Abstract
Two cDNA clones encoding cytochrome P450 enzymes belonging to the CYP79 family have been isolated from Triglochin maritima. The two proteins show 94% sequence identity and have been designated CYP79E1 and CYP79E2. Heterologous expression of the native and the truncated forms of the two clones in Escherichia coli demonstrated that both encode multifunctional N-hydroxylases catalyzing the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of the two cyanogenic glucosides taxiphyllin and triglochinin in T. maritima. This renders CYP79E functionally identical to CYP79A1 from Sorghum bicolor, and unambiguously demonstrates that cyanogenic glucoside biosynthesis in T. maritima and S. bicolor is catalyzed by analogous enzyme systems with p-hydroxyphenylacetaldoxime as a free intermediate. This is in contrast to earlier reports stipulating p-hydroxyphenylacetonitrile as the only free intermediate in T. maritima. l-3,4-Dihydroxyphenyl[3-14C]Ala (DOPA) was not metabolized by CYP79E1, indicating that hydroxylation of the phenol ring at the meta position, as required for triglochinin formation, takes place at a later stage. In S. bicolor, CYP71E1 catalyzes the subsequent conversion of p-hydroxyphenylacetaldoxime to p-hydroxymandelonitrile. When CYP79E1 from T. maritima was reconstituted with CYP71E1 and NADPH-cytochrome P450 oxidoreductase from S. bicolor, efficient conversion of tyrosine to p-hydroxymandelonitrile was observed.
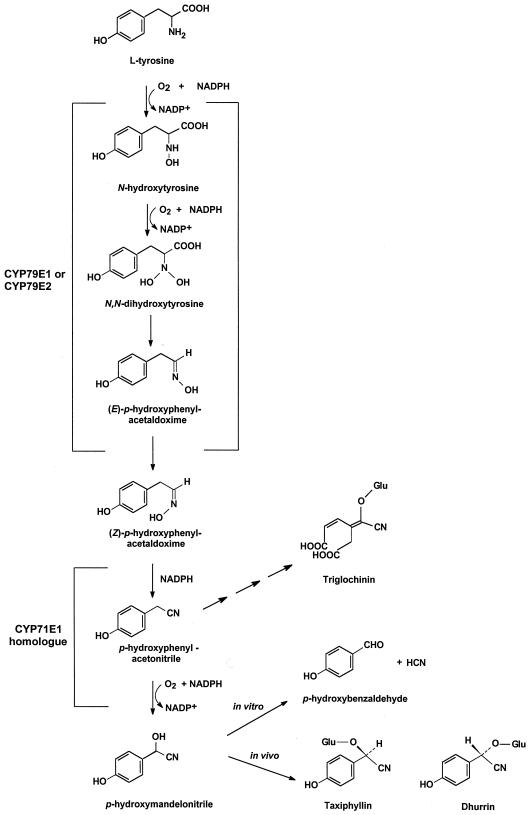
Triglochin maritima (seaside arrow grass) contains two Tyr-derived cyanogenic glucosides, triglochinin (4-carboxy-methyl-5-cyano-5-β-d-glucopyranosyloxy-cis-penta-2, 4-dienoic acid) and taxiphyllin (β-d-glucopyranosyloxy-(R)-p-hydroxymandelonitrile) (Sharples et al., 1972; Conn, 1973). The biosynthesis of taxiphyllin in T. maritima has been shown to proceed with N-hydroxy-Tyr, (Z)-p- hydroxyphenylacetaldoxime, p-hydroxyphenylacetonitrile, and p-hydroxymandelonitrile as intermediates (Hösel and Nahrstedt, 1980; Cutler et al., 1981; Nielsen and Møller, 1999) (Fig. 1) and to be catalyzed by cytochrome P450 (Cyt P450) enzymes (Nielsen and Møller, 1999). In Sorghum bicolor, the same intermediates had previously been shown to be involved in the synthesis of β-d-glucopyranosyloxy-(S)-p-hydroxymandelonitrile (dhurrin), the epimer of taxiphyllin, and to be catalyzed by two multifunctional Cyt P450 enzymes, CYP79A1 (P450tyr) (Sibbesen et al., 1994, 1995; Koch et al., 1995) and CYP71E1 (P450ox) (Kahn et al., 1997; Bak et al., 1998a).
Figure 1.
Putative biosynthetic pathway for taxiphyllin and triglochinin, the two cyanogenic glucosides in T. maritima. In the present study, CYP79E1 and CYP79E2 were isolated and shown to catalyze the conversion of l-Tyr to (Z)-p-hydroxyphenylacetaldoxime. A CYP71E1 homolog is thought to catalyze the conversion of (Z)-p-hydroxyphenylacetaldoxime to p-hydroxymandelonitrile. p-Hydroxyphenylacetonitrile is thought to constitute the branch point between taxiphyllin and triglochinin synthesis. Dhurrin is shown for comparison.
Additional gene sequences encoding Cyt P450s belonging to the CYP79 family are available (http://drnelson. utmem.edu/CytochromeP450.html) from glucosinolate-producing plant species and are thought to catalyze a similar set of reactions in glucosinolate biosynthesis (Bak et al., 1998b). However, except for CYP79A1 from S. bicolor, the precise catalytic properties of the CYP79 members remain elusive, because neither reconstitution of isolated proteins nor functional expression has been achieved. In S. bicolor, CYP79A1 catalyzes the conversion of Tyr to p-hydroxyphenylacetaldoxime, whereas CYP71E1 catalyzes the conversion of (Z)-p-hydroxyphenylacetaldoxime to p-hydroxymandelonitrile. The involvement of two multifunctional enzymes explains why this pathway is highly channeled in S. bicolor with p-hydroxyphenylacetaldoxime as the only intermediate in equilibrium with the exogenously added substrate (Møller and Conn, 1980; Halkier et al., 1989).
Based on a common biosynthetic route for cyanogenic glucoside biosynthesis in a number of different plant species (Cutler and Conn, 1981; Collinge and Hughes, 1982; Koch et al., 1992), it is tempting to infer the involvement of analogous multifunctional Cyt P450s. However, this may not be so in T. maritima, since in this plant, p- hydroxyphenylacetaldoxime and p-hydroxyphenylacetonitrile are free to equilibrate (Cutler et al., 1981; Nielsen and Møller, 1999). The Cyt P450-catalyzed conversion of aldoxime to nitrile is a dehydration reaction and, as such, is non-classical. In T. maritima it could be carried out as an additional enzyme activity associated with the first multifunctional Cyt P450 enzyme instead of being the first catalytic event catalyzed by the second Cyt P450 involved. If so, the second Cyt P450 in T. maritima would be a normal C-hydroxylase.
To discriminate between these possibilities, cloning of a putative Cyt P450 enzyme from T. maritima that is analogous to CYP79A1 from S. bicolor was initiated based on a PCR approach. We report the isolation and functional reconstitution of two cDNA clones from T. maritima, designated CYP79E1 and CYP79E2, encoding two multifunctional Cyt P450s catalyzing the conversion of Tyr to p-hydroxyphenylacetaldoxime in taxiphyllin and triglochinin biosynthesis. Reconstitution of the complete conversion of Tyr into p-hydroxymandelonitrile was achieved upon insertion of native or truncated forms of CYP79E1 into liposomes in the presence of CYP71E1 and NADPH-Cyt P450 oxidoreductase isolated from S. bicolor.
MATERIALS AND METHODS
PCR Approach to Generate cDNA Fragments of a CYP79 Homolog in Triglochin maritima
A unidirectional plasmid cDNA library was made by Invitrogen (Carlsbad, CA) from flowers and fruits (schizocarp) of T. maritima, using the expression vector pcDNA2.1, which contains the lacZ promoter. Plant material was collected at Aflandshage on Southern Amager, at the coast of Øresund, Denmark, frozen directly in liquid N2, and stored at −80°C.
