Abstract
In this study, we report that antigen-specific CD19+CD27+CD21lo (CD21lo) B cells are transiently induced 14-28 days after immunization, at the time germinal centers (GCs) peak. Although clonally related to memory B cells and plasmablasts, CD21lo cells form distinct clades within phylogenetic trees based on accumulated variable gene mutations, supporting exit from active GCs. CD21lo cells express a transcriptional program suggesting they are primed for plasma cell differentiation and are refractory to GC differentiation, although they do not spontaneously secrete antibody. Additionally, CD21lo cells differentially express multiple cell surface markers and have elevated intracellular levels of Blimp-1 and T-bet protein compared to memory B cells. Together, these data support a model in which CD21lo cells are recent GC graduates that represent a distinct population from CD27+ classical memory cells, are refractory to GC reentry and are predisposed to differentiate into long-lived plasma cells.
Introduction
Immunological memory is the ability to generate rapid, effective responses to previously encountered pathogens and is a hallmark of adaptive immunity. Germinal centers play a major role in B cell memory development where somatic hypermutation of the immunoglobulin genes allows for the rapid adaptation to antigens. Competition for cognate T cells between limited numbers of B cell clones within each GC allows high affinity memory and plasma cells to emerge, forming the basis of humoral memory1,2. When memory B cells encounter their cognate antigen, they are rapidly reactivated and can differentiate into plasmablasts that secrete large amounts of protective antibodies into the bloodstream, or they can return to GC reactions where further affinity maturation occurs3,4. In contrast, long lived plasma cells continuously secrete antibody over extended periods of time, providing continual serum-level protection5. The secreted antibodies from both plasmablasts and plasma cells can bind pathogens and protect by directly inhibiting receptor-ligand interactions or by facilitating the phagocytosis or lysis of the pathogen6,7.
Although memory B cells and long lived plasma cells are relatively well characterized, a growing literature describes various memory-like B cell subsets that are phenotypically distinct from classical memory populations. They are typically characterized by elevated levels of negative regulators of BCR signaling, like FCRL4 or FCRL58–11, or decreased levels of positive regulators, like CD2112–17. Memory B cells with decreased levels of CD21 are further separated into subsets that express8,9,13,16,18, or do not express14,15,17,19,20 the canonical human memory B cell marker, CD2721,22, or have heterogeneous CD27 expression12. These subsets are likely not mutually exclusive. The subsets defined by elevated FCRL4 or FCRL5 expression all show decreased levels of CD218–11. Additionally, multiple studies of cells defined by decreased CD21 expression also show higher levels of FCRL4 or FCRL513–15,17,20. The immunological role of these different populations, as well as their relationship to other B cell subsets, remains unclear. They have largely been identified in the context of chronic infection11,14–17,19,20, but have also been documented in autoimmunity13,23, and in healthy tonsils and peripheral blood8,9,12. The variety of contexts in which non-classical memory B cells have been identified suggests that the memory B cell compartment is highly heterogeneous and that these non-classical cells may have distinct functional roles in humoral immunity.
These various non-classical memory B cells share many characteristics, despite variations in how various investigators have chosen to define their identifying cell surface markers. One common characteristic is evidence of GC experience. Many studies have found direct evidence of this by demonstrating that these subsets have undergone isotype switching10,11,17,20,24 and somatic hypermutation11,17. Additionally, non-classical memory B cells identified in chronic infection settings are enriched for antigen specific cells, which suggests they have undergone affinity maturation in GCs10–12,17,20.
Another common observation is that non-classical B cells are functionally distinct from classical memory B cells. Multiple studies have found elevated levels of CD95 (Fas) expression and an increased propensity for apoptosis both with and without stimulus in CD27+FCRL4+, CD27+CD21lo and CD27-CD21lo cells9,12,13,16. These subsets have a reduced capacity for BCR signaling compared to memory B cells: they have elevated levels of BCR inhibitory molecules like SIGLEC6 and SIGLEC109,10,13,14,17, decreased calcium flux after BCR stimulation12,13,16,17, a decreased ability to proliferate after BCR specific and nonspecific stimulation8,12,13,15,17, and a diminished potential to differentiate into antibody secreting cells10,16,17.
Many non-classical memory B cell subsets downregulate receptors required to participate in GC reactions, including L-selectin, CCR7, CXCR4 and CXCR58,10,13–17,20. Furthermore, many of these populations also upregulate FGR, a gene that negatively regulates chemokine signaling14,16. There is also limited evidence that the Blimp-1 pathway, the master regulator of plasma cell differentiation and an antagonist to the BCL-6 driven GC program is also upregulated. One study found an increase in Blimp-1 by RNASeq in FCRL5+ cells in individuals exposed to malaria10, while another showed that Bach2, a Blimp-1 inhibitor, was significantly decreased in CD21lo cells in HCV patients16.
The FCRL4/5+, CD27+CD21lo, and CD27-CD21lo subsets also share characteristics with another recently described memory population that is expanded in aging mice and humans25,26. These cells have been named age-associated B cells (ABCs) and lack CD21 expression, upregulate Fas and CD11c, and are GC experienced. They overexpress CD80, CD86, and MHC class II, are enriched for IgG2a+ cells in mice and the phenotype of these cells is dependent on their expression of the transcriptional factor, T-bet25,27.
However, there is a growing appreciation that ABCs may not simply be induced by age, but are generated in response to a combination of BCR, TLR7, and IFNGR signaling, that would occur during anti-viral responses25,27. Furthermore, ABCs in mice with viral infections were found to be antigen specific and their depletion results in a dampened antiviral B cell response27. Together, the data suggests that ABCs may be a similar and overlapping population with the FCRL4/5+, CD27+CD21lo, or CD27-CD21lo subsets.
The diverse context in which these non-classical populations have been observed strongly suggests that they are a critical component of the humoral immune system, but because the majority of previous studies characterizing these cells involved individuals with chronic infection or autoimmunity, the role of these cells in healthy acute immune responses is unknown. We set out to identify and comprehensively characterize one of these populations, the CD19+CD27+CD21lo (CD21lo) cells, in a cohort of healthy individuals who received the seasonal influenza vaccine. We tested the antigen specificity of CD21lo cells, analyzed their phylogenetic relationships to other B cell populations, and compared their transcriptional profile to memory B cells, the cells they most phenotypically resemble. We found substantial differences between CD21lo cells and classical memory B cells, which lead us to conclude that the CD21lo population is a developmentally distinct stage of B cell differentiation. In addition, we found evidence that CD21lo cells are recent GC graduates that are inhibited from re-entering the GC and are upregulating the Blimp-1 driven plasma cell program, indicating that CD21lo cells are a potential long lived plasma cell pre-cursor.
RESULTS
CD27+CD21lo B cells are enriched for antigen specific cells after influenza immunization
Previous studies have found that the various CD21lo populations described previously are expanded in individuals with chronic viral infection or autoimmunity and are enriched for antigen-specific cells13–16. To evaluate whether the expansion of either CD27+CD2lo or CD27-CD21lo cells occurs in an acute immune response, we determined the proportion of the four B cell subsets based on CD27 and CD21 expression within the CD19+CD38lo gate at four time points after seasonal influenza vaccination (Fig 1A). We found that the proportion of each cell type remain constant throughout the vaccine response (Fig 1B).
Fig. 1. CD21lo cells are enriched for influenza specific cells.
