Abstract
Glioblastoma is a universally lethal cancer with a median survival of approximately 15 months1. Despite substantial efforts to define druggable targets, there are no therapeutic options that meaningfully extend glioblastoma patient lifespan. While previous work has largely focused on in vitro cellular models, here we demonstrate a more physiologically relevant approach to target discovery in glioblastoma. We adapted pooled RNA interference (RNAi) screening technology2–4 for use in orthotopic patient-derived xenograft (PDX) models, creating a high-throughput negative selection screening platform in a functional in vivo tumour microenvironment. Using this approach, we performed parallel in vivo and in vitro screens and discovered that the chromatin and transcriptional regulators necessary for cell survival in vivo are non-overlapping with those required in vitro. We identified transcription pause-release and elongation factors as one set of in vivo-specific cancer dependencies and determined that these factors are necessary for enhancer-mediated transcriptional adaptations that enable cells to survive the tumour microenvironment. Our lead hit, JMJD6, mediates the upregulation of in vivo stress and stimulus response pathways through enhancer-mediated transcriptional pause-release, promoting cell survival specifically in vivo. Targeting JMJD6 or other identified elongation factors extends survival in orthotopic xenograft mouse models, supporting targeting the transcription elongation machinery as a therapeutic strategy for glioblastoma. More broadly, this study demonstrates the power of in vivo phenotypic screening to identify new classes of ‘cancer dependencies’ not identified by previous in vitro approaches, which could supply untapped opportunities for therapeutic intervention.
Chromatin regulators have emerged as a promising class of ‘druggable’ targets for cancer therapy3,5,6. Chromatin regulation is often context-specific7–11, suggesting the microenvironment mediates cancer cell response to inhibition of specific chromatin regulators. Therefore, we developed an in vivo RNAi screening strategy to enable identification of chromatin regulators critical for survival of glioblastoma cells within a functional tumour microenvironment (Fig. 1a). Utilizing an advanced shRNA delivery vector2–4 (Extended Data Fig. 1a–c), glioblastoma PDX cells (Supplemental Table 1) were transduced with a pooled library containing 1,586 inducible shRNAs targeting 406 known chromatin and transcriptional regulators (2–4 shRNAs per gene) and controls, at efficiencies to achieve a single shRNA per cell. Transduced cells were selected by a constitutive green fluorescent reporter using fluorescence activated cell sorting (FACS) and employed in concurrent in vivo and in vitro screens. In each screen, transduced cells were split into an induced arm and an uninduced control arm. Cells in the induced arm were treated with doxycycline, which induced shRNA expression and a second fluorescent reporter, dsRED. After 3 weeks, induced cells (dsRED+) or uninduced control cells were sequenced and shRNA representation was quantified. For the in vivo screen, 61 mice were implanted with cells and randomly assigned to the control or induced arm. Multiple mice were grouped as single biological replicates, providing the fold coverage necessary to obtain reproducible results (Extended Data Fig. 2a), and enabling successful in vivo negative-selection screening in a solid tumour model (see Methods for full screening methods).
Figure 1. Parallel in vivo and in vitro screen identifies environment-specific cancer dependencies and reveals transcriptional pause-release and elongation as an in vivo-specific target.
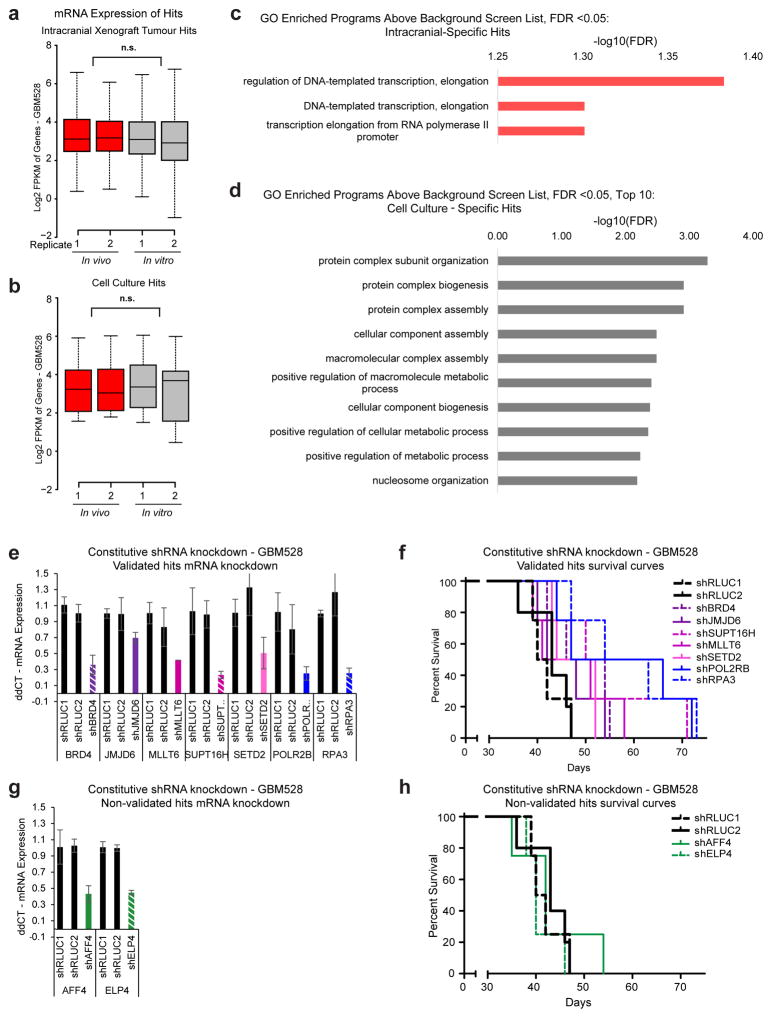
a, Schematic diagram depicting screen. b, Plot of score of 2nd best shRNA targeting each gene in each screen as calculated by RIGER28. Boxes indicate target gene ‘hits’ that caused depletion of the cell population when inhibited. c, Venn diagram of hits from each screen. d, Enrichment of hits in GO gene sets, using the screened library gene list as background to control for bias toward chromatin modifiers. Significance calculated by Benjamini-Hochberg FDR. e, Schematic of transcription elongation machinery, highlighting in vivo-specific hits.
Genes critical for glioblastoma cell survival were prioritized by calculating depletion scores for each shRNA, based on the decrease in shRNA frequency in the induced arm compared to the uninduced control arm. Positive hits were defined as expressed genes that were targets of at least two non-overlapping shRNAs that effectively mediated cellular depletion (Fig. 1b and Supplementary Table 2). In vivo hits outnumbered in vitro hits and, surprisingly, there was almost no overlap between hits that caused cell depletion in vitro versus in vivo (Fig. 1c). Genes that caused cell depletion in both screens were restricted to the positive control gene, replication protein A3 (RPA3) (Extended Data Fig. 2b) and two genes essential for transcription and maintenance of DNA methylation, POLR2B and DNMT1. Differences in molecular dependencies were not explained by expression of the hits, as there were no significant differences in the in vivo expression of the hits compared to their in vitro expression (Extended Data Fig. 3a, b). Collectively, these primary screen results reveal unique molecular dependencies for glioblastoma cells in vivo.
Gene ontology (GO) analysis of the hits revealed significantly enriched molecular classes that were microenvironment-specific. In vitro-specific hits were enriched for genes that promote cellular metabolism and macromolecule biogenesis, whereas in vivo-specific hits were enriched for genes controlling transcriptional elongation (Fig. 1d and Extended Data Fig. 3c, d). Nearly all mediators of transcriptional pause-release and elongation included in the screen scored as in vivo-specific hits, including recently annotated regulators, Jumonji C-domain-containing Protein 6 (JMJD6)12, the DOT1 Like Histone Lysine Methyltransferase (DOT1L) complex13, and Ring Finger Protein 20 (RNF20)14 (Fig. 1e). Several primary hits were validated in secondary in vivo survival assays (Extended Data Fig. 3e–h).
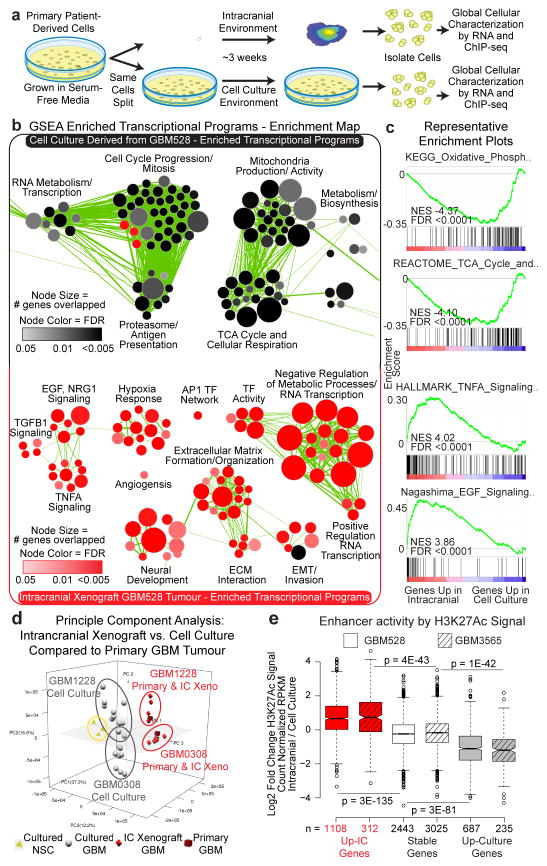
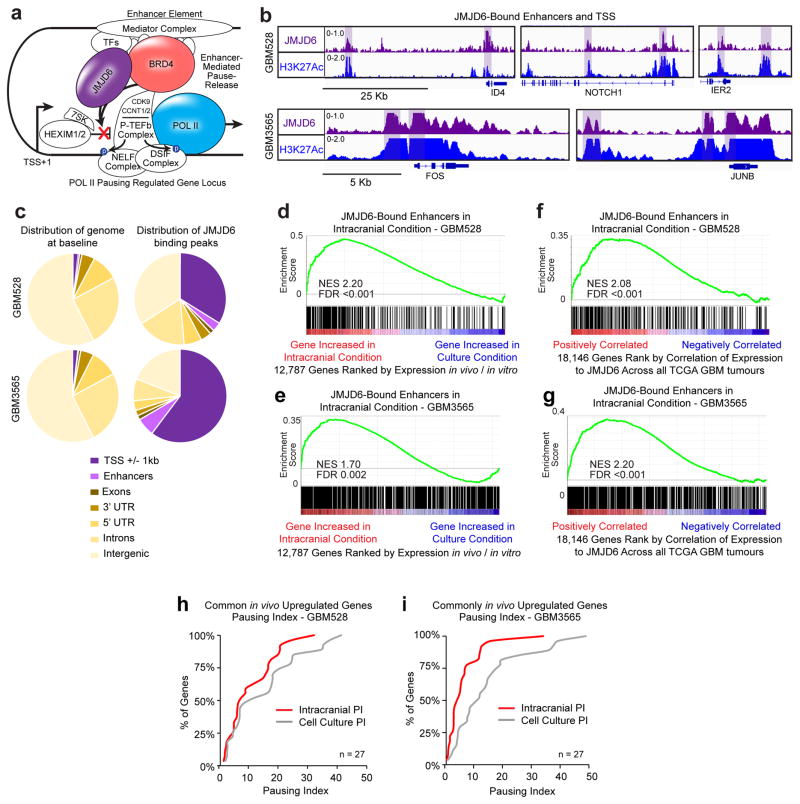
To investigate the mechanisms underlying the selective dependency of glioblastoma cells on transcriptional pause-release and elongation in vivo, we derived and analysed global transcriptional and chromatin landscapes from cells grown in both the intracranial and cell culture environments (Fig. 2a). Multiple PDX models representing different molecular glioblastoma subtypes from our lab were included in the analysis (Supplemental Table 1), as were data generated by an independent lab15. Gene expression profiles were strikingly different when cells were grown concurrently for ~3 weeks in an intracranial xenograft model as compared to cell culture conditions (Extended Data Fig. 4a, b, and Supplementary Table 3).
