Abstract
Introduction
Genetic variants associated with nicotine dependence have previously been identified, primarily in European-ancestry populations. No genome-wide association studies (GWAS) have been reported for smoking behaviors in Hispanics/Latinos in the United States and Latin America, who are of mixed ancestry with European, African, and American Indigenous components.
Methods
We examined genetic associations with smoking behaviors in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) (N = 12 741 with smoking data, 5119 ever-smokers), using ~2.3 million genotyped variants imputed to the 1000 Genomes Project phase 3. Mixed logistic regression models accounted for population structure, sampling, relatedness, sex, and age.
Results
The known region of CHRNA5, which encodes the α5 cholinergic nicotinic receptor subunit, was associated with heavy smoking at genome-wide significance (p ≤ 5 × 10–8) in a comparison of 1929 ever-smokers reporting cigarettes per day (CPD) > 10 versus 3156 reporting CPD ≤ 10. The functional variant rs16969968 in CHRNA5 had a p value of 2.20 × 10–7 and odds ratio (OR) of 1.32 for the minor allele (A); its minor allele frequency was 0.22 overall and similar across Hispanic/Latino background groups (Central American = 0.17; South American = 0.19; Mexican = 0.18; Puerto Rican = 0.22; Cuban = 0.29; Dominican = 0.19). CHRNA4 on chromosome 20 attained p < 10–4, supporting prior findings in non-Hispanics. For nondaily smoking, which is prevalent in Hispanic/Latino smokers, compared to daily smoking, loci on chromosomes 2 and 4 achieved genome-wide significance; replication attempts were limited by small Hispanic/Latino sample sizes.
Conclusions
Associations of nicotinic receptor gene variants with smoking, first reported in non-Hispanic European-ancestry populations, generalized to Hispanics/Latinos despite different patterns of smoking behavior.
Implications
We conducted the first large-scale genome-wide association study (GWAS) of smoking behavior in a US Hispanic/Latino cohort, and the first GWAS of daily/nondaily smoking in any population. Results show that the region of the nicotinic receptor subunit gene CHRNA5, which in non-Hispanic European-ancestry smokers has been associated with heavy smoking as well as cessation and treatment efficacy, is also significantly associated with heavy smoking in this Hispanic/Latino cohort. The results are an important addition to understanding the impact of genetic variants in understudied Hispanic/Latino smokers.
Introduction
Cigarette smoking continues to create a heavy public health burden both globally and in the United States.1,2 In US adults overall, the prevalence of smoking has declined from 20.9% in 2005 to 15.1% in 2015.3,4 However, this reduced prevalence still corresponds to 40 million smokers in the United States alone, and globally the number of smokers is increasing,5 leading to predictions of over 1 billion smokers worldwide by 2025.6
Much progress has been made in identifying genetic variants associated with nicotine dependence and other smoking behaviors through genome wide association studies (GWAS) that test large numbers of genetic variants, such as single nucleotide polymorphisms (SNPs), across the genome. The earliest and largest genome-wide studies of smoking were performed in populations of European ancestry (excluding Hispanics/Latinos in the United States and Latin America),7–13 hereafter referred to as “European ancestry.” GWAS results are also available for other racial/ethnic populations including East Asians14 and African Americans.15 The most prominent, well-replicated findings are variants in the CHRNA5/A3/B4 cluster of cholinergic nicotinic receptor subunit genes that are associated with risk for nicotine dependence and heavy smoking. This finding was initially discovered in European-ancestry populations11–13,16–18 and has also been reported in African Americans and East Asians.15,19,20 Multiple correlated variants represent this locus, and the SNP rs16969968 in CHRNA5 has gained particular attention because it is a non-synonymous coding change (D398N), is consistently associated across continental populations,19 and demonstrates nicotine-related functional effects in vitro21,22 and in transgenic mouse models.23
Variation has been found across racial and ethnic groups in patterns of tobacco use, nicotine metabolism, and risk of smoking-related diseases.24 To date, no large-scale genome-wide genetic study of smoking has examined Hispanic/Latino populations, who represent the largest minority group living in the United States25 and are of mixed ancestry with European, African, and American Indigenous components.26 In 2015, the prevalence of smoking among US Hispanics/Latinos was 10.1%, lower than among non-Hispanic Whites (16.6%), African Americans (16.7%) and American Indians/Alaska Natives (21.9%), and higher than that among Asian Americans (7%).27 Other evidence suggests less intensive tobacco use (measured by cigarettes per day [CPD]) among Hispanic/Latino smokers compared with other populations.28–31
The prevalence of nondaily smoking is markedly higher in Hispanics/Latinos than in other population groups, making this trait particularly interesting to examine in genetic studies of Hispanics/Latinos.28,29,31,32 Social phenomena, and acculturation-related factors particularly among Hispanics/Latinos, likely contribute to this higher rate of nondaily smoking,33,34 but biological factors, perhaps involved in lower addiction susceptibility or differing response to tobacco constituents, may also play a role. Of note, American Indigenous populations have a uniquely long history of tobacco use: tobacco has been cultivated in the Americas for more than 5000 years and used across the Americas for at least the last 2000 years.35 Still, overall metabolism of nicotine does not appear to differ between Hispanics/Latinos and non-Hispanic Whites.36 Daily and nondaily smoking may differ in their effects on target organs,37 further increasing our interest in this phenotype.
Here we report the first large-scale genome-wide association study of smoking behaviors in Hispanic/Latino individuals. A unique aspect of this study is the assessment of nondaily versus daily smoking and the large number of nondaily smokers in this Hispanic/Latino cohort, making it possible to seek genetic loci influencing this smoking behavior, which has not previously been examined in any GWAS. We also analyzed more commonly assessed smoking traits, such as heavy smoking as defined by CPD, to look for novel associations and also to evaluate the degree to which known genetic associations identified in other population groups generalize to this Hispanic/Latino cohort.
Materials and Methods
Study Samples
Participants in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) included 16 415 adults aged 18 to 74 years old recruited as a population sample drawn from four US communities (Bronx, NY, Chicago, IL, Miami, FL, and San Diego, CA), of which 12 803 were genotyped genome-wide as described below.28,38 All participants identified themselves as Hispanic, Latino, or as a member of a specific Latin American group. A household-based recruitment approach was used, with an average of 1.8 members per household enrolled. Standardized questionnaires were administered by in person interview using either Spanish or English. Venous blood samples were drawn, separated and frozen on site. DNA was prepared from thawed buffy coats at the HCHS/SOL Central Laboratory. All participants included in this report provided consent for their data and biosamples to be used for genetic aspects of the HCHS/SOL research aims. Table 1 shows demographics of the genetic study sample for whom smoking data were available (N = 12 741).
Table 1.