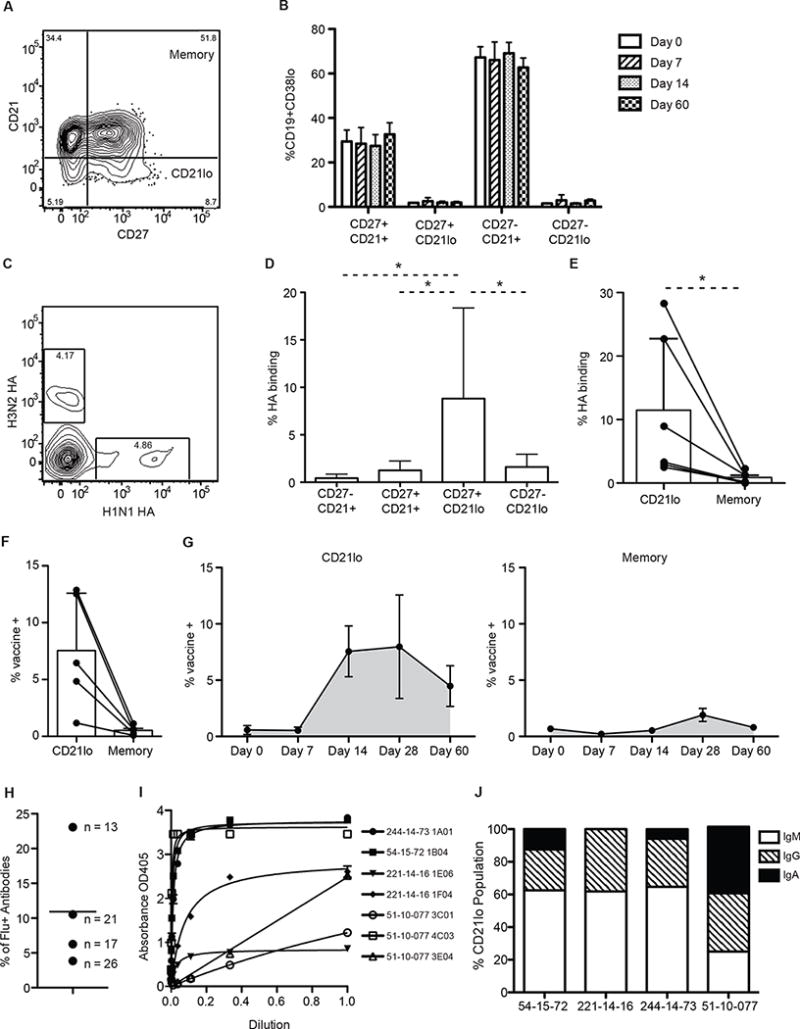
The proportion of influenza specific cells in the CD21lo population compared to the memory B cell population was evaluated from cells isolated from peripheral blood. (A) Representative FACS gating of CD19+ B cells by CD27 and CD21. (B) Proportion of CD19+CD38lo cells in the CD27−CD21+, CD27+CD21+, CD27+CD21lo, and CD27−CD21lo compartments at 0, 7, 14, and 60 days after vaccination (n=3). Bar graphs show mean (+/− SD) (C) Representative gating of hemagglutinin (HA) staining 14 days post-immunization in the CD19+CD38−CD27+CD21lo compartment. (D) Percentage of H1N1 and H3N2 HA binding cells in the CD27−CD21+, CD27+CD21+, CD27+CD21lo, and CD27-CD21lo subsets as determined by FACS analysis 14 days after immunization (n=6). Significance was determined using the Wilcoxon matched-pairs test. A CD19+CD38lo gate was included in the FACS analysis to remove plasmablasts from the analysis. Bar graphs show mean (+/− SD) (E) Percentage of H1N1 and H3N2 HA binding cells in the CD21lo or memory compartment as determined by FACS analysis (n=6). Lines link data points from the same individual. Significance was determined using the Wilcoxon matched-pairs test. Bar graphs show mean (+/− SD) (F) Percentage of vaccine specific IgM, IgG and IgA+ CD21lo and classical memory B cells as measured by ELISPOT 14 days post-immunization (n=5). Significance was determined using a two-tailed paired t-test. Bar graphs show mean (+/− SD) (G) Percentage of vaccine specific IgM, IgG and IgA+ CD21lo or classical memory B cells as measured by ELISPOT from 0 (n=4), 7 (n=3), 14 (n=5), 28 (n=5), and 60 (n=3) days post-immunization. Cells were sorted and stimulated for 5 days with CpG, PWM and SAC before being plated on ELISPOT plates coated with the 2012–2013 or 2014–2015 vaccine. Line graphs show mean (+/− SE) (H) Percentage of vaccine binding monoclonal antibodies generated from each individual. Values beside each data point represent the number of antibodies assayed for that person. (I) Scatchard plots of binding of specific antibodies to the 2014–2015 seasonal influenza vaccine (244-014-73, 54-15-72, 221-14-16) or inactivated H1N1 A/California/07/09 virus (51-10-077). (J) Isotype of the CD21lo cells isolated for mAb generation was determined using blast on sequences generated during the PCR steps of cloning. Bar graphs show the proportion of CD21lo cells that were cloned into mAbs that express IgM, IgG and IgA antibodies for each individual.
We also determined whether any of the four B cell subsets based on CD27 and CD21 expression were enriched for antigen specificity during the vaccine response using flow cytometry to measure the proportion of hemagglutinin (HA) specific cells present after seasonal influenza. HA is a glycoprotein on the surface of influenza and is the predominant target of the antibody response28. Fluorescently tagged HA from the A/California/04/09 H1N1 and A/Perth/16/09 H3N2 influenza vaccine strains were used to stain B cells to determine the proportion of antigen specific cells generated within the different subsets. We verified the specificity of the HA staining by single cell sorting CD19+ CD27+ HA+ B cells and generating monoclonal antibodies (mAbs) of which 92% were confirmed to bind HA by ELISA (Fig S1A-B). We found that 14 days after vaccination, the CD27+CD21lo population contained a significantly larger proportion of HA+ cells compared to the CD27+CD21+ classical memory, the CD27-CD21+ naïve, and the CD27-CD21lo “tissue-like” memory populations (Fig 1C–D). This enrichment was consistent across all six subjects we sampled (Fig 1E). On average, 11.45% of CD21lo cells from each donor bound HA while only 0.85% of classical memory B cells did.
The enrichment of antigen-specific CD21lo B cells 14 days after immunization was further verified in 5 additional subjects using a polyclonal stimulation ELISPOT assay29. On average, 7.5% of CD21lo cells bound vaccine, while only 0.52% of the memory B cells did (Fig 1F). This assay was also performed 0, 7, 14, 28, and 60 days after vaccination. Notably, the percentage of vaccine specific cells peaks at different times for CD21lo and classical memory B cells (Fig 1G). While the peak of vaccine specific CD21lo B cells began at 14 days post-vaccination and was waning at day 28, the peak of vaccine specific classical memory B cells did not begin until 28 days post-vaccination. Furthermore, unlike classical memory B cells, the CD21lo population contained virtually no influenza-specific cells before vaccination, demonstrating that antigen specificity in this population is transient.
In a third assay, mAbs were expressed recombinantly from the variable genes of single cell sorted CD21lo B cells from 4 individuals. Similar to the findings observed by flow cytometry and ELISPOT, 11% (7/74) of CD21lo B cells isolated 14 days after influenza vaccination were reactive to either the 2014–2015 quadrivalent vaccine (antibodies isolated during the 2014–2015 season) or the H1N1 A/California influenza virus (antibodies isolated during the 2010–2011 season), as measured by ELISA (Fig 1H–I). Notably, a sizeable proportion (average of 53%) of the CD21lo cells we isolated for mAb cloning express IgM BCRs, and notably all contained somatic mutations (Fig 1J). This supports a GC origin of CD21lo cells, as these IgM+ CD21lo cells are likely derived from naïve cells that were activated and underwent affinity maturation for the first time. In total, these results demonstrate that the CD21lo subset is a ready source for high affinity mAbs, as antigen specific cells are present in this population for a 2-week period following immunization and that these cells can also be isolated from frozen samples (Fig S2A). Similar to what is found in freshly isolated blood, CD21lo cells make up about 5% of the circulating CD19+CD27+ B cell population after thawing (Fig S2B).
Based on the three assays described above, we estimate that approximately 10% of CD21lo cells were influenza specific 14 days after vaccination, which is after the formation of germinal centers30,31. The specificity of the remaining CD21lo cells is unknown but likely includes additional vaccine-specific cells that bind epitopes not detected by our assays, or non-native forms of vaccine antigens32,33. Further, as the CD21lo subset remains enriched with antigen-specific cells for several weeks, this population should represent all specificities induced by recent immune responses besides the vaccine antigens. The antigen specific CD21lo population is rapidly induced and then decays following immunization, with kinetics consistent with a transient post-GC stage.
CD21lo B cells are clonally related to but phylogenetically distinct from plasmablasts and classical memory B cells
The differential kinetics of antigen specific CD21lo cells from plasmablasts and memory B cells suggest they are developmentally distinct. To further investigate this hypothesis, we performed high throughput sequencing of PCR amplified heavy chain genes from the plasmablasts (CD19+ CD38+ CD27+), memory B cells (CD19+ CD38lo CD27+ CD21+), and CD21lo cells (CD19+ CD38lo CD27+ CD21lo). The plasmablasts were isolated from peripheral blood 7 days post-immunization and the other populations were isolated at 14 and 90 days post-immunization.
The majority of VH genes used across the various B cell compartments belonged to the IGHV1, IGHV3, and IGHV4 families, consistent with previous observations34–36 (S3A). However, there was great diversity in which VH gene and VH and JH pairings were used as measured by Simpson’s Index (Fig S3B), indicating high levels of heterogeneity in the VH and JH usage between the different B cell subpopulations. To further characterize broad patterns in the genetic structure of these populations, neighbor joining trees were generated using a distance matrix generated from the number of sequences that used each VH gene or VH-JH combination. The samples in the tree clustered by individual, showing that each person utilized a diverse set of immunoglobulin genes in responding to influenza vaccination and possessed a unique gene usage signature (Fig S3C–D).
We then analyzed the patterns of somatic mutation within expanded clones to determine the phylogenetic relationships between the plasmablast, memory, and CD21lo sequences. We grouped the sequences into clonotypes based on VH and JH gene usage, CDR3 length, and 85% nucleotide similarity in the CDR335,37. Clonal prevalence was normalized across subjects for the number of sequences in each sample by downsampling those with more sequences. The majority of clones (average 80%) were present at a low frequency with less than 5 unique sequences (Fig 2A). Numerous large clonal expansions were also found in each sample. We found that CD21lo cells were clonally related to both plasmablasts and memory B cells. On average, about 20% of clones containing CD21lo-encoded sequences also had clonal plasmablasts or memory B cell sequences, or both (Fig 2B). This high degree of clonal relatedness suggests that these cells share a common ancestor that underwent affinity maturation and generated multiple daughter lineages with varied differentiation fates.
