Abstract
The interaction between SStp, the transit peptide of the precursor protein to the small subunit of Rubisco (prSSU) and two Hsp70 molecular chaperones, Escherichia coli DnaK and pea (Pisum sativum) CSS1, was investigated in detail. Two statistical analyses were developed and used to investigate and predict regions of SStp recognized by DnaK. Both algorithms suggested that DnaK would have high affinity for the N terminus of SStp, moderate affinity for the central region, and low affinity for the C terminus. Furthermore, both algorithms predicted this affinity pattern for >75% of the transit peptides analyzed in the chloroplast transit peptide (CHLPEP) database. In vitro association between SStp and these Hsp70s was confirmed by three independent assays: limited trypsin resistance, ATPase stimulation, and native gel shift. Finally, synthetic peptides scanning the length of SStp and C-terminal deletion mutants of SStp were used to experimentally map the region of greatest DnaK affinity to the N terminus. CSS1 displayed a similar affinity for the N terminus of SStp. The major stromal Hsp70s affinity for the N terminus of SStp and other transit peptides supports a molecular motor model in which the chaperone functions as an ATP-dependent translocase, committing chloroplast precursor proteins to unidirectional movement across the envelope.
The semi-autonomous chloroplast, which contains its own genome, acquires the vast majority of its proteins as nuclear-encoded, larger Mr precursors synthesized in the cytosol and transported across the envelope membranes. These precursors contain an amino-terminal extension known as a transit peptide, which is both necessary and sufficient to direct the targeting and translocation of precursors with high fidelity (for review, see Bruce and Keegstra, 1994). Specifically, transit peptides enable the productive interaction of precursors with two distinct membrane protein complexes: components of the translocon at the outer membrane of the chloroplast (Toc) and components of the translocon at the inner membrane of the chloroplast (Tic) (Schnell et al., 1997). Recent progress has been made in identifying many of the individual components associated with Tic (Lubeck et al., 1996; Kouranov et al., 1998) and Toc (Hirsch et al., 1994; Kessler et al., 1994; Seedorf et al., 1995). However, with the exception of the Hsp70 homologs, none of the components identified to date is related to proteins identified in the other membrane translocation systems, such as bacterial secretion, mitochondria, and the endoplasmic reticulum (Schatz and Dobberstein, 1996).
In contrast, much less progress has been reported on the molecular analysis of the functional properties of the transit peptide itself. Despite >250 transit peptides sequences in the CHLPEP database (von Heijne et al., 1991) and hundreds of more recently identified transit peptides, few in-depth structural or functional analyses of these sequences are reported (Lancelin et al., 1994; Krimm et al., 1999; Wienk et al., 1999). However, arguments have recently been made to suggest that transit peptides are modular, with discrete domains providing different functional roles. Although early sequence analysis suggested the existence of three semi-conserved domains (Karlin Neumann and Tobin, 1986), only recent work combining both in vitro (Pilon et al., 1995; Pinnaduwage and Bruce, 1996; Bruce, 1998) and in vivo (Kindle, 1998; Rensink et al., 1998) approaches demonstrates that different regions of the transit peptide perform different functions in the import process.
Although these analyses have only been performed for a few transit peptides, the emerging consensus is that transit peptides contain three functional domains. The N-terminal domain appears to perform an essential, as-yet-undefined role in the initiation and commitment of the precursor to translocation; the central region is more dispensable, functioning as a flexible hinge region between the N and C termini, and, finally, the C terminus may be involved both in lipid interaction and in correct processing by the stromal processing peptidase. An obvious problem with this modular organization is that transit peptides vary widely in length and share very limited sequence homology. This sequence degeneracy is particularly difficult to explain in light of the essential role that the N terminus performs in chloroplast import (Pilon et al., 1995; Kindle, 1998). Therefore, either a common, as-yet-unknown secondary structure or the involvement of an interaction that intrinsically requires low sequence specificity would provide the best hypothesis for the mechanism of transit peptide function.
Most of the current models of protein translocation include a peripherally attached molecular motor (Schatz and Dobberstein, 1996). In the mitochondria and ER, this motor is believed to be a Hsp70 molecular chaperone. The involvement of Hsp70 as the molecular motor assumes a direct interaction between the incoming precursor and the peptide binding domain of the molecular chaperone, and most current models show the targeting sequence as the region of the precursor that is recognized by the chaperone (Gray and Row, 1995; Keegstra et al., 1995; Heins et al., 1998). Indeed, a recent study shows significant interaction between mitochondrial presequences and DnaK (Zhang et al., 1999). Consistent with these models, chloroplast transit peptides have been suggested to serve as substrates for Hsp70 chaperones (von Heijne and Nishikawa, 1991). Although this proposal is supported by statistical analyses indicating that transit peptides are enriched in sequences predicted to exist as random coils, to date only one report has demonstrated a direct interaction between a transit peptide and Hsp70 (Ivey and Bruce, 2000). In any event, no clear agreement exists for the involvement of Hsp70s in the chloroplast protein import process (Soll and Waegemann, 1992; Nielsen et al., 1997).
In this report we have investigated the chloroplast transit peptide sequences that enable it to function as a substrate for the Hsp70 class of molecular chaperones. Based on the results of two independent statistical algorithms that calculate an index of DnaK affinity, we predict that SStp, the transit peptide for the precursor protein to the small subunit of Rubisco (prSSU) contains two regions that should be recognized by DnaK. When these algorithms are applied to the transit peptides in the CHLPEP database, >95% of the transit peptides analyzed contained at least one potential DnaK recognition domain; furthermore, these sequences occurred predominantly at the N terminus of the transit peptide. We have verified this interaction for SStp by three in vitro assays: a proteolysis protection assay with DnaK, substrate stimulation of ATPase activity with CSS1, and a native gel shift assay with both Hsp70s. Finally, we have confirmed the algorithms' predictions by partially mapping the transit peptide regions responsible for DnaK/CSS1 interaction using both co-affinity precipitation of DnaK and the in vitro native gel shift assay. The results of these observations are discussed in the context of both transit peptide design and the potential involvement of Hsp70s as the chloroplast translocation molecular motor.