Figure 2. Transcription of pause-controlled programs is upregulated in the in vivo tumour microenvironment.
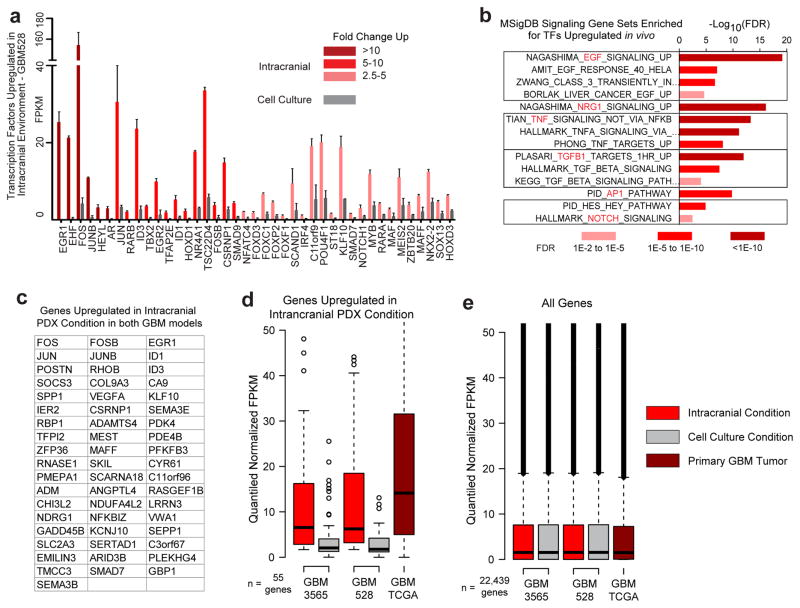
a, Workflow for global analysis of glioblastoma cells. b, Cellular programs enriched by GSEA in cells grown in each condition represented using Enrichment Map. c, Example GSEA plots. FDR calculated by GSEA software. d, Principle component analysis of matched glioblastoma cells in primary tumours, intracranial tumours and cell culture. e, Fold change of H3K27Ac signal at enhancers of genes with >2.5 fold mRNA expression change between conditions, or 0.9–1.1 fold change (stable). P-values by 2-sided Mann-Whitney (M-W) U Test.
Gene set enrichment analysis (GSEA)16 coupled to Enrichment Map17 visualization was used to annotate differentially enriched biological pathways (Fig. 2b, c, Extended Data Fig. 4, and Supplementary Table 4a–f). Cancer cells cultured in serum-free conditions, where nutrients and space are in abundant supply, were enriched for transcriptional programs of proliferation. In contrast, transcriptional programs associated with stress response, signalling response, and other stimulus response pathways were enriched in intracranial tumours, where nutrients and space are less abundant. The stimulus response pathways upregulated in the intracranial tumour environment include pause-controlled pathways comprised of genes with a strong reliance on transcription pause-release and elongation for their expression13,18–22. Of the 55 genes that were upregulated more than 2.5 fold in tumour cells grown intracranially in both the proneural GBM528 and mesenchymal GBM3565 models, many were important transcription factors and signalling molecules regulated by Pol II pausing, including pause-controlled genes, such as early growth response protein 1 (EGR1) and FOS13,23 (Extended Data Fig. 5a–c). These 55 genes were also highly expressed in primary glioblastoma patient tumours (Extended Data Fig. 5d, e). Principal component analysis of expression profiles of matched primary tumours and derived models revealed that cells grown as intracranial tumours more accurately modelled the transcriptome of the primary patient tumour from which they were derived, as compared with cells grown in culture (Fig. 2d). Collectively, these data suggest that the upregulated pause-controlled gene programs that occur in vivo may allow the tumour cells to interact with and adapt to their complex microenvironment, supporting their increased dependency on transcription pause-release and elongation factors for in vivo survival.
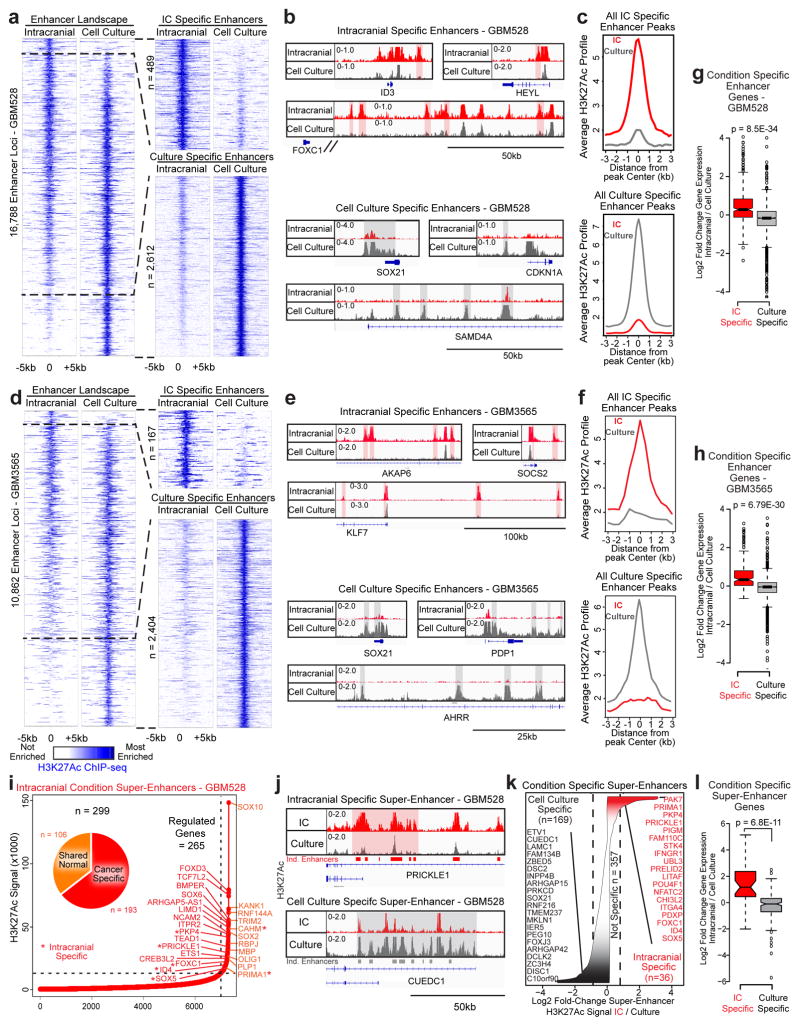
Enhancers act in conjunction with transcription factors to drive transcriptional changes through transcriptional pause-release and elongation. Leveraging chromatin immunoprecipitation (ChIP)-sequencing of the dynamic enhancer mark lysine-27 acetylation of histone 3 (H3K27Ac), transcriptional changes in tumour cells grown intracranially were reflective of genome-wide alterations in enhancers in two glioblastoma models (Fig. 2e). Nearly 20% of all enhancer elements, including “super-enhancer” loci, were condition-specific, and expression of their putative target genes, as determined by nearest expressed gene, corresponded to the condition-dependent changes in enhancer signal (Extended Data Fig. 6 and Supplementary Table 5). Together, these data show that the microenvironment regulates the epigenome to transform the glioblastoma cell state by differentially activating enhancers and their target genes.
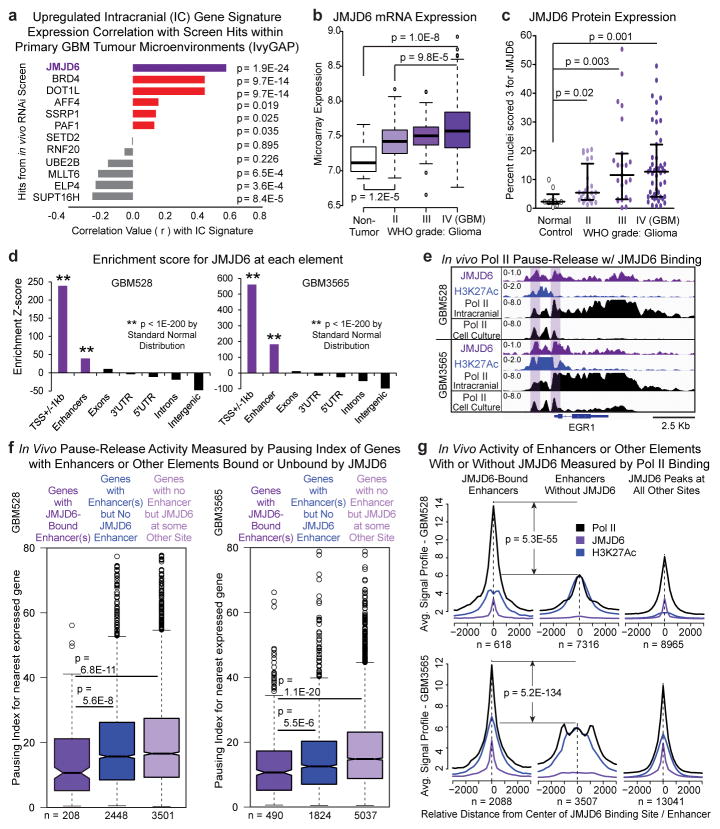
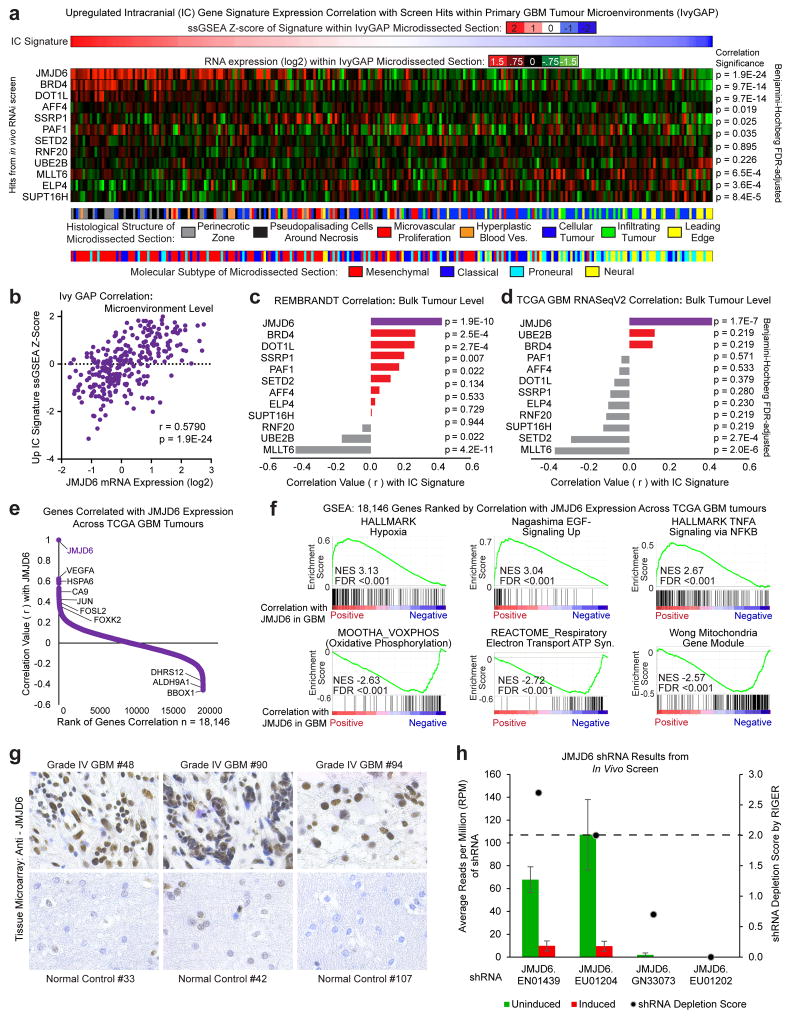
Of the in vivo-specific hits, we prioritized the 12 genes encoding transcription elongation factors for further consideration as therapeutic targets. Using large, independent datasets of primary patient tumours, we correlated expression of each of the 12 transcription elongation factors with the expression of the 55 genes that were consistently upregulated in vivo across both PDX tumour models, which we hypothesized are main drivers of the transcriptional programs needed by tumours to adapt and survive in the stressful and dynamic in vivo microenvironment. JMJD6 was the most positively correlated hit across all datasets, including the Ivy Glioblastoma Atlas Project dataset, which provides intratumour microenvironment-specific expression24 (Fig. 3a and Extended Data Fig. 7a–d), suggesting JMJD6 regulates many of the genes important for survival of glioblastoma cells in vivo.