Fig. 2. CD21lo cells are clonally related to plasmablasts and memory B cells but form distinct clades.
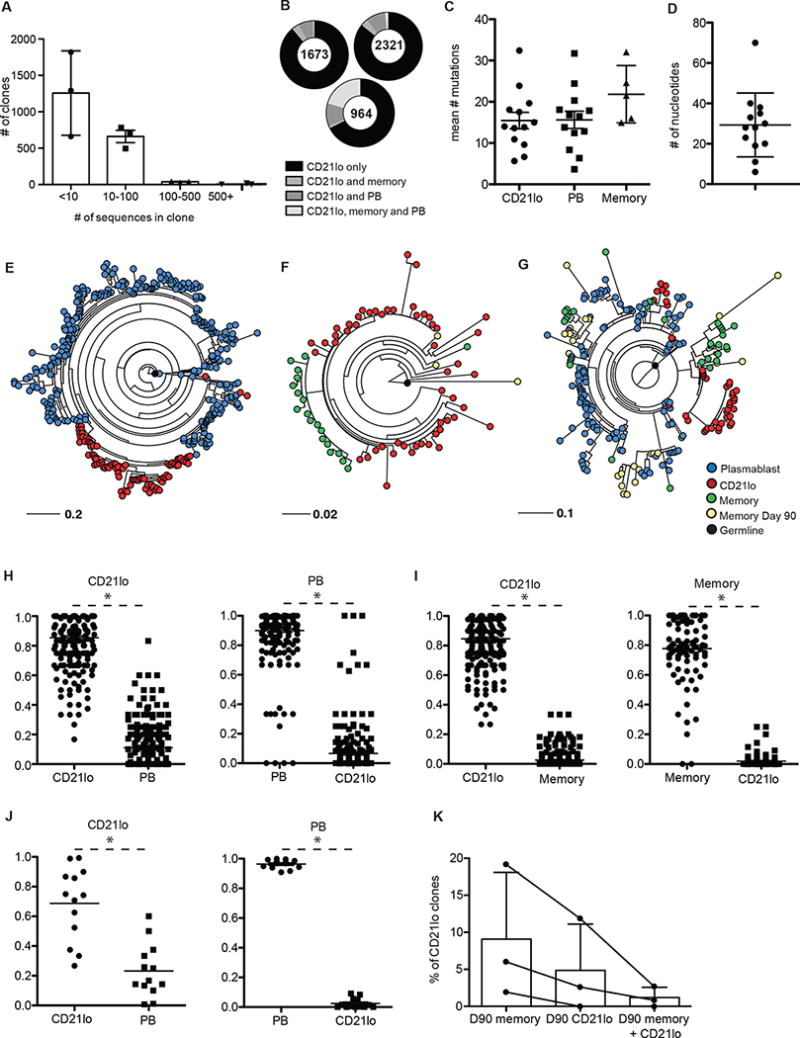
454 sequencing was performed on cDNA libraries generated from PCR amplified antibody genes from plasmablasts, memory B cells, and CD21lo cells isolated from 3 subjects that received the 2010–2011 seasonal influenza vaccine. (A) Number of clonal families of each size (n=3). Clone numbers were normalized for sequencing sample size by rarefaction. (B) Pie charts showing the proportion of Day 14 CD21lo containing clones that also contain PB or Day 14 memory sequences. (C) Mean number of mutations in the V region for Day 14 CD21lo and plasmablast sequences for each influenza specific clone (n=13). (D) Difference in V region nucleotide mutation number between the most and least mutated sequences within each influenza specific clone (n=13). (E–G) Representative maximum likelihood trees. Trees are rooted on the germline VH-JH sequence of the clone. Empirical probability of a Day 14 CD21lo sequence will have another Day 14 CD21lo sequence as its nearest neighbor in the phylogenetic tree or vice versa for plasmablast (n=219) (H) or Day 14 memory B cell (n=87) (I) or sequences in experimentally verified flu specific clones (n=12) (J). Significance was determined with a two-tailed paired t- test. Each dot represents the probability calculated for an individual clone. The line denotes the mean. (K) Percentage of clones that contain Day 14 CD21lo sequences that also contain sequences from Day 90 CD21lo, Day 90 memory, or both.
Due to the frequency of exposure to influenza, the common ancestor of cells responding to the vaccine is likely from the reactivated memory B cell pool that dominates the immune response to the vaccine in most individuals38. Reactivated memory B cells with high affinity for the immunizing antigen tend to differentiate directly into plasmablasts, while lower affinity cells re-enter the GC to undergo further affinity maturation39. The extensive GC experience of these two populations was evident when we analyzed 13 clones that contained experimentally verified flu binding antibodies (Table S1, Fig S4). While there were wide ranges in the number of mutations within each clone, there was no significant difference in the mean number of V region nucleotide mutations between CD21lo, plasmablasts, and memory B cell populations (Fig 2C,D Fig S5). The small number of memory B cells in these influenza binding clones is consistent with the kinetics identified by assays measuring antigen specificity (Fig 1G).
The de novo affinity maturation of the CD21lo cells was evident when we modeled the GC evolution of each clone using maximum likelihood trees that were rooted on the germline VH-JH sequence. The presence of distinct CD21lo clades within each clone was obvious upon visual inspection (Fig 2E–G). Using those trees, we empirically determined the probability that the nearest neighbor in the phylogenetic tree for each CD21lo sequence was another CD21lo sequence, a plasmablast sequence (Fig 2H) or a memory B cell sequence (Fig 2I). CD21lo sequences had a significantly higher probability of being neighbors with other CD21lo sequences versus plasmablast or memory B cell sequences. The same analysis was performed for plasmablast and memory B cell sequences and revealed there is a greater probability that the nearest neighbor is a sequence from the same B cell subset than from a CD21lo sequence (Fig 2H–I). The same segregation of CD21lo cells was observed in the influenza-binding clones mentioned above (Fig 2J). The distinct CD21lo clades in these trees show that while this population may have shared a common GC ancestor with plasmablasts and memory B cells, they evolve separately during affinity maturation. This unique pattern of somatic mutations in CD21lo cells supported an origin of these cells from ongoing GC reactions.
There were very few clones that contained day 14 and day 90 CD21lo cells as well as day 90 CD21lo cells. In one individual, we identified no CD21lo cells from day 14 that were clonally related to any CD21lo cells from day 90. However, a larger number of clones had both day14 CD21lo cells and day 90 memory cells (Fig 2G, 2K). This data, coupled with the ELISPOT experiments (Fig 1G) showing that the enrichment of antigen specific cells in the CD21lo compartment decays over time, supports the notion that CD21lo cells are in a post-GC transitional stage.
Analysis of the transcriptional program of CD21lo cells
The phylogenetic segregation of CD21lo cells from classical memory B cells within the same clone suggested that the two populations are functionally distinct. To further explore the functional capacities of these cells and better understand the transcriptional programs of the different post-GC lineages, we performed RNASeq analysis on CD21lo and memory B cells. The two populations were sorted from the peripheral blood of four healthy subjects 14 days after receiving the 2014–15 seasonal influenza vaccine. Due to the small number of antigen specific cells, we chose to analyze the entire CD21lo and memory B cells populations in this assay. RNA was extracted using TRIzol and libraries were prepared using the SmartSeq2 protocol that we modified slightly for bulk RNASeq40. Single end 50 base pair sequencing was performed on an Illumina HiSeq2000 machine by the University of Chicago Functional Genomics core and sequence analysis was done as previously described41. Differential expression analysis was performed using Cuffdiff and genes with a q value < 0.05 were considered to have significantly different expression between the two groups. We identified 260 genes that were differentially expressed between CD21lo and classical memory B cells (Fig 3A–C, S7A), many of which correlate with functional differences found in CD27+CD21lo B cell subsets previously characterized during chronic infection14,16. As detailed below, differentially expressed genes fell into various functional categories, including the promotion of recirculation, inhibition of activation or GC differentiation, increased susceptibility to peripheral tolerance, and differentiation into a plasma cell phenotype.
Fig. 3. CD21lo cells are transcriptionally distinct from memory B cells.