MATERIALS AND METHODS
Predictive DnaK Affinity Algorithms
Phage Display Based Algorithm
In work by Gragerov et al. (1994), an index of each amino acid's appearance in high-affinity versus low-affinity peptides in a random peptide phase display (RPPD) library was calculated. For example, the value determined for Leu was 2, reflecting the ratio of frequency of occurrence of Leu found in selected versus unselected phage. Using these values, we developed a simple algorithm using a sliding six-amino acid window in one-amino acid steps to predict DnaK's affinity to transit peptides in the CHLPEP database. For each window, we multiplied the indices of six adjacent amino acids in SStp:
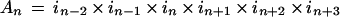 |
where An is an index of the six-amino acid window's affinity for DnaK, in is the third residue in the window, in+1 is the fourth residue, and so on. Met, Cys, and Glu were statistically underrepresented in the display library, so we assigned them values of 1 for equal probability of being in a strongly or weakly selected peptide. Based on values obtained when the algorithm was applied to several peptides described in the original study, we designated a “cut-off” index value of 2.0. Therefore, a six-amino acid window with an index value greater than 2.0 is predicted to have an affinity to bind DnaK that is similar to a strongly selected peptide from the original study.
Cellulose Display Based Algorithm
A second, more recent report utilized 3,725 synthetic, cellulose-bound peptides (13 mers) that span the length of 37 naturally occurring proteins to develop a cellulose-bound peptide scanning (CBPS) algorithm for DnaK affinity (Rudiger et al., 1997). In this study, each amino acid is assigned a Δ Δ Go value that reflects the change in free energy of the DnaK/peptide complex when that amino acid is present. Because of the apparent preferences of different amino acids to accommodate different positions relative to the center of the DnaK peptide-binding pocket, three Δ Δ Go values are assigned to each amino acid. As described in Rudiger et al. (1997):
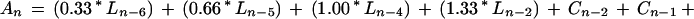 |
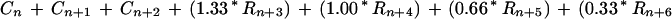 |
where An is an index of the 13 amino acid window's affinity for DnaK, n describes the amino acid's position relative to the center of DnaK's binding site, and L, C, and R are experimentally derived values for each amino acid left-of-center, center, and right-of-center, respectively. The weighting values (0.33, 0.66, 1.00, and 1.33) were statistically determined by Rudiger et al. (1997) to maximize the accuracy of the algorithm. We applied this algorithm via a sliding 13-amino acid window in one-amino acid steps to predict DnaK's affinity to transit peptides in the CHLPEP database. Based on values obtained when the algorithm was applied to two peptides described in the original study, we designated a “cut-off” index value of −4.0. Therefore, a 13-amino acid window with a Δ Δ Go value less than −4.0 is predicted to bind DnaK similarly to a strongly selected peptide from the original study.
SStp Fusion Proteins
SStp fusions with glutathione S-transferase (GST) (pGEX vector, Pharmacia Biotech, Piscataway, NJ) and the dual His affinity tag fused to the RNase S peptide epitope tag (His-S) (pET vector, Novagen, Madison, WI) as well as DnaK were expressed and purified as described previously (Ivey and Bruce, 2000). SStp derived from the His-S Tag system was used for all in vitro analyses involving purified components.
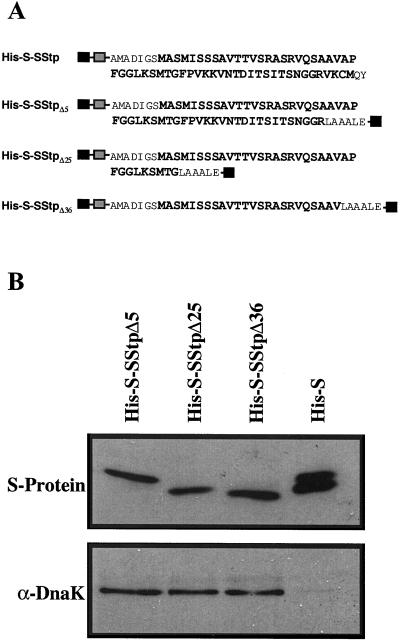
C-terminal deletions of His-S-SStp were generated via Exonuclease III digestion using the Erase-a-Base kit (Promega, Madison, WI). pET30a-SStp was linearized with HindIII, and the 5′ overhangs were filled in with α-phosphorylthiolates to generate blunt, exonuclease-resistant ends. This linear plasmid was then restricted with EcoRI to generate a single 5′ overhang for ExoIII digestion. The exonuclease reaction was stopped at timed intervals, and the samples were treated with S1 nuclease and Klenow fragment to generate blunt, ligatable ends. The mixed plasmid species were recircularized, transformed into Escherichia coli cells, and screened using direct colony PCR with the forward and reverse T7 promoter primers. Isolated DNA from appropriate transformants was sequenced using an automated sequencer (PE-Applied Biosystems, Foster City, CA). Three deletion mutants—His-S-SStpΔ5, His-S-SStpΔ25, and His-S-SStpΔ36—lacking 5, 25, and 36 amino acids, respectively, from the C terminus of the full-length His-S-SStp were selected for use in these studies. Coincidentally, all of the deletions were in frame with the optional, C-terminal His-Tag engineered into the pET30a vector.
Limited Trypsin Proteolysis of DnaK
DnaK was subjected to trypsin digestion (5 ng/mg Hsp70), as in Freeman et al. (1995) in 50-μL reactions for 60 min at 37°C in buffer C (20 mm Tris-HCl, pH 6.9, 100 mm EDTA, and 100 mm NaCl). Subsequent assays also contained α-lactalbumin, reduced, carboxymethylated α-lactalbumin (RCMLA), or SStp as potential DnaK substrates. Aliquots were removed during the course of the digestion and immediately boiled in SDS sample buffer (100 mm Tris-HCl, pH 6.8 containing 10% [v/v] glycerol, 0.04% [w/v] bromphenol blue, and 0.1% [w/v] SDS). The samples were run on SDS-PAGE, with protein visualization by Coomassie Brilliant Blue staining. Quantitative scanning densitometry was performed using a computing densitometer (model 3000, Molecular Dynamics, Sunnyvale, CA).
Purification of Native CSS1
CSS1 was purified from 14-d-old pea (Pisum sativum) cotyledons applying an affinity chromatography method similar to that used to purify Bovine taurus Hsc70 (Schlossman et al., 1984). Fractionated stroma from pea was prepared as described previously (Bruce et al., 1994) and diluted 1:10 with buffer M (20 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.5, 20 mm NaCl, 2.5 mm Mg[OAc]2, 1 mm dithiothreitol [DTT], and 0.1% [v/v] Triton X-100). Next, the entire sample was loaded onto an ATP-agarose column (Sigma-Aldrich, St. Louis) pre-equilibrated with buffer M at 4°C. The column was washed exhaustively with buffer M, then buffer M containing 1 m NaCl, and finally buffer M again. CSS1 was then eluted with 10 mm ATP in buffer M titrated to pH 7.5. Authenticity of CSS1 was demonstrated via cross-reactivity on a western blot probing with a polyclonal DnaK antiserum. Eluted fractions containing CSS1 were dialyzed exhaustively and concentrated into buffer M by ultrafiltration against a 30-kD molecular mass cutoff membrane, aliquoted, and stored at −85°C.