Figure 3. JMJD6 is top hit and regulates enhancer mediated pause-release in GBM.
a, Correlation across all Ivy GAP samples of elongation factor hits with gene signature of genes upregulated in vivo in both PDX models. P-value by FDR-adj. B-H procedure. b, JMJD6 mRNA expression across all gliomas in REMBRANDT database. c, Tissue microarray analysis of JMJD6 protein expression in over 100 gliomas. d, Global enrichment Z-scores of JMJD6 binding by JMJD6 ChIP-seq. e, Browser track example for (f). Pausing index of target genes (nearest expressed gene) (f) or aggregate plots of ChIP-seq signal (g) of JMJD6-bound and unbound enhancers, and of JMJD6-bound sites outside of enhancers. P-values (b–c, f–g) by 2-sided M-W U Test.
To further assess the regulatory role of JMJD6 in patient tumours, we correlated the expression of all genes in TCGA glioblastoma tumours individually with the expression of JMJD6 (Extended Data Fig. 7e and Supplementary Table 6). Genes positively correlated with JMJD6 were enriched in pause-controlled programs, similar to those upregulated in intracranial tumours, whereas genes negatively correlated with JMJD6 were enriched in metabolic programs, similar to those upregulated in cell culture (Extended Data Fig. 7f and Supplementary Table 4g). These findings indicate a potential mechanism for the in vivo specificity of JMJD6 in the screen, and provide evidence that JMJD6 may control transcriptional pause-release in primary glioblastoma tumours.
To explore the clinical significance of JMJD6, we analysed gene expression in primary tumours and found that JMJD6 mRNA (Fig. 3b) and protein (Fig. 3c and Extended Data Fig. 7g) were highly expressed in gliomas, and increased with tumour grade. These data, along with the robust in vivo depletion of tumour cells harbouring JMJD6 shRNA in the primary screen (Extended Data Fig. 7h), provide further evidence that JMJD6 constitutes a strong lead target for further evaluation.
In HEK293T and HeLa cells, JMJD6 acts with bromodomain containing 4 (BRD4) as a key activator of enhancer-mediated pause-release at genes controlled by Pol II pausing12 (Extended Data Fig. 8a). To determine if JMJD6 localizes to enhancers in glioblastoma in vivo, we performed JMJD6 ChIP-seq of tumour cells in intracranial tumours. Globally, JMJD6 was distributed throughout the genome, but strongly enriched at enhancers and promoters (Extended Data Fig. 8b, c and Fig. 3d). Furthermore, in the PDX models, target genes of JMJD6-bound enhancers were enriched for genes upregulated in vivo (Extended Data Fig. 8d, e) and for genes that positively correlated with JMJD6 in primary patient tumours (Extended Data Fig. 8f, g). These results indicate that JMJD6 may regulate the expression of genes targeted by JMJD6-bound enhancers through enhancer-mediated pause-release, both in the intracranial tumour environment in our PDX models, as well as in patient tumours.
To investigate, we conducted ChIP-seq of RNA Pol II in GBM528 and GBM3565 cells both in vivo and in vitro. Transcription pause-release and elongation was measured by the Pausing Index (PI), which is the ratio of Pol II density surrounding the transcriptional start sites (TSSs) to the density of Pol II over the gene body22. The higher the PI, the more paused the gene transcript. Genes that were consistently upregulated in vivo in both PDX tumours models, such as EGR1, were transcriptionally paused in vitro and released in vivo (Fig. 3e and Extended Data Fig. 8h, i). To interrogate globally whether JMJD6 activity at enhancers promotes pause-release in vivo, we calculated the in vivo PI for all expressed genes with or without enhancers. Genes regulated by JMJD6-bound enhancers had lower PIs, or increased levels of pause-release activity, compared to genes regulated by enhancers not bound by JMJD6 (Fig. 3f). This effect was specific to JMJD6 binding at enhancers, as genes bound by JMJD6 at locations other than an enhancer did not demonstrate increased pause-release.
We then evaluated the activity of JMJD6-bound enhancers relative to enhancers not bound by JMJD6 in vivo. Building upon recent evidence that active enhancers are transcribed and that activity can be estimated by RNA Pol II binding25, we measured relative RNA Pol II binding at JMJD6-bound enhancers versus enhancers not bound by JMJD6, and found that JMJD6-bound enhancers had significantly higher levels of Pol II binding (Fig. 3g). Measurements of Pol II binding at JMJD6 sites, including outside of enhancers, revealed that Pol II binding was highest at JMJD6-bound enhancers. Collectively, these data provide evidence that JMJD6 binding is associated with increased enhancer activity and promotes pause-release in human glioblastoma cells within the tumour microenvironment.
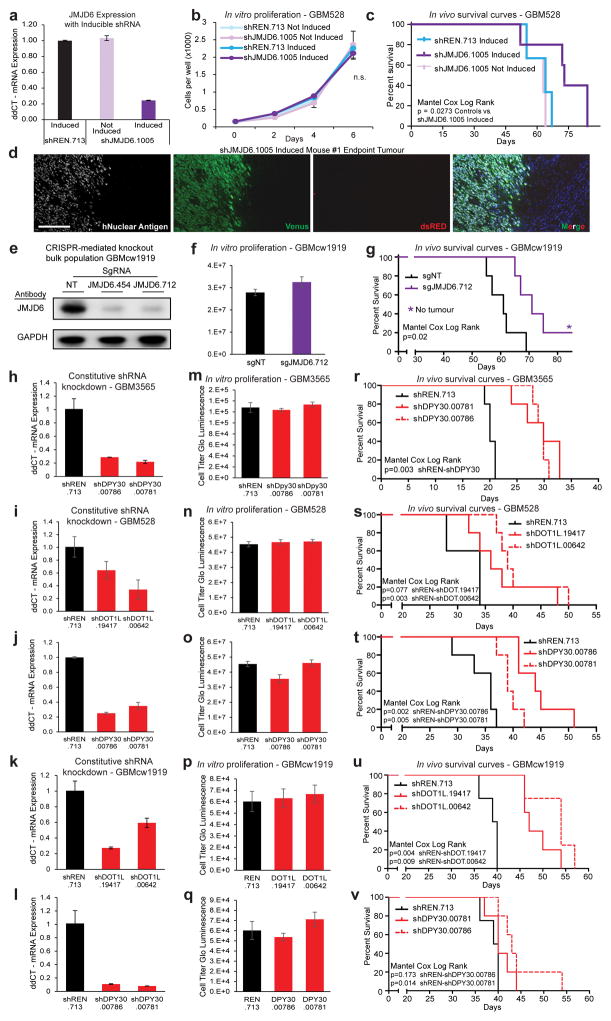
We next sought to determine the potential preclinical value of JMJD6 as a therapeutic target. Targeting JMJD6 with an inducible shRNA distinct from the primary screen again did not alter glioblastoma cell growth or survival in vitro, but did extend survival in an orthotopic xenograft mouse model (Extended Data Fig. 9a–c). Furthermore, the inducible shRNA transgene was silenced in tumours that grew in the knockdown arm (Extended Data Fig. 9d), suggesting that achieving sustained JMJD6 inhibition may provide an even more striking therapeutic benefit.
These data prompted us to use CRISPR-Cas9 technology to create multiple clonal populations of glioblastoma cells in vitro in which JMJD6 expression was completely knocked out by expression of one of two independent guide RNAs (sgRNAs) (Fig. 4a). Glioblastoma cells lacking JMJD6 formed clonal populations and proliferated with little or no growth defect in vitro when compared to non-targeting sgRNA control cell populations (Fig. 4b). However, mice bearing JMJD6-deficient cells showed a striking survival advantage compared to mice with control cells implanted, with over 25% of mice in the JMJD6-deficient group tumour-free after 100 days (Fig. 4c). Similar results were found using an independent PDX model (Extended Data Fig. 9e–g).
Figure 4. JMJD6 and other hits are potential therapeutic targets in GBM.
a, CRISPR mediated knockout of JMJD6 in GBM528 cells. b–c, Parallel in vitro proliferation assay (b) and in vivo survival assay (c) of cells from (a). d, Constitutive shRNA knockdown of DOT1L. e–f, Parallel in vitro proliferation assay (e) and in vivo survival assay (f) of cells from (d). Error bars of bar graphs +/− s.d. of at least triplicates.
To validate other screen hits using in vivo survival studies, we performed similar experiments targeting DOT1L, a recently discovered mediator of transcription elongation13 and top scoring hit from the primary screen, and DPY30, another top scoring hit from the primary screen that is not known to be involved in transcription pause-release or elongation. Constitutive shRNA-mediated knockdown of these genes in glioblastoma cells from three independent PDX models caused no proliferation defect in vitro (Fig. 4d–e and Extended Data Fig. 9h–q). However, mice bearing cells with DOT1L or DPY30 knockdown survived significantly longer than mice implanted with cells expressing non-targeting shRNAs (Fig. 4f and Extended Data Fig. 9r–v). These results further demonstrate the power of in vivo screening to identify in vivo-specific cancer dependencies and indicate that many of the identified hits from the primary screen are strong candidates for future investigation as potential therapeutic targets in glioblastoma.
Pharmaceutical approaches to cancer drug discovery typically involve high-throughput screening of established cell lines cultured in vitro to reveal individual targetable oncogenes that predominantly regulate cancer cell proliferation. However, drugs developed to modulate these targets have thus far achieved limited success in patients, especially for glioblastoma. This therapeutic roadblock prompted us to develop and validate a novel in vivo functional screening strategy for glioblastoma that recapitulates the majority of the stressors and stimuli of the tumour microenvironment. Our approach revealed a number of in vivo-specific biological targets for glioblastoma, including JMJD6, indicating that cancer cells are dependent on unique molecular effectors for growth and survival depending on the extrinsic factors in their microenvironment. Specifically, we found that glioblastoma cells in vivo, but not in vitro, were dependent on Pol II pause-release and transcription elongation machinery for survival (Extended Data Fig. 10). This machinery is necessary to upregulate pause-controlled stress and signalling response pathways that promote cell survival13,18,20,21. This finding unexpectedly revealed that in the primary tumour, targeting the microenvironment-induced stress response mechanisms of the cancer cell may be a more effective therapeutic strategy than targeting cell growth, the main target of traditional chemotherapies. Overall, our results demonstrate feasibility for direct target identification in vivo in solid tumours and suggest new avenues for therapeutic development.