RNASeq was performed on CD21lo and memory B cells isolated 14 days post-immunization. Sequence alignment and differential gene expression was analyzed using the Tuxedo suite on Galaxy. (A) Scatter plot showing the mean expression (log2 RPKM) plotted against log fold change of the 260 genes that are significantly differentially expressed between CD21lo and memory B cells (FDR < 0.05). Red dots represent genes highlighted in the text. (B) Functional pathway analysis of differentially expressed genes. (C) Heatmap representing the expression (RPKM) of selected differentially expressed genes. (D) Expression fold change of CD21lo over memory from QPCR validation of key plasma cell associated genes (n=11). The line represents the mean. To measure protein expression of Blimp-1, CD21lo and memory B cells were isolated 21 days post-immunization. (E) Representative histogram from FACS analysis of intracellular Blimp-1 expression in HA+ CD21lo, HA+ memory B cells, and plasmablasts. (F) Proportion of HA+ CD21lo, HA+ memory B cells, and plasmablasts that are Blimp-1lo, Blimp-1int, and Blimp-1hi (n=3). Statistical significance is calculated using paired student t-test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
CD21lo cells downregulate tissue homing molecules and recirculate in the peripheral blood
Compared to recirculating classical memory B cells, CD21lo cells differentially express many trafficking related molecules. The chemokine receptors, CCR7, CXCR4, and CXCR5, which control trafficking to and within the GC, were all downregulated on CD21lo cells and FGR, which inhibits chemokine signaling, was upregulated in CD21lo cells (Fig 3C). This inhibition of trafficking is further seen in the decreased RNA expression of L-selectin (SELL, CD62L) (Fig 3C) and decreased protein expression of CD73 (Fig 4A–B) in CD21lo cells. L-selectin is required for migration of B cells into peripheral lymph nodes via the high endothelial venules, while CD73 regulates adhesion and transmigration of lymphocytes to the endothelium42,43. Downregulation of L-selectin, CXCR4 and CXCR5 was further verified at the protein level by flow cytometry (Fig 4A–B). In addition, CD21lo cells also express elevated levels of CD11c (ITGAX) at both the transcript and protein levels (Fig 3C, 4A–B). CD11c is an integrin glycoprotein that assists cells in adhesion to the endothelium44,45 and increased expression of CD11c has been identified in almost every non-classical memory B cell population in humans9,10,12–16,20. Overall, the expression profile of CD21lo cells suggests that they are less likely to re-enter GCs.
Fig. 4. Differential expression of surface markers by CD21lo cells compared to memory B cells.
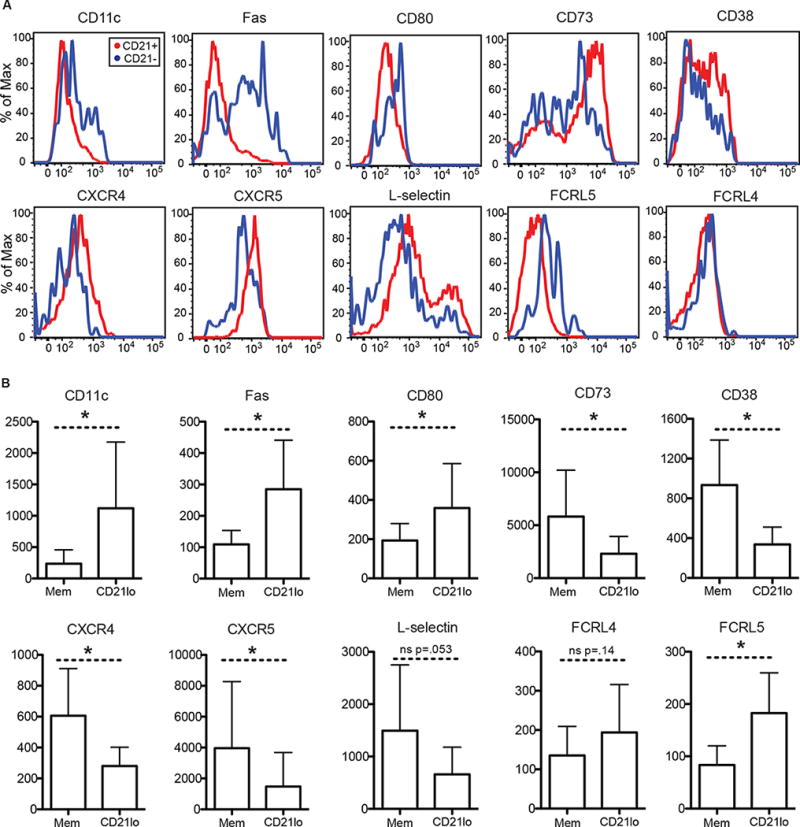
Flow cytometry analysis was performed on PBMCs isolated 14 days after immunization with the 2013–2014 or 2014–2015 seasonal influenza vaccine. PBMCs were frozen after isolation and thawed prior to antibody staining. (A) Representative histograms from FACS analysis of CD21lo and memory B cells for a panel of markers. (B) Surface expression as represented by the median fluorescence intensity (MFI) for a panel markers (n>= 6). Statistical significance is calculated using paired student t-test (*, p < 0.05). Bar graphs show mean (+/− SD).
CD21lo cells have potential for increased T-B interactions and are more susceptible to Fas mediated apoptosis
We also found that proteins related to T-B cell interactions were differentially expressed. As mentioned earlier, CD11c expression was significantly higher in CD21lo cells compared to classical memory (Fig 3C, 4A–B). CD11c+ B cells in mice localize to the T-B border in the spleen and form more stable interactions with T cells than other B cells46. CD80, which binds CD28 and CTLA4, and is required for T cell interaction, is also upregulated on CD21lo cells, as reported on CD11c+ B cells in mice46 (Fig 4A–B). Interestingly, CD21lo cells also showed significantly higher expression level of Fas compared to classical memory B cells, a phenotype that has been reported in other non-classical populations9,12,13,15,16. Furthermore, the elevated levels of Fas were independent of HA specificity (Fig S7A–B). Higher levels of are associated with a greater susceptibility to Fas mediated apoptosis47. We speculate that CD21lo cells are testing their newly generated BCRs for self-reactivity and are subject to Fas mediated apoptosis if they are activated by self-antigen and are unable to find T cell help.
CD21lo B cells upregulate negative regulators of BCR signaling but can signal in response to strong BCR stimulus
CD21lo B cells possessed elevated levels of negative regulators of BCR signaling compared to classical memory B cells. BCR signaling and amplification involves a cascade of phosphorylation events, that can be modulated by various co-receptors48. FCRL5, SIGLEC6, and SIGLEC10, are all BCR negative regulators that were upregulated on CD21lo cells (Fig 3C, Fig 4A–B). The increased expression of these inhibitory molecules, combined with the decreased levels of CD21, suggests that CD21lo cells may be more difficult to activate compared to classical memory B cells. However, we found that CD21lo cells can mobilize calcium in response to strong BCR stimulus. We measured the change in free calcium concentration after stimulation with ionomycin or anti-IgG and anti-IgK by flow cytometry using the calcium dye Fluo-4. There was no significant difference in the ability of CD21lo cells to mobilize calcium compared to memory B cells (Fig. 5C–D). This suggests that while negative regulators of BCR signaling are upregulated and the BCR co-receptor, CD21, is downregulated on CD21lo cells, these cells are still capable of responding to strong BCR stimuli. Their ability to flux intracellular calcium is perhaps unsurprising because in the vaccine response, CD21lo cells are exposed to antigen transiently, in contrast to the high levels of persistent antigen associated with chronic infection or autoimmune stimulation13,16.
Fig. 5. CD21lo cells are functionally distinct from memory and plasma cells.
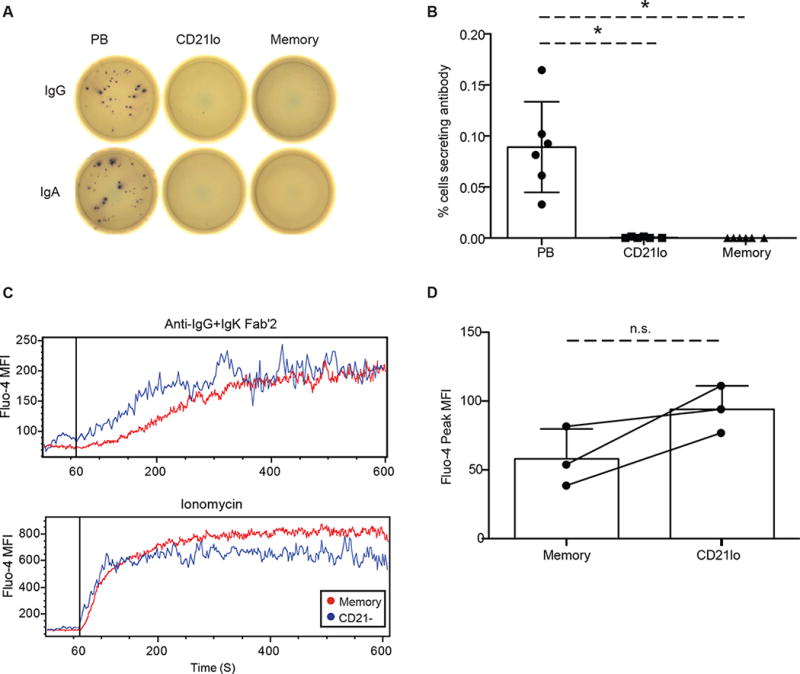
CD21lo and memory B cells were isolated form the peripheral blood from healthy individuals during steady state. Cells were incubated on ELISPOT plates for 5 hours or overnight with no stimulation. (A) Representative ELISPOT image showing plasmablasts producing both IgG and IgA antibodies while memory and CD21lo cells produce neither. (B) Summary of the frequency of IgG (n=3) or IgA (n=3) antibody secreting cells in the plasmablast, CD21lo and memory populations. Additionally, CD21lo and memory B cells were isolated 0 (n=4), 7 (n=3), 14 (n=5), 28 (n=5), and 60 (n=3) days post-immunization and stimulated for 5 days with CpG, PWM and SAC before being plated ELISPOT plates coated with vaccine. Bar graphs represent mean (+/− SD). (C) Intracellular calcium levels were measured in a flow cytometry assay using Fluo-4 AM in memory and CD21lo cells 21 days after immunization. Cells loaded with Fluo-4 AM were run on the flow cytometer for 60 seconds and then stimulated with Anti-IgG Fab’2 (25 μg/ml) and Anti-IgK Fab’2 (25 μg/ml) or 20 uM ionomycin. Representative traces show the mean fluorescence intensity (MFI) levels of Fluo-4 before and after stimulus. (D) Maximum free calcium concentration for memory and CD21lo cell types is represented by the peak Fluo-4 MFI for each cell type (n=3). Quantification of intracellular calcium levels was adjusted by basal levels of calcium, which were established before stimulation. Statistical significance is calculated using paired student t-test (*, p < 0.05). Bar graphs represent mean (+/− SD).