CSS1 ATPase Activity
ATPase activity assays were performed with [γ-32P]ATP in buffer M as previously described (Sadis and Hightower, 1992). Each 50-μL reaction contained CSS1, unlabeled ATP, and [γ-32P]ATP, and was incubated at 37°C. Peptide substrates were provided at 100-fold molar excess relative to the chaperone. Aliquots of the reaction mixture were withdrawn at regular intervals and mixed with 1 mL of 50 mm HCl/5 mm H3PO4 containing 7% (w/v) activated charcoal. After microcentrifugation, 200-μL aliquots of the free 32Pi-containing supernatants were removed and counted via scintillation. Observation of spontaneous ATP hydrolysis was necessary because Hsp70 ATPase activities are extremely low.
Co-Precipitation of DnaK with Ni-Sepharose
His-S-SStp and His-S-SStp truncation mutants were expressed in E. coli as described previously for His-S-SStp (Ivey and Bruce, 2000). Small cell cultures (10 mL) were grown and induced normally but lysed by sonication in native lysis buffer (Novagen). The crude lysates were spun at 16,000g; then each of the supernatants was added to microfuge tubes containing 100 μL of Ni-Sepharose (Pharmacia) and mixed gently for 5 min at 4°C. After washing the Ni-Sepharose in batch three times with 1 mL of the same lysis buffer, SDS sample buffer was added directly to the Ni-Sepharose to elute the bound proteins. After SDS-PAGE and electroblotting onto polyvinylidene difluoride membrane, the blot was divided laterally. The upper half was probed with α-DnaK antiserum, the lower half with S-protein conjugated to alkaline phosphatase.
Native Gel Shift Competition Assay with 125I-RCMLA
DnaK and CSS1 binding competition studies with 125I-RCMLA were performed as described by Freeman and co-workers (Freeman et al., 1995). Hsp70 was incubated with 125I-RCMLA and the competing peptide/protein for 30 min at 37°C in buffer A. Native sample buffer (100 mm Tris-HCl, pH 6.8, containing 10% [v/v] glycerol and 0.04% [w/v] bromphenol blue) was added and the samples were resolved by electrophoresis on a 6% native acrylamide gel. The proteins were then fixed with acetic acid, the gels dried, and autoradiograms were developed. Quantitative scanning densitometry was performed with a computing densitometer (model 3000 Series, Molecular Dynamics, Sunnyvale, CA).
RESULTS
The N Terminus of Transit Peptides Is Predicted to Interact with DnaK
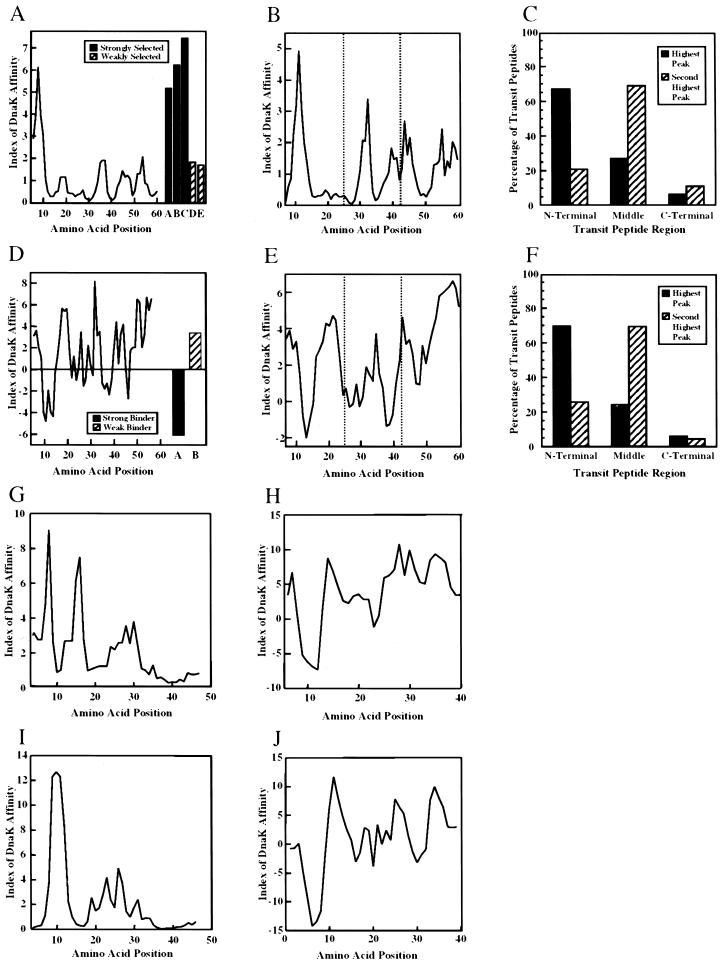
To investigate more precise region(s) within SStp responsible for chaperone association, we used previously published data in two novel algorithms, the first of which was derived from a RPPD analysis (Gragerov et al., 1994). The second algorithm was obtained from data in a CBPS analysis (Rudiger et al., 1997). The RPPD and CBPS algorithms calculate a DnaK affinity index for a sliding window of six and 13 amino acids in length, respectively. Analysis of the entire SStp sequence by both algorithms implicated the same regions as those interacting with DnaK (Fig. 1, A and D). To compare the algorithms' predictive indices against the experimentally determined DnaK binding activity to given peptide substrates, five peptides reported from the RPPD library and two from the CBPS study were analyzed with their respective algorithms (Fig. 1, A and D). In both cases, the “control” peptides defined experimental limits for DnaK affinity. Utilizing the RPPD algorithm, an index value greater than 2.0 should be strongly selected by DnaK; for the CBPS algorithm, which describes free energy changes, high-affinity regions have index values less than −4.0. Based on these criteria, DnaK is predicted to exhibit a strong association for the region of SStp centered at amino acid position 10, and may interact with a second site centered at position 37. Interestingly, these two regions of SStp align well with one another when calculated from either algorithm.