Degenerate PCR primers were designed based on conserved amino acid sequences in CYP79A1 derived from Sorghum bicolor (GenEMBL no. u32624; Koch et al., 1995), CYP79B1 from Sinapis alba (GenEMBL no. AF069494; Bak et al., 1998b), CYP79B2 from Arabidopsis (GenEMBL no. AF069495; Bak et al., 1998b), and PCR fragment of CYP79D1 from Manihot esculenta (GenEMBL no. AF140613; Andersen et al., 2000) (Fig. 2). Two rounds of PCR amplification reactions (total volume: 50 μL) were carried out using 100 pmol of each primer, 5% (v/v) dimethyl sulfoxide, 200 μm dNTPs, and 2.5 units of Taq DNA polymerase in PCR buffer (50 mm KCl, 10 mm Tris-HCl, pH 8.8, 1.5 mm MgCl2, and 0.1% [v/v] Triton X-100). Thermal cycling parameters were 2 min at 95°C, 30× (5 s at 95°C, 30 s at 45°C, and 45 s at 72°C), and finally 5 min at 72°C. The first PCR reaction was performed using primer 1F and 1R (Table I) on 100 ng of template DNA prepared from the cDNA library, or genomic DNA prepared using an extraction kit (Nucleon Phytopure Plant DNA Extraction Kit, Amersham, Buckinghamshire, UK).
Figure 2.
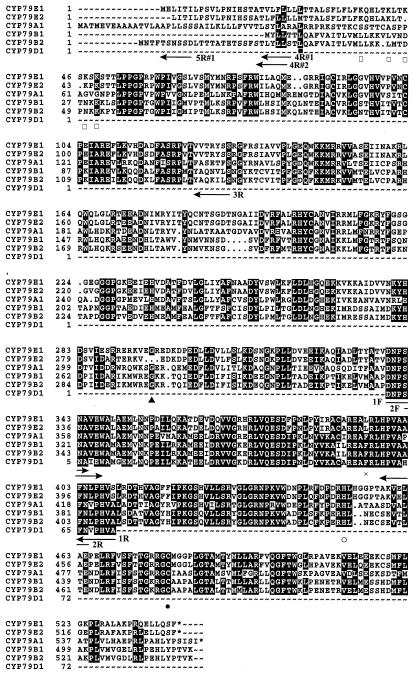
Alignment of CYP79E1, CYP79E2, CYP79A1, CYP79B1, CYP79B2, and a PCR fragment of CYP79D1. Position of degenerate and gene-specific primers used to isolate the two CYP79s from T. maritima is indicated by arrows. Vector primers and the gene-specific primer for CYP79E1 5′-UTR are not shown. ●, Heme-binding Cys residue. ○, In the PERF region Phe is replaced by a His residue and Glu by a Asp residue. ×, In the conserved KETLR region, Leu is restored in CYP79Es. ▪, Met present in CYP79E2 but not in CYP79E1. □, Positively charged region in CYP79Es. ▴, Deletion in CYP79E2.
Table I.
Compilation of primers used
| Primer Name | Amino Acid Sequence | Nucleotide Sequencea | Restriction Site |
|---|---|---|---|
| 1Fb | DNPSNAc | GCGGAATTCGAYAAYCCIWSIAAYGC | EcoRI |
| 1Rb | FNV/LPHVAc | GCGGATCCGCIACRTGIGGIAHRTTRAA | BamHI |
| 2F | SNAVEWc | GCGGAATTCWSIAAYGCIRTIGARTGG | EcoRI |
| 2R | HPVAXFNc | GCGGATCCRTTRAAIIINGCIACIGGRTG | BamHI |
| 3F | d | GCGGAATTCCACACAGGAAACAGCTATGAC | EcoRI |
| 3Re | VVTRYSS | GCGGATCCAGACGAGTAGCGAGTCACAAC | BamHI |
| 4R#1f | TVLFLL | GCGGATCCAAGAGGAACAGTACT | BamHI |
| 4R#2f | ATLFLL | GCGGATCCAAGAGGAACAATGTG | BamHI |
| 5F#1f | g | GCGAATGCATTGCTCCCACTAGCC | |
| 5R#1f | MELITI | GCGATGGTTATGAGTTCCATTTTG | |
| 6F#1 (na) | MELITIL | GCGCATATGGAACTAATAACAATTCTT | NdeI |
| 6R | LLQSF*h | GCGAAGCTTATTAGAAGCTCTGGAGCAG | HindIII |
| 6F#1 [Δ(1–31)17α(8aa)] | MALLLAVFFLFLFKQ | GCGCATATGGCTCTGTTATTAGCAGTTTTTTTCC | |
| TCTTCCTCTTCAAACAA | NdeI | ||
| 6F#1 [Δ(1–52)2E1(10aa)] | MARQVHSSWNLPPGP | GCGCATATGGCTCGTCAAGTTCATTCTTCTTGG | |
| AATTTACCACCAGGCCCC | NdeI |
The sequence is shown from 5′ end to 3′ end.
F, Forward primer; R, reverse primer.
Amino acid consensus sequence based on alignment in Fig. 2 and used for primer design.
A specific primer for pcDNA2.1 placed just upstream the insertion site of the 5′ end of the cDNA library.
Covers a sequence that is identical in the two clones #1 and #2.
Covers a sequence that is specific for either of the two clones #1 and #2.
A specific primer for the 5′-UTR in #1.
The asterisk indicates a stop codon.
The PCR products were purified using a kit (QIAquick, Qiagen, Valencia, CA), eluted in 30 μL of 10 mm Tris-HCl, pH 8.5, and used as template (1 μL) for the second round of PCR reactions carried out using PCR fragments derived from both cDNA and genomic DNA and using the two degenerate primers 2F and 2R (Table I). An aliquot (5 μL) of the PCR reaction was applied to a 1.5% (w/v) agarose/Tris-borate/EDTA (TBE) gel, and a band of the expected size (approximately 200 bp) was observed using both cDNA and genomic DNA as template. The rest of the PCR reaction was purified using the QIAquick kit, and eluted in 30 μL of 10 mm Tris-HCl, pH 8.5. The purified PCR fragments (5 μL) were digested with EcoRI and BamHI, excised from a 1.5% (w/v) agarose/TBE gel, purified using an agarose gel extraction kit (QIAEX II, Qiagen), and ligated into an EcoRI- and BamHI-digested pBluescript II SK vector (Stratagene, La Jolla, CA). Seven clones derived from the cDNA library and three clones derived from genomic DNA were sequenced (ALF Express, Pharmacia, Piscataway, NJ) using the fluorescent-labeled primer cycle sequencing kit with 7-deaza dGTP (Thermo Sequenase, Amersham). All sequence analyses were performed using programs in the GCG Wisconsin Sequence Analysis package (Genetics Computer Group, Madison, WI).
Screening of a Plasmid cDNA Library Made from Flowers and Fruits of T. maritima
Both cDNA and genomic DNA produced an identical PCR fragment with high sequence resemblance to the other known CYP79s. The PCR fragment was used as template to generate a 350-bp digoxigenin-11-dUTP-labeled (Boehringer Mannheim, Basel) probe (TRI1) by PCR using T3 and T7 primers. The labeled probe was employed to screen 660,000 colonies of the pcDNA2.1 cDNA library. Hybridizations were carried out overnight at 68°C in 5× SSC (0.75 m NaCl and 75 mm sodium citrate, pH 7.0), 0.1% (w/v) N-lauroylsarcosine, 0.02% (w/v) SDS, and 1% (w/v) blocking reagent (Boehringer Mannheim). Membranes were washed twice under high-stringency conditions (65°C, 0.1× SSC, and 0.1% [w/v] SDS), incubated with anti-digoxigenin-AP, and developed using 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium according to the manufacturer's (Boehringer Mannheim) instructions. Positive colonies were rescreened under the same conditions, and single positive colonies were sequenced and analyzed.