METHODS
Human glioblastoma specimen culture conditions
All human glioblastoma tissues were obtained from excess surgical materials from consented patients after review from a neuropathologist and used in accordance with an approved protocol by the Institutional Review Board at Cleveland Clinic. As previously described26, glioblastoma cells were derived immediately after dissociation of primary patient tumour or after transient xenograft passage. For all in vitro studies, glioblastoma cells were cultured in Neurobasal medium (Gibco) with B27 (without vitamin A, ThermoFisher), basic fibroblast growth factor (20ng/ml, R&D) and epidermal growth factor (20ng/ml, R&D).
Primary glioblastoma models were validated to be unique by short tandem repeat (STR) analysis at multiple xenograft passage numbers (STR conducted by ATCC and Duke University, Cell Line Authentication Service). STR results are available upon request. All cells used were derived from primary patient tissues, and are not included in the database of commonly misidentified cell lines maintained by ICLAC. All lines are routinely tested for mycoplasma contamination and were negative.
Patient-derived orthotopic xenografts
All animal procedures were performed in accordance with Cleveland Clinic Institutional Animal Care and Use Committee (IACUC) approved protocols. For intracranial tumours, size cannot be measured directly. Therefore, specific neurological signs indicating the presence of brain tumours (seizures, ataxia, lethargy) are monitored, in accordance with the IACUC approved protocol, and mice were always euthanized when these signs became present. The number of animals included in each of the described studies was based on extensive past experience in the development and use of glioblastoma xenograft models by our group. Each study was designed to minimize unnecessary animal use, optimize statistical power, and account for known variance in each model system. For all in vivo experiments, human glioblastoma cells were intracranially implanted into NOD SCID gamma (NSG, JAX Charles River) mice 4–6 weeks of age. Within this age window, mice were age-matched across groups. For the primary screen, mice were randomized by cage into control or induced group. The xenograft implantations were performed blinded to group. Researchers were not blinded during the rest of the experiment as no subjective measurements were used. 31 female mice and 30 male mice were used in the screen, with an even split between control and induced populations. For individual shRNA or CRISPR survival analysis, xenograft implantations were performed blinded to group, and the gender of the mice in a given study were kept identical to maintain consistency across experimental and control groups. See individual methods for number and gender of mice used. Mice were euthanized at the onset of neurological signs, and tumours were removed for further analysis.
In vivo and in vitro inducible shRNA screens
Summary (more technical details in sections following summary)
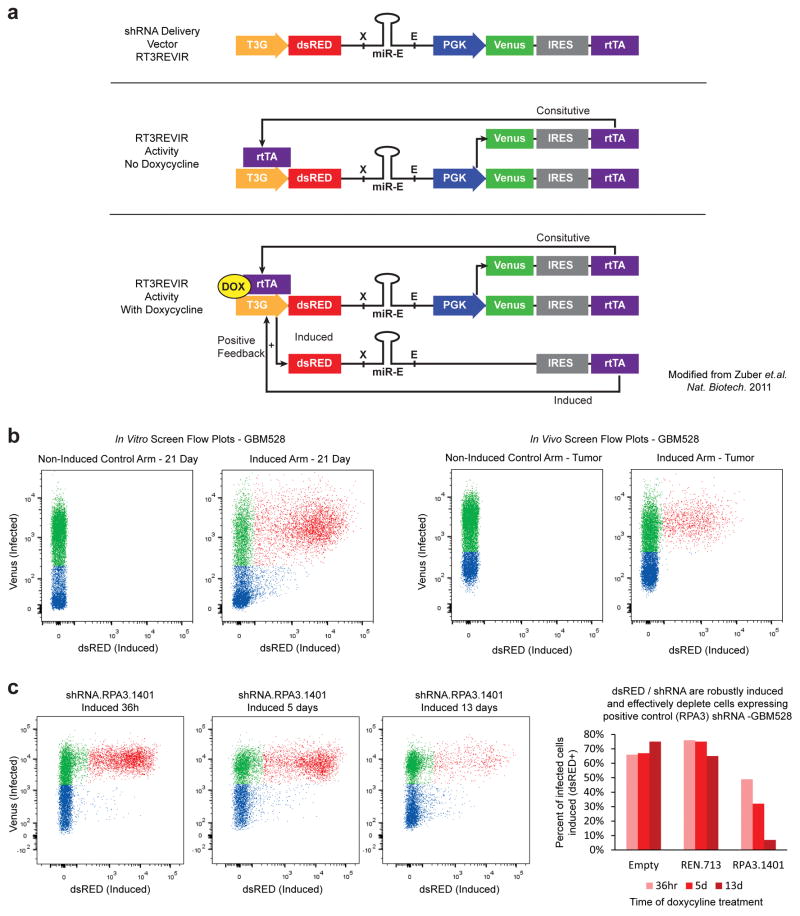
To investigate the effects of targeting epigenetic regulators in glioblastoma cells within the tumour microenvironment, we modified an inducible in vivo RNAi screening system, previously used for screening in hematologic malignancies2,3, for use in solid tumours (Fig. 1a). The advanced inducible shRNA delivery vector has high fidelity expression and dual fluorescent reporters to reduce the noise and bias that plague traditional shRNA screens2,4 (Extended Data Fig. 1a–c). Our shRNA library contained 1,586 shRNAs targeting 406 known chromatin and transcriptional regulator genes (2–4 shRNAs per gene), with positive and negative control shRNAs.
Well-characterized, PDX glioblastoma cells (Supplementary Table 1) were transduced with the shRNA library pool such and cells with genomic integration of shRNAs, as monitored by expression of a constitutive green fluorescent reporter, were selected using FACS. With the selected pool of cells, we performed an in vitro screen in previously defined serum-free cell culture conditions15 concurrent with an in vivo screen in intracranial mouse xenografts. Each screen used the same population of cells distributed across two arms: 1) control – cells were left uninduced (no shRNA expression), and 2) experimental – cells were treated with doxycycline to induce shRNA expression. Induction also led to expression of a second fluorescent reporter, dsRED, which allowed isolation of cells actively expressing an shRNA at the endpoint of the screen (Fig. 1a and Extended Data Fig. 1a, b). The in vitro screen consisted of 3 independent replicates per arm, and cells were collected after 3 weeks. In the parallel in vivo screen, single animals could not be used as independent replicates as the maximum number of engrafted cells per animal provided insufficient fold coverage of the shRNA library. Therefore, 61 mice were implanted with cells and randomly assigned to the control (20 mice) or induced arm (41 mice). Multiple mice were then grouped together as a single biological replicate in order obtain 3 independent replicates for the induced arm and 2 for the uninduced arm. This provided the fold coverage necessary to obtain reproducible results (Extended Data Fig. 2a), and enabled successful in vivo negative-selection screening in a solid tumour model. Tumours developed for 2–3 weeks until mice exhibited signs of neurologic dysfunction. Mice were then sacrificed, tumours excised and dissociated, and uninduced or induced tumour cells from the control or experimental arms, respectively, were collected by FACS. The representation of shRNAs from each cell population was quantified by high depth sequencing of lentivirally integrated shRNA barcodes.
For both screens, genes critical for glioblastoma cell survival were prioritized by calculating depletion scores for each shRNA, based on the decrease in shRNA frequency in the induced arm compared to the uninduced control arm. Positive hits were defined as expressed genes that were targets of at least two non-overlapping shRNAs that effectively mediated cellular depletion (Fig. 1b, c and Supplementary Table 2).
shRNA library generation
An all-in-one inducible retroviral vector with an optimized “miR-E” shRNA design (RT3REVIR)4 was used to deliver shRNAs. Briefly, this construct contains an optimized 3rd generation Tet-responsive element and rtTA3 to potentiate a positive feedback loop upon addition of doxycycline, enhancing expression of the construct upon induction and reducing construct leakiness. It also contains an enhanced design of the miR30 backbone to allow for more efficient shRNA processing. In addition, the second reporter is inducible and is expressed in conjunction with the shRNA, which allows for selection of cells actively expressing an shRNA at the time of collection, and significantly reduces the number of cells included in the final analysis that contained silenced shRNAs.
We shuttled the library of 1,568 shRNAs (shERWOOD Epigenetics-related genes library, transOMIC) into the RT3REVIR vector using previously published methods4. To produce virus, retroviral plasmid library was transfected into HEK293T Phoenix packaging cells as previously described2. Chloroquine (25 μM, Sigma-Aldrich) was added to enhance plasmid stability.
Functional screening assays
60 million patient-derived glioblastoma cells were transduced with the pooled inducible shRNA library at 1.1% transduction efficiency to ensure that cells were transduced with only 1 shRNA per cell. Protamine sulfate (Sigma-Aldrich) was used for transduction. Successfully transduced cells were selected by a constitutive Venus fluorescent reporter using Fluorescence-activated cell sorting (FACS). Cells were allowed to recover and expand for three passages. The same population of cells at the same time was used for a screen completed in triplicate in standard serum free cell culture conditions and for a screen completed in an in vivo intracranial xenograft mouse model.
For the in vivo screen, each mouse was intracranially injected with 500,000 cells to maximize number of engrafted cells per mouse. Fold coverage of the shRNA library is determined by the number of engrafted cells (not number implanted). Based upon GFP spike-in dilution assays, we estimated between 10–20% of cells implanted are engrafted (50,000 – 100,000 cells per mouse). To overcome this issue of low-fold coverage when using a single animal, typically the limiting factor when screening in solid tumours in vivo, we injected 61 mice and randomly assigned them to the induced arm (41 mice) or the control arm (20 mice). Mice in the induced arm were maintained on doxycycline containing water (2 mg/ml Doxycycline and 2% sucrose, Sigma-Aldrich) for the duration of the screen. Cells were not exposed to doxycycline prior to being injected into the mouse, allowing us to screen for factors important for tumour growth rather than engraftment efficiency. When a mouse began to show overt neurological signs, which occurred between 2 to 3 weeks, it was euthanized, the tumour was harvested, macroscopically dissected, dissociated to single cells (Tumor Dissociation Kit, human and GentleMACS™ Octo Dissociator with Heaters, Miltenyi Biotech) and depleted of any remaining mouse cells using magnetic-activated cell sorting (MACS, Mouse Cell Depletion Kit, Miltenyi Biotech). Each mouse was processed independently. Tumour cells from the mice in the induced arm were sorted by FACS to collect Venus+dsRED+ cells (denoting shRNA expression). The average number of double positive cells collected across 41 mice was 229,964 (range − 12,440 to 758,143 cells). 26/41 mice had greater than 100,000 cells collected (see source data). Mice were combined after sequencing into “biological replicate” groups of 13–14 mice (see source data), which provided, on average, about 3 million cells per biological replicate or “n”. This represented a fold coverage of ~2,000 per replicate. Tumour cells from mice in the uninduced control arm were sorted by FACS to collect Venus+dsRED− cells (shRNA integrated but not expressed). The average number of single positive control cells collected across 20 mice was 851,916 (range − 144,184 to 1.8 million cells). 16/20 mice had over 500,000 cells collected. Control mice were combined after sequencing into two biological replicate groups of 10 mice (see source data), which provided on average about 8 million cells per “n”. This represented a fold coverage of ~5,000 per replicate. The baseline induction rate of the inducible system in vivo is 30–40%, which limits the number of cells that can collected from mice in the induced arm compared to the control arm (from which we collected all cells). For this reason, we used more than double the number of mice in the induction arm compared to the control arm in order obtain similar fold coverage rates.