Figure 1.
Predictive DnaK affinity algorithms. A through C utilize data found in the RPPD study (Gragerov et al., 1994); D through F utilize data found in the CBPS study (Rudiger et al., 1997). A and D, DnaK affinity algorithms (see “Materials and Methods” for details) applied to SStp from pea. Also shown in right of A is the analysis of five peptides reported from the original study. The black bars indicate values obtained using peptides from the original phage display study that were “strongly selected”: A, NRLLLT; B, ARLLLT; and C, NRLLLA. The hatched bars indicate values obtained using “weakly selected” peptides: D, KWVHLF and E, LLTNRG. In the right of D are values from two peptides from the original study: AKTLILSHLRFVV, a strongly selected peptide, and VVHIARNYAGYG, a weakly selected peptide. B and E, The algorithms (Legend continues on facing page.)applied to all angiosperm prSSU transit peptides in the CHLPEP database. The average values at each position in the sequence are plotted. C and F, Distribution by thirds of predicted highest and second highest affinity regions in 115 angiosperm, stromally targeted transit peptides in the CHLPEP database. G and I, RPPD analyses of nitrite reductase and Fru-1,6-bisphosphatase, respectively. H and J, CBPS analyses of nitrite reductase and Fru-1,6-bisphosphatase, respectively.
The profile of pea SStp shown in Figure 1, A and D, is typical of prSSU transit peptides from other organisms (data not shown) and transit peptides of other stromally targeted precursors. Figure 1, B and E, represents the average of all angiosperm prSSU transit peptides found in the CHLPEP database (von Heijne et al., 1991). Again, both algorithms strongly agree that these transit peptides display a major peak of predicted DnaK affinity at the N terminus and exhibit a similar periodicity of successive peaks whose affinity for DnaK diminishes toward the C-terminal cleavage site. The apparent small peak values, especially in Figure 1E, reflect averaging of “misaligned” peaks.
Furthermore, when these analyses were performed for 115 transit peptides from stromally localized angiosperm precursors in the CHLPEP database, the domain with the highest predicted DnaK affinity occurred in the N-terminal region of approximately 70% of the transit peptides analyzed (Fig. 1, C and F). Moreover, both algorithms predicted that approximately 70% of the transit peptides contained a second peak of lower strength positioned in the central region. Finally, only approximately 5% of the peptides contained a prominent peak in the C-terminal region, indicating that this domain was largely devoid of sequences that would function as good substrates for DnaK recognition. Both algorithms predicted at least one high-affinity site for >95% of transit peptides tested and were in good alignment agreement approximately 80% of the time (data not shown). Two unrelated transit peptides from precursors for nitrite reductase from spinach (Fig. 1, G and H) and Fru-1,6-bisphosphatase from pea (Fig. 1, I and J) also show DnaK affinity profiles similar to that of prSSU using both algorithms.
SStp Association with DnaK Provides Protease Protection
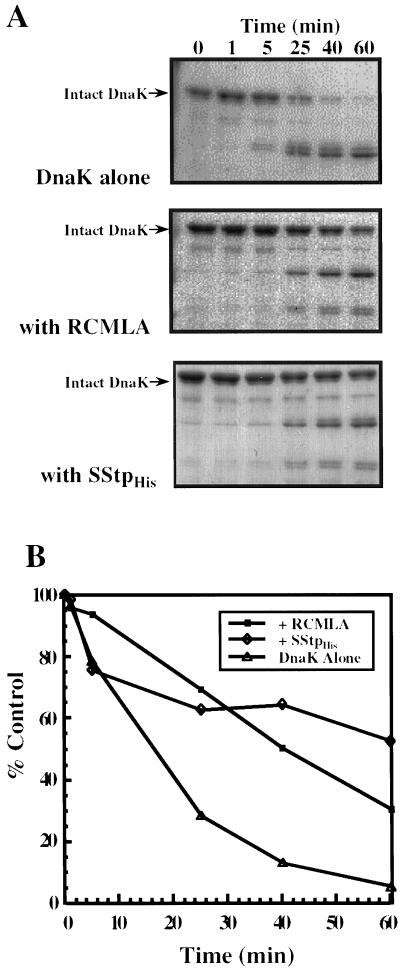
To experimentally assess the validity of the algorithms' predictions, three in vitro assays were performed that explored the consequences of an Hsp70/SStp interaction. Previous studies have shown that Hsp70s undergo a change in conformation upon binding to a peptide substrate such that the substrate-bound form is more resistant to trypsin digestion (Cyr et al., 1992; Palleros et al., 1992; Freeman et al., 1995). Using the components purified above, we investigated whether a similar interaction could be demonstrated in vitro by employing the limited trypsin proteolysis technique. RCMLA, a permanently unfolded protein and model Hsp70 substrate, and SStp were also utilized to protect DnaK from trypsin degradation. When incubated with trypsin alone, DnaK was readily digested, yielding a 43-kD fragment (Fig. 2A, top). This fragment probably corresponded to the N-terminal ATPase domain of DnaK (DeLuca Flaherty et al., 1988). However, in the presence of 10-fold molar excesses of RCMLA (Fig. 2A, middle) or SStp (Fig. 2A, bottom), DnaK was protected substantially from proteolysis. As previously shown (Freeman et al., 1995), this protection was not simply the result of unfolded substrates competing for the protease, because the control substrate, native α-lactalbumin, failed to protect DnaK from trypsin digestion (data not shown). The amount of intact DnaK remaining at each time point was quantitated via scanning densitometry and plotted in Figure 2B. Whereas approximately 40% to 60% of the DnaK remained intact after 1 h of trypsin digestion in the presence of RCMLA or SStp, DnaK alone was almost completely digested (<5% remaining) by trypsin in the same time frame. These results support previous observations that in vitro binding of a substrate, either RCMLA or SStp, induces a significant, substrate-dependent conformational change, rendering the DnaK much more resistant to trypsin digestion. These results confirm that SStp contains at least one sequence that serves as a good substrate for Hsp70 binding, as was predicted from earlier secondary structural analyses (von Heijne and Nishikawa, 1991).
Figure 2.
Substrate protection of DnaK from trypsin degradation. A, 0.7 μm DnaK was treated with trypsin after a 5-min incubation in the presence of no unfolded protein substrate (top); 7 μm RCMLA (middle); and 7 μm SStp (bottom). Aliquots were removed at the given times and immediately boiled in SDS sample buffer. All samples were examined by SDS-PAGE and stained with Coomassie Brilliant Blue. B, Quantitation of the amount of intact DnaK remaining at each time point in A for each treatment.