CD21lo cells upregulate the plasma cell transcriptional program but do not secrete antibody
The transcriptional profile of CD21lo cells indicates that they are primed for differentiation into a long-lived plasma cell fate. In classical memory B cells, there was no or minimal expression of Blimp-1 (PRDM1), the transcriptional regulator of the plasma cell program, and high expression of Bach-2 (BACH2), a Blimp-1 repressor, as expected (Fig 3C)49,50. However, CD21lo cells significantly upregulated Blimp-1 and downregulated Bach-2 (Fig 3C).
In addition to Blimp-1 and Bach-2, several direct targets of Blimp-1 were also differentially regulated in CD21lo cells (Fig 3C). These genes include XBP-1, which controls the unfolded protein response in plasma cells, c-Myc (MYC), a GC associated proliferation marker, the J-chain (IGJ), which links secretory multimeric immunoglobulins, and MZB1, an ER protein required for heavy chain synthesis51,52. Other plasma cell associated genes not regulated by Blimp-1 were also differentially expressed. Notably, ZBTB32, an early repressor of CIITA and MHCII in long lived plasma cells, was upregulated in CD21lo cells53. BCMA (TNFRSF17) and IL-6R (IL6R), genes associated with the plasma cell niche in the bone marrow, were also significantly upregulated in CD21lo cells compared to classical memory. The shift in functional capabilities of these cells suggested that these cells might be a precursor stage to the long-lived plasma cell population. The differential expression of key plasma cell related genes including PRDM1, XBP1, and BCMA was verified using QPCR (Fig 3D).
We also found an upregulation of Blimp-1 in CD21lo cells at the protein level. Using flow cytometry, we observed that a greater proportion of HA+ CD21lo cells were Blimp-1int compared to HA+ memory B cells (Fig 3E-F). Both populations had significantly fewer Blimp-1hi cells compared to plasmablasts, which is consistent with the CD21lo cells’ lack of antibody secretion. Furthermore, histograms show that the elevated levels of Blimp-1 are not driven by a few outlier cells that have very high level of Blimp-1 expression, but are reflective of the population has a whole (Fig 3E–F).
It is important to emphasize that despite the upregulation of plasma cell associated genes, CD21lo cells are not yet plasma cells. Surface expression of CD38, a plasma cell marker, was significantly lower on CD21lo cells than on memory B cells (Fig 4A–B). Additionally, the CD21lo population did not secrete antibody. CD21lo cells, along with plasmablasts as a positive control and memory B cells as a negative control, were sorted from peripheral blood and then immediately plated on anti-IgG or anti-IgA coated ELISPOT plates. Despite the upregulation of the Blimp-1 gene network, the CD21lo cells do not actively secrete antibodies of either isotype (Fig 5A, B). This finding was consistent with past studies showing no influenza specific antibody secreting cells in the peripheral blood at day 14 post-vaccination33,54. Because CD21lo cells peak after all detectable antigen-specific plasmablasts have declined, it is unlikely these cells are pre-plasmablasts. Together, these results suggest that the CD21lo compartment contains cells that are primed for long lived plasma cell differentiation, but have not yet begun the process.
CD21lo cells have elevated levels of T-bet mRNA and protein
Recent studies have demonstrated transcription factor T-bet dependent differentiation of a memory B cell subset after TLR7 and IFNGR signaling, which occur during influenza responses25,27. Further, T-bet sequesters BCL-6, the transcriptional regulator of both Tfh cell and GC B cell programs55. This sequestration blocks the initiation of the Tfh program and could potentially have a similar effect on the GC B cell program. We hypothesized that CD21lo cells upregulate T-bet, which could serve to block GC differentiation and competitive invasion of ongoing GC reactions. We found that CD21lo cells consistently had greater levels of T-bet transcripts compared to memory B cells by RNASeq analysis (Fig 6A). Furthermore, intracellular levels of T-bet protein were significantly elevated in CD21lo cells compared to memory B cells, as measured by flow cytometry (Fig 6B–C). Interestingly, a greater percentage of cells that bound HA expressed T-bet in both the CD21lo and classical memory subsets. The HA+ CD21lo, HA− CD21lo, and HA+ memory B cell populations all had significantly greater percentages of cells expressing T-bet compared to the HA− memory B cell population (Fig 6D–E). Notably, not all CD21lo and HA+ cells expressed T-bet. Thus, T-bet induction after influenza vaccination is associated with both memory and CD21lo (pre-plasma cell) differentiation.
Fig. 6. CD21lo cells have elevated levels of T-bet compared to memory B cells.

(A) T-bet RNA expression, as determined by RNASeq experiments described in Figure 3, in CD21lo and memory B cells. Statistical significance is calculated using paired student t-test (*, p < 0.05). T-bet protein expression was measured using flow cytometry. CD21lo and memory B cells were isolated form the peripheral blood from healthy individuals 14 days after immunization (n=4). (B) Protein expression is represented by the MFI (median fluorescence intensity). Statistical significance is calculated using paired student t-test (*, p < 0.05) (n=7). Bar graphs represent mean (+/− SD). (C) Representative histograms from FACS analysis of T-bet expression in CD21lo and memory B cells. (D) The CD21lo and memory populations were divided into HA+ and HA− subsets. The percent of T-bet+ cells in each population was determined by measuring the proportion of cells that had greater levels of staining than the isotype control. Statistical significance is calculated using paired student t-test (*, p < 0.05) (n=4). Bar graphs represent mean (+/− SD). (E) Representative histograms from FACS analysis of T-bet expression in CD21lo HA+, CD21lo HA−, memory HA+ and memory HA− cells.
Discussion
A growing literature that includes studies of both mice and humans has documented a variety of memory-like B cell subsets whose role in a healthy immune system remains unknown. The vast majority of these cells have been identified in patients with either chronic viral infection or autoimmune disease10,11,13–17,19,20. Although these non-classical memory B cells may have a similar phenotype to exhausted cells resulting from chronic inflammation, we find it is an unlikely explanation for the CD21lo population that we observe to be consistently present after vaccination in healthy subjects. A recent report from Ellebedy and colleagues described an activated CD71hi B cell subset that appears to be an early memory population and has clonal relationships to both plasmablasts and long-lived memory cells. Although they also display low expression of CD21, transcriptome analysis of these cells at day 7 after vaccination (as opposed to day 14) did not indicate upregulation of the Blimp1 gene program. We suspect that CD71hi cells include precursors for both the CD21lo subset described herein as well as newly generated classic memory cells18. The CD21lo population characterized herein bears some similarities CD21lo cells isolated by Thorarinsdottir et al., from peripheral blood of healthy individuals12. However, the CD21−/lo cells examined in that study were heterogeneous in CD27 expression, while the cells studied here are CD27+. We focused our study on the CD27+CD21lo population because it is the subset that is the most enriched for HA specific cells after vaccination. We suggest that CD21lo cells represent recent GC emigrants that are in a transitional stage where they are refractory to further stimulation, do not participate in further GC reactions, and are well-suited for post-GC negative selection. Thus, we propose that these cells are precursors of long-lived plasma cells, with a selective checkpoint to remove potentially pathological autoreactivities acquired during somatic hypermutation prior to long-term antibody secretion.
We determined that the CD21lo compartment is enriched for antigen specific cells after immunization compared to classical memory B cells. The two populations follow a distinct temporal pattern of enrichment with antigen-specific CD21lo cells peaking earlier and rapidly declining. The relative abundance of antigen specific cells in the CD21lo population compared to classical memory B cells marks them as potential targets for monoclonal antibody discovery. As we have shown, CD21lo cells can be used as an enriched source of antigen specific antibodies from peripheral blood isolated at time points outside of the narrow plasmablast burst that is now commonly used for mAb production. A comprehensive comparison of plasmablast to CD21lo mAbs will be of significant interest. CD21lo B cells appear to arise as the product of ongoing GC reactions compared to plasmablasts that are likely from the direct activation of memory B cells. Therefore, there may be important qualitative differences in mAbs from CD21lo cells, such as adaptation to de novo epitopes or improved binding affinity.