For the in vitro screen, 1.5 million cells were placed in serum-free sphere culture in a 15cm plate for each replicate. 3 replicates were used for induced arm and 3 replicates were used for the control arm. Doxycycline (1 μg/ml, Sigma-Aldrich) was added to cells in the induced arm at time of plating and cells were maintained on doxycycline for 21 days, which is the same amount of time that the last of the intracranial tumours were harvested for the in vivo screen. Cells in the uninduced control arm were also maintained for 21 days in culture with no doxycycline. Cells had a doubling time of 1.5 days and were passaged every 3–4 days to prevent cells from becoming over-confluent. During passage, at least 2 million cells were re-seeded in the plate to maintain library representation. At the conclusion of the screen Venus+dsRED+ cells (1.5 million for each replicate) were collected by FACS from the induced arm, while Venus+dsRED− cells (1.5 million for each replicate) were collected by FACS for the undinduced arm.
shRNA amplification and sequencing
Each mouse was processed and analysed separately. Genomic DNA was isolated and sequenced as described2 with slight modification. Genomic DNA was isolated by two rounds of phenol extraction using PhaseLock tubes (5prime) followed by isopropanol precipitation. Deep sequencing libraries were generated by PCR amplification of shRNA guide strands using barcoded primers that tag the product with standard Illumina adapters (p7+loop, CAAGCAGAAGACGGCATACGA-NNNN (4 nt barcode)-TAGTGAAGCCACAGATGTA; p5+miR3′, AATGATACGGCGACCACCGATGGATGTGGAATGTGTGCGAGG). Libraries were sequenced on the HiSeq 2500 platform at the Cleveland Clinic Genomics Core Facility. Libraries were sequenced using a primer that reads in reverse into the guide strand (miR30EcoRISeq, TAGCCCCTTGAATTCCGAGGCAGTAGGCA).
Sequencing analysis and shRNA scoring
Sequence processing was performed using two custom workflows using usegalaxy.org27. Annotated workflows are stored in the usegalaxy.org published workflows repository and can be accessed and utilized using the following links:
Workflow 1 can be found here: https://usegalaxy.org/u/tyleremiller/w/shrna-pipeline1. It is used first to generate an output needed for workflow 2.
Workflow 2 can be found here: https://usegalaxy.org/u/tyleremiller/w/shrnastep2.
Raw read counts were converted to RPM (Reads Per Million) to control for variations in total shRNA reads in each sample.
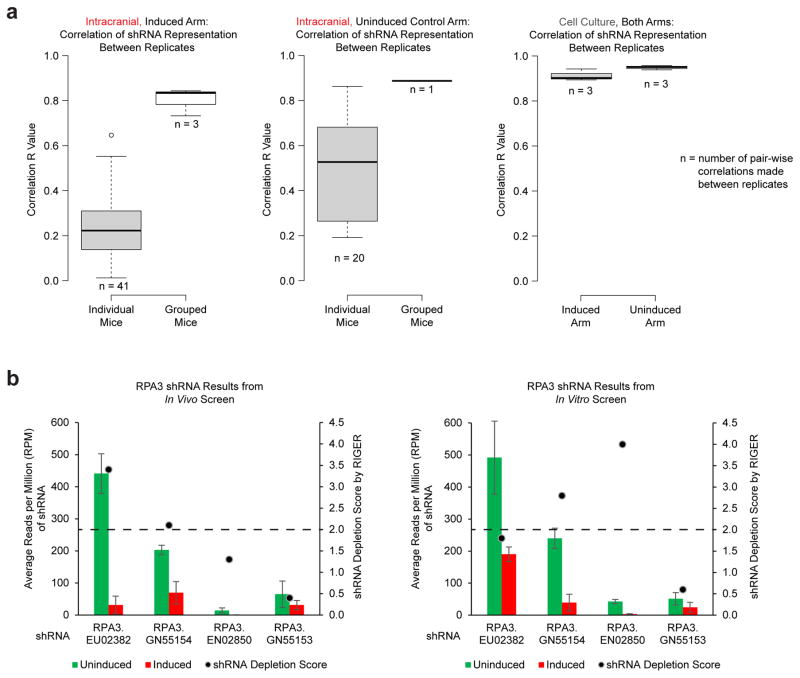
Results from multiple mice were randomly pooled together to form replicates (n = 3 for the induced arm, with each replicate containing 13–14 mice, while n = 2 for the control arm, with each replicate containing 10 mice). To provide a sufficient baseline for detecting shRNA depletion (negative selection) in experimental samples, we aimed to acquire an average of >1,000 reads per shRNA in each replicate, which would require 1.6×106 reads per replicate. In practice, we achieved >1.5×107 reads for all replicates (nearly 10,000-fold coverage). Pooling the mice was essential for achieving proper fold coverage and highly correlated replicates. The average correlation score between individual mice from the induced groups was R = 0.24, while the average correlation value between the 3 combined replicates was R = 0.80.
shRNAs were scored using RIGER, and extension of the GENE-E package (Broad Institute)28. Median RPM value for each replicate was used for analysis. Signal to Noise of replicates was used to calculate individual shRNA score based on their ability to deplete cells in the induced arm compared to the control arm and 2nd-Best shRNA score was used to rank genes. Expressed genes with a RIGER depletion score ≥ 2 were considered hits.
Gene ontology enrichment analysis and visualization
Analysis for screen hits
Gene ontology analysis for screen hits (Fig. 1 and Extended Data Fig. 3) was conducted using background correction in order to understand what is enriched above what would be expected for a list comprised of chromatin modifiers. Specifically, the gene list of in vivo or in vitro hits was imported into gProfiler29 to generate enrichment scores for all gene ontology (GO) gene sets according to recommended settings for gProfiler http://baderlab.org/Software/EnrichmentMap/GProfilerTutorial, with the exception that we used the full list of genes included in the screen as a background list (“Gene list as a stat. background” feature) to control for bias towards chromatin modifiers.
Analysis when starting with whole transcriptome data
For gene ontology analysis when analysing whole transcriptome data (Fig. 2, and Extended Data Fig. 4), data were imported into Gene Set Enrichment Analysis (GSEA)16 software to generate enrichment scores for gene sets in Hallmark, C2.all and C5.all MSigDB datasets. Exact settings are listed in Supplemental Table 4. Cytoscape (v3.2.1) and the Enrichment Map17 plug-in was used to generate networks for gene sets enriched with an FDR cut-off of < 0.05. For GSEA analysis in Extended Data Fig. 7, ranked lists were generated from whole transcriptome expression data of GBM528 or GBM3565 cells grown in vivo versus in vitro (for Extended Data Fig. 8d, e) or from the correlations of genes with JMJD6 in glioblastoma tumours from TCGA database (for Extended Data Fig. 8f, g). These ranked lists were run against a Gene Set comprised of genes targeted by JMJD6-bound enhancers in GBM528 cells or GBM3565 cells. Normalized Enrichment scores and False Discovery Rate was generated by GSEA software.
Global characterization of glioblastoma cells grown in vivo and in vitro
To characterize the cell state of human tumour cells grown in vivo and compare it to those grown in culture, we used the same experimental setup as was done in the screen (schematic in Fig. 2a). We took patient-derived glioblastoma cells growing in vitro and split them in half. We injected half into 20 mice per glioblastoma model and maintained the other half in culture in independently maintained replicates. Multiple PDX models representing different molecular glioblastoma subtypes were included in the analysis, including our primary screen model, the proneural model GBM528, and the mesenchymal model GBM3565 (Supplemental Table 1). We also analysed data obtained from an independent lab from two other PDX models, GBM1228 and GBM0308, which were grown similarly in vivo and in vitro15. In addition, these two models had matched data from the corresponding primary tumour15. For the in vivo arm, each mouse was intracranially implanted with 500,000 cells. When a mouse began to show overt neurological signs, which occurred between 2 to 3 weeks, it was euthanized, the tumour was harvested, macroscopically dissected, dissociated to single cells (Tumor Dissociation Kit, human and GentleMACS™ Octo Dissociator with Heaters, Miltenyi Biotech) and depleted of any remaining mouse cells using magnetic-activated cell sorting (MACS, Mouse Cell Depletion Kit, Miltenyi Biotech). We used two rounds of mouse cell depletion to ensure only human tumour cells were used in the analysis. We consistently were able to obtain greater than 98% purity of human cells with this method. Each mouse was processed independently. For RNA-seq samples, cells from 2–3 mice were pooled together to form each replicate. For ChIP-seq samples, many mice were pooled together for ChIP-seq as cell number was a limiting factor. Cells grown in culture were maintained for 3 weeks and then harvested for RNA-seq and ChIP-seq.
RNA-sequencing
RNA was extracted from human glioblastoma cells with TRIzol (ThermoFisher), separated using Phase Lock Gel tubes (5 Prime), and purified using the miRNAeasy kit (Qiagen) according to the manufacturer’s protocol. Total RNA was prepared for sequencing by Beckman-Coulter Genomics using the Illumina TruSeq Stranded Total RNA Library Prep Kit according to the manufacturer’s protocol. RNA-seq libraries were sequenced on the Illumina HiSeq 2500 platform by Beckman-Coulter Genomics. For gene expression analysis, reads were aligned to the hg19 genome build (retrieved from http://cufflinks.cbcb.umd.edu/igenomes.html), using Tophat30 v2.0.6. The distribution of alignments was analysed using the CollectRnaSeqMetrics module of Picard v1.89 (http://picard.sourceforge.net/). FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values for known genes were calculated using Cufflinks31 v2.0.2 provided with the GTF file via the −G (known genes only) option. FPKM values were quantile normalized. Differential expression testing was performed using Cuffdiff v2.0.2; however, all FPKM values provided are those calculated by Cufflinks. To generate an expressed genes list, an average of replicates for each condition was calculated and genes with FPKM value >0.25 in either intracranial sample or culture sample were considered expressed. Genes that did not meet this expression cut-off (Replicate average FPKM <0.25 in Intracranial and in Culture conditions) were removed as not expressed. Expressed genes were tabled by converting FPKMs <0.25 to 0.25. Private link to raw and processed RNA-seq data: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sfopmemwjxgzhyb&acc=GSE74529
Exome-sequencing
DNA was harvested from GBM528, GBM3565 and GBMcw1919 cells using standard techniques. The Case Genomics Core processed the DNA using the Illumina Nextera Rapid Capture Exome kit and sequenced on the Illumina HiSeq 2500 platform at greater than 100X coverage. Data was processed by the Case Genomics Core to generate variant call format (VCF) files and clinical variant calls were made using Omicia Opal Software (http://www.omicia.com/).
ChIP-sequencing
ChIP-seq
Chromatin Immunoprecipitation (ChIP) was performed as previously described32. For histone modification and transcription factor ChIP-Seq, 2 million cells (H3K27Ac), 15–20 million cells (JMJD6), or 5 million cells (Pol2) were crosslinked in PBS + 1% fresh formaldehyde for 10 minutes at 25 C, quenched for 5 minutes with 125 mM glycine, washed twice in cold PBS with protease inhibitors (complete PI, Roche), and stored at −80 C. Briefly, formaldehyde-fixed cells were lysed and sheared (Branson S220) on wet ice. The sheared chromatin was cleared and incubated overnight at 4°C with the following antibodies: H3K27ac (Active Motif, 39133), JMJD6 (Abcam, ab64575, lot GR54735-1), and total Pol II (Santa Cruz, sc-899-X, lot H0510). Antibody-chromatin complexes were immunoprecipitated with protein G magnetic Dynal beads (Life Technologies), washed, eluted, reverse crosslinked, and treated with RNAse A followed by proteinase K. ChIP DNA was purified using Ampure XP beads (Beckmann Coulter) and then used to prepare sequencing libraries for sequencing with the Next-Seq Illumina genome analyser.