SStp Stimulates the ATPase Activity of CSS1
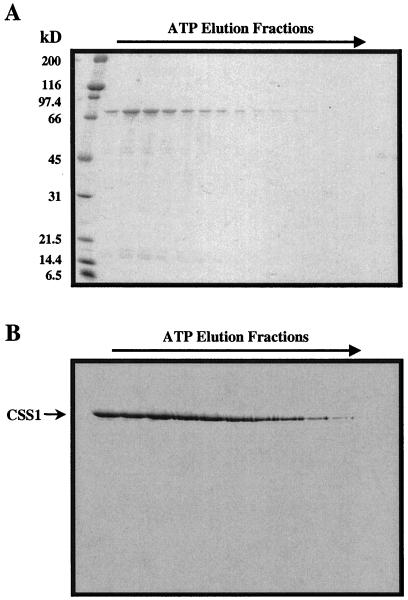
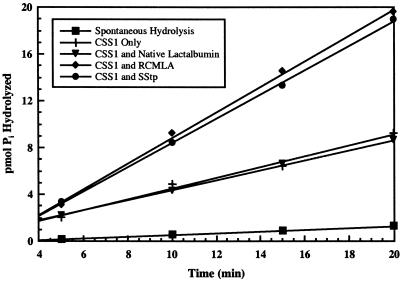
A second indicator of a protein or peptide's interaction with Hsp70/DnaK as a substrate is the stimulation of the intrinsic ATPase activity of the chaperone (Sadis and Hightower, 1992; Ziegelhoffer et al., 1995). We used this assay to evaluate the ability of both RCMLA and full-length SStp to stimulate the intrinsic ATPase activity of purified CSS1. First, CSS1 was purified from intact pea chloroplasts via ATP affinity chromatography. Figure 3A shows the ATP elution profile from the ATP-Sepharose affinity column. Western blotting indicated that the major band at 73 kD was CSS1 (Fig. 3B). Subsequent CSS1 ATPase assay results, as shown in Figure 4, showed a basal activity of approximately 2.6 pmol ATP min−1 pmol−1 enzyme. This rate was stimulated to approximately 5.1 pmol ATP min−1 pmol−1 enzyme when a 100-fold molar excess of either RCMLA or SStp was added to the reaction. This effect was dependent on the specific interaction of the chaperone with a substrate, since an equal addition of native α-lactalbumin did not result in ATPase stimulation. Both the substrate levels required and the level of stimulation observed were quite similar to those observed for DnaK (Liberek et al., 1991).
Figure 3.
Biochemical purification of CSS1 from pea. A, Coomassie Brilliant Blue-stained SDS gel of the elution profile from an ATP-agarose column after incubation with 10 mm ATP. B, Western blot of the fractions in A probing with α-DnaK.
Figure 4.
SStp stimulation of CSS1 ATPase activity. CSS1 (0.7 μm) was incubated with 50 μm cold ATP and [γ-32P]ATP (3,000 Ci/mmol; molar ratio of unlabeled:32P-labeled, 5,250:1) in the absence and presence of 70 μm native α-lactalbumin, RCMLA, or SStp. Spontaneous hydrolysis indicates liberated 32Pi in absence of chaperone.
Mapping of SStp Region Recognized by Dnak in E. coli
The in vivo association of SStp and DnaK described by Ivey and Bruce (2000) was used to experimentally map the region of SStp responsible for DnaK binding and provide support for the algorithms' predictions. First, C-terminal deletion mutants of SStp fused to the dual His-S Tag (Fig. 5A) were expressed in E. coli. Then, co-affinity precipitations of DnaK using Ni-Sepharose were performed for each deletion and the His-S Tag alone. Figure 5B, top, shows equal loadings of the three deletions and the His-S Tag visualized by far-western blotting. The bottom panel is a western blot of the same samples showing DnaK binding to every deletion containing at least part of the SStp sequence. However, DnaK did not bind to the His-S Tag itself, which serves as a negative control. Therefore, sequences in the first 24 residues of SStp must contain a DnaK recognition motif allowing this interaction in vivo. Because the experiment did not include N-terminal SStp deletions, these data do not exclude the possibility of other DnaK-binding sites C-terminal to amino acid position 24.
Figure 5.
Affinity precipitations of DnaK with C-terminal deletions of SStp. A, Amino acid sequences of His-S-SStp and C-terminal deletions. Residues in bold correspond to SStp. Black boxes indicate the N- and C-terminal His-tags, while gray boxes indicate N-terminal S-tags. B, Top, Far-western analysis of His-S-SStp deletions and the His-S tag alone using the S-protein conjugated to alkaline phosphatase; bottom, western blot of the samples in the top using α-DnaK antiserum.
In Vitro Interaction of DnaK and CSS1 with the N Terminus of SStp
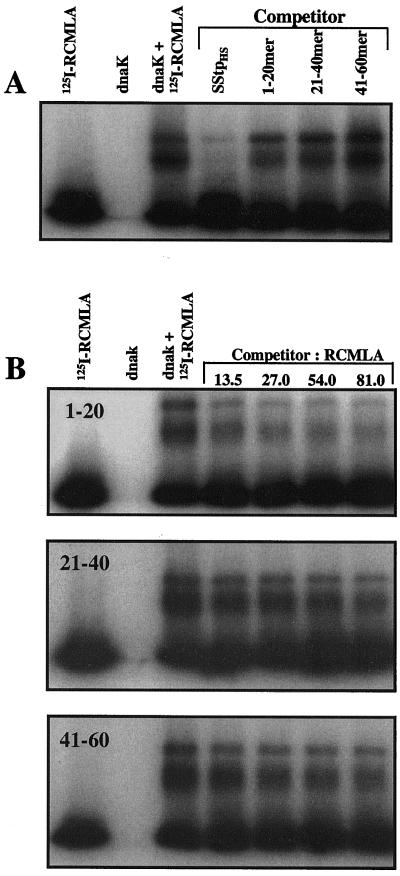
To further test the algorithms' predictions for SStp, a competitive native gel shift assay was used. Incubation of 125I-RCMLA with purified DnaK results in the formation of at least two stable complexes whose electrophoretic mobilities on native polyacrylamide gels are retarded relative to 125I-RCMLA alone (Fig. 6A, lanes 1–3). The two discrete complexes may represent 125I-RCMLA associated with a monomeric and a dimeric form of DnaK, since several Hsp70s exist in an equilibrium between a monomeric and dimeric species (Palleros et al., 1991; Azem et al., 1997). To determine the relative affinity of DnaK for different peptide substrates, synthetic, unlabeled peptides were added to the 125I-RCMLA-DnaK incubation as competitors for binding to 125I-RCMLA DnaK binding. Competitive association between the unlabeled peptide and DnaK displaces 125I-RCMLA, thus reducing the amount of 125I-RCMLA-DnaK complexes. Figure 6A shows an autoradiogram of a competitive gel shift assay in which 125I-RCMLA-DnaK complexes formed in the presence of a 10-fold molar excess of SStp or synthetic 20 amino acid peptides spanning the SStp sequence. SStp effectively competed with 125I-RCMLA for DnaK binding, as previously shown (Ivey and Bruce, 2000). Synthetic peptides (20-mers) corresponding to the N-terminal (1–20), middle (21–40), and C-terminal (41–60) thirds of SStp clearly competed with an activity very similar to their predicted values of DnaK affinity (Fig. 1, A and D). This difference in activity was confirmed when the unlabeled SStp 20-mers were titrated relative to 125I-RCMLA (Fig. 6B). The N-terminal sequences (peptide 1–20) competed best with 125I-RCMLA for DnaK, whereas the central region (peptide 21–40) was considerably less effective, and the C-terminal region (peptide 41–60) displaced 125I-RCMLA even less than the central region (Fig. 6B).