We also observed that the kinetics of the appearance of antigen specific CD21lo cells were different from that of plasmablasts and memory B cells. A distinct developmental pathway for CD21lo cells, likely from reactivated memory B cells that re-entered a GC, was also reflected in the phylogenetic organization of the clonal families of these varied populations. While the three different cell types were clonally related, they segregated distinctly within the phylogenies of each clonal family, forming definitive clades.
This division became more apparent when we examined their transcriptional programs. CD21lo cells have a gene program that inhibits their ability to be activated via their B cell receptor and traffic back to the GC. Of particular note, CD21lo cells have upregulated the Blimp-1 program but are not plasma cells and do not secrete antibody. The increased expression of Blimp-1 may play a role in preventing the induction of the BCL-6 controlled germinal center program. Furthermore, CD21lo cells also upregulate the transcription factor T-bet, which has been shown to sequester BCL-6 in T cells. These two mechanisms of BCL-6 inhibition may be in place because the functional consequences of recently graduated cells re-joining the GC reaction are great. These cells would likely have a selective advantage in affinity maturation compared to the still evolving cells in the GC. The recent graduates would outcompete the still evolving GC cells for survival, leading to a severe loss of BCR diversity and a less protective humoral response. Such a mechanism would be advantageous for effective protection from pathogens such as influenza that undergo antigenic drift mainly for key protective epitopes, resulting in low affinity B cell responses that would need to competitively mature for protection. Thus a tendency for viral antigens to upregulate T-bet through TLR7 and IFNGR stimulation, as previously described25,27, may have evolved as a mechanism to ensure diversity of GC responses to viral antigens. Future studies on the significance of T-bet upregulation should provide important insight into the regulation of anti-viral B cell immunity.
We also noted that CD21lo cells have increased levels of Fas, which modulates apoptosis, and CD11c and CD80, which play roles in T-B interactions. This suggests that CD21lo cells may be more susceptible to peripheral tolerance. While autoreactive GC B cells will not receive T cell help and will undergo apoptosis, B cells that are cross reactive to self and the antigen (in this case, the vaccine) can emigrate from the GC. We propose that as recent emigrants from GC reactions that are both recirculating and activation-inhibited, CD21lo cells represent an ideal transient stage for post-GC peripheral tolerance. If CD21lo cells encounter immunizing antigens, cognate T cell help will rescue them. However, if a CD21lo cell binds self-antigen, they will not be able receive T cell help and will undergo apoptosis given the elevated levels of Fas observed.
An alternative but not mutually exclusive model focuses primarily on our observation that CD21lo cells upregulate a number of plasma cell associated genes, as well as Blimp-1 protein. The precursor to the long-lived plasma cell compartment has long been mysterious, despite the clear importance of that population to providing long term serum level protection. The transcriptional program of CD21lo cells suggests that it may be that transitional stage that is subjected to a selective checkpoint where they are susceptible to Fas mediated apoptosis. A fate mapping study, though likely not possible in human subjects, would be able to definitively address this hypothesis. A selective checkpoint allowing recirculation and selection would be most important prior to differentiation to long-lived antibody secreting plasma cells, but not for memory cells that do not secrete antibodies. This transient selective phase of differentiation may account for the developmental delay noted for long-lived plasma cell differentiation56.
In summary, we report that in normal, healthy immune responses, CD21lo cells are potential plasma cell precursors that represent the earliest immigrants to the peripheral blood from GC reactions. A variety of phenotypic and functional characteristics support a model in recent GC graduates enter circulation as CD21lo cells and will transiently recirculate while being inhibited from adopting a GC phenotype but maintaining their BCR signaling competence. This phenotype will make CD21lo cells both ideally suited as the stage for post-GC tolerance induction and would avoid invasion of other GC reactions to maximize memory cell diversity. Upregulation of the Blimp1 program, as well as plasma cell markers like BCMA, IL-6R and ZBTB32, by CD21lo cells would also prime them to differentiate into long-lived plasma cells.
Materials and Methods
Study Design
This study was initiated to identify and characterize populations of potential long-lived plasma cell precursors in the peripheral blood after influenza vaccination. To study these cells, we isolated peripheral blood mononuclear cells (PBMCs) at 0, 14, 28 and 60 days after vaccination and monitored the enrichment of vaccine specific cells in the populations of interest. Once we identified the CD21lo population as the main focus of our study, we collected additional samples for immunoglobulin repertoire sequencing and whole transcriptome profiling. Surface protein expression was determined by flow cytometry analysis performed on thawed frozen PBMC samples isolated in our laboratory. Subjects in our study were healthy adults (over 18) and had varying histories of previous exposure to influenza.
PBMC Collection and Isolation
Peripheral blood samples were collected in accordance with the University of Chicago Institutional Review Board (#09-043-A). Peripheral blood mononuclear cells were isolated from 40 mL of venous blood from healthy adults 0, 14, 28, and/or 60 days after vaccination with the 2010–2011 or 2011–2012 trivalent or 2013–2014, 2014–2015 or 2016–2017 quadrivalent seasonal influenza vaccine. B cells were first enriched using the RosetteSep Human B Cell Enrichment Cocktail (STEMCELL Technologies) and washed in PBS/0.2% BSA and then isolated using a Lymphoprep gradient. For frozen PBMC, cells were frozen in FBS/10%DMSO before the B cell enrichment step and stored in liquid nitrogen. Frozen PBMCs were thawed in a 37C degree water bath and diluted in warm RPMI/10%FBS. The cells were washed in media and then PBS prior to staining for flow cytometry.
Flow Cytometry and Cell Sorting
Flow cytometry analysis was performed on fresh or frozen purified human PBMCs from day 14 or 21 after vaccination using a BD LSRFortessa. Freshly isolated PBMCs were sorted for RNASeq analysis and for monoclonal antibody generation using a BD FACSAriaII machine. Plasmablasts were identified as CD19+ CD38hi CD27+, classical memory cells as CD19+CD38loCD27+CD21+, and CD21lo cells as CD19+CD38loCD27+CD21lo. The following antibodies (from BioLegend, unless noted) were used for cell sorting: anti-CD19 Pacific Blue, anti-CD38 PECy7, anti-CD27 BV605, anti-CD21 FITC. The following antibodies were used for assaying surface expression of a panel of proteins (from BioLegend, unless noted): anti-CD80 FITC (Caltag), anti-CD95 BV421 (BD), anti-CD11c PE, anti-FCRL4 PE, anti-FCRL5 APC, anti-CXCR4 APC, anti-CXCR5 PE, and anti-CD73 PE-Dazzle.
Intracellular T-bet and Blimp-1 staining were performed on freshly isolated PBMCs 14 days after immunization. Cells were treated with human Fc blocking antibody (BD) and then stained with the CD19, CD27, CD38 antibodies described above as well as with AF647-conjugated anti HA-biotin. Then the cells were fixed with IC Fixation buffer (eBioscience) or 30 min at room temperature (RT) and intracellularly stained with anti–T-bet (Biolegend) or anti-Blimp-1 (BD) antibodies at RT for 30 min in the dark. Cells were washed in a permeabilization buffer (BD) and analyzed using a BD LSRFortessa. CD21lo HA-specific cells were defined as CD19+CD27+CD38neg. Percentages and median florescence intensities (MFI) were adjusted using a T-bet isotype control, (mouse IgG1-PE (Biolegend)) or Blimp-1 isotype control (Rat IgG2a-PE, (BD)).
Recombinant HA Production
Recombinant influenza HA was produced in 293T cells transfected with plasmids donated by the Mascola lab. Supernatant of the cells was collected and washed through a Ni-NTA column. Eluted protein was then further purified using size exclusion chromatography. The purified protein was then biotinylated using a BirA biotinylation kit (Avidity) and conjugated to SA-PE or SA-AF647.
Monoclonal Antibody Cloning
Cells were single cell sorted into a catching solution of 2 uL of cell lysis buffer (0.2% Triton X-100, RNase Inhibitor), 1uL oligo-dT (10 uM), and 1uL dNTP (10mM) and cDNA was generated as per the SmartSeq2 protocol32. The cDNA was then used to produce monoclonal antibodies as previously described48. In brief, 1 uL of the cDNA was further amplified for sequencing and cloning using nested PCRs. Sanger sequencing of the single cells was performed, and analyzed using IMGT V-Quest. These sequences were used to determine the isotype of the cells with Blast+57. Restriction enzyme sites were incorporated onto the amplicons by PCR and the genes were cloned into IgG1, IgK, or IgL expression vectors. Antibodies were expressed in HEK293T cells and purified using protein A beads (Pierce).
ELISA
Vaccine or IgG ELISAs were performed with a 1:20 dilution of the 2014-2015 seasonal influenza vaccine, 8HAU inactivated H1N1 A/California/07/09 virus or 2ug/uL of anti-IgG antibody per well. Antibodies were serially diluted threefold seven times with starting concentrations of 10 ug/mL. Microtiter plates were coated with diluted vaccine or anti-IgG antibody. The well- characterized Cr9114 antibody was used as a positive control for the vaccine ELISAs, recombinant human IgG was used for the IgG ELISAs. Intermediate washes were performed with PBS/0.05% Tween. A horseradish peroxidase-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories) was used as a secondary and plates were developed with SuperAquaBlue (EBioscience) until the positive controls reached an OD405 of 3.0.