PCR Approach to Design 5′ End Probes to Screen for Full-Length Clones
Library screening resulted in two very similar partial clones designated #1 and #2, that particularly differ in their N-terminal sequence (Fig. 2). To isolate the corresponding full-length clones from the pcDNA2.1 library, two consecutive PCR reactions were performed using the same PCR conditions as above, with the exception that the annealing temperature was set at 55°C. The first PCR reaction was performed with primers 3F and 3R (Table I) using 100 ng of DNA from the cDNA library as template. The purified PCR products from the first PCR reaction were used as template (1 μL) for a second round of PCR reactions using primer 4R#1 or 4R#2 against primer 3F (Table I). The PCR fragments from the second round were separated on a 2% (w/v) agarose/TBE gel, and the slowest migrating bands were excised from the gel, purified on the agarose gel extraction kit, digested with EcoRI and BamHI, cloned in pBluescript II SK, and sequenced. Using primer 4R#1 together with primer 3F in the second round of PCR, a PCR fragment with a putative start Met 26 amino acids downstream of the EcoRI cloning site was found. The PCR reaction with primers 4R#2 and 3F produced a PCR fragment of exactly the same length as the partial cDNA clone already isolated using the TRI1 probe. As a consequence, the PCR fragment cloned with 4R#1 and 3R was used as a template to generate a digoxigenin-11-dUTP-labeled probe, TRI2, using primers 5F#1 and 5R#1. Using the same conditions as above, TRI2 covering partly the 5′ untranslated region (UTR) and 5′ end of the open reading frame of #1 was employed to screen the pcDNA2.1 library together with the TRI1 probe. The first lifts were hybridized with TRI2 and the second with TRI1. Two individual cDNA clones of exactly the same length as the PCR fragment were isolated screening 1,000,000 colonies (Fig. 2).
Expression Constructs
Three different constructs of clone #1 were generated with PCR, using Pwo polymerase (Boehringer Mannheim) to introduce a NdeI restriction site at the start codon and a HindIII restriction site immediately after the stop codon. A full-length construct was synthesized using primers 6F#1(na) and 6R#1. Two truncated constructs were made using primers 6F#1[Δ(1–31)17α(8aa)] and 6R#1 or primers 6F#1[Δ(1—52)2E1(10aa)] and 6R#1 (Table I). The PCR fragments were digested with NdeI and HindIII and ligated into NdeI- and HindIII-digested pSP19g10L expression vector (Barnes, 1996). The unique restriction sites NcoI and PmlI were used to cut out a fragment (1,045 bp) of the middle part of the PCR clones, which was then replaced with the analogous fragment from the cDNA clone. The remaining portions of the constructs that derived from PCR were sequenced to exclude PCR errors. Clone #2 was found to be in frame with the first 24 amino acids of the lacZ gene. A similar clone of #1, in frame with lacZ at same nucleotide position as in #2, was also expressed in pcDNA2.1 for comparison.
Expression in Escherichia coli
All expression constructs were transformed into the E. coli strains JM109 (Stratagene) and XL-1 Blue (Stratagene). The native and Δ(1–52)2E1(10aa) constructs of clone #1 and the construct of clone #2 were co-transformed with pSBET (Schenk et al., 1995), which encodes a tRNA gene for rare Arg codons, into JM109. Single colonies were grown overnight in Luria-Bertani medium (50 μg/mL ampicillin, 37°C, 225 rpm) and used to inoculate 100× volume of modified TB medium (50 μg/mL ampicillin, 1 mm thiamine, 75 μg/mL δ-amino-levulinic acid, 1 mm isopropyl β-d-thiogalactopyranoside [IPTG], 28°C, 125 rpm, 48 h).
Measurements of Expression Levels and Biosynthetic Activities
Expression levels of the different constructs were determined by CO difference spectroscopy and quantified using an extinction coefficient ε450–490 of 91 mm−1 cm−1 (Omura and Sato, 1964). Spectra were made from 100 or 500 μL of whole E. coli cells or using the rich phases from Triton X-114 phase partitioning solubilized in 50 mm KH2PO4/K2HPO4, pH 7.5, 2 mm EDTA, 20% (v/v) glycerol, and 0.2% (v/v) Triton X-100 (total volume: 1 mL). E. coli cells for in vivo studies were prepared by centrifugation (2 min and 30 s at 7,000g) of 1 mL of cell culture and resuspension in 100 μL of 50 mm N-[2-hydroxy-1,1-Bis(hydroxymethyl)ethyl]glycine (Tricine), pH 7.9, and 1 mm phenylmethylsulfonyl fluoride. For in vitro studies, spheroblasts were made from E. coli (JM109) cells expressing native or Δ(1–52)2E1(10aa) constructs of clone #1 or the construct of clone #2, followed by temperature-induced phase partitioning (0.6% [v/v] Triton X-114 and 30% [v/v] glycerol) as previously described (Halkier et al., 1995).
Measurements of in vivo catalytic activity were carried out by the administration of [U-14C]Tyr (0.35 μCi, 7.39 μm), p-hydroxyphenylacetaldoxime (0 or 0.1 mm), or p- hydroxyphenylacetonitrile (0 or 0.1 mm) to resuspended E. coli cells (100 μL). In vitro activities were measured in reconstitution experiments using the rich phase from phase partitioning. A standard reaction mixture (total volume: 50 μL) contained 5 μL of rich phase, 0.375 unit of S. bicolor NADPH-Cyt P450 oxidoreductase, 5 μL of L-α-dilauroyl phosphatidylcholine (DLPC), 0.6 mm NADPH, and 14 mm KH2PO4/K2HPO4, pH 7.9. The following substrates were tested: l-[U-14C]Tyr (0.20 μCi, 9.04 μm), l-[U-14C]Phe (0.20 μCi, 8.8 μm), and l-3,4-dihydroxyphenyl[3-14C]Ala (0.20 μCi, 400 μm). l-[U-14C]Tyr (0.20 μCi, 9.04 μm) was also tested in reconstitution experiments that included purified CYP71E1 (Kahn et al., 1997; Bak et al., 1998a) from S. bicolor.
Incubations (1 h, 30°C, shaking water bath) were started by the addition of substrate (in vivo experiments) or NADPH (in vitro experiments), and were stopped by the addition of ethyl acetate. Biosynthetic activity was monitored by the formation of radioactive products using thin-layer chromatography (TLC) analysis, as previously described (Møller and Conn, 1979), and detection and quantification using a phosphor imager (Storm 840, Molecular Dynamics, Sunnyvale, CA). Before TLC application, the sample was extracted with ethyl acetate. During this step, the surplus of radiolabeled Tyr remains in the aqueous phase, thus preventing overexposure at the origin. The total ethyl acetate phase was applied to the TLC. In some experiments, inevitable carry-over of small amounts of the aqueous phase results in the appearance of a Tyr band at the origin. Unlabeled reference compounds (p-hydroxyphenylacetaldoxime, p-hydroxyphenylacetonitrile, and p-hydroxybenzaldehyde) were prestreaked on the TLC plates to permit visual detection under UV light.
RESULTS
Cloning of CYP79E1 and CYP79E2
A sequence alignment of CYP79A1 and putative N-hydroxylases belonging to the CYP79 family is shown on Figure 2. Based on this alignment, four degenerate oligonucleotide primers covering two CYP79-specific regions were designed (1F, 2F, 1R, and 2R) (Fig. 2) and employed in nested PCR reactions using genomic DNA and cDNA made from flowers and fruits of T. maritima as templates. A PCR fragment of the expected size (approximately 200 bp) and showing 62% to 70% identity to CYP79 sequences at the amino acid level was isolated using both templates and employed to screen the cDNA library. Two clones, denoted #1 and #2, were isolated and verified by sequence comparison to share high sequence identity with the CYP79 family. Using clone-specific PCR primers, a full-length clone corresponding to #1 was isolated. The open reading frame encodes a protein with a molecular mass of 60.8 kD. A comparison of the full-length sequence of clone #1 with that of clone #2 demonstrated that clone #2 is 6 bp shorter at the 5′ end, but contains a Met codon not found in clone #1 at a position corresponding to amino acid residue 26 specified by clone #1. The surrounding sequence of this Met codon did not fit the general context sequence for a start codon in a monocotyledonous plant (Joshi et al., 1997). Most likely, clone #2 lacks 6 bp to be full-length.