Peak calling
Reads were aligned to hg19 using Burrows-Wheeler Aligner (BWA)33 and identical ChIP-seq sequence reads were collapsed to a single read to avoid PCR duplicates. Peaks were called using HOMER v4.634 using matched inputs with the following parameters:
H3K27ac, –histone –tagThreshold 50 was used for enhancer analysis.
JMJD6, –factor.
Private link to raw and processed ChIP-seq data: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=sfopmemwjxgzhyb&acc=GSE74529
Enhancer Landscape Analysis
To generate enhancer loci lists for each condition, H3K27Ac ChIP-seq peak files were filtered to remove all peaks overlapping ENCODE blacklisted regions for functional genomics analysis (https://sites.google.com/site/anshulkundaje/projects/blacklists) as well as peaks with any overlap occurring within +/−1kb from transcription start sites (TSSs) of all annotated RefSeq genes to exclude promoters. To call target genes of enhancers, enhancer loci were mapped to the nearest expressed gene within the glioblastoma cells. An expressed gene had to be within 200kb for an enhancer loci to be mapped to a gene. The expressed gene list was the same as described in the RNA-seq methods section above. Peaks were visualized with the Integrative Genomics Viewer (IGV, Broad Institute).
To call condition-specific enhancers
H3K27Ac ChIP-seq enhancer lists from in vivo intracranial xenograft and in vitro cultured cells were merged to create a single peak file. RPKM (Reads Per Kilobase per Million) values within merged peaks were calculated. In vivo-specific or in vitro-specific enhancers were called as peaks with 3-fold increased or decreased RPKM values in vivo relative to in vitro, respectively.
Enhancer data presentation
Heatmaps, Aggregate plots and motif analysis were conducted using the Cistrome35 galaxy instance. Heatmaps were created using the Heatmap tool, with 5000 bp upstream and downstream, 200 bp step and saturation of 0.05. Aggregative plots were created using the Aggregation Plot tool, using a Span of 3000 bp and resolution of 200 bp.
Super-enhancers (SEs)
In vivo and in vitro SEs were identified using the dynamicEnhancer software (retrieved from https://github.com/BradnerLab/pipeline). H3K27Ac peaks identified as enhancers and separated by less than 12.5 kb were stitched together. All stitched peaks were then ranked by the density of H3K27Ac minus input. Peaks higher than the inflection point on the density curve were designated SEs. To call condition-specific SEs, SEs from in vivo and in vitro conditions were merged and H3K27Ac signals for all merged SEs in each cell line were calculated. To be considered specific to in vivo or in vitro, SEs had to be only called a SE in a single condition and had to have at least 1.5-fold change in H3K27Ac signal between conditions.
JMJD6 binding analysis
Genomic distribution of JMJD6 (Extended Data Fig. 8c)
To call the genomic distribution of JMJD6 binding, each JMJD6 ChIP-seq binding peak was assigned to the underlying genomic element. Enhancers were defined as above, areas surrounding TSS’s were defined as +/− 1Kb of all annotated TSS’s in hg19, and other elements were defined by hg19 annotation. Each peak was assigned to a single element. In the cases in which a JMJD6 binding peak overlapped with two elements (e.g., enhancer and intron), priority was assigned in the following order: Enhancer > TSS +/− 1Kb > 5′ UTR > 3′ UTR > Exons > Introns > Intergenic.
Enrichment analysis of JMJD6 binding (Fig. 3d)
To determine enrichment scores of JMJD6 peaks at a particular genomic element (e.g. exons), a null binding distribution of JMJD6 peaks for each element was generated by randomizing the peaks throughout the genome 1000 times using bedtools shuffle. A z-score enrichment value at each genomic element was calculated using the actual number of JMJD6 peaks assigned to the genomic element compared to the average number of JMJD6 peaks and standard deviation from the null distribution. P-values were obtained from this z-score using the standard normal distribution.
JMJD6, Enhancer and Pol II binding data presentation (Fig. 3g)
Aggregate plots were constructed using the Cistrome35 galaxy instance with the Aggregation Plot tool, using a Span of 3000 bp and resolution of 200 bp. P-values were calculated using Mann-Whitney-U test of RPKM values of Pol II at each set of loci.
Pausing Index Calculation
RPKM values were generated for all hg19 TSS loci (from 50 bp upstream to 200 bp downstream of TSS (−50, +200)) and for the gene body of these transcripts (from 500 bp downstream of TSS to transcription termination site (TTS) (+500, TTS)) by first using the Galaxy 27 tool “Count intervals in one file overlapping intervals in another file” to obtain total read counts for each TSS loci and gene body. Total Pol II reads were aligned to hg19 as above to create the necessary BAM file. RPKM values were then calculated using the formula: ((number_of_reads +1)/(size_of_peak_in_bp/1000)) /number_of_aligned_reads_in_millions.
Results were filtered as follows to ensure only high confidence calls were considered in the downstream analysis:
Transcripts <750 bp were removed
Transcripts overlapping any of the ENCODE blacklisted regions were removed
Transcripts with Pol II density < 3.0 RPKM in the TSS region in either in vivo OR in vitro condition were removed
Transcripts with Pol II density < p0.4 RPKM in the BODY region in both in vivo AND in vitro conditions were removed
For genes with multiple TSS’s, only the transcript with the highest occupied TSS (by RPKM) was used
Any gene that was not expressed at 0.25 FPKM by RNA-seq was removed
Any transcript that overlapped other transcripts at this step was removed to avoid potential artefacts that occur by including reads from other TSS regions within the gene BODY region
These filtering steps resulted in ~9,000 transcripts for GBM528 and ~8,000 transcripts for GBM3565. Pausing Index (PI) was then calculated for each transcript using the formula: PI = (RPKM TSS)/(RPKM Body).
Individual gene knockdown studies by shRNA
For inducible shRNA knockdown studies of JMJD6 (Extended Data Fig. 9a–d), LT3REVIR, a lentiviral version of the same inducible RT3REVIR delivery vector used in the primary screen was used4. Glioblastoma cells were infected in culture and Venus+ cells harbouring the shRNA were selected by FACS. Cells were induced with doxycycline for 48 hr and dsRED+ cells expressing the shRNA were then selected by FACS. For in vitro studies, dsRED+ cells were plated in 96-well plates on Geltrex™ LDEV-Free hESC-qualified Reduced Growth Factor Basement Membrane Matrix (ThermoFisher) and maintained in doxycycline containing media. Cells were imaged using an Operetta High Content Imaging System (Perkin Elmer) on indicated days. Number of dsRED+ cells for each well was calculated using Harmony High Content Image Analysis (Perkin Elmer). For in vivo studies, dsRED+ cells were intracranially implanted into age-matched female NSG mice. Animals in the induced arm were maintained on doxycycline containing water (2mg/ml Doxycycline and 2% sucrose, Sigma) for the duration of the experiment. Mice in the uninduced control arm were maintained on water containing 2% sucrose. The induced shJMJD6 group contained 5 mice, while non-targeting (shREN) and uninduced shJMJD6 control groups each contained 3 mice, for a total of 6 control mice. All mice were monitored daily until development of neurological signs, at which time they were euthanized.
shRNA sequences:
JMJD6.1005: TGCTGTTGACAGTGAGCGACAGCAAGACGAAGCTATTACATAGTGAAGCCACAGATGTATGTAATAGCTTCGTCTTGCTGGTGCCTACTGCCTCGGA
REN.713: TGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGA
Validation studies of in vivo screen hits
We used the constitutive shRNA vector SGEP4, which is a constitutive version of the lentiviral vector used above. It allows for puromycin selection and has a constitutive GFP fluorescent reporter. For validation of 9 primary screen hits (Extended Data Fig. 3e–h), including the positive control, patient-derived glioblastoma GBM528 cells were transduced with individual shRNAs targeting primary screen hits, or 2 independent negative controls (totalling 11 independent transduced populations, each transduced with a single shRNA). Infected populations of GBM528 cells were selected by 1 ug/ml puromycin for 3 days and immediately intracranially implanted into age-matched female NSG mice; 4 mice were used for each experimental and control group. All mice were monitored daily until development of neurological signs, at which time they were euthanized.
In parallel, we conducted more in-depth validation of a top scoring transcription elongation factor in the primary screen, DOT1L, as well as another top screen hit that was not a transcription elongation factor, DPY30 (Fig. 4c, d and Extended Data Fig. 9i–x). We used the top two scoring shRNAs from the screen for each gene and used three independent glioblastoma models, GBM3565, GBM528 and GBMcw1919. As above, we transduced cells with shRNAs targeting DOT1L, DPY30, or a non-targeting shRNA. Infected populations were selected by 1 ug/ml puromycin for 5 days and knockdown of target was confirmed by qRT-PCR. To determine the effect of target knockdown on cells in vitro and in vivo, the same populations of cells harvested at the same time point were used in in vitro proliferation assays and for in vivo survival studies in intracranial PDX models. For in vitro studies, cells were plated in 96 well plates on Geltrex™ as above and maintained in standard serum free media. Cell proliferation was measured using Cell-Titer Glow (Promega). For in vivo studies, cells were intracranially implanted into age-matched NSG mice. All mice were monitored daily until development of neurological signs, at which time they were euthanized.
shRNA sequences:
DPY30.00786: TGCTGTTGACAGTGAGCGACAGAAAAGTCATCAAAGCAGATAGTGAAGCCACAGATGTATCTGCTTTGATGACTTTTCTGCTGCCTACTGCCTCGGA
DPY30.00781: TGCTGTTGACAGTGAGCGCCAACGTTGAGAGAATAGTAGATAGTGAAGCCACAGATGTATCTACTATTCTCTCAACGTTGTTGCCTACTGCCTCGGA
DOT1L.19417: TGCTGTTGACAGTGAGCGACGCGAGTTCAGGAAGTGGATATAGTGAAGCCACAGATGTATATCCACTTCCTGAACTCGCGGTGCCTACTGCCTCGGA
DOT1L.00642: TGCTGTTGACAGTGAGCGCTCAGAATAAACAGTAGAAACATAGTGAAGCCACAGATGTATGTTTCTACTGTTTATTCTGAATGCCTACTGCCTCGGA
CRISPR-mediated JMJD6 knockout studies
CRISPR design and construction
The CRISPR design tool from Broad institute (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) was used to design the guide-RNA (gRNA). Oligonucleotides were purchased from Fisher, annealed and cloned into LentiCRISPR v2 plasmid, which was a gift from Dr. Feng Zhang (Addgene plasmid #52961). The oligonucleotides used were as follows:
JMJD6.454 Forward:CACCGTTAATTCCCTCAGGATAACG
JMJD6.454 Reverse:AAACCGTTATCCTGAGGGAATTAAC
JMJD6.712 Forward:CACCGCTGCTGTCAAAGATGTAAAG
JMJD6.712 Reverse:AAACCTTTACATCTTTGACAGCAGC
Experimental design for complete knockout in clonal populations
For complete knockout studies (Fig. 4a–c), GBM528 cells were transduced with 1 of 2 independent CRISPR-Cas9 constructs targeting JMJD6 or a non-targeting (NT) control and selected for integration of the lentiviral construct by puromycin. Single cells were expanded in vitro to obtain clonal populations and knockout was confirmed by Western-blot (see below for methods). Two clonal populations per sgRNA were subjected to parallel in vitro proliferation assays and in vivo survival assays. For in vitro studies, cells were plated in 96 well plates on Geltrex™ as above and maintained in standard serum free media. Cell proliferation was measured using AlamarBlue® cell viability reagent (ThermoFisher). For in vivo studies, cells were intracranially implanted into age-matched female NSG mice. 5 mice for each clone, or 10 mice for each sgRNA construct were used. All mice were monitored daily until development of neurological signs, at which time they were euthanized.