ELISPOT
Vaccine and immunoglobulin ELISPOTs were performed as previously described46. In short, ELISPOT filter plates were coated overnight with influenza vaccine, anti-IgG or anti-IgA at 4C. Plates were sorted and blocked with RPMI with 10% FCS for 2 hours at 37C. Sorted cells in RPMI with 10% FCS, 1% HEPES, 1% L-Glutamine, and 1% penicillin/streptomycin were added to the plates in a 1:2 dilution series. Cells were incubated for 5 hours or overnight. Plates were then washed with PBS with .05% Tween and biotinylated anti-IgG or anti-IgA (Southern Biotech) was added at a 1:1000 dilution. Streptavidin-AP (Southern Biotech) was used as a secondary antibody and the ELISPOT was revealed using NBT/BCIP (Thermo Scientific).
Memory B Cell Stimulation
B cells were activated to differentiate into antibody secreting cells as previously described25,49. In short, CD21lo and memory B cells were sorted from peripheral blood and cultured for 5 days with SAC (Sigma-Aldrich), CpG (InvivoGen) and PWM (gift from Shane Crotty). The cells were then placed on vaccine coated ELISPOT plates in a 1:2 dilution series and an ELISPOT was performed as described above.
Calcium flux
Fresh human PBMCs were isolated after 21 days of immunization, and resuspended in HBSS/1%BSA and supplemented with calcium and magnesium. Cells were loaded with Fluo-4 AM Calcium Indicators (Invitrogen) at final concentration of 5 uM and incubated at 37°C for 45 min in dark. Cells were washed with HBSS/1%BSA and rested for 20 min at room temperature in the dark. Loaded cells were stained with anti-CD19, anti-CD27 and anti-CD21 antibodies as described above, but were kept at room temperature. Prior to analysis, cells were incubated at 37°C for 10 minutes. Flow cytometry analysis was performed on a BD LSRFortessa. Baseline calcium levels were read for 60 s, after which the cells were stimulated with 25 ug/uL of anti-IgG Fab’2 (Jackson ImmunoResearch Laboratories) and anti-IgK Fab’2 (SouthernBiotech) antibodies or for a positive control, 20 uM of ionomycin (Calbiochem). Calcium curves were read for 10 mins. Kinetics curves for each calcium response were generated using FlowJo software.
Library Preparation and Sequencing
For RNASeq, RNA was extracted for sequencing using TRIzol. Libraries were prepared using the SmartSeq2 protocol that was lightly modified to make low input bulk RNASeq libraries. 250 pg of input RNA was used to generate cDNA that was transcribed from mRNA primed with oligo-dTs and then purified with Ampure XP beads. The cDNA was then tagmented and adapters were added using the Illumina Nextera kit. Library preparation for all RNASeq samples was performed in parallel. 50 bp single end sequencing was performed on an Illumina HiSeq2000 at the University of Chicago Functional Genomics core and all samples were run in the same lane.
For repertoire sequencing, RNA was extracted using a Qiagen RNeasy Micro kit. cDNA was prepared by PCR amplifying the antibody transcripts using degenerate primers for the V region and specific primers for the constant region. The cDNA libraries were prepared and sequenced by the Georgiou lab or iRepertoire.
RNASeq Analysis
Differential gene expression was performed on Galaxy (usegalaxy.org) using the Tuxedo suite as previously described33. In short, sequences were aligned to the hg19 version of the human genome using Tophat and Bowtie2. Cuffmerge was used to combine transcript assemblies generated by Cufflinks and differential gene expression was determined using Cuffdiff. A differentially expressed gene is defined in our study as one with a q value less than 0.05 as calculated in Cuffdiff using the Benjamini-Hochberg correction. Additionally, the RPKM values for each sample were determined using Cuffquant and Cuffnorm on the transcript assemblies.
Quantitative PCR
QPCR was performed on cDNA made from using RNA extracted using Trizol as described above for RNASeq. The PRDM1, XBP1,TNFRSF17, and B2M Taqman Gene Expression Assays were used for QPCR and were run on an Applied Biosystems 7300 machine. The delta-delta Ct method was used for analysis.
Antibody Repertoire Analysis
Alignment and Gene Usage Analysis: Sequences were aligned to the reference IMGT database to using IMGT High V-Quest to identify the V, D, and J genes, the CDR3 nucleotide and amino acid sequence and the number of mutations in each region.
Gene-usage trees: IGHV gene frequency vectors and IGHV-IGHJ gene pair frequency vectors were generated after the alignment step. The number of occurrences of each gene was tabulated in the vectors, which were then scaled and used to generate a distance matrix. The similarity between the samples from the 3 subjects was then determined by generating a neighbor-joining tree using the APE package in R58.
Clonal assignments: Sequences were segregated into clones, which we define as a group of sequences that have the same IGHV gene, IGHJ gene, and CDR3 length, and are at least 85% similar in the CDR3 region, similar to previous studies. Sequences were segregated into subgroups based on IGHV gene, IGHJ gene and CDR3 length and a hierarchical clustering tree was generated for each subgroup (code adapted from Laserson et al35). The trees were then cut at the Levenshtein edit distance of 0.15*CDR3 length, rounded to the nearest integer. The clusters generated by this cut were then defined as B cell clones.
Maximum Likelihood Trees: We generated a multiple alignment for the sequences in each clone using MUSCLE59 and then estimated a maximum likelihood tree for each clone using RAxML with the GTR+G+I model60. We analyzed the probability that the neighboring sequence would belong to each cell type by calculating the empirical probability. We excluded small clones (less than 10 sequences) and clones with less than 3 sequences of each B cell subset from the analysis. Rooted trees were generated using the germline IGHV-IGHJ sequence. The rooted trees were visualized using the APE package in R58.
Flu Positive Clones: Flu positive clones were identified using sequencing data from experimentally verified flu binding antibodies generated from Day 7 plasmablasts from the subjects in the study. Clones containing sequences with the identical CDR3 to any of the flu binding antibodies were considered flu positive clones for analysis.
Supporting code for repertoire analysis can be found on at: https://github.com/denilau17/RepSeq3
Statistical Analyses
All statistical analyses, except for RNASeq and immunoglobulin repertoire sequencing experiments, were performed using GraphPad Prism. Statistical significance was determined using a two-tailed non-parametric unpaired Mann-Whitney test, Wilcoxon matched pair test or a two-tailed paired Student’s T-test. P values less than 0.05 were considered significant.
Supplementary Material
Fig. S1. Validation of the HA binding FACS assay.
Fig. S2. CD21lo cells can be isolated from frozen PBMC.
Fig. S3. PB, CD21lo, and memory cells are diverse in their immunoglobulin gene usage.
Fig. S4. Phylogenetic trees of experimentally validated influenza binding clones.
Fig. S5. Mutational load in influenza-reactive clones.
Fig. S6. Differential gene expression between CD21lo and memory B cells
Fig. S7. CD21o cells have elevated protein expression of Fas, independently of HA specificity.
Table S1. Specificity of influenza-reactive antibodies
One-sentence summary for SPi.
A distinct population of B cells that respond to vaccination serve as potential plasma cell precursors.
Editor’s summary.
Plasma Cell Precursors
Memory B cells are critical players in the rapid secondary immune response to pathogens; however, little is known about B cell subsets that are phenotypically different from classical memory populations. Now Lau et al. report that CD21lo B cells in healthy humans are predisposed to differentiate into long-lived plasma cells. CD21lo B cells were induced during the peak of germinal center activity after influenza vaccination, but formed distinct clades from memory B cells and plasmablasts. These cells were primed for plasma cell differentiation, but resistant to further GC differentiation. These data suggest that CD21lo cells are a distinct population of memory B cells that may contribute to plasma cell formation.
Acknowledgments
We would like to thank all study participants and lab members Will Taylor, Xinyan Qu, and Samuel Lim for their contributions to this work. We would also like to thank Sarah Cobey and Marcos Viera for their feedback on our repertoire analysis and the Mascola lab for generously sharing their modified hemagglutinin plasmids.
Funding: This project was funded in parts with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under CEIRS contract No HHSN272201400006C (P.W.); and NIH grant Nos NIH U19AI109946 (P.W.), P01AI097092 (P.W.), and U19AI057266 (P.W.), UIH U19AI05766-11 (P.W.).
Footnotes
Author contributions: DL designed and performed experiments, analyzed data and wrote the manuscript; LL and SA designed and performed experiments, analyzed data and revised the manuscript; CH, KTR, KN, MH, YH, BD, AKEP, BD, GI, and GG performed experiments and revised the manuscript; PW oversaw the project.
Competing interests: There are no competing interests.