Cyt P450 enzymes are heme-containing enzymes constituting a supergene family. In plants, they are divided into two distinct groups (Durst and Nelson, 1995): The A-group probably derived from a common ancestor and is involved in the biosynthesis of secondary plant products such as cyanogenic glucosides and glucosinolates. The non-A-group is heterogeneous and clusters near to animal, fungal, and microbial Cyt P450s. Cyt P450s showing amino acid sequence identities above 40% are grouped within the same family (Nelson et al., 1993). Cyt P450s showing more than 55% identity belong to the same subfamily. The Cyt P450s encoded by clones #1 and #2 showed 45% to 49% identity, respectively, to the previously known members of the CYP79 family (Table II) and accordingly were identified as the first two members of a new subfamily and assigned as CYP79E1 (accession no. AF140609) and CYP79E2 (accession no. AF140610), respectively, by the International Cytochrome P450 Nomenclature Committee. The sequence identity between CYP79E1 and CYP79E2 is 94%.
Table II.
Percent identity and similarity between six members of the CYP79 family
| Identity | Similarity
|
|||||
|---|---|---|---|---|---|---|
| CYP79E1 | CYP79E2 | CYP79A1 | CYP79B1 | CYP79B2 | CYP79D1 | |
| CYP79E1 | 95.2 | 61.7 | 58.1 | 58.9 | 60.0 | |
| CYP79E2 | 94.1 | 61.5 | 57.6 | 58.5 | 59.2 | |
| CYP79A1 | 48.8 | 48.8 | 65.5 | 67.1 | 65.8 | |
| CYP79B1 | 44.9 | 44.9 | 51.3 | 92.3 | 65.1 | |
| CYP79B2 | 44.5 | 44.6 | 52.6 | 89.3 | 67.3 | |
| CYP79D1 | 46.4 | 46.5 | 51.5 | 49.1 | 50.7 | |
Expression of CYP79E1 and CYP79E2 in E. coli
The expression vector pSP19g10L was used for expression of CYP79E1 and CYP79E2 constructs in E. coli. This expression vector contains the lacZ promoter fused with the short leader sequence of gene 10 from T7 bacteriophage (g10L) and has been shown effective for heterologous protein expression in E. coli (Olins and Rangwala, 1990). In Cyt P450s, increased expression levels have been obtained by modifying the 5′ end of the open reading frame to increase the content of A′s and T′s (Stormo et al., 1982; Schauder and McCarthy, 1989; Barnes et al., 1991) and by replacement of a number of codons at the 5′ end with codons specifying the N-terminal sequence of bovine P45017α (Barnes et al., 1991) or human P4502E1 or 2D6 (Gillam et al., 1994, 1995). To take advantage of this knowledge, a number of different constructs were made (Fig. 3). The first construct, CYP79E1na, encoded native CYP79E1 with silent mutations introduced at codons 3 and 5 to increase the AT content. The second construct, CYP79E1Δ (1–31)17α(8aa), encoded a truncated form of CYP79E1 in which 31 codons of the native 5′sequence had been replaced by eight AT-enriched codons of P45017α (Barnes et al., 1991; Halkier et al., 1995). In the third construct, CYP79E1Δ (1–52)2E1(10aa), the first 52 codons of the native 5′sequence were replaced by 10 AT-enriched codons of P4502E1, and silent mutations were introduced in codons 53 and 55. Because the CYP79E2 clone was isolated in frame with the first 24 codons of the lacZ gene in the vector pcDNA2.1, this was tested as a fourth expression construct designated CYP79E2lacZ(24aa) (Fig. 3). For comparison, an equivalent fifth construct, CYP79E1Δ (1–2)lacZ(24aa), was also prepared.
Figure 3.
N-Terminal amino acid sequences of the four different constructs of CYP79E1 and the construct of CYP79E2. Native sequence from CYP79E is underlined.
All constructs contained the original stop sequence, TAAT, which is found in most highly expressed E. coli genes (Tate and Brown, 1992). All constructs made using the vector pSP19g10L had their 3′-UTR removed, because inclusion of the 3′-UTR has been reported to prevent or reduce expression of some genes (Richardson et al., 1995). In constructs based on pcDNA2.1, the 3′-UTR was retained. To further optimize the expression, all constructs were tested in two E. coli strains, XL-1 Blue and JM109. The expression level was monitored in dependence of time by carbon monoxide difference spectroscopy. In all cases, the JM109 strain was most efficient. CYP79E1 and CYP79E2 contain 19 and 17 AGA or AGG Arg codons, which are rare in E. coli genes. A strong positive correlation between the occurrence of codons and tRNA content has been established (Ikemura, 1981). Accordingly, co-transformation with pSBET, which encodes a tRNA for rare Args, was also pursued.
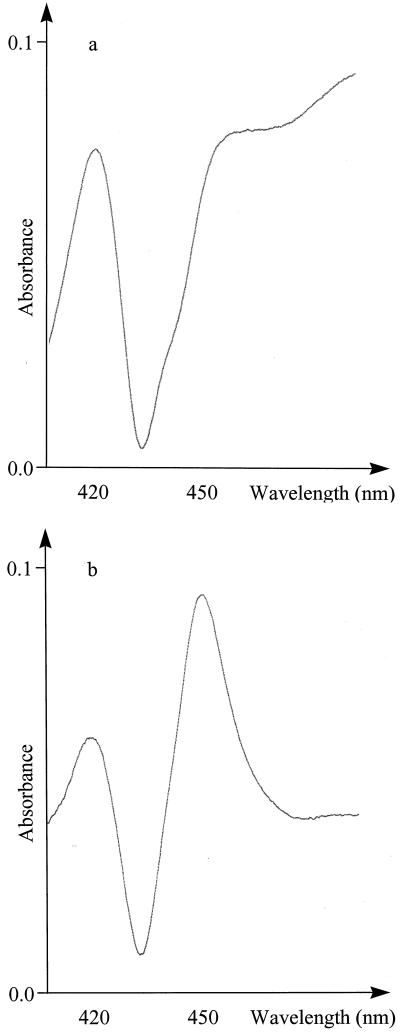
Carbon monoxide binding spectra using intact E. coli cells showed the absorption maximum at 450 nm, which is diagnostic for the formation of functional Cyt P450 with the following three constructs: CYP79E1na, CYP79E1Δ (1–52)2E1(10aa), and CYP79E2lacZ(24aa). The spectra were obtained without and with co-transformation with pSBET, but in all cases the Cyt P450 content was too low to permit quantification. To obtain an accurate determination, the Cyt P450s were enriched by isolation of E. coli spheroblasts, followed by temperature-induced Triton X-114 phase partitioning (Werck-Reichart et al., 1991; Halkier et al., 1995). The highest expression level (JM109 cells, 48 h) was 56 nmol/L culture and was obtained using CYP79E2lacZ(24aa) (Fig. 4). This level is comparable to the expression level of 62 nmol/L culture obtained with the S. bicolor construct CYP79A1Δ (1–33)17α(8aa) (Halkier et al., 1995), which was included as a positive control. CYP79E1Δ (1–31)17α(8aa) with a modified P45017α N terminus and the empty vector control never revealed any detectable spectrum.
Figure 4.
CO difference spectra of rich phase from temperature-induced Triton X-114 phase partitioning of (a) CYP79E1na and (b) CYP79E2lacZ(24aa).