Demographics of the HCHS/SOL Genetic Sample With Smoking Data
| Current daily smokers | Current nondaily smokers | Former smokers | Never-smokers | Total | |
|---|---|---|---|---|---|
| Total N | 1786 | 778 | 2555 | 7616 | 12 741 |
| Male: N (%) | 892 (49.94) | 453 (58.23) | 1393 (54.52) | 2485 (32.63) | 5227 (41.03) |
| Age: Mean (SD) | 46.6 (12.1) | 41.0 (13.2) | 51.4 (11.9) | 44.8 (14.4) | 46.1 (13.9) |
| Recruitment site: N (%) | |||||
| Bronx, NY | 551 (30.85) | 152 (19.54) | 584 (22.86) | 1946 (25.55) | 3233 (25.37) |
| Chicago, IL | 304 (17.02) | 236 (30.33) | 562 (22.00) | 1971 (25.88) | 3075 (24.13) |
| Miami, FL | 676 (37.85) | 167 (21.47) | 730 (28.57) | 1869 (24.54) | 3445 (27.04) |
| San Diego, CA | 255 (14.28) | 223 (28.66) | 679 (26.58) | 1830 (24.03) | 2988 (23.45) |
| Self-reported Hispanic/Latino heritage: N (%) | |||||
| Central American | 106 (5.94) | 83 (10.67) | 250 (9.78) | 884 (11.61) | 1325 (10.40) |
| South American | 67 (3.75) | 49 (6.30) | 177 (6.93) | 546 (7.17) | 839 (6.59) |
| Mexican | 366 (20.49) | 367 (47.17) | 992 (38.83) | 3009 (39.51) | 4736 (37.17) |
| Puerto Rican | 557 (31.19) | 140 (17.99) | 455 (17.81) | 1033 (13.56) | 2186 (17.16) |
| Cuban | 514 (28.78) | 82 (10.54) | 432 (16.91) | 1006 (13.21) | 2035 (15.97) |
| Dominican | 103 (5.77) | 27 (3.47) | 165 (6.46) | 906 (11.90) | 1201 (9.43) |
| Missing | 6 (0.34) | 0 (0.00) | 4 (0.16) | 15 (0.20) | 25 (0.20) |
| More than one heritage | 53 (2.97) | 24 (3.08) | 59 (2.31) | 173 (2.27) | 309 (2.43) |
| Other | 14 (0.78) | 6 (0.77) | 21 (0.82) | 44 (0.58) | 85 (0.67) |
| Genetic analysis group: N (%) | |||||
| Central American | 106 (5.94) | 87 (11.18) | 276 (10.80) | 925 (12.15) | 1396 (10.96) |
| Cuban | 558 (31.24) | 95 (12.21) | 464 (18.16) | 1139 (14.96) | 2257 (17.71) |
| Dominican | 96 (5.38) | 23 (2.96) | 165 (6.46) | 895 (11.75) | 1179 (9.25) |
| Mexican | 372 (20.83) | 378 (48.59) | 991 (38.79) | 3004 (39.44) | 4747 (37.26) |
| Puerto Rican | 581 (32.53) | 139 (17.87) | 461 (18.04) | 1059 (13.90) | 2241 (17.59) |
| South American | 73 (4.09) | 56 (7.20) | 198 (7.75) | 594 (7.80) | 921 (7.23) |
| Cigarettes per day (CPD): N | Other smokers with CPD (current/former unspecified) | Total | |||
| CPD ≤ 10 (light smokers) | 946 | 675 | 1531 | 4 | 3156 |
| 10 < CPD ≤ 20 | 664 | 73 | 641 | 2 | 1380 |
| 20 < CPD ≤ 30 | 82 | 7 | 123 | 0 | 212 |
| CPD > 30 | 93 | 17 | 227 | 0 | 337 |
| CPD missing | 1 | 6 | 33 | 0 | 40 |
| Total | 1786 | 778 | 2555 | 6 | 5125 |
| Total with CPD available | 1785 | 772 | 2522 | 5085 | |
Smoking Behavior: Defining Phenotypic Traits for Analysis
Ever-smokers were defined as having smoked at least 100 cigarettes in their lifetime. Ever-smokers were asked whether they currently smoked daily, some days (ie, nondaily smokers), or not at all (ie, former smokers). Ever-smokers reported the average number of CPD smoked across their lifetime during the time that they smoked. In addition, current daily smokers reported their current number of CPD, and current nondaily smokers reported both the number of smoking days during the past month, and the average number of CPD on smoking days.
We used two binary traits in our primary genome-wide analyses. First, with analysis limited to ever-smokers, we defined heavy smoking cases as CPD > 10 and compared them to light smokers defined as CPD ≤ 10. Among former smokers we used the average lifetime CPD to define heavy versus light smoking, and among current smokers we used either the average lifetime CPD or the current CPD, whichever value was higher. The threshold of 10 CPD to define our primary heavy/light variable was selected a priori based on the empiric distribution of CPD in the HCHS/SOL cohort and other data on Hispanic/Latino populations that report an average number of CPD of 10–12 among current daily smokers.28,31 The proportion of lighter smokers in HCHS/SOL is higher than is reported in European-ancestry cohorts (Supplementary Figure S1). The second trait used in primary analysis, in models that were limited to current smokers, contrasted nondaily smokers as “cases” versus daily smokers.
We conducted several secondary analyses of other smoking traits. These included two alternative CPD-based traits, both analyzed among ever-smokers: (1) an alternative heavy (CPD > 20) versus light (CPD ≤ 10) binary smoking trait, and (2) a 4-level CPD variable (CPD ≤ 10; 10 < CPD ≤ 20; 20 < CPD ≤ 30; CPD > 30). Both these traits have been used in prior genetic studies of non-Hispanic smokers.16,19 These alternative phenotypes allowed us to examine the robustness of the primary heavy smoking trait by comparing the strength of association across these alternative CPD-based phenotypes in the known smoking-associated region of CHRNA5/A3/B4. Finally, we examined the binary traits of current smoker (N = 2564) versus former smoker (N = 2555), and ever-smoker (N = 5119) versus never-smoker (N = 7616).
Table 1 summarizes smoking traits, including CPD within ever-smokers. Of the 5085 ever-smokers with CPD data, 3156 (62.1%) reported light smoking (CPD ≤ 10) and 1929 (37.9%) reported heavy smoking (CPD > 10). Of the 2564 current smokers, 778 were nondaily smokers (30.3%) and 1786 (69.7%) were daily smokers. A total of six ever-smokers reported CPD but did not report their current or nondaily smoking status. Heavy/light smoking and daily/nondaily smoking were related: among nondaily smokers with CPD data, the vast majority (87.4%) reported CPD ≤ 10, and the 1df chi-square was 275.43 (p < .001); the correlation was moderate (r2 = .117) due in part to the differing marginal frequencies of these two traits.