References and Notes
- 1.Victora GD, Nussenzweig MC. Germinal Centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 2.Tas JMJ, et al. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 4.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-Specific Memory B Cell Development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 5.Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral Immunity Due to Long-Lived Plasma Cells. Immunity. 1998;8:363–372. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 6.Hart SP, Smith JR, Dransfield I. Phagocytosis of opsonized apoptotic cells: roles for ‘old-fashioned’ receptors for antibody and complement. Clin Exp Immunol. 2004;135:181–185. doi: 10.1111/j.1365-2249.2003.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz P, Simone CB, Henkart PA, Fauci AS. Mechanisms of antibody-dependent cellular cytotoxicity: the use of effector cells from chronic granulomatous disease patients as investigative probes. J Clin Invest. 1980;65:55–63. doi: 10.1172/JCI109660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrhardt GRA, et al. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J Exp Med. 2005;202:783–791. doi: 10.1084/jem.20050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrhardt GRA, et al. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med. 2008;205:1807–1817. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan RT, et al. FCRL5 Delineates Functionally Impaired Memory B Cells Associated with Plasmodium falciparum Exposure. PLoS Pathog. 2015;11:e1004894. doi: 10.1371/journal.ppat.1004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muellenbeck MF, et al. Atypical and classical memory B cells produce Plasmodium falciparum neutralizing antibodies. J Exp Med. 2013;210:389–399. doi: 10.1084/jem.20121970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorarinsdottir K, et al. CD21−/low B cells in human blood are memory cells. Clin Exp Immunol. 2016 doi: 10.1111/cei.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isnardi I, et al. Complement receptor 2/CD21− human naive B cells contain mostly autoreactive unresponsive clones. Blood. 2010;115:5026–5036. doi: 10.1182/blood-2009-09-243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kardava L, et al. Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals. J Clin Invest. 2014;124:3252–3262. doi: 10.1172/JCI74351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moir S, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008;205:1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charles ED, et al. Clonal B cells in patients with hepatitis C virus–associated mixed cryoglobulinemia contain an expanded anergic CD21low B-cell subset. Blood. 2011;117:5425–5437. doi: 10.1182/blood-2010-10-312942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Portugal S, et al. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. eLife. 2015;4:e07218. doi: 10.7554/eLife.07218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellebedy AH, et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol. 2016;17:1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramaniam KS, et al. HIV Malaria Co-Infection Is Associated with Atypical Memory B Cell Expansion and a Reduced Antibody Response to a Broad Array of Plasmodium falciparum Antigens in Rwandan Adults. PLoS ONE. 2015;10:e0124412. doi: 10.1371/journal.pone.0124412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss GE, et al. Atypical Memory B Cells Are Greatly Expanded in Individuals Living in a Malaria-Endemic Area. J Immunol. 2009;183:2176–2182. doi: 10.4049/jimmunol.0901297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein U, Rajewsky K, Küppers R. Human Immunoglobulin (Ig)M+IgD+ Peripheral Blood B Cells Expressing the CD27 Cell Surface Antigen Carry Somatically Mutated Variable Region Genes: CD27 as a General Marker for Somatically Mutated (Memory) B Cells. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tangye SG, Liu YJ, Aversa G, Phillips JH, de Vries JE. Identification of Functional Human Splenic Memory B Cells by Expression of CD148 and CD27. J Exp Med. 1998;188:1691–1703. doi: 10.1084/jem.188.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas MW, et al. A novel subset of memory B cells is enriched in autoreactivity and correlates with adverse outcomes in SLE. Clin Immunol. 2008;126:189–201. doi: 10.1016/j.clim.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Borrego F, Nagata S, Tolnay M. Fc Receptor–like 5 Expression Distinguishes Two Distinct Subsets of Human Circulating Tissue–like Memory B Cells. J Immunol. 2016:1501027. doi: 10.4049/jimmunol.1501027. [DOI] [PubMed] [Google Scholar]
- 25.Rubtsov AV, et al. Toll-like receptor 7 (TLR7)–driven accumulation of a novel CD11c+ B-cell population is important for the development of autoimmunity. Blood. 2011;118:1305–1315. doi: 10.1182/blood-2011-01-331462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Y, O’Neill P, Naradikian MS, Scholz JL, Cancro MP. A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood. 2011;118:1294–1304. doi: 10.1182/blood-2011-01-330530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubtsova K, Rubtsov AV, Dyk LF, van Kappler JW, Marrack P. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc Natl Acad Sci. 2013;110:E3216–E3224. doi: 10.1073/pnas.1312348110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur K, Sullivan M, Wilson PC. Targeting B cell responses in universal influenza vaccine design. Trends Immunol. 2011;32:524–531. doi: 10.1016/j.it.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crotty S, Aubert RD, Glidewell J, Ahmed R. Tracking human antigen-specific memory B cells: a sensitive and generalized ELISPOT system. J Immunol Methods. 2004;286:111–122. doi: 10.1016/j.jim.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 30.DeFranco AL. The germinal center antibody response in health and disease. F1000Research. 2016;5:999. doi: 10.12688/f1000research.7717.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Havenar-Daughton C, et al. CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci U S A. 2016;113:2702–2707. doi: 10.1073/pnas.1520112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuraoka M, et al. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 35.Laserson U, et al. High-resolution antibody dynamics of vaccine-induced immune responses. Proc Natl Acad Sci. 2014;111:4928–4933. doi: 10.1073/pnas.1323862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu YC, et al. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tipton CM, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16:755–765. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrews SF, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192–316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan TG, et al. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203:2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 41.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salmi M, Jalkanen S. Cell-surface enzymes in control of leukocyte trafficking. Nat Rev Immunol. 2005;5:760–771. doi: 10.1038/nri1705. [DOI] [PubMed] [Google Scholar]
- 43.Tang MLK, Steeber DA, Zhang XQ, Tedder TF. Intrinsic Differences in L-Selectin Expression Levels Affect T and B Lymphocyte Subset-Specific Recirculation Pathways. J Immunol. 1998;160:5113–5121. [PubMed] [Google Scholar]
- 44.Bilsland CA, Diamond MS, Springer TA. The leukocyte integrin p150,95 (CD11c/CD18) as a receptor for iC3b. Activation by a heterologous beta subunit and localization of a ligand recognition site to the I domain. J Immunol. 1994;152:4582–4589. [PubMed] [Google Scholar]
- 45.López-Rodríguez C, Chen HM, Tenen DG, Corbí AL. Identification of Sp1-binding sites in the CD11c (p150,95 alpha) and CD11a (LFA-1 alpha) integrin subunit promoters and their involvement in the tissue-specific expression of CD11c. Eur J Immunol. 1995;25:3496–3503. doi: 10.1002/eji.1830251243. [DOI] [PubMed] [Google Scholar]
- 46.Rubtsov AV, et al. CD11c-Expressing B Cells Are Located at the T Cell/B Cell Border in Spleen and Are Potent APCs. J Immunol. 2015:1500055. doi: 10.4049/jimmunol.1500055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith KG, Nossal GJ, Tarlinton DM. FAS is highly expressed in the germinal center but is not required for regulation of the B-cell response to antigen. Proc Natl Acad Sci U S A. 1995;92:11628–11632. doi: 10.1073/pnas.92.25.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scharenberg AM, Humphries LA, Rawlings DJ. Calcium signalling and cell-fate choice in B cells. Nat Rev Immunol. 2007;7:778–789. doi: 10.1038/nri2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaffer AL, et al. Blimp-1 Orchestrates Plasma Cell Differentiation by Extinguishing the Mature B Cell Gene Expression Program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 50.Ochiai K, et al. Plasmacytic Transcription Factor Blimp-1 Is Repressed by Bach2 in B Cells. J Biol Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 51.Johansen FE, Braathen R, Brandtzaeg P. Role of J chain in secretory immunoglobulin formation. Scand J Immunol. 2000;52:240–248. doi: 10.1046/j.1365-3083.2000.00790.x. [DOI] [PubMed] [Google Scholar]
- 52.Rosenbaum M, et al. MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev. 2014;28:1165–1178. doi: 10.1101/gad.240762.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon HS, et al. ZBTB32 Is an Early Repressor of the CIITA and MHC Class II Gene Expression during B Cell Differentiation to Plasma Cells. J Immunol. 2012;189:2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brokstad KA, Cox RJ, Olofsson J, Jonsson R, Haaheim LR. Parenteral influenza vaccination induces a rapid systemic and local immune response. J Infect Dis. 1995;171:198–203. doi: 10.1093/infdis/171.1.198. [DOI] [PubMed] [Google Scholar]
- 55.Szabo SJ, et al. A Novel Transcription Factor, T-bet, Directs Th1 Lineage Commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 56.Weisel FJ, Zuccarino-Catania GV, Chikina M, Shlomchik MJ. A Temporal Switch in the Germinal Center Determines Differential Output of Memory B and Plasma Cells. Immunity. 2016;44:116–130. doi: 10.1016/j.immuni.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madden T. The BLAST Sequence Analysis Tool. National Center for Biotechnology Information; US: 2003. [Google Scholar]
- 58.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 59.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014:btu033. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Validation of the HA binding FACS assay.
Fig. S2. CD21lo cells can be isolated from frozen PBMC.
Fig. S3. PB, CD21lo, and memory cells are diverse in their immunoglobulin gene usage.
Fig. S4. Phylogenetic trees of experimentally validated influenza binding clones.
Fig. S5. Mutational load in influenza-reactive clones.
Fig. S6. Differential gene expression between CD21lo and memory B cells
Fig. S7. CD21o cells have elevated protein expression of Fas, independently of HA specificity.
Table S1. Specificity of influenza-reactive antibodies