Determination of the Enzymatic Activity of CYP79E1 and CYP79E2
Radiolabeled Tyr was administered to E. coli cells containing all five CYP79E1 and CYP79E2 constructs described above as well as the S. bicolor construct CYP79A1Δ (1–33)17α(8aa) as a positive control. In all cases, except for CYP79E1Δ (1–31)17α(8aa), p-hydroxyphenylacetaldoxime was obtained as the sole product (data not shown). This demonstrates that CYP79E1 and CYP79E2 are functionally equivalent to CYP79A1 isolated from S. bicolor, in spite of earlier reports of p-hydroxyphenylacetonitrile as the only free intermediate in T. maritima (Hösel and Nahrstedt, 1980; Cutler et al., 1981). A detailed analysis of the expression levels obtained with the different constructs provided the following information: (a) The CYP79E1Δ (1–2)lacZ(24aa) and CYP79E2lacZ(24aa) constructs provided the highest activity; (b) for all constructs, JM109 cells were superior to XL-1 Blue; (c) in time course studies (0–72 h), the highest activity was obtained at 24 to 48 h; (d) omission of IPTG reduced the expression level to approximately 50%, as documented using the CYP79E1na construct; (e) co-transformation with pSBET further increased the high expression level obtained with CYP79E2lacZ(24aa), whereas the expression level obtained with all other constructs diminished.
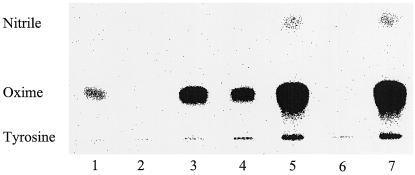
For in vitro studies, the recombinant Cyt P450s obtained by expression of CYP79E1na, CYP79E1Δ (1–52)2E1(10aa), and CYP79E2lacZ(24aa) were partially purified by temperature-induced Triton X-114 phase partitioning (Werck-Reichart et al., 1991; Halkier et al., 1995) and reconstituted using S. bicolor NADPH-Cyt P450 oxidoreductase and DLPC (Fig. 5). As demonstrated above using intact E. coli cells, Tyr is metabolized to p-hydroxyphenylacetaldoxime by reconstituted recombinant CYP79E1 and CYP79E2. No p-hydroxyphenylacetaldoxime accumulates using an empty vector control or in reconstitutions without the addition of reductase. In the experiment without administration of NADPH, a small amount of p-hydroxyphenylacetaldoxime accumulated (Fig. 5, lane 1). Using the constructs CYP79E2lacZ(24aa) and S. bicolor CYP79A1Δ (1–33)17α(8aa), the turnover of Tyr per nanogram of Cyt P450 obtained was about the same.
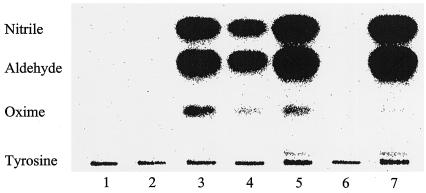
Figure 5.
Reconstitution of recombinant CYP79E1 and CYP79E2 using radiolabeled Tyr as substrate. Spheroblasts were isolated from E. coli cells expressing different constructs of CYP79E1 and CYP79E2, followed by temperature-induced Triton X-114 phase partitioning and reconstitution with S. bicolor NADPH-Cyt P450 oxidoreductase. Lanes 1 to 6 contain 20 μg protein/reaction mixture. Lane 7 contains 10 μg protein/reaction mixture. Lane 1, CYP79E1na, no NADPH; lane 2, CYP79E1na, no NADPH-Cyt P450 oxidoreductase; lane 3, CYP79E1na; lane 4, CYP79E1Δ (1–52)2E1(10aa); lane 5, CYP79E2lacZ(24aa); lane 6, expression vector pSP19g10L; lane 7, S. bicolor CYP79A1Δ (1–33)17α(8aa). Reaction mixtures were analyzed by TLC and the products formed monitored and quantified using a phosphor imager. The position of authentic standards is indicated. Oxime, (E)- and (Z)-p-Hydroxyphenylacetaldoxime; nitrile, p-hydroxyphenylacetonitrile.
Because of the lack of accurate CO difference spectra of CYP79E1na and CYP79E1Δ (1–52)2E1(10aa) from the in vitro and in vivo experiments, it was not possible to measure the turnover of Tyr per nanogram of Cyt P450. The relative values for turnover of Tyr per milligram of protein are: CYP79E1Δ (1–52)2E1(10aa) = 1; CYP79E1na = 1.5; CYP79E2lacZ(24aa) = 22; S. bicolor CYP79A1Δ (1–33)17α(8aa) = 46. As already stated above, CYP79E1Δ (1–31)17α(8aa) showed no activity at all. In CYP79E2lacZ(24aa) and CYP79A1Δ (1–33)17α(8aa), accumulation of p-hydroxyphenylacetonitrile could be detected (Fig. 5). No catalytic activity could be detected using DOPA or l-[U-14C]Phe as the substrate in reconstitution experiments (data not shown). Administration of DOPA to microsomal preparations from T. maritima flowers and fruits also did not show any measurable activity.
Reconstitution of CYP79E with CYP71E1
Reconstitution of the membrane-associated pathway of cyanogenic glucoside synthesis resulting in the formation of p-hydroxymandelonitrile (the aglycon of dhurrin, seen as p-hydroxybenzaldehyde in vitro) was achieved using enzymes from the two species S. bicolor and T. maritima (Fig. 6). In all reconstitutions, including Tyr, NADPH, NADPH-Cyt P450 oxidoreductase, CYP79E1 or CYP79E2, and CYP71E1, considerable amounts of p-hydroxyphenylacetonitrile and p-hydroxybenzaldehyde accumulated (Fig. 6).
Figure 6.
Reconstitution of recombinant CYP79E1 and CYP79E2 from T. maritima with CYP71E1 from S. bicolor using radiolabeled Tyr as substrate. Spheroblasts were isolated from E. coli cells expressing different constructs of CYP79E1 and CYP79E2, followed by temperature-induced Triton X-114 phase partitioning. Lanes 1 to 6 contain 20 μg protein/reaction mixture. Lane 7 contains 10 μg of protein/reaction mixture. Reconstitution was performed with isolated CYP71E1 and NADPH-Cyt P450 oxidoreductase from S. bicolor. All reconstitutions contain CYP71E1. Lane 1, CYP79E1na, no NADPH; lane 2, CYP79E1na, no NADPH-Cyt P450 oxidoreductase; lane 3, CYP79E1na; lane 4, CYP79E1Δ (1–52)2E1(10aa); lane 5, CYP79E2lacZ(24aa); lane 6, expression vector pSP19g10L; lane 7, S. bicolor CYP79A1Δ (1–33)17α(8aa). Reaction mixtures were analyzed by TLC and the products formed monitored and quantified using a phosphor imager. The position of authentic standards is indicated. Oxime, (E)- and (Z)-p-Hydroxyphenylacetaldoxime; Nitrile, p- hydroxyphenylacetonitrile; Aldehyde, p-hydroxybenzaldehyde.
DISCUSSION
The biosynthesis from amino acids of the aglycon part of cyanogenic glucosides has been shown to involve Cyt P450 enzymes in S. bicolor (Halkier and Møller, 1991), M. esculenta (Koch et al., 1992), and T. maritima (Nielsen and Møller, 1999) (Fig. 1). In S. bicolor, two multifunctional Cyt P450 enzymes have been shown to catalyze all of the membrane-associated steps leading to the aglycon of dhurrin. Tyr is converted to (Z)-p-hydroxyphenylacetaldoxime by CYP79A1 (Sibbesen et al., 1994, 1995; Koch et al., 1995), and (Z)-p-hydroxyphenylacetaldoxime is subsequently converted to p-hydroxymandelonitrile by CYP71E1 (Kahn et al., 1997; Bak et al., 1998a). In M. esculenta, a CYP79 homolog, CYP79D1, has recently been identified and characterized (Andersen et al., 2000). Also, the biosynthetic pathway of at least some glucosinolates involves Cyt P450 enzymes in the conversion of the parent amino acid to the corresponding aldoxime (Du et al., 1995). CYP79 homologs have been identified in the two glucosinolate-producing plants, S. alba (CYP79B1) and Arabidopsis (CYP79B2), but so far no successful expression of the isolated clones has been achieved (Bak et al., 1998b).
