery of free energy landscapes via diffusion-map-directed molecular dynamics. Phys Chem Chem Phys. 2014;16:19181–91. doi: 10.1039/c3cp54520b. [DOI] [PubMed] [Google Scholar]
- 51.Risken H, Frank T. The Fokker-Planck Equation: Methods of Solution and Applications. 2 Berlin: Springer-Verlag; 1996. [Google Scholar]
- 52.Rojnuckarin A, Kim S, Subramaniam S. Brownian dynamics simulations of protein folding: access to milliseconds time scale and beyond. PNAS. 1998;95:4288–92. doi: 10.1073/pnas.95.8.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rojnuckarin A, Livesay DR, Subramaniam S. Bimolecular reaction simulation using weighted ensemble Brownian dynamics and the University of Houston Brownian dynamics program. Biophys J. 2000;79:686–93. doi: 10.1016/S0006-3495(00)76327-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubino G, Tuffin B, editors. Rare Event Simulation using Monte Carlo Methods. Chichester, UK: Wiley; 2009. [Google Scholar]
- 55.Saglam AS, Chong LT. Highly efficient computation of the basal kon using direct simulation of protein–protein association with flexible molecular models. J Phys Chem B. 2016;120:117–22. doi: 10.1021/acs.jpcb.5b10747. Demonstrates the power of WE for even highly coarse-grained simulations of protein–protein association. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51:91–97. [Google Scholar]
- 57.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–46. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 58.Shi Y, Xia Z, Zhang J, Best R, Wu C, et al. The polarizable atomic multipole-based AMOEBA force field for proteins. J Chem Theory Comput. 2013;9:4046–63. doi: 10.1021/ct4003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spiriti J, Zuckerman DM. Tabulation as a high-resolution alternative to coarse-graining protein interactions: initial application to virus capsid subunits. J Chem Phys. 2015;143:243159. doi: 10.1063/1.4938479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stone JE, Hardy DJ, Ufimtsev IS, Schulten K. GPU-accelerated molecular modeling coming of age. J Mol Graph Model. 2010;29:116–25. doi: 10.1016/j.jmgm.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suarez E, Adelman JL, Zuckerman DM. Accurate estimation of protein folding and unfolding times: beyond Markov state models. J Chem Theory Comput. 2016;12:3473–81. doi: 10.1021/acs.jctc.6b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suarez E, Lettieri S, Zwier MC, Stringer CA, Subramanian SR, et al. Simultaneous computation of dynamical and equilibrium information using a weighted ensemble of trajectories. J Chem Theory Comput. 2014;10:2658–67. doi: 10.1021/ct401065r. Introduction of the non-Markovian analysis of WE trajectories, which is essential for unbiased kinetics when continuous trajectories are mapped to a discrete bin space. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suarez E, Pratt AJ, Chong LT, Zuckerman DM. Estimating first-passage time distributions from weighted ensemble simulations and non-Markovian analyses. Protein Sci. 2016;25:67–78. doi: 10.1002/pro.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tse MJ, Chu BK, Roy M, Read EL. DNA-binding kinetics determines the mechanism of noise-induced switching in gene networks. Biophys J. 2015;109:1746–57. doi: 10.1016/j.bpj.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Erp TS, Moroni D, Bolhuis PG. A novel path sampling method for the calculation of rate constants. J Chem Phys. 2003;118:7762. doi: 10.1063/1.1644537. [DOI] [PubMed] [Google Scholar]
- 66.Vanden-Eijnden E, Venturoli M. Exact rate calculations by trajectory parallelization and tilting. J Chem Phys. 2009;131:7. doi: 10.1063/1.3180821. [DOI] [PubMed] [Google Scholar]
- 67.Villen-Altamirano M, Villen-Altamirano J. Proc Winter Simul Conf. New York: IEEE; 1994. RESTART:a straightforward method for fast simulation of rare events; pp. 282–89. [Google Scholar]
- 68.Wall FT, Erpenbeck JJ. New method for the statistical computation of polymer dimensions. J Chem Phys. 1959;30:634–37. [Google Scholar]
- 69.Zhang BW, Jasnow D, Zuckerman DM. Efficient and verified simulation of a path ensemble for conformational change in a united-residue model of calmodulin. PNAS. 2007;104:18043–48. doi: 10.1073/pnas.0706349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang BW, Jasnow D, Zuckerman DM. The “weighted ensemble” path sampling method is statistically exact for a broad class of stochastic processes and binning procedures. J Chem Phys. 2010;132:054107. doi: 10.1063/1.3306345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–46. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimmerman MI, Bowman GR. FAST conformational searches by balancing exploration/exploitation trade-offs. J Chem Theory Comput. 2015;11:5747–57. doi: 10.1021/acs.jctc.5b00737. [DOI] [PubMed] [Google Scholar]
- 73.Zuckerman DM. Statistical Physics of Biomolecules: An Introduction. Boca Raton, FL: CRC Press; 2010. [Google Scholar]
- 74.Zwier MC, Adelman JL, Kaus JW, Pratt AJ, Wong KF, et al. WESTPA: an interoperable, highly scalable software package for weighted ensemble simulation and analysis. J Chem Theory Comput. 2015;11:800–9. doi: 10.1021/ct5010615. A freely available, highly scalable WE software package that interfaces with any dynamics engine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwier MC, Kaus JW, Chong LT. Efficient explicit-solvent molecular dynamics simulations of molecular association kinetics: methane-methane, Na+/Cl−, methane/benzene, and K+/18-crown-6 ether. J Chem Theory Comput. 2011;7:1189–97. doi: 10.1021/ct100626x. [DOI] [PubMed] [Google Scholar]
- 76.Zwier MC, Pratt AJ, Adelman JL, Kaus JW, Zuckerman DM, Chong LT. Efficient atomistic simulation of pathways and calculation of rate constants for a protein-peptide binding process: application to the MDM2 protein and an intrinsically disordered p53 peptide. J Phys Chem Lett. 2016;7:3440–45. doi: 10.1021/acs.jpclett.6b01502. The largest-scale atomistic WE simulation to date, yielding the first atomistic simulations of complete pathways for a protein–peptide binding process with rigorous rate constants. [DOI] [PMC free article] [PubMed] [Google Scholar]