Experimental design for knockout in bulk populations without clonal selection
Due to knockout efficiency being extremely high in the experiment above, we used CRISPR-mediated knockout of JMJD6 on a population of cells without clonal selection to confirm our results in another PDX glioblastoma model (Extended Data Fig. 9e–g). GBMcw1919 cells were transduced as above, selected by puromycin, subjected to Western blot to confirm population based knockdown, and then assayed in vitro and in vivo as above. For in vivo survival studies, 5 age-matched female NSG mice were used for each sgRNA construct.
Western Blotting for JMJD6 to determine knockout
Cells were collected and lysed in hypotonic buffer with nonionic detergent (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% NP-40; 50 mM NaF with protease inhibitors), incubated on ice for 15 minutes, and cleared by centrifugation at 10,000 g at 4°C for 10 minutes. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories). Equal amounts of protein were mixed with reducing Laemmli loading buffer, boiled, and electrophoresed in NuPAGE Gels (ThermoFisher), and then transferred to PVDF membranes (Millipore). Blocking was performed for 30 minutes with 5% nonfat dry milk in TBST and blotting performed with primary JMJD6 antibody (Active Motif, 61494) for 16 hours at 4°C.
Tissue processing, histology and imaging
Tissues for histological sections was performed as reported previously36. Primary antibodies used: GFP to stain for Venus (1:250; Aves Labs Cat# GFP-1020), mCherry to stain for dsRED (1:250; Abcam Cat# Ab167453) and human nuclear antigen to stain for human cells (1:250; NovusBio clone 235-1 Cat# NBP2-34525V3). Species-specific Alexa-Fluor-conjugated secondary antibodies were used for detection (1:500; ThermoFisher).
Tissue microarray immunohistochemistry and analysis
Briefly, deidentified tissue microarrays (TMAs) were constructed from gliomas after obtaining University of Kentucky Institutional Review Board Approval. Three 2-mm diameter cores per tumour were obtained, with each core embedded in a separate TMA block. A total of 104 cases comprised the TMAs, including 9 nonneoplastic controls (cortical dysplasias), 9 grade II astrocytomas, 11 grade III astrocytomas, 12 anaplastic oligodendrogliomas, 16 grade II oligodendrogliomas, and 47 grade IV glioblastomas (GBMs). Immunohistochemistry was performed for each core as described previously37, but using an antibody towards JMJD6 (Abcam, ab64575). Briefly, each core was semiquantified on a relative scale from 0 to 3, with 0 = negative and 3 = strongest. Results from all 3 cores were averaged together to produce a final score for a tumour. Results were plotted based on WHO grade and differences were calculated via Mann-Whitney-Wilcoxon Test.
Retrospective analysis of gene expression in human gliomas
Gene expression correlations across primary patient glioblastoma tumours, expression of individual genes or gene signatures in primary patient gliomas, and patient survival were determined through analysis of the Allen Institute’s Ivy Glioblastoma Atlas Project (IvyGAP; http://glioblastoma.alleninstitute.org), the National Cancer Institute’s Repository for Molecular Brain Neoplasia Data (REMBRANDT, https://caintegrator.nci.nih.gov/rembrandt/), or The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/). Normalized Ivy Glioblastoma Atlas Project (Ivy GAP; http://glioblastoma.alleninstitute.org) and REMBRANDT datasets were downloaded from GlioVis (http://gliovis.bioinfo.cnio.es/), and TCGA GBM RNAseqV2 dataset was downloaded from the Broad Institute GDAC Firehose via the TCGA2STAT package on R38.
Correlation analysis of gene expression and gene signature scores (Fig. 3a and Extended Data Fig. 7a–d)
We created a gene signature of the 55 genes upregulated in vivo >2.5 fold in both GBM528 and GBM3565 models. Gene signature scores within Ivy GAP samples were analysed via single-sample GSEA on GenePattern and normalized as Z-scores across all samples in the given dataset16,39. Correlation between gene signature Z-scores and median-centred gene expression was determined as Pearson coefficient (r)-values. Associated FDR-adjusted p-values were calculated using the Benjamini-Hochberg procedure. MatLab (MathWorks, Natick, MA, USA) was used to determine correlation values and p-values.
Individual gene correlation with JMJD6 (Extended Data Fig. 7e)
To test correlation of all genes with JMJD6 across glioblastoma tumours, TCGA RNAseqV2 data were downloaded and analysed using R (www.r-project.org). The function ggpairs, available through the GGally package, was utilized to generate plots and determine pairwise correlation coefficients (http://CRAN.R-project.org/package=GGally). All glioblastoma tumours in the TCGA with RNA-seq data available were used.
Survival Analysis
For gene expression changes, high and low groups were defined as above and below the median, respectively. Kaplan–Meier curves were generated and log-rank (Mantel-Cox) analysis was performed using GraphPadPrism software (GraphPad Software, La Jolla, CA, USA).
Direct comparison of TCGA RNAseq gene expression data to our RNAseq data (Extended Data Fig. 5d, e)
In order to directly evaluate the expression/activity level of genes in an existing glioblastoma gene expression databases, we utilized the RNAseqV2 TCGA database of glioblastoma tumours that were characterized by mRNA-sequencing. FPKM data for RNAseqV2 were downloaded from https://gdc-portal.nci.nih.gov/. We compared this data to our mRNA-sequencing data from both intracranial and culture conditions in two glioblastoma tumour models. In order to directly compare values, we took the raw FPKM values from the 166 TCGA tumours and our data and quantile normalized them together. We evaluated expression of the 55 genes that were upregulated in vivo in both GBM3565 and GBM528 or all genes.
Statistical Analysis
Analysis for each plot is listed in figure legend and/or in corresponding methods above. Briefly, all grouped data are presented as mean +/− S.D. All box and whisker plots of expression data are presented as median (middle line of box) +/− 25 percentile (top and bottom line of box, respectively). P-values presented are calculated by 2-sided Mann-Whitney-U Test. Kaplan–Meier curves were generated and log-rank (Mantel-Cox) analysis was performed to generate P-values using GraphPadPrism software (GraphPad Software, La Jolla, CA, USA). Sample sizes for each experiment are given in corresponding figures and/or methods above. Sizes were chosen based on previous experience with given experiments, or in the case of retrospective analysis, all available samples were included.
Data Availability
The datasets generated during and analysed during the current study are included within the published article (and supplemental information tables) or have been deposited in Gene Expression Omnibus (RNA-seq and ChIP-seq Data) under accession number GSE74529 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74529
Extended Data
Extended Data Figure 1. shRNA delivery vector performance.
a, Schematic of the RT3REVIR shRNA delivery vector (Top). Once integrated into cells, a constitutive PGK promoter drives Venus-fluorescent reporter and rtTA through an IRES element, creating an All-in-One inducible vector (Middle). When doxycycline is introduced to cells, it binds to rtTA and drives activity of the 3rd generation TET-inducible promoter. This drives dsRED-fluorescent reporter and shRNA expression. In addition, it drives higher rtTA transcription through the IRES element, creating a positive-feedback loop that increases rtTA expression in the cell resulting in higher expression of inducible elements (Bottom). b, The inducible shRNA delivery vector displays almost no unintended induction. Representative FACS plots from the parallel screen of cells infected with RT3REVIR with and without doxycycline treatment in vitro (Left) and in vivo (Right). c, RT3REVIR robustly expresses shRNAs and depletes cells expressing cell-lethal shRNAs in a competitive proliferation assay. Representative FACS plots over time of cells infected with a positive control shRNA against RPA3 and induced (Left). Quantification of fluorescent cells in the representative competitive proliferation assay. Empty = cells with vector that had no shRNA. REN.713 = cells with vector containing a negative control shRNA targeting the Renilla protein (not expressed in human cells). Bars represent percent of cells actively expressing the shRNA within the total infected population from a single dish (Right).
Extended Data Figure 2. Importance of combining multiple mice to achieve increased reproducibility.
a, Correlation (r) values between individual mice (41 pair-wise comparisons for induced mice and 20 pair-wise comparisons for control uninduced mice) or of grouped replicates containing multiple mice (3 pair-wise comparisons for triplicate induced arm replicates and 1 pair-wise comparison for the duplicate control uninduced replicates). b, Positive control gene RPA3 was effectively depleted from cell populations in both in vivo and in vitro screens using grouped replicates for the in vivo screen. Four shRNAs targeting RPA3 were included in the shRNA screening library. At least 2 of 4 shRNAs achieved a RIGER depletion score of 2.0 or greater.
Extended Data Figure 3. Validation of in vivo screen results.
a, b, Average mRNA expression of intracranial-specific (a) or cell culture-specific hits (b) in vivo and in vitro. P-values calculated by 2-sided Mann-Whitney U Test and were >0.05. c, d, Screen hits were analysed for enrichment of GO gene sets, using the screened library gene list as background to control for bias toward chromatin modifiers. All results with an FDR of 0.05 or lower for in vivo-specific hits are presented (c). Top 10 results with an FDR of 0.05 or lower for in vitro-specific hits are presented (d). Significance calculated by Benjamini-Hochberg FDR. e–h, Validation of elongation factor hits by in vivo survival assays. shRNAs from the primary screen were used to transduce GBM528 cells using a constitutive expression vector. Primary screen hits that led to an increase survival with knockdown, with at least 2/4 mice surviving longer than all 9 negative control mice, were considered validated. e, f, Target mRNA knockdown by qRT-PCR (e) and Kaplan-Meier survival curve (f) of validated hits. Black lines and bars represent two independent negative control shRNAs. Purple/pink lines/bars represent validated in vivo-specific hits and blue lines/bars represent validated common hits found in both in vivo and in vitro screen. g, h, Target mRNA knockdown by qRT-PCR (g) and Kaplan-Meier survival curve (h) of primary screen hits that did not validate (green lines/bars).
Extended Data Figure 4. Independent models confirm stimulus-controlled and stress response programs upregulated in vivo and in primary tumours.
a, b, Genes with average of >2.5 fold expression change between conditions in GBM528 (a) GBM3565 (b) cells, as determined by RNA-seq. GBM528 heatmap associated with Main Figure 2b, c. c, d, Cellular programs enriched by GSEA in cells grown in each condition. Representation of all enriched programs in vitro and in vivo using Enrichment Map (c). Example GSEA plots for cells grown in vitro and in vivo (d). FDR calculated by GSEA software. d–g, In the same manner as in (b–d), GSEA was performed using data generated from 2 independent models by Lee and colleagues15 on cells grown in vitro vs. in vivo intracranial xenograft tumours (e, g or in vitro vs. the primary glioblastoma from which the cells were derived (f, h). Cellular programs enriched in cells grown in each condition are presented using Enrichment Map. FDR calculated by GSEA software.
Extended Data Figure 5. Transcription factors and signalling molecules that drive stimulus-controlled programs consistently upregulated in vivo.
a, Transcription factors upregulated in GBM528 cells upon growth in vivo. Values are mean FPKM +/− s.d from biological duplicates. b, Cell signalling programs regulated by pause-control that are enriched for upregulated transcription factors in (a). FDR calculated by MSigDB for enrichment against all genes. c, 55 genes upregulated more than 2.5-fold in both GBM528 and GBM3565 upon growth in vivo. d, Expression of those 55 genes in vivo and in vitro in GBM528 and GBM3565 compared to expression in primary glioblastoma tumours from the TCGA RNAseqV2 database. Data was FPKM quantile normalized across all datasets before plotting. e, All genes in datasets shown to confirm normalization.