Figure 6.
Mapping the high-affinity DnaK binding site(s) within SStp. A, Autoradiogram demonstrating 125I-RCMLA/DnaK complex stability and subsequent competition by unlabeled competitors in a native gel system. DnaK was added to a final concentration of 0.7 μm, while the concentrations of 125I-RCMLA and SStp were 7 and 70 μm, respectively. B, Autoradiograms of 125I-RCMLA/DnaK complexes in the presence of the N-terminal (top), middle (middle panel), and C-terminal (bottom) thirds of SStp from pea. The DnaK and 125I-RCMLA concentrations were the same as in A. Molar ratios of the unlabeled competitor to 125I-RCMLA are shown in lanes 5 through 8.
In previous work, three peptides of similar length, which are not predicted by either algorithm to bind DnaK and are thus clear negative controls, were tested and failed to disrupt the 125I-RCMLA-DnaK complex (Ivey and Bruce, 2000). Thus, SStp bound DnaK with higher affinity than any of the 20-mers, but the greatest local affinity for DnaK was detected in the N-terminal 20 amino acids, and progressively less affinity was found toward the middle and C-terminal thirds of SStp.
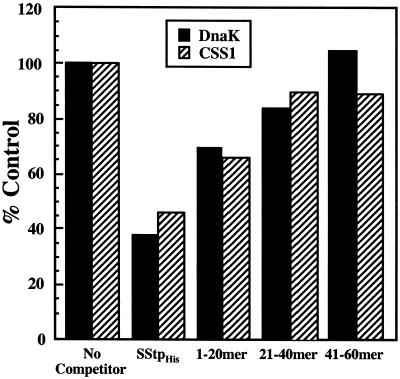
Thus far, we have shown that SStp functions in vivo and in vitro as an effective substrate for DnaK. The major chloroplast Hsp70 homolog, CSS1, is most similar to prokaryotic homologs of DnaK, with 55% amino acid sequence identity and 74% similarity (Marshall and Keegstra, 1992). Therefore, one would predict similar peptide binding properties for both chaperones. Using purified CSS1 instead of DnaK, competitive gel shift assays were performed as described above. The results for both DnaK and CSS1 are represented graphically in Figure 7. The trend observed for DnaK binding to SStp was almost identical to that observed for CSS1, suggesting that both chaperones preferred sequences found at the N-terminal region of SStp.
Figure 7.
Comparison of SStp binding by DnaK and CSS1. Native gel shift competition assays were used to compare the binding of SStp by DnaK and CSS1 and to map the highest affinity site for both Hsp70s. The black bars were quantified from the autoradiogram in Figure 6A. Using the same 125I-RCMLA, chaperone, and competitor concentrations as before, the hatched bars were obtained similarly using CSS1 instead of DnaK.
DISCUSSION
Although several recent studies have attempted to identify the linear peptide sequences recognized by Hsp70s (Blond-Elguindi et al., 1993; Gragerov et al., 1994; Rudiger et al., 1997), only one has directly analyzed targeting sequences (Zhang et al., 1999); that study also used the same CBPS algorithm used in this study to predict interactions between mitochondrial presequences and DnaK. However, the current study demonstrates, for the first time to our knowledge, a direct interaction between an organellar molecular chaperone and a physiologically relevant precursor targeting sequence. Specifically, our data confirm that the transit peptide of prSSU contains one or more sequences that are recognized by two Hsp70 chaperones, DnaK and CSS1.
Transit Peptide Design
Both the x-ray crystal structure of the DnaK peptide-binding domain and peptide binding studies of a eukaryotic Hsp70 indicate a preference for binding substrates six to eight amino acids in length (Flynn et al., 1991; Zhu et al., 1996). Therefore, the full-length SStp may contain up to seven contiguous Dnak/CSS1 binding sites. To identify the number and position of potential DnaK binding sites in SStp, we utilized data from two extensive studies (Gragerov et al., 1994; Rudiger et al., 1997), which provide statistical data on the probability of individual amino acids to occur in DnaK selected peptides. Both algorithms predict potential DnaK binding sites in SStp. Moreover, both algorithms predicted that the strongest peptide-DnaK interactions were restricted primarily to the N terminus. When these analyses were applied to all prSSU transit peptides from angiosperms in the CHLPEP database (von Heijne et al., 1991), N-terminal bias was observed by both algorithms. This suggests that the Hsp70 binding site(s) at the N terminus of prSSU is a conserved trait, independent of phylogeny.
Although many stromal proteins, such as the precursors to Fru-1,6-bisphosphatase from pea and nitrite reductase from spinach, have patterns very similar to SStp, analyses of 115 angiosperm transit peptides for stromally localized precursors indicate that the placement of high-affinity binding sites is not absolutely conserved. However, both algorithms indicate that >95% of these transit peptides contain at least one potential Hsp70 recognition domain and that approximately 70% of these transit peptides contain sequences at their N terminus predicted to most favorably interact with Hsp70s. In addition, both analyses indicate additional site(s) with lower affinity found within the central third of the transit peptide and that, for most transit peptides, the C-terminal third is largely devoid of Hsp70 binding sites.