The CYP79B1 clone was isolated by screening an S. alba cDNA library with an expressed sequence tag (CYP79B2) probe from Arabidopsis (Bak et al., 1998b). Both plants are members of the Brassicaceae family, but contain glucosinolates derived from different substrates (Hogge et al., 1988; Du et al., 1995). Despite this difference, the two full-length clones showed 89% identity at the amino acid level (Table II). Accordingly, the first attempts to isolate a CYP79 homolog from T. maritima was carried out using heterologous probes from S. bicolor to take advantage of the fact that both plants are monocotyledonous and use Tyr as substrate. However, no CYP79 homolog could be isolated using this strategy. Instead, we used a PCR approach, taking advantage of the presence of some highly conserved CYP79-specific regions covering part of the I-helix (primers 1F and 2F), β6-1 and β1-4 (primers 1R and 2R) (Fig. 2). These regions are involved in substrate recognition (SRS) (Gotoh, 1992) as well as heme binding, and accordingly were thought to be less variable than other SRSs (Hasemann et al., 1995) and therefore suitable for the design of degenerate primers. Conserved regions within groups and families of Cyt P450s have previously been used successfully to design degenerate primers for the amplification of specific Cyt P450 clones (Holton et al., 1993; Meijer et al., 1993; Frank et al., 1996; Bak et al., 1998b).
The major PCR product obtained using cDNA and genomic DNA revealed 62% to 70% identity to other members of the CYP79 family. This confirmed that the primers were highly specific toward the CYP79 family, as illustrated with primer 1F (DNPSNA) covering a region that in the vast majority of other Cyt P450s contains a highly conserved Thr residue (Hasemann et al., 1995) instead of Asn. Only one of the degenerate primers (1R), covering FN(V/L)PHVA, did not anneal perfectly with the sequences subsequently obtained from T. maritima, having a single nucleotide difference at the 5′ end of the primer. Here the Ala codon in the primer was replaced by a Ser codon in the T. maritima sequences. The PCR fragment was used as a probe to screen the cDNA library made from T. maritima flowers and fruits. Microsomal preparations isolated from these tissues had previously been shown to have high catalytic activity compared with other tissues (Cutler et al., 1981; Nielsen and Møller, 1999), thus raising the abundance of the putative mRNA. Two partial CYP79 homologs were isolated and used to isolate a full-length and a nearly full-length clone designated CYP79E1 and CYP79E2, respectively, as the first members of a new subfamily (Fig. 2). These clones show 94% identity (Table II).
CYP79E1 showed 45% to 49% identity to other members of the CYP79 family (Table II). In sequence alignment to other members of the CYP79 family, CYP79E1 showed the highest identity and similarity to CYP79A1 (Table II). In contrast, CYP79A1 showed higher identity/similarity toward the three other sequences, CYP79B1, CYP79B2, and CYP79D1, that were all isolated from dicotyledonous plants (Table II). This is surprising considering that CYP79B1 and CYP79B2 are putatively involved in the biosynthesis of glucosinolates, whereas CYP79D1 catalyzes the conversion of aliphatic amino acids. The very high degree of identity between CYP79B1 and CYP79B2 (89%) is not seen between the three clones involved in the biosynthesis of cyanogenic glucosides, explaining the lack of success using a heterologous probe from S. bicolor for screening the T. maritima library.
Cyt P450s are thought to possess the same tertiary structure (Hasemann et al., 1995; Peterson and Graham, 1998); nevertheless, they only exhibit one highly conserved sequence, FXXGXRXCXG (X being any amino acid), harboring the heme-binding Cys residue (Nelson et al., 1993). When restricted groups or families of Cyt P450s are aligned, additional conserved amino acid residues are found, such as the proposed highly conserved A-group heme-binding consensus sequence, PFGXGRRXCXG (Durst and Nelson, 1995; Bak et al., 1998b), and the CYP79-specific sequence, SFSTG(K/R)RGC(A/I)A (Bak et al., 1998b). The CYP79 family diverges from the A-group heme-binding consensus sequence and within the CYP79 family, CYP79E diverges in several positions from otherwise conserved CYP79 residues (Fig. 2). A notable difference is that CYP79E1, CYP79E2, and CYP79A2 (GenEMBL no. AB010692 comp [11,000–13,200 region]) contain the generally highly conserved Gly residue positioned two amino acids downstream from the heme-binding Cys residue (amino acid 479 in CYP79E1), whereas all the other CYP79s contain Ala at this position. This Gly residue is positioned closely to the heme plain and allows a sharp turn from the Cys-pocket into the l-helix (Hasemann et al., 1995).
In the highly conserved PERF region within microsomal Cyt P450s (448–451 in CYP79E1), the Phe residue has been replaced by His, as is also observed in other CYP79s (Bak et al., 1998b). The K-helix contains a partly conserved region, KETLR (392–396 in CYP79E1), in many Cyt P450s, with the Glu and the Arg residues being particularly conserved (Hasemann et al., 1995). In the CYP79 family, Lys has been replaced by Arg, Thr has been replaced by Ala, and Leu has been replaced with Phe, with the exception that CYP79A2 still contains Lys and CYP79Es still contains Leu. Only minor differences are observed between the two different clones isolated from T. maritima. The region of positive charges preceding the Pro-rich region contains a repeat of 3× KS in CYP79E1, whereas CYP79E2 contains the sequence KPKS in the same region. Finally, the amino acids EGR (296–298 in CYP79E1) are deleted in CYP79E2, even though the Gly is conserved in all other CYP79s. The differences both within the CYP79 family and in relation to residues that are more or less conserved within the A-group or microsomal P450s in general illustrate how difficult it is, based on sequence alignments, to predict amino acids important for different functions.
All eukaryotic Cyt P450 enzymes found so far, except CYP55A (Park et al., 1997), are membrane bound. Some are localized in the mitochondrial membranes, while most, including plant Cyt P450s, are localized in the endoplasmic reticulum. The N terminus of the microsomal Cyt P450s functions as a hydrophobic signal sequence directing the proteins to the endoplasmic reticulum (Sakaguchi et al., 1987). The CYP79Es from T. maritima contains the four structurally conserved domains in the N terminus (Halkier et al., 1995). CYP79Es have a short stretch of four uncharged amino acids between the positively charged region and the Pro-rich region, as opposed to CYP79A1, which has a long stretch of 18 uncharged amino acids at this position. It has previously been suggested that a long stretch of uncharged residues serves to facilitate the access of the rather hydrophilic substrate Tyr to the active site (Koch et al., 1995). The fact that the cyanogenic glucosides in T. maritima are derived from Tyr contradicts this suggestion.
To verify the catalytic activity of the two CYP79 homologs from T. maritima, they were functionally expressed in E. coli. Codon usage in pro- and eukaryotic genes is different, and most highly expressed E. coli genes have a high AT content in the 5′ end of the gene. Therefore, modification of the 5′ end of most eukaryotic Cyt P450 genes is necessary to achieve high expression (Barnes et al., 1991; Halkier et al., 1995; Gillam, 1998). Chimeric CYP79E2, containing the first 24 amino acids of the bacterial lacZ gene, showed a dramatically increased expression level (Fig. 4) and turnover of Tyr per milligram of protein (Figs. 5 and 6) compared with CYP79E1 expressed with a few silent mutations or in a truncated form containing a modified P4502E1 N terminus (Gillam et al., 1994, 1995) devoid of the membrane-spanning anchor (Fig. 4). A partial CYP79E1 clone in-frame with lacZ at the same nucleotide position as CYP79E2 showed the same high expression level as lacZ-modified CYP79E2, indicating the importance of the N-terminal sequence and choice of expression vector for efficient expression. Surprisingly, the CYP79E1 construct, containing a modified P45017α N terminus with a stretch of 13 hydrophobic amino acids for membrane insertion, resulted in no detectable expression and no activity. The same type of construct [TYRΔ(1–25)bov] resulted in the highest level of expression of CYP79A1 (Halkier et al., 1995). All constructs showed higher expression level and activity when expressed in the E. coli strain JM109 compared with XL-1 Blue.