Genotyping
HCHS/SOL participants were genotyped at 2.4 million SNPs using a custom Illumina array including the HumanOmni2.5-8v1-1 array content plus approximately 150 000 investigator-chosen SNPs.39 We followed standard quality-assurance and quality-control (QA/QC) procedures: samples were checked for annotated or genetic sex, gross chromosomal anomalies, relatedness and population structure, missing call rates, batch effects, and duplicate-sample discordance.39–42 At the SNP level, checks were performed for Hardy-Weinberg equilibrium, minor allele frequency (MAF), duplicate-probe discordance, Mendelian errors, and missing call rate. QA/QC steps were performed using the R/Bioconductor package GWASTools.43
Genotypes were pre-phased using SHAPEIT244 and imputed using IMPUTE245,46 with the 1000 Genomes Phase 3 cosmopolitan (“ALL”) reference panel,47 resulting in 49 million imputed variants.48 For each imputed SNP, we calculated R2 for imputation quality (the ratio of observed to expected variance of imputed dosage, also called “oevar”) and excluded SNPs with R2 < .3. We filtered variants using the minimum effective number of copies of the minor allele (effN), which is approximately its minor allele count. We estimated effN as 2 × MAF × (1−MAF) × N × R2, where MAF is the minor allele frequency, N is the number of participants, and R2 is set to one for genotyped variants. For binary traits, SNPs with effN < 50 in either the cases or the controls were filtered out. For quantitative traits, SNPs with effN < 30 in all participants were filtered out.
Statistical Analyses
Association Testing
Genetic variants were tested for association using a penalized pseudolikelihood-based score test49 to fit a logistic mixed model for binary traits, and using mixed linear regression for quantitative traits.50 All models accounted for population structure and admixture by including fixed effects for the first five genetic eigenvectors and the “genetic analysis group,”39 a classification of study individuals into the population groups Mexican, Puerto Rican, Cuban, Dominican, South American, and Central American. These genetic analysis groups mostly overlap with the corresponding self-reported ethnicities, yet are refined based on genotyping data, and provide classification in instances where self-reported ethnicities were unavailable.39 Other covariates (fixed effects) were included for sex, age, study center, and sample design (using an AIC-determined function of the sampling weights). Both models included random effects for genetic relatedness (kinship), household, and correlation among individuals due to shared community (block group).
Quantile-quantile (Q-Q) plots and the genomic inflation factor (λ) were used to evaluate control of Type I error. LocusZoom51 was used to plot regions harboring genome-wide significant signals (p < 5 × 10–8), using correlations obtained from the HCHS/SOL cohort to visualize linkage disequilibrium patterns. Statistical analyses were performed at the University of Washington’s Genetic Analysis Center (GAC). All analyses and metadata were saved and tracked in the GAC’s HCHS/SOL analysis database and GAC analysis IDs for each GWAS are documented in Supplementary Table S1.
Replication Tests for Daily/Nondaily Smoking
For replication testing we focused on the novel daily/nondaily phenotype, which had not been previously examined for genetic association in any population including European-ancestry cohorts. Three studies that have assessed daily/nondaily smoking evaluated our top signals for this trait in their data: the BioMe Biobank,52 The Collaborative Genetic Study of Nicotine Dependence (COGEND),8,17 and The Genetic Study of Atherosclerosis Risk (GeneSTAR).53,54 Only BioMe included Hispanic/Latino participants. Supplementary Text S1 details these studies’ recruitment, genotyping, phenotyping, and analysis models. Each study fit logistic regression models for the binary daily/nondaily trait and adjusted for sex, age, and appropriate study-specific variables. For the replication analyses, each study contributed samples as follows.
The BioMe sample contributed 428 Hispanic/Latino current smokers (343 daily, 85 nondaily), and 405 African American current smokers (336 daily, 69 nondaily). BioMe participants were asked the same question used in HCHS/SOL: “Do you NOW smoke daily, some days, or not at all.” The replication analysis compared participants reporting “daily” to those reporting “some days” (nondaily).
COGEND contributed 1252 European–American current smokers (202 nondaily and 1050 daily) and 587 African American current smokers (56 nondaily and 531 daily current smokers). In COGEND current smokers, daily/nondaily smoking was defined using the question “About how many days out of the last 30 did you smoke at least one cigarette?” with a threshold of ≤ 20 cigarettes for nondaily and > 20 cigarettes for daily.
GeneSTAR contributed 296 European-American current smokers (30 nondaily and 266 daily) and 272 African American current smokers (33 nondaily and 239 daily). In GeneSTAR the daily and nondaily smoking trait for replication testing was defined using the question “On how many days a week do you smoke cigarettes.” Nondaily smokers were defined as those who responded “usually on one day or less” or “usually on 2 to 4 days,” and daily smokers as those who responded “almost every day.”
Results
Chromosome 15 CHRNA5/A3/B4 Results for Heavy/Light Smoking and Other Smoking Traits
We first focused on the functional SNP rs16969968 in CHRNA5 on chromosome 15, which is known to be associated with nicotine dependence and heavy smoking in European-, African-, and East Asian-ancestry populations. The primary heavy/light smoking trait (CPD > 10 vs. CPD ≤ 10) was associated with rs16969968 with nearly genome-wide significance: p = 2.2 × 10–7, odds ratio (OR) = 1.32 for the minor allele A. The MAF overall was 0.22, which varied only slightly by genetic analysis group (Central American = 0.17; South American = 0.19; Mexican = 0.18; Puerto Rican = 0.22; Cuban = 0.29; Dominican = 0.19). This association is supported by 13 other genotyped and imputed SNPs that attained genome-wide significance (p < 5 × 10–8) and appear to represent the same signal (Supplementary Figure S2); after conditional analysis with rs16969968 as a covariate, none remain associated even at p < .005 (Table 2). More broadly, conditional analysis found no genome-wide significant signals anywhere in the region. These results demonstrate that the locus represented by rs16969968 in CHRNA5 has an effect on heavy smoking that generalizes to Hispanic/Latino smokers.
Table 2.