These predictions were confirmed experimentally for SStp. The trypsin resistance and the ATPase stimulation data reported here support previous observations that in vitro binding of a substrate, either RCMLA or SStp, induces a significant substrate-dependent conformational change. Binding of the peptide substrate renders DnaK more resistant to trypsin digestion and more active as a ATPase. These results confirm that SStp contains at least one sequence that serves as a good substrate for Hsp70s, as was predicted from earlier secondary structural analyses (von Heijne and Nishikawa, 1991).
These results suggest that chloroplast transit peptides have one or more regions that may function as a high-affinity substrate for Hsp70s. A shared peptide sequence preference among chaperones is expected, since a recent study with three Hsp70 homologs (Hsc70, BiP, and DnaK) shows several common peptide binding tendencies (Fourie et al., 1994). Interestingly, the full-length transit peptide appeared to display greater interaction than any of the 20-mers. These data argue strongly that CSS1-peptide substrate interactions are generally governed by the same rules as those for DnaK.
Our observation that CSS1 exhibits high affinity for the N terminus of transit peptides and diminishing affinity toward the C terminus has several important implications. First, if the translocation proceeds with the N terminus first, as most models depict, the suggested design would enable the emerging transit peptide to encounter the chaperone at the earliest possible point in the translocation process. This initial interaction may represent the first committed step of protein translocation. Second, most transit peptides may contain additional secondary Hsp70-binding sites that would enable multiple chaperone molecules to concurrently bind the precursor and drive translocation. The observation that full-length SStp was a better substrate than any of the 20-mers suggests that multiple potential binding sites may promote some level of cooperativity, possibly by interaction with both substrate binding domains of the DnaK dimer. This multiplicity of binding sites could be the reason that certain precursors are translocated more efficiently. Third, these observations may confirm the hypothesis of a modular organization of transit peptides.
Chloroplast Hsp70s as Molecular Motors and “Unfoldases”
The most widely accepted generic protein import model describes an active, energy-dependent “molecular motor” model that is bound to the membrane and directly utilizes a conformational change in the chaperone to unidirectionally drive translocation (Glick, 1995; Gisler et al., 1998). Our data directly support the hypothesis that an individual transit peptide contains one or more Hsp70 recognition domains, enabling a precursor to simultaneously engage more than one chaperone molecule concurrently during translocation. Analyses of prSSU transit peptides in CHLPEP indicate at least two such sites. Interestingly, the two sites are separated by approximately 26 amino acids, which would be sufficient to span a bilayer. Therefore, this spacing could allow one transit peptide to simultaneously engage two chaperones on either side of a membrane. If both high-affinity sites are active in recruiting Hsp70s, the ability of SStp to interact concurrently with two Hsp70 molecules may synergistically promote a much stronger “unfoldase” activity than might be expected from the sum of their individual contributions. Differences in amino acid sequences of transit peptides from different precursors could then affect their translocation efficacies, which would constitute a novel form of post-transcriptional/post-translational regulation of gene expression.
Although most investigators agree that isolated translocation complexes contain one or more Hsp70s (Waegemann and Soll, 1991; Schnell et al., 1994), recent reports also describe a second potential molecular chaperone, ClpC, which is associated with the translocation complex (Akita et al., 1997; Nielsen et al., 1997). However, no evidence of transit peptide or precursor binding has been presented, and the presence of ClpC in a translocation complex is independent of the presence of a precursor protein (Nielsen et al., 1997). Furthermore, organellar Clp homologs have been implicated in the degradation of misfolded precursor proteins after import (Schmitt et al., 1995; Halperin and Adam, 1996).
CONCLUSIONS
Interestingly, recent work has shown that when a small N-terminal region is deleted from the ferredoxin or plastocyanin transit peptide, transport into chloroplasts is reduced to an undetectable level both in vitro and in vivo (Pilon et al., 1995; Kindle, 1998). Although these reports conclude that the N-terminal region of the transit peptide is required for efficient protein translocation, neither provides an explanation for this effect. Analyses of these two transit peptides by the above algorithms indicate that the characterized deletions remove potentially critical Hsp70 binding sites.
To our knowledge, our study utilizes the first statistical analyses that predict a common biochemical activity associated with chloroplast transit peptides. This general profile defines a novel property intrinsic to the design of stromally targeted transit peptides whose primary sequences are otherwise unrelated. It will be interesting to extend these analyses to transit peptides of precursors destined to other chloroplast compartments and other organelles to determine the universality of this observation.
ACKNOWLEDGMENTS
We thank Drs. R. Moore and K. Keegstra for the generous gifts of peptides, Dr. C. Georgopoulos for the generous gifts of antibodies, Dr. S. Perry for the gift of the pGEX-2T-SStp construct, and Drs. J. Churchich and R. Miltenberger for many helpful discussions.
Footnotes
This work was supported by the Cell Biology Program at the National Science Foundation (grant nos. MCB–9401840 and MCB–9604535 to B.D.B.) and by The Science Alliance Program at the University of Tennessee-Knoxville.