T. maritima contains the two cyanogenic glucosides triglochinin and taxiphyllin, which are both derived from Tyr (Sharples et al., 1972; Conn, 1973). Triglochinin biosynthesis requires a ring cleavage step supposedly taking place after an additional hydroxylation of the phenolic side chain. To further study this pathway, the catalytic activity of the two isolated CYP79Es from T. maritima were tested using other putative substrates. Neither Phe nor DOPA were metabolized by the Cyt P450s, strongly suggesting that the ring cleavage reaction leading to triglochinin formation proceeds after p-hydroxyphenylacetonitrile formation. The ring cleavage reaction could be dependent on hydroxylation carried out by a 2-oxoglutarate-dependent dioxygenase (Prescott and John, 1996).
(Z)-p-Hydroxyphenylacetaldoxime is converted to p-hydroxymandelonitrile by CYP71E1 in S. bicolor (Kahn et al., 1997; Bak et al., 1998a). Oximes have not been found to accumulate in cyanogenic plants (Tapper and Butler, 1972). The membrane-bound enzymes involved in the biosynthesis of cyanogenic glucosides are therefore believed to be closely associated (Møller and Seigler, 1998). Reconstitution experiments including native or 2E1 constructs of CYP79E1 or the CYP79E2 construct, all from T. maritima, together with isolated CYP71E1 from S. bicolor, indeed revealed formation of considerable amounts of p-hydroxybenzaldehyde and the presence of low amounts of p-hydroxyphenylacetaldoxime (Fig. 6). CYP71E1 is a labile enzyme and, in its isolated state, releases considerable amounts of p-hydroxyphenylacetonitrile from the active site before the subsequent C-hydroxylation reaction proceeds (Kahn et al., 1997; Bak et al., 1998a). Accordingly, p-hydroxyphenylacetonitrile is also observed to accumulate in the reaction mixtures, including the reaction mixture solely composed of S. bicolor CYP79A1 and CYP71E1.
In biosynthetic experiments using S. bicolor microsomes, no p-hydroxyphenylacetonitrile is observed (Møller and Conn, 1980) while p-hydroxyphenylacetonitrile accumulates in T. maritima (Nielsen and Møller, 1999). In the reconstitution experiments (Fig. 6), the relative levels of p-hydroxyphenylacetonitrile and p-hydroxybenzaldehyde were the same in all experiments, indicating that the activity of CYP71E1 is not differentially affected by the presence of CYP79A1 from S. bicolor or by native and truncated forms of CYP79E1 and CYP79E2 from T. maritima. This indicates that other membrane-associated regions of CYP79E1 are sufficient to retain correct orientation and association with CYP71E1. The efficient interaction between enzymes from the two species, T. maritima and S. bicolor, indicates a high conservation of the enzymes involved in the biosynthesis of cyanogenic glucosides, and suggest the presence of a CYP71E1 homolog in T. maritima. In vivo, it is conceivable that, in contrast to CYP71E1 from S. bicolor, the CYP71E1 homolog present in T. maritima releases sufficient amounts of p-hydroxyphenylacetonitrile to permit the formation of triglochinin, the major cyanogenic glucoside in T. maritima.
ACKNOWLEDGMENTS
Dr. Søren Bak (Department of Entomology, University of Arizona, Forbes 410) is thanked for helpful discussions and Karina Juul Peitersen for technical assistance. Dr. Peter Busk kindly provided a PCR fragment of CYP79D1, and Dr. Mette Dahl Andersen has kindly provided CYP79D1 for sequence comparison studies. Dr. Rachel Alice Kahn (Département d'Enzymologie Cellulaire et Moléculaire, Institut de Biologie Moléculaire des Plantes, Centre National de la Recherche Scientifique, Strasbourg, France) is acknowledged for kindly providing the NADPH-Cyt P450 oxidoreductase and CYP71E1 from S. bicolor. Dr. Hans-Henning Steinbiss (Max-Planck-Institut für Züchtungsforschung, Köln, Germany) is acknowledged for kindly providing the pSBET vector.
Footnotes
Financial support from the Danish Agricultural and Veterinary Research Council, Danish Biotechnology Program, and the Danish National Research Foundation is gratefully acknowledged.
LITERATURE CITED
- Andersen MD, Busk PK, Svendsen I, Møller BL. Cytochromes P450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin: cloning, functional expression in Pichia pastoris and substrate specificity of the isolated recombinant enzymes. J Biol Chem. 2000;275:1966–1975. doi: 10.1074/jbc.275.3.1966. [DOI] [PubMed] [Google Scholar]
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA. Cloning of three A-type cytochrome P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol. 1998a;36:393–405. doi: 10.1023/a:1005915507497. [DOI] [PubMed] [Google Scholar]
- Bak S, Nielsen HL, Halkier BA. The presence of CYP79 homologous in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol. 1998b;38:724–734. doi: 10.1023/a:1006064202774. [DOI] [PubMed] [Google Scholar]
- Barnes HJ. Maximizing expression of eukaryotic cytochrome P450s in Escherichia coli. Methods Enzymol. 1996;272:3–14. doi: 10.1016/s0076-6879(96)72003-7. [DOI] [PubMed] [Google Scholar]
- Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc Natl Acad Sci USA. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge DB, Hughes MA. In vitro characterization of the Ac locus in white clover (Trifolium repens L.) Arch Biochem Biophys. 1982;218:38–45. doi: 10.1016/0003-9861(82)90318-6. [DOI] [PubMed] [Google Scholar]
- Conn EE. Biosynthesis of cyanogenic glucosides. Biochem Soc Symp. 1973;38:277–302. [PubMed] [Google Scholar]
- Cutler AJ, Conn EE. The biosynthesis of cyanogenic glucosides in Linum usitatissimum (linen flax) in vitro. Arch Biochem Biophys. 1981;212:468–474. doi: 10.1016/0003-9861(81)90389-1. [DOI] [PubMed] [Google Scholar]
- Cutler AJ, Hösel W, Sternberg M, Conn EE. The in vitro biosynthesis of taxiphyllin and the channeling of intermediates in Triglochin maritima. J Biol Chem. 1981;256:4253–4258. [PubMed] [Google Scholar]
- Du L, Lykkesfeldt J, Olsen CE, Halkier BA. Involvement of a cytochrome P450 in glucosinolate biosynthesis as demonstrated by an in vitro microsomal enzyme system isolated from jasmonic acid-induced seedlings of Sinapis alba L. Proc Natl Acad Sci USA. 1995;92:12505–12509. doi: 10.1073/pnas.92.26.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst F, Nelson DR. Diversity and evolution of plant P450 and P450-reductases. Drug Metabol Drug Interact. 1995;12:189–206. doi: 10.1515/dmdi.1995.12.3-4.189. [DOI] [PubMed] [Google Scholar]
- Frank MR, Deyneka JM, Schuler MA. Cloning of wound-induced cytochrome P450 monooxygenases expressed in pea. Plant Physiol. 1996;110:1035–1046. doi: 10.1104/pp.110.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam EMJ. Human cytochrome P450 enzymes expressed in bacteria: reagents to probe molecular interactions in toxicology. Clin Exp Pharmacol Physiol. 1998;25:877–886. doi: 10.1111/j.1440-1681.1998.tb02338.x. [DOI] [PubMed] [Google Scholar]