Association of rs16969968 and Chromosome 15 CHRNA5/A3/B4 SNPs With Heavy/Light Smoking in HCHS/SOL
| rsID | bp | Gene | Function | Qualitya | Effect allele/ref allele | Effect allele freq | Primary heavy/light (>10 vs. ≤ 10) | Primary heavy/light (> 10 vs ≤ 10) conditioned on rs16969968 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |||||||
| rs16969968 | 78882925 | CHRNA5 | Non-syn | 1 | A/G | 0.219 | 1.32 (1.19–1.46) | 2.20E-7 | — | — |
| rs1051730 | 78894339 | CHRNA3 | Syn | 1 | A/G | 0.231 | 1.34 (1.21–1.49) | 1.74E-08 | 1.64 (1.10–2.45) | .014739 |
| rs4243084 | 78911672 | CHRNA3/B4 | Intronic | 0.998 | C/G | 0.269 | 1.32 (1.20–1.45) | 1.78E-08 | 1.26 (1.03–1.54) | .022103 |
| rs58365910 | 78849034 | CHRNA5 | Intergenic | 0.994 | C/T | 0.245 | 1.33 (1.21–1.47) | 2.00E-08 | 1.27 (1.03–1.57) | .025894 |
| rs8040868 | 78911181 | CHRNA3 | Syn | 1 | C/T | 0.317 | 1.30 (1.19–1.43) | 2.16E-08 | 1.21 (1.04–1.40) | .011916 |
| — | 78898675 | 0.887 | Indel | 0.385 | 1.31 (1.19–1.44) | 2.21E-08 | 1.21 (1.06–1.38) | .006382 | ||
| — | 78913353 | CHRNA3 | 0.89 | Indel | 0.363 | 1.30 (1.19–1.43) | 2.66E-08 | 1.20 (1.05–1.37) | .76013 | |
| — | 78871382 | CHRNA5 | Intronic | 0.796 | Indel | 0.281 | 1.32 (1.20–1.46) | 3.12E-08 | 1.21 (1.04–1.41) | .657196 |
| rs55676755 | 78898932 | CHRNA3 | Intronic | 0.999 | G/C | 0.237 | 1.33 (1.20–1.48) | 3.24E-08 | 1.38 (1.00–1.69) | .052231 |
| rs12914385 | 78898723 | CHRNA3 | Intronic | 1 | T/C | 0.263 | 1.32 (1.20–1.45) | 3.32E-08 | 1.24 (1.01–1.54) | .042405 |
| — | 78859605 | CHRNA5 | Intronic | 0.988 | Indel | 0.241 | 1.33 (1.20–1.47) | 4.01E-08 | 1.27 (0.99–1.64) | .062451 |
| rs190065944 | 78859610 | CHRNA3 | Intronic | 1 | A/G | 0.239 | 1.33 (1.20–1.47) | 4.15E-08 | 1.25 (0.99–1.58) | .058969 |
| rs2036527 | 78851615 | CHRNA5 | Intergenic | 1 | A/G | 0.237 | 1.33 (1.20–1.47) | 4.29E-08 | 1.25 (0.99–1.57) | .059432 |
| — | 78899560 | CHRNA3 | Intronic | 0.986 | Indel | 0.246 | 1.32 (1.20–1.46) | 4.94E-08 | 1.28 (0.97–1.69) | .084076 |
CI = confidence interval; HCHS/SOL = Hispanic Community Health Study/Study of Latinos; OR = odds ratio.
aImputation quality score (R2) is given (value 1 if genotyped).
Alternative CPD-based smoking traits (CPD > 20 vs. CPD ≤ 10, and 4-level CPD) gave no genome-wide significant signals in the chromosome 15 region. In general, results were similar but less significant than for the primary heavy/light trait. For example, at rs16969968, testing CPD > 20 versus CPD ≤ 10 gave p = 6.8 × 10–7 and 4-level CPD gave p = 1.6 × 10–5.
We also tested rs16969968 for association with other smoking traits in this Hispanic/Latino cohort. We saw no evidence for association with current/former (p = .388) or ever-/never-smoking (p = .340), in agreement with prior analyses in European-ancestry samples.12 The novel daily/nondaily smoking trait also showed no association with rs16969968 (p = .192), which suggests important differences between this trait and heavy smoking, despite some phenotypic relationship.
Heavy/Light Smoking GWAS
The primary heavy/light smoking trait was defined by CPD > 10 versus CPD ≤ 10. Quantile-quantile (Q-Q) plots show no evidence of inflation, with λ = 1.002 for the set of ~11.4 million filtered SNPs (Supplementary Figure S3). Two genomic regions reached GWAS significance (p ≤ 5 × 10–8): the known CHRNA5/A3/B4 region on chromosome 15 described above, and an intergenic region on chromosome 20, as shown in the Manhattan plot (Supplementary Figure S4). Top associated variants are listed in Supplementary Table S2. The chromosome 20 signal, near HAO1, is supported only by three imputed, low-frequency variants: rs117372249 (MAF = 0.015), rs117253780 (MAF = 0.014), and rs143072330 (MAF = 0.015). Though the R2 estimates for these imputed variants are > .9, imputation quality can be poor for low frequency variants even when R2 is high.55,56 Moreover, these associations are not supported by the genotyped SNPs in the region (denoted by circles in Supplementary Figure S5).
In European-ancestry populations, other genes besides CHRNA5/A3/B4 are known from GWAS to be associated with nicotine dependence or CPD: CHRNB3/A6,8,13,17,57CHRNA4,10 and CYP2A6.12 Therefore we looked up HCHS/SOL results for the primary heavy/light trait, for top SNPs previously reported in GWAS of European-ancestry smokers10 (Supplementary Table S3). For CHRNA4 on chromosome 20, results were strong with p = 9 × 10–5 at rs45497800 and p < .001 for several additional SNPs, and the direction of effect agrees with non-Hispanic reports,10 for example, OR = 1.13 for the minor allele C at rs2273500. For CHRNB3/A6 on chromosome 8, several variants had p < .05, again with direction of effect consistent with non-Hispanic results. Finally, the examined CYP2A6 SNPs on chromosome 19 were not significant.
We also examined an alternative heavy/light smoking trait defined by CPD > 20 (N = 549) versus CPD ≤ 10 (N = 3156), which has been used in prior genetic studies of non-Hispanic smokers.16,19 No SNPs reached genome-wide significance. Variants attaining p < 5 × 10–7 are all in the CHRNA5/A3/B4 region on chromosome 15 (Supplementary Table S4).
Daily/Nondaily Smoking GWAS
Analysis of daily/nondaily smoking identified two novel genomic regions, on chromosomes 2 and 4, that surpassed genome-wide significance (p ≤ 5 × 10–8) (Supplementary Figures S6 and S7; Supplementary Table S5). Both signals are supported by genotyped and imputed SNPs (Figures 1 and 2). Neither region has been identified in prior GWAS of smoking traits.