Extended Data Figure 6. Epigenomic regulation of glioblastoma cells is microenvironment-specific.
a–h, Global enhancer landscape of GBM528 (a) and GBM3565 (d) cells in both conditions and microenvironment-specific enhancers. b–h, Browser track examples (b, e), aggregate plots (c, f), and gene expression fold change of target genes (nearest expressed gene) (g, h) of microenvironment-specific enhancer loci from (a, d). i, Super-enhancers identified in vivo in GBM528. j, Browser track examples of condition-specific SEs. k, Super-enhancers specific to each condition were identified. l, Expression of condition-specific super-enhancer target genes. Boxplot P-values calculated by 2-sided Mann-Whitney U Test.
Extended Data Figure 7. Prioritization of JMJD6 as lead target.
a, Expression across all Ivy GAP samples of gene signature of 55 genes upregulated in vivo in both PDX models (top), and of elongation factor hits (middle). Corresponding histological tumour structure and TCGA molecular subtype of each sample represented below (bottom). The signature and JMJD6 expression is highest in hypoxic regions, which also corresponds to more Mesenchymal-like regions of the tumour. b, Expression correlation dotplot for JMJD6 data represented in (a). c–d, Expression correlation of each elongation factor hit with gene signature of 55 genes upregulated in vivo in both PDX models across all REMBRANDT (c) and TCGA (d) Glioblastoma tumours (bulk tumour expression. Ivy Gap data in (a) and (Main Figure 3a) is microenvironment-specific expression). P-value (a–d) by FDR-adj. B–H procedure. e, JMJD6 mRNA-seq correlation with each gene across TCGA GBM tumours. f, Example plots from GSEA of using the gene correlations in (b) and a pre-rank list. e, Representative images from tissue microarray analysis of JMJD6 protein expression in (Main Fig. 3c). h, Primary screen results for the 4 shRNAs targeting JMJD6. Only 2 of the 4 shRNAs were represented in the library at appreciable levels, and both led to a RIGER depletion score of greater than 2. Values are median RPM +/− s.d. of 3 biological replicates for induced populations and 2 biological replicates for the uninduced population.
Extended Data Figure 8. JMJD6 regulates enhancer mediated pause-release in GBM.
a, Known role of JMJD6 in transcription pause-release. In HEK293T and HeLa cells, JMJD6 acts with bromodomain containing 4 (BRD4) as a key activator of enhancer-mediated pause-release at genes controlled by Pol II pausing12. Upon enhancer activation, JMJD6 demethylates 7SK RNA releasing positive transcription elongation factor (P-TEFb) inhibition from the 7SK/HEXIM complex12 b, Browser tracks of JMJD6 at enhancers and TSS’s. c, Global distribution of genomic elements as determined by Hg19 reference genome and H3K27Ac ChIP-seq (left) and global distribution of JMJD6 binding peaks per genomic element as determined by JMJD6 ChIP-seq (right). Enrichments shown in (Main Fig. 3d). d–g, GSEA enrichment plots of genes with JMJD6-bound enhancers in the GBM528 (d) or GBM3565 (e) PDX model against differential expression of genes between in vivo and in vitro conditions (expression from Main Fig. 2b for GBM528 and Ext. Data Fig. 4a for GBM3565). GSEA enrichment plots of genes with JMJD6-bound enhancers in the GBM528 (f) or GBM3565 (g) PDX model against gene correlations with JMJD6 in TCGA tumours (correlations from Ext. Data Fig. 7e). h, i, Distribution of pausing-index of the common in vivo upregulated genes from Extended Data Fig. 5c for which pausing index could be determined in GBM528 (h) and GBM3565 (i). All P-values calculated by 2-sided Mann-Whitney U Test.
Extended Data Figure 9. Validation of JMJD6 and other hits in multiple PDX models of glioblastoma.
a, JMJD6 mRNA expression by qRT-PCR after inducible shRNA knockdown of JMJD6. b, In vitro proliferation and c, in vivo survival compared to uninduced and induced non-targeting controls. Values are mean +/− s.d. of 3 technical replicates. d, Endpoint tumours harvested from the induced arm of (c) stained to show human tumour cells (human nuclear antigen) that harbour a JMJD6 shRNA (Venus+) or harbour and express a JMJD6 shRNA (Venus+dsRED+). The vast majority of tumour cells at endpoint had silenced the shRNA (Venus+dsRED−). Scale bar: 200 μM. e, CRISPR mediated knockout of JMJD6 in a bulk population of GBMcw1919 cells in vitro. f–g, Parallel in vitro proliferation assay (f) and in vivo survival assay (g) of cells from (e). h–l, Constitutive shRNA knockdown of DOT1L and DPY30 in vitro. m–v, Parallel in vitro proliferation assays (m–q) and in vivo survival assays (r–v) of cells from (h–l), respectively. Error bars of bar graphs +/− s.d. of at least triplicates.
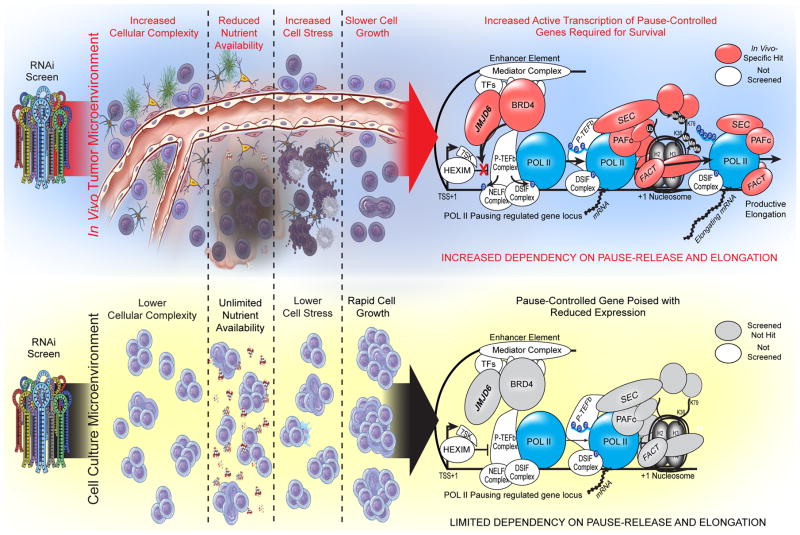
Extended Data Figure 10. Summary Figure.
Overview summary of results. The in vivo tumour microenvironment, both in primary glioblastoma tumours and intracranial xenograft tumours, is complex and stressful for cells. Tumour cells must appropriately interact with and respond to a large number of other cells, both cancerous and non-cancerous, in order to survive. They also must activate response pathways to survive in the face of reduced nutrient availability, including hypoxic and low glucose conditions, and in the face of increased cell stress due to immune regulators, and debris and signalling from apoptotic cells. Thus, slower growth is seen as the cells expend energy on responding to these microenvironmental stimuli in order to survive. Due to the large number of pause-controlled genes needed to appropriately respond to the cell stresses in vivo, cells are dependent on transcriptional pause-release and elongation. In contrast, cell culture conditions are optimized to reduce cell stress and drive growth by providing a surplus of all required nutrients for cell growth. Cells are largely homogenous and cancerous. Together, these in vitro conditions lead to rapid cell growth and little need for pause-controlled pathways that respond to environmental stimuli and stress. Therefore, in vitro cells are not as dependent on transcriptional pause-release and elongation for growth and survival.
Supplementary Material
Acknowledgments
The authors thank the Hemann, Paddison, and Zuber labs, along with Kareem Alazem for technical help with RNAi screening. We thank Bjarne Kristensen and Ann Mari Rosager for input on histological analysis and Alina Saiakhova, E. Ricky Chan, Massimo Squatrito and Wen Liu for bioinformatic support. Additional support was provided by the Cytometry & Imaging Microscopy and Genomics core facilities of the Case Comprehensive Cancer Center at Cleveland Clinic and Case Western (P30CA043703), respectively. We also thank members of the Rich, Tesar and Scacheri labs for input on the project and discussion about the manuscript. This work was supported by Velosano (J.N.R.); New York Stem Cell Foundation-Robertson Investigator Award (P.J.T.); CIHR Banting Fellowship (S.C.M.); National Institutes of Health grants CA183510 (T.E.M.); GM007250 (T.E.M., A.R.M., L.J.Y.K., J.J.M.); CA189647 (C.G.H.); CA154130, CA169117, CA197718, CA171652, NS087913, and NS089272 (J.N.R.).
Footnotes
Author Contributions
T.E.M., P.J.T. and J.N.R designed the overall screening strategy. T.H., J.Z., P.J.P. and M.T.H provided technical expertise, unpublished reagents and methods for the screen. T.E.M., S.C.M., C.G.H. and L.W. performed the screen. T.E.M., L.C.W. and S.C.M. produced and sequenced screening libraries. T.E.M., P.J.T. and J.N.R. processed and analysed screening results. T.E.M., L.C.W. and D.C.F. generated and analysed RNA-seq data. T.E.M., B.B.L., S.M.G., R.T., B.E.B., L.C.W. and S.C.M. generated ChIP-seq data. T.E.M., B.B.L, A.R.M., B.E.B., J.J.M. and P.C.S. analysed and interpreted ChIP-seq data. T.E.M., L.K., and W.A.F. performed bioinformatic analysis of published expression datasets. T.E.M, D.D., Q.X., L.C.W. and L.W. performed functional knockdown and CRISPR knockout studies. C.H. created and analysed tissue microarray for JMJD6 expression. T.E.M., P.J.T. and J.N.R. analysed all data and wrote the paper. All authors provided intellectual input, edited and approved the final manuscript.
The authors declare no competing financial interests.
LITERATURE CITED
- 1.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The lancet oncology. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature biotechnology. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell reports. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 5.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 7.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adam RC, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JD, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Molecular cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akhtar-Zaidi B, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nature reviews. Molecular cell biology. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L, Li L, Zhou B, Qin Z, Dou Y. H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Molecular cell. 2014;54:920–931. doi: 10.1016/j.molcel.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams LH, et al. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Molecular cell. 2015;58:311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews. Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilchrist DA, et al. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes & development. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galbraith MD, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nature genetics. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroder S, et al. Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Molecular cell. 2013;52:314–324. doi: 10.1016/j.molcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nameeta Shah PD, Feng Xu, Ph.D, Lankerovich Michael, Ph.D, Puchalski Ralph B, Ph.D, Keogh Bart., M.D., Ph.D 2016 http://dx.doi.org/10.7937/K9/TCIA.2016.XLwaN6nL.
- 25.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 27.Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reimand J, Arak T, Vilo J. g:Profiler--a web server for functional interpretation of gene lists (2011 update) Nucleic acids research. 2011;39:W307–315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suva ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome biology. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller TE, et al. Lgr5 Marks Post-Mitotic, Lineage Restricted Cerebellar Granule Neurons during Postnatal Development. PloS one. 2014;9:e114433. doi: 10.1371/journal.pone.0114433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert MR, et al. Autophagy and oxidative stress in gliomas with IDH1 mutations. Acta neuropathologica. 2014;127:221–233. doi: 10.1007/s00401-013-1194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan YW, Allen GI, Liu Z. TCGA2STAT: simple TCGA data access for integrated statistical analysis in R. Bioinformatics. 2016;32:952–954. doi: 10.1093/bioinformatics/btv677. [DOI] [PubMed] [Google Scholar]
- 39.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analysed during the current study are included within the published article (and supplemental information tables) or have been deposited in Gene Expression Omnibus (RNA-seq and ChIP-seq Data) under accession number GSE74529 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74529