- Gillam EMJ, Guo Z, Guengerich FP. Expression of modified human cytochrome P450 2E1 in Escherichia coli, purification, and spectral and catalytic properties. Arch Biochem Biophys. 1994;312:59–66. doi: 10.1006/abbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- Gillam EMJ, Guo Z, Martin MV, Jenkins CM, Guengerich FP. Expression of cytochrome P450 2D6 in Escherichia coli, purification, and spectral and catalytic characterization. Arch Biochem Biophys. 1995;319:540–550. doi: 10.1006/abbi.1995.1329. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- Halkier BA, Møller BL. Involvement of cytochrome P-450 in the biosynthesis of dhurrin in Sorghum bicolor (L.) Moench. Plant Physiol. 1991;96:10–17. doi: 10.1104/pp.96.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Nielsen HL, Koch B, Møller BL. Purification and characterization of recombinant cytochrome P450TYR expressed at high levels in Escherichia coli. Arch Biochem Biophys. 1995;322:369–377. doi: 10.1006/abbi.1995.1477. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Olsen CE, Møller BL. The biosynthesis of cyanogenic glucosides in higher plants: the (E)- and (Z)-isomers of p-hydroxyphenylacetaldehyde oxime as intermediates in the biosynthesis of dhurrin in Sorghum bicolor (L.) Moench. J Biol Chem. 1989;264:19487–19494. [PubMed] [Google Scholar]
- Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- Hogge RL, Reed DW, Underhill EW. HPLC separation of glucosinolates from seeds and leaves of Arabidopsis thaliana and their identification using thermospray liquid chromatography-mass spectrometry. J Chromatogr Sci. 1988;26:551–560. [Google Scholar]
- Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu C-Y, Farcy E, Stevenson TW, Cornish EC. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature. 1993;366:276–279. doi: 10.1038/366276a0. [DOI] [PubMed] [Google Scholar]
- Hösel W, Nahrstedt A. In vitro biosynthesis of cyanogenic glucoside taxiphyllin in Triglochin maritima. Arch Biochem Biophys. 1980;203:753–757. doi: 10.1016/0003-9861(80)90235-0. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Mol Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Kahn RA, Bak S, Svendsen I, Halkier BA, Møller BL. Isolation and reconstitution of cytochrome P450ox and in vitro reconstitution of the entire biosynthetic pathway of the cyanogenic glucoside dhurrin from sorghum. Plant Physiol. 1997;115:1661–1670. doi: 10.1104/pp.115.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B, Nielsen VS, Halkier BA, Olsen CE, Møller BL. The biosynthesis of cyanogenic glucosides in seedlings of cassava (Manihot esculenta Crantz) Arch Biochem Biophys. 1992;292:141–150. doi: 10.1016/0003-9861(92)90062-2. [DOI] [PubMed] [Google Scholar]
- Koch BM, Sibbesen O, Halkier BA, Svendsen I, Møller BL. The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of l-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys. 1995;323:177–186. doi: 10.1006/abbi.1995.0024. [DOI] [PubMed] [Google Scholar]
- Meijer AH, Souer E, Verpoorte R, Hoge JHC. Isolation of cytochrome P-450 cDNA clones from the higher plant Catharanthus roseus by a PCR strategy. Plant Mol Biol. 1993;22:379–383. doi: 10.1007/BF00014944. [DOI] [PubMed] [Google Scholar]
- Møller BL, Conn EE. The biosynthesis of cyanogenic glucosides in higher plants: N-hydroxytyrosine as an intermediate in the biosynthesis of dhurrin in Sorghum bicolor (Linn) Moench. J Biol Chem. 1979;254:8575–8583. [PubMed] [Google Scholar]
- Møller BL, Conn EE. The biosynthesis of cyanogenic glucosides in higher plants: channelling of intermediates in dhurrin biosynthesis by a microsomal enzyme system from Sorghum bicolor (Linn.) Moench. J Biol Chem. 1980;255:3049–3056. [PubMed] [Google Scholar]
- Møller BL, Seigler DS. Biosynthesis of cyanogenic glycosides, cyanolipids and related compounds. In: Singh BK, editor. Plant Amino Acids: Biochemistry and Biotechnology. New York: Marcel Dekker; 1998. pp. 563–609. [Google Scholar]
- Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, Gonzales FJ, Coon MJ, Gunsalus IC, Gotoh O, Okuda K, Nebert DW. The P450 superfamilies: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- Nielsen JS, Møller BL. Biosynthesis of cyanogenic glucosides in Triglochin maritima and the involvement of cytochrome P450 enzymes. Arch Biochem Biophys. 1999;368:121–130. doi: 10.1006/abbi.1999.1258. [DOI] [PubMed] [Google Scholar]
- Olins PO, Rangwala SH. Vector for enhanced translation of foreign genes in Escherichia coli. Methods Enzymol. 1990;185:115–119. doi: 10.1016/0076-6879(90)85012-d. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes: I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- Park S-Y, Shimizu H, Adachi S-I, Nakagawa A, Tanaka I, Nakahara K, Shoun H, Obayashi E, Nakamura H, lizuka T, Shiro Y. Crystal structure of nitric oxide reductase from denitrifying fungus Fusarium oxysporum. Nat Struct Biol. 1997;4:827–832. doi: 10.1038/nsb1097-827. [DOI] [PubMed] [Google Scholar]
- Peterson JA, Graham SE. A close family resemblance: the importance of structure in understanding cytochromes P450. Structure. 1998;6:1079–1085. doi: 10.1016/s0969-2126(98)00109-9. [DOI] [PubMed] [Google Scholar]
- Prescott AG, John P. Dioxygenases: structure and function. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:245–271. doi: 10.1146/annurev.arplant.47.1.245. [DOI] [PubMed] [Google Scholar]
- Richardson TH, Jung F, Griffin KJ, Wester M, Raucy JL, Kemper B, Bornheim LM, Hassett C, Omiecinski CJ, Johnson EF. A universal approach to the expression of human and rabbit cytochrome P450s of the 2C subfamily in Escherichia coli. Arch Biochem Biophys. 1995;323:87–96. doi: 10.1006/abbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Mihara K, Sato R. A short amino-terminal segment of microsomal cytochrome P-450 functions as an insertion signal and as a stop-transfer sequence. EMBO J. 1987;6:2425–2431. doi: 10.1002/j.1460-2075.1987.tb02521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder B, McCarthy JEG. The role of bases upstream of the Shine-Dalgarno region and in the coding sequence in the control of gene expression in Escherichia coli: translation and stability of mRNAs in vivo. Gene. 1989;78:59–72. doi: 10.1016/0378-1119(89)90314-4. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss H-H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- Sharples D, Spring MS, Stroker JR. Biosynthesis of the major cyanogenic glycoside of Thalictrum aquilegifolium. Phytochemistry. 1972;11:2999–3002. [Google Scholar]
- Sibbesen O, Koch B, Halkier BA, Møller BL. Isolation of the heme-thiolate enzyme cytochrome P-450TYR, which catalyzes the committed step in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Proc Natl Acad Sci USA. 1994;91:9740–9744. doi: 10.1073/pnas.91.21.9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbesen O, Koch B, Halkier BA, Møller BL. Cytochrome P450TYR is a multifunctional heme-thiolate enzyme catalyzing the conversion of l-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. J Biol Chem. 1995;270:3506–3511. doi: 10.1074/jbc.270.8.3506. [DOI] [PubMed] [Google Scholar]
- Stormo GD, Schneider TD, Gold LM. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapper BA, Butler GW. 2-Hydroxyaldoximes as possible precursors in the biosynthesis of cyanogenic glucosides. Phytochemistry. 1972;11:1041–1046. [Google Scholar]
- Tate WP, Brown CM. Translational termination: “stop” for protein synthesis or “pause” for regulation of gene expression. Biochemistry. 1992;31:2443–2450. doi: 10.1021/bi00124a001. [DOI] [PubMed] [Google Scholar]
- Werck-Reichart D, Benveniste I, Teutsch H, Durst F, Gabriac B. Glycerol allows low-temperature phase separation of membrane proteins solubilized in Triton X-114: application to the purification of plant cytochromes P-450 and b5. Anal Biochem. 1991;197:125–131. doi: 10.1016/0003-2697(91)90367-3. [DOI] [PubMed] [Google Scholar]