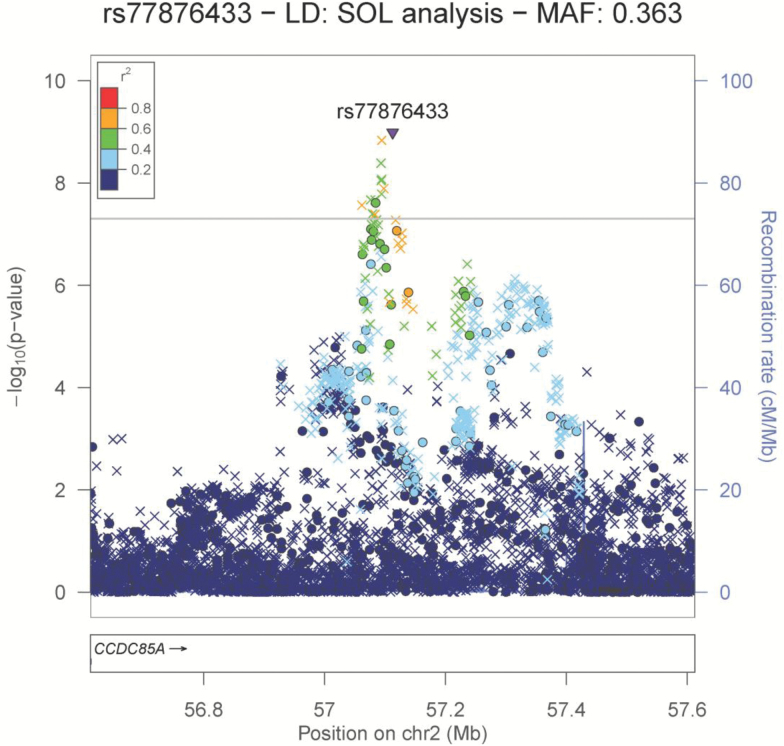
Figure 1.
Locus zoom plot of the chromosome 2 region associated with daily/nondaily smoking. Genotyped SNPs are denoted by circles; imputed SNPs are denoted by triangles (refsnp) and × symbols.
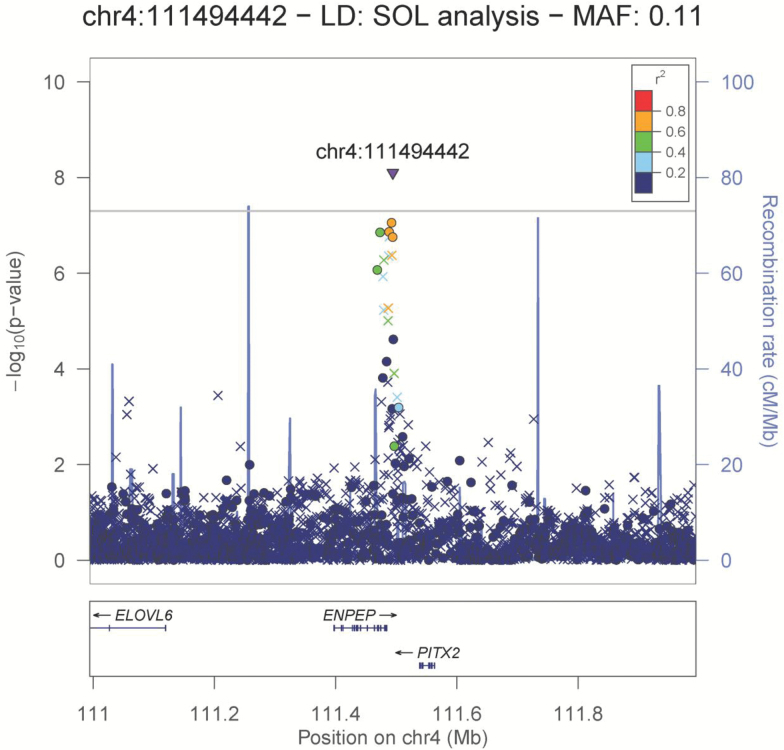
Figure 2.
Locus zoom plot of the chromosome 4 region associated with daily/nondaily smoking. Genotyped SNPs are denoted by circles; imputed SNPs are denoted by triangles (refsnp) and × symbols.
On chromosome 2, both genotyped and imputed SNPs attained p ≤ 5 × 10–8 in an intergenic region downstream of CCDC85A (Figure 1). The lead signal was an imputed SNP, rs77876433 (p = 1.03 × 10–9, OR = 1.57, MAF = 0.363 [allele C]). The most significant genotyped SNP was rs1989725 (p = 2.45 × 10–8, OR = 1.49, MAF = 0.38 [allele G]).
On chromosome 4, a single imputed insertion variant at 111494442 Mb, in the region of ENPEP and PITX2, surpassed genome-wide significance (no rsID available, p = 7.94 × 10–9, OR = 1.87, MAF = 0.11 [allele GA]). Several additional genotyped SNPs were in linkage disequilibrium and had low p values supporting this signal, down to p = 8.83 × 10–8 for rs1562640 (OR = 1.95, MAF = 0.074 for allele G).
Replication Testing of Daily/Nondaily Smoking
We provided the independent studies BioMe, COGEND, and GeneSTAR with lists of the top SNPs (p < 5 × 10–6) in the two regions that surpassed GWAS significance for daily/nondaily smoking, on chromosomes 2 and 4. Available SNPs were tested, stratifying by race/ethnicity. Results were obtained for one Hispanic/Latino cohort (BioMe, N = 428), three African American cohorts (BioMe, N = 405; COGEND, N = 587; GeneSTAR, N = 272), and two European-American cohorts (COGEND, N = 1252; GeneSTAR, N = 96).
Supplementary Table S6 shows replication and meta-analysis results for the best available SNP in each region, namely the SNP that had the most significant association in HCHS/SOL and was also available in all three replication studies: BioMe, COGEND, and GeneSTAR. We found no evidence for replication, either in the Hispanic/Latino cohort from BioMe or in the additional cohorts of African or European ancestry. On chromosome 2, the SNP with the lowest p value in HCHS/SOL, rs77876433, was available in all three studies, and showed no evidence for replication. Specifically, in the BioMe Hispanic/Latino cohort, rs77876433 was nonsignificant with an opposite direction of effect (OR = 0.77 for the minor allele C, p = .27). The African American and European–American cohorts also yielded nonsignificant results, and the meta-analysis of all replication cohorts across race/population gives OR = 0.93 and p value .31. On chromosome 4, the best available SNP rs1562640 (OR = 1.95, p = 3.43 × 10–8 in HCHS/SOL) also did not replicate in the BioMe Hispanic/Latino sample (OR = 1.02, p = .96); the meta-analysis of all replication cohorts was nonsignificant (OR = 1.13, p = .36).
Secondary GWAS of Ever/Never and Current/Former Smoking
No variants reached genome-wide significance for ever-/never-smoking or current/former smoking. The Q-Q plot for ever-/never-smoking showed no evidence of bias across ~15.4M variants, with λ = 0.981. For current/former smoking, the Q-Q plot across ~11.9M variants and λ = 0.926 suggested some downward bias.
Discussion and Conclusions
Genetic analyses of non-European and mixed-ancestry populations are important for several reasons. Identifying genetic effects that generalize across populations paves the way to ensure that eventual “personalized medicine” benefits will extend to understudied, underserved populations, and also nominates likely causal variants that exert effects despite differences in linkage disequilibrium and population history.58–60 Studying smoking genetics in diverse populations also can identify novel genetic associations due to enhanced power from population differences in smoking behaviors and allele frequencies.