LITERATURE CITED
- Akita M, Nielsen E, Keegstra K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol. 1997;136:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azem A, Oppliger W, Lustig A, Jeno P, Feifel B, Schatz G, Horst M. The mitochondrial hsp70 chaperone system: effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mhsp70. J Biol Chem. 1997;272:20901–20906. doi: 10.1074/jbc.272.33.20901. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Bruce BD. The role of lipids in plastid protein transport. Plant Mol Biol. 1998;38:223–246. [PubMed] [Google Scholar]
- Bruce BD, Keegstra K. Translocation of proteins across chloroplast membranes. In: Barber J, editor. Advances in Molecular and Cell Biology: Molecular Processes of Photosynthesis. Greenwich, CT: Jai Press; 1994. pp. 389–430. [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. In vitro import of proteins into chloroplasts; pp. 1–15. [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- DeLuca Flaherty C, Flaherty KM, McIntosh LJ, Bahrami B, McKay DB. Crystals of an ATPase fragment of bovine clathrin uncoating ATPase. J Mol Biol. 1988;200:749–750. doi: 10.1016/0022-2836(88)90487-1. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Fourie AM, Sambrook JF, Gething MJ. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J Biol Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler SM, Pierpaoli E V, Christen P. Catapult mechanism renders the chaperone action of Hsp70 unidirectional. J Mol Biol. 1998;279:833–840. doi: 10.1006/jmbi.1998.1815. [DOI] [PubMed] [Google Scholar]
- Glick BS. Can Hsp70 proteins act as force-generating motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- Gray J, Row P. Protein translocation across chloroplast envelope membranes. Trends Cell Biol. 1995;5:243–247. doi: 10.1016/s0962-8924(00)89018-2. [DOI] [PubMed] [Google Scholar]
- Halperin T, Adam Z. Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol. 1996;30:925–933. doi: 10.1007/BF00020804. [DOI] [PubMed] [Google Scholar]
- Heins L, Collinson I, Soll J. The protein translocation apparatus of chloroplast envelopes. Trends Plant Sci. 1998;3:56–61. [Google Scholar]
- Hirsch S, Muckel E, Heemeyer F, von Heijne G, Soll J. A receptor component of the chloroplast protein translocation machinery. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- Ivey RA, Bruce BD. In vivo and in vitro interaction of DnaK and a chloroplast transit peptide. Cell Stress Chaperones. 2000;5:62–71. doi: 10.1043/1355-8145(2000)005<0062:IVAIVI>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin Neumann GA, Tobin EM. Transit peptides of nuclear-encoded chloroplast proteins share a common amino acid framework. EMBO J. 1986;5:9–13. doi: 10.1002/j.1460-2075.1986.tb04170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Bruce BD, Hurley M, Li H-M, Perry S. Targeting of proteins into chloroplasts. Physiol Plant. 1995;93:157–162. [Google Scholar]
- Kessler F, Blobel G, Patel HA, Schnell DJ. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Kindle KL. Amino-terminal and hydrophobic regions of the Chlamydomonas reinhardtii plastocyanin transit peptide are required for efficient protein accumulation in vivo. Plant Mol Biol. 1998;38:365–377. doi: 10.1023/a:1006025606330. [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm I, Gans P, Hernandez JF, Arlaud GJ, Lancelin JM. A coil-helix instead of a helix-coil motif can be induced in a chloroplast transit peptide from Chlamydomonas reinhardtii. Eur J Biochem. 1999;265:171–180. doi: 10.1046/j.1432-1327.1999.00701.x. [DOI] [PubMed] [Google Scholar]
- Lancelin JM, Bally I, Arlaud GJ, Blackledge M, Gans P, Stein M, Jacquot JP. NMR structures of ferredoxin chloroplastic transit peptide from Chlamydomonas reinhardtii promoted by trifluoroethanol in aqueous solution. FEBS Lett. 1994;343:261–266. doi: 10.1016/0014-5793(94)80568-7. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck J, Soll J, Akita M, Nielsen E, Keegstra K. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, Keegstra K. Isolation and Characterization of a cDNA clone encoding the major Hsp70 of the pea chloroplastic stroma. Plant Physiol. 1992;100:1048–1054. doi: 10.1104/pp.100.2.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleros DR, Reid KL, McCarty JS, Walker GC, Fink AL. DnaK, hsp73, and their molten globules: two different ways heat shock proteins respond to heat. J Biol Chem. 1992;267:5279–5285. [PubMed] [Google Scholar]
- Palleros DR, Welch WJ, Fink AL. Interaction of hsp70 with unfolded proteins: effects of temperature and nucleotides on the kinetics of binding. Proc Natl Acad Sci USA. 1991;88:5719–5723. doi: 10.1073/pnas.88.13.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Wienk H, Sips W, de Swaaf M, Talboom I, van ‘t Hof R, de Korte-Kool G, Demel R, Weisbeek P, de Kruijff B. Functional domains of the ferredoxin transit sequence involved in chloroplast import. J Biol Chem. 1995;270:3882–3893. doi: 10.1074/jbc.270.8.3882. [DOI] [PubMed] [Google Scholar]
- Pinnaduwage P, Bruce BD. In vitro interaction between a chloroplast transit peptide and chloroplast outer envelope lipids is sequence-specific and lipid class-dependent. J Biol Chem. 1996;271:32907–32915. doi: 10.1074/jbc.271.51.32907. [DOI] [PubMed] [Google Scholar]
- Rensink WA, Pilon M, Weisbeek P. Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 1998;118:691–699. doi: 10.1104/pp.118.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadis S, Hightower LE. Unfolded proteins stimulate molecular chaperone Hsc70 ATPase by accelerating ADP/ATP exchange. Biochemistry. 1992;31:9406–9412. doi: 10.1021/bi00154a012. [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schlossman DM, Schmid SL, Braell WA, Rothman JE. An enzyme that removes clathrin coats: purification of an uncoating ATPase. J Cell Biol. 1984;99:723–733. doi: 10.1083/jcb.99.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M, Neupert W, Langer T. Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 1995;14:3434–3444. doi: 10.1002/j.1460-2075.1995.tb07349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Keegstra K, Kessler F, Ko K, Soll J. A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol. 1997;7:303–304. doi: 10.1016/S0962-8924(97)01111-2. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Waegemann K, Soll J. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 1995;7235:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- Soll J, Waegemann K. A functionally active protein import complex from chloroplasts. Plant J. 1992;2:253–256. [Google Scholar]
- von Heijne G, Hirai T, Klösgen RB, Steppuhn J, Bruce BD, Keegstra K, Herrmann R. CHLPEP: a database of chloroplast transit peptides. Plant Mol Biol Rep. 1991;9:104–126. [Google Scholar]
- von Heijne G, Nishikawa K. Chloroplast transit peptides: the perfect random coil? FEBS Lett. 1991;278:1–3. doi: 10.1016/0014-5793(91)80069-f. [DOI] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1991;1:149–158. [Google Scholar]
- Wienk HLJ, Czisch M, de Kruijff B. The structural flexibility of the preferredoxin transit peptide. FEBS Lett. 1999;453:318–326. doi: 10.1016/s0014-5793(99)00653-5. [DOI] [PubMed] [Google Scholar]
- Zhang XP, Elofsson A, Andreu D, Glaser E. Interaction of mitochondrial presequences with DnaK and mitochondrial hsp70. J Mol Biol. 1999;288:177–190. doi: 10.1006/jmbi.1999.2669. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhoffer T, Lopez-Buesa P, Craig EA. The dissociation of ATP from hsp70 of Saccharomyces cerevisiae is stimulated by both Ydj1p and peptide substrates. J Biol Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]