This study is the first GWAS of smoking behaviors in a Hispanic/Latino cohort. We demonstrated that the involvement of CHRNA5 in smoking risk generalizes to Hispanic/Latino smokers. The non-synonymous, functional SNP rs16969968 and its correlates were associated with heavy smoking in HCHS/SOL. The dichotomous heavy/light trait provided stronger evidence for association at CHRNA5/A3/B4 than 4-level semi-quantitative CPD. Furthermore, in this Hispanic/Latino cohort we observed stronger association by defining heavy smokers as CPD > 10, rather than CPD > 20, when compared against light smokers with CPD ≤ 10. This strengthening of evidence with a lower threshold may reflect not only the larger number of cases, but also the differing distribution of CPD in Hispanics/Latinos, who tend to report lower CPD than European-ancestry smokers (Supplementary Figure S1). This finding may gain added relevance as the trend in smoking behavior of all US smokers continues towards lower CPD.3 In HCHS/SOL, the heavy/light trait was also associated with imputed variants in a region of chromosome 20 near HAOI. We did not pursue replication for this signal because no genotyped SNPs supported it and the associated imputed SNPs were all of low frequency, diminishing imputation reliability55,56 and replication power. These SNPs also lack validation of allele frequency estimates from other reference databases besides 1000 Genomes, as documented at dbSNP, www.ncbi.nlm.nih.gov/projects/SNP/.
Analyses of variants known to be associated in non-Hispanic populations supported association of heavy/light smoking with CHRNA4 in Hispanics/Latinos. Multiple SNPs attained p < .0001 in targeted tests of this gene, which was recently associated with nicotine dependence in European-ancestry smokers.10 We observed modest evidence (p < .05) in the CHRNAB3/A6 region and no evidence with CYP2A6, though this latter region is complex and difficult to query due to numerous duplications and deletions.
For the a priori loci at CHRNA5/A3/B4, CHRNB3/A6, and CHRNA4, our results for heavy/light smoking are in line with expectations from power calculations61–63 for this HCHS/SOL cohort of 1929 heavy smokers and 3156 light smokers. Assuming a stringent GWAS-significant threshold of alpha = 5 × 10–8, the predicted detection power would be 47% for rs16969968 and its correlates in CHRNA5/A3/B4, assuming an effect size of 1.3 as reported in non-Hispanic cohorts16–18 and a MAF of ~0.2 based on the 1000G AMR (Ad Mixed American) population; in contrast, predicted power to detect CHRNB3/A6 and CHRNA4 at this stringent alpha is negligible (<2%), assuming an effect size of 1.15 for both loci (reflecting non-Hispanic reports10,13), and MAF = 0.3 and MAF = 0.1 respectively (from 1000G AMR).61–63 Our findings are consistent with these predictions, as CHRNA5/A3/B4 was the only one of these three gene regions that attained p < 5 × 10–8. Moreover, using the relaxed alpha level of .05, the predicted power would be 93% for CHRNB3/A6 SNPs and 67% for CHRNA4 SNPs under the above assumptions; hence it is not surprising that both CHRNB3/A6 and CHRNA4 yielded confirmatory p values < .05.
This study presented strong but not conclusive evidence for two novel genetic loci influencing daily versus nondaily smoking. Nondaily smoking is more prevalent in Hispanic/Latino smokers than in European- and African Americans.28,30,31 In HCHS/SOL, we found genome-wide significant evidence that daily/nondaily smoking is associated with two loci, on chromosome 2 downstream of CCDC85A, and on chromosome 4 near ENPEP and PITX2. Some evidence suggests that the chromosome 2 region may be relevant to gene function in the brain. Median expression levels of CCDC85A are relatively high across brain tissues in data from the Genotype-Tissue Expression (GTEx) project (www.gtexportal.org, Supplementary Figure S8),64 and the lead SNP rs77876433 shows nominal evidence for association (uncorrected p = 9 × 10–3) as an eQTL of CCDC85A in caudate (basal ganglia) samples (N = 100) and significant evidence for association (p = 3.8 × 10–5, surpassing the false discovery rate threshold) with the overlapping transcript RP11-481J13.1 in a larger set of esophageal samples (N = 241). The chromosome 4 signal has less suggestive evidence for brain relevance in GTEx. ENPEP and PITX2 show low median expression levels across brain tissues (Supplementary Figures S9 and S10). The chr4:111494442 indel was not available in GTEx for testing as an eQTL; however the correlated SNP rs1562640 showed nominal association with ENPEP expression in hippocampus (p = .046) and hypothalamus (p = .045).
We were unable to replicate either of the daily/nondaily loci in a small sample of additional Hispanic/Latino smokers, or in independent samples of European- and African Americans. Lack of replication suggests that these signals may be false positives, or may possibly be true effects that are most detectable in Hispanic/Latinos and would require a larger cohort from this population to replicate. Prior to our replication analyses, we calculated that for common SNPs (MAF ~ 35% for the chromosome 2 locus), we could expect 80% power to detect an effect size of ~ 1.5 in a sample of 400 to 500 Hispanic/Latino smokers,61–63 comparable to the BioMe Hispanic/Latino sample size. However, differences in the distribution of national background and a lower frequency of nondaily smokers in BioMe compared to HCHS/SOL, together with the “winner’s curse” effect,65,66 likely reduced our replication power.
A better understanding of the health implications of nondaily versus daily smoking is needed to assess the impact of any genetic findings for this trait. Nondaily smoking appears to differ in its effects on some organs compared to daily smoking,37 and a study of Mexican smokers showed that nondaily smokers are more likely to quit than heavy daily smokers.67 Still, adverse health effects remain for smokers who are nondaily smokers.68 It remains unclear what health benefits might be conferred by reducing from daily to nondaily smoking, compared to actual cessation which greatly improves health outcomes.69–71
In closing, we emphasize that the influence of CHRNA5 variation on heavy smoking extends to Hispanic/Latino smokers, and this genetic association is most evident when accounting for the differing distribution of smoking in this population by using a lower CPD threshold to define heavy smokers. To translate this finding to patient care, it will be important to understand the impact of these genetic variants on smoking cessation and treatment in Hispanics/Latinos. Importantly, CHRNA5 variants have already been associated with cessation and efficacy of pharmacologic treatment in European-American smokers.72,73 Our evidence that CHRNA5 is associated with heavy smoking in Hispanic/Latino smokers suggests that cessation and pharmacogenetic effects may similarly generalize to this population. Genetics studies and clinical trials must include understudied populations such as Hispanics/Latinos to ensure that the benefits of precision medicine extend to all.
Supplementary Material
Supplementary data are available at Nicotine & Tobacco Research online.
Funding
The baseline examination of the Hispanic Community Health Study / Study of Latinos (HCHS/SOL) was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Genotyping efforts were supported by NHLBI HSN 26220/20054C, NCATS CTSI grant UL1TR000124, and NIDDK Diabetes Research Center (DRC) grant DK063491. The Mount Sinai BioMe Biobank is supported by The Andrea and Charles Bronfman Philanthropies and genotyping has been in part supported by a grant from NHGRI (HG007417). COGEND is supported by P01CA89392 from the National Cancer Institute; COGEND genotyping was funded by 1 X01 HG005274-01 and performed at Center for Inherited Disease Research (CIDR) which is funded through a federal contract from NIH to JHU (HHSN268200782096C). GeneSTAR was supported by grants (U01 HL72518 and HL087698) from the National Institutes of Health/National Heart, Lung, and Blood Institute and by a grant from the National Institutes of Health/National Center for Research Resources (M01-RR000052) to the Johns Hopkins General Clinical Research Center. We would like to acknowledge the following additional funding support. From the National Institute on Drug Abuse (NIDA): R01 DA026911; R01 DA036583; R01DA035825; R01 DA019963; R01 DA013423; R21 DA038241. From the National Institute on Aging: P30 AG15272. This research was supported in part by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Declaration of Interests
LJB is listed as an inventor on Issued US Patent 8080371 “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction, and served as a consultant for Pfizer in 2008. The spouse of NLS is also listed as an inventor on the above patent. The other authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the participants and staff of the HCHS/SOL study for their contributions to this study. This manuscript has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications. We thank Erin Rice for administrative and communications support. Disclaimer: The findings and conclusions in this article are those of the authors and do not necessarily represent the views or the official position(s) of the National Institutes of Health or any of the sponsoring organizations and agencies of the US government.
References
- 1. Jha P, Peto R. Global effects of smoking, of quitting, and of taxing tobacco. N Engl J Med. 2014;370(1):60–68. [DOI] [PubMed] [Google Scholar]
- 2. US Department of Health and Human Services, The Health Consequences of Smoking - 50 Years of Progress: a Report of the Surgeon General. Atlanta, GA: US Department of Health and Human Services, CDC; 2014. [Google Scholar]
- 3. Jamal A, Homa DM, O’Connor E et al. . Current cigarette smoking among adults - United States, 2005-2014. MMWR Morb Mortal Wkly Rep. 2015;64(44):1233–1240. [DOI] [PubMed] [Google Scholar]
- 4. Ward BW, Clarke TC, Nugent CN, Schiller JS,. Early release of selected estimates based on data from the 2015 National Health Interview Survey, National Center for Health Statistics; May 2016. www.cdc.gov/nchs/nhis.htm. Accessed November 7, 2016. [Google Scholar]
- 5. Ng M, Freeman MK, Fleming TD et al. . Smoking prevalence and cigarette consumption in 187 countries, 1980-2012. JAMA. 2014;311(2):183–192. [DOI] [PubMed] [Google Scholar]
- 6. Bilano V, Gilmour S, Moffiet T et al. . Global trends and projections for tobacco use, 1990-2025: an analysis of smoking indicators from the WHO Comprehensive Information Systems for Tobacco Control. Lancet. 2015;385(9972):966–976. [DOI] [PubMed] [Google Scholar]
- 7. Berrettini W, Yuan X, Tozzi F et al. . Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bierut LJ, Madden PA, Breslau N et al. . Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caporaso N, Gu F, Chatterjee N et al. . Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4(2):e4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hancock DB, Reginsson GW, Gaddis NC et al. . Genome-wide meta-analysis reveals common splice site acceptor variant in CHRNA4 associated with nicotine dependence. Transl Psychiatry. 2015;5:e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu JZ, Tozzi F, Waterworth DM et al. ; Wellcome Trust Case Control Consortium. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. The Tobacco and Genetics Consortium, Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thorgeirsson TE, Gudbjartsson DF, Surakka I et al. ; ENGAGE Consortium. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumasaka N, Aoki M, Okada Y et al. . Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PLoS One. 2012;7(9):e44507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. David SP, Hamidovic A, Chen GK et al. . Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saccone NL, Culverhouse RC, Schwantes-An TH et al. . Multiple Independent Loci at Chromosome 15q25.1 Affect Smoking Quantity: a Meta-Analysis and Comparison with Lung Cancer and COPD. PLoS Genet. 2010;6(8):e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saccone SF, Hinrichs AL, Saccone NL et al. . Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thorgeirsson TE, Geller F, Sulem P et al. . A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen LS, Saccone NL, Culverhouse RC et al. . Smoking and genetic risk variation across populations of European, Asian, and African American ancestry–a meta-analysis of chromosome 15q25. Genet Epidemiol. 2012;36(4):340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saccone NL, Wang JC, Breslau N et al. . The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69(17):6848–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bierut LJ, Stitzel JA, Wang JC et al. . Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165(9):1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) α5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (α4β2)₂α5 AChR function. Mol Pharmacol. 2011;79(1):119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frahm S, Slimak MA, Ferrarese L et al. . Aversion to nicotine is regulated by the balanced activity of β4 and α5 nicotinic receptor subunits in the medial habenula. Neuron. 2011;70(3):522–535. [DOI] [PubMed] [Google Scholar]
- 24. Pérez-Stable EJ, Herrera B, Jacob P III, Benowitz NL. Nicotine metabolism and intake in black and white smokers. JAMA. 1998;280(2):152–156. [DOI] [PubMed] [Google Scholar]
- 25. Ennis SR, Rios-Vargas M, Albert NG,. The Hispanic Population: 2010. Suitland, MD: E.a.S.A. U.S. Department of Commerce, U.S. Census Bureau, Editor; 2011. [Google Scholar]
- 26. Banda Y, Kvale MN, Hoffmann TJ et al. . Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jamal A, King BA, Neff LJ, Whitmill J, Babb SD, Graffunder CM. Current Cigarette Smoking Among Adults - United States, 2005-2015. MMWR Morb Mortal Wkly Rep. 2016;65(44):1205–1211. [DOI] [PubMed] [Google Scholar]
- 28. Kaplan RC, Bangdiwala SI, Barnhart JM et al. . Smoking among U.S. Hispanic/Latino adults: the Hispanic community health study/study of Latinos. Am J Prev Med. 2014;46(5):496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reyes-Guzman CM, Pfeiffer RM, Lubin J et al. . Determinants of light and intermittent smoking in the United States: results from three pooled National Health Surveys. Cancer Epidemiol Biomarkers Prev. 2017;26(2):228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodriquez EJ, Oh SS, Perez-Stable EJ, Schroeder SA. Changes in smoking intensity over time by birth cohort and by Latino national background, 1997–2014. Nicotine Tob Res. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trinidad DR, Pérez-Stable EJ, Emery SL, White MM, Grana RA, Messer KS. Intermittent and light daily smoking across racial/ethnic groups in the United States. Nicotine Tob Res. 2009;11(2):203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hassmiller KM, Warner KE, Mendez D, Levy DT, Romano E. Nondaily smokers: who are they?Am J Public Health. 2003;93(8):1321–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levy DE, Biener L, Rigotti NA. The natural history of light smokers: a population-based cohort study. Nicotine Tob Res. 2009;11(2):156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodriquez EJ, Stoecklin-Marois MT, Hennessy-Burt TE, Tancredi DJ, Schenker MB. Acculturation-related predictors of very light smoking among Latinos in California and nationwide. J Immigr Minor Health. 2015;17(1):181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilbert J. Tobacco and Shamanism in South America. New Haven, CT: Yale University Press; 1987. [Google Scholar]
- 36. Benowitz NL, Pérez-Stable EJ, Herrera B, Jacob P III. Slower metabolism and reduced intake of nicotine from cigarette smoking in Chinese-Americans. J Natl Cancer Inst. 2002;94(2):108–115. [DOI] [PubMed] [Google Scholar]
- 37. Franceschini N, Deng Y, Flessner MF et al. . Smoking patterns and chronic kidney disease in U.S. Hispanics: the Hispanic Community Health Study/ the Study of Latinos. Nephrology Dialysis Transplantation. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parrinello CM, Isasi CR, Xue X et al. . Risk of cigarette smoking initiation during adolescence among US-Born and Non-US-Born Hispanics/Latinos: the Hispanic Community Health Study/Study of Latinos. Am J Public Health. 2015;105(6):1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Conomos MP, Laurie CA, Stilp AM et al. . Genetic diversity and association studies in US Hispanic/Latino populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 2016;98(1):165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39(4):276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free estimation of recent genetic relatedness. Am J Hum Genet. 2016;98(1):127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laurie CC, Doheny KF, Mirel DB et al. ; GENEVA Investigators. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34(6):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gogarten SM, Bhangale T, Conomos MP et al. . GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28(24):3329–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. [DOI] [PubMed] [Google Scholar]
- 45. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abecasis GR, Auton A, Brooks LD et al. . An integrated map of genetic variation from 1,092 human genomes. Nature, 2012; 491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nelson SC, Stilp AM, Papanicolaou GJ et al. . Improved imputation accuracy in Hispanic/Latino populations with larger and more diverse reference panels: applications in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Hum Mol Genet. 2016;25(15):3245–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen H, Wang C, Conomos MP et al. . Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98(4):653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Conomos MP, Thornton T.. GENESIS: GENetic EStimation and Inference in Structured samples (GENESIS): Statistical methods for analyzing genetic data from samples with population structure and/or relatedness. R package version 2.4.0. 2016. [Google Scholar]
- 51. Pruim RJ, Welch RP, Sanna S et al. . LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tayo BO, Teil M, Tong L et al. . Genetic background of patients from a university medical center in Manhattan: implications for personalized medicine. PLoS One. 2011;6(5):e19166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Becker DM, Becker LC, Pearson TA, Fintel DJ, Levine DM, Kwiterovich PO. Risk factors in siblings of people with premature coronary heart disease. J Am Coll Cardiol. 1988;12(5):1273–1280. [DOI] [PubMed] [Google Scholar]
- 54. Vaidya D, Yanek LR, Moy TF, Pearson TA, Becker LC, Becker DM. Incidence of coronary artery disease in siblings of patients with premature coronary artery disease: 10 years of follow-up. Am J Cardiol. 2007;100(9):1410–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin P, Hartz SM, Zhang Z et al. ; COGA Collaborators COGEND Collaborators, GENEVA. A new statistic to evaluate imputation reliability. PLoS One. 2010;5(3):e9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ramnarine S, Zhang J, Chen LS et al. . When does choice of accuracy measure alter imputation accuracy assessments?PLoS One. 2015;10(10):e0137601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rice JP, Hartz SM, Agrawal A et al. ; GENEVA Consortium. CHRNB3 is more strongly associated with Fagerström test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107(11):2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saccone NL, Saccone SF, Goate AM et al. . In search of causal variants: refining disease association signals using cross-population contrasts. BMC Genet. 2008;9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teo YY, Ong RT, Sim X, Tai ES, Chia KS. Identifying candidate causal variants via trans-population fine-mapping. Genet Epidemiol. 2010;34(7):653–664. [DOI] [PubMed] [Google Scholar]
- 60. Zaitlen N, Paşaniuc B, Gur T, Ziv E, Halperin E. Leveraging genetic variability across populations for the identification of causal variants. Am J Hum Genet. 2010;86(1):23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155(5):478–484. [DOI] [PubMed] [Google Scholar]
- 62. Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21(1):35–50. [DOI] [PubMed] [Google Scholar]
- 63. Gauderman WJ, Morrison JM. QUANTO 1.2.3: A computer program for power and sample size calculations for genetic-epidemiology studies 2007. http://hydra.usc.edu/gxe. Accessed February 13, 2015.
- 64. Lonsdale J, Thomas J, Salvatore M et al. . The Genotype-Tissue Expression (GTEx) project. Nat Genet, 2013;45(6): 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhong H, Prentice RL. Correcting “winner’s curse” in odds ratios from genomewide association findings for major complex human diseases. Genet Epidemiol. 2010;34(1):78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zollner S, Pritchard JK. Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80(4):605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Swayampakala K, Thrasher J, Carpenter MJ, Shigematsu LM, Cupertio AP, Berg CJ. Level of cigarette consumption and quit behavior in a population of low-intensity smokers–longitudinal results from the International Tobacco Control (ITC) survey in Mexico. Addict Behav. 2013;38(4):1958–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schane RE, Ling PM, Glantz SA. Health effects of light and intermittent smoking: a review. Circulation. 2010;121(13):1518–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anthonisen NR, Connett JE, Murray RP. Smoking and lung function of Lung Health Study participants after 11 years. Am J Respir Crit Care Med. 2002;166(5):675–679. [DOI] [PubMed] [Google Scholar]
- 70. Doll R, Peto R. Cigarette smoking and bronchial carcinoma: dose and time relationships among regular smokers and lifelong non-smokers. J Epidemiol Community Health. 1978;32(4):303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doll R, Peto R, Boreham J, Sutherland I. Mortality in relation to smoking: 50 years’ observations on male British doctors. BMJ. 2004;328(7455):1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen LS, Baker TB, Piper ME et al. . Interplay of genetic risk factors (CHRNA5-CHRNA3-CHRNB4) and cessation treatments in smoking cessation success. Am J Psychiatry. 2012;169(7):735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen LS, Hung RJ, Baker T. et al. . CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis-a meta-analysis. J Natl Cancer Inst. 2015;107(5):djv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.