Abstract
Despite their evolutionary, developmental, and functional importance the origin of vertebrate paired appendages remains uncertain. In mice, a single enhancer termed ZRS is solely responsible for Shh expression in limbs. Here, zebrafish and mouse transgenic assays trace the functional equivalence of ZRS across the gnathostome phylogeny. CRISPR/Cas9-mediated deletion of the medaka-ZRS and enhancer assays reveal the existence of ZRS shadow enhancers in both teleost and human genomes. Deletion of both ZRS and shadow ZRS abolish shh expression and completely truncate pectoral fin formation. Strikingly, deletion of ZRS results in an almost complete ablation of the dorsal fin. This finding indicates that a ZRS-Shh regulatory module is shared by paired and median fins, and that paired fins likely emerged by the co‐option of developmental programs established in the median fins of stem gnathostomes. Shh function was later reinforced in pectoral fin development with the recruitment of shadow enhancers, conferring additional robustness.
The emergence of paired fins, either pectoral or pelvic, constituted a major morphological transformation of the vertebrate body plan. Despite this, their evolutionary origin remains a mystery. Classical ideas center on three hypotheses: that paired fins relate to gill arches, are derived from a fin fold, or that median fins evolved first and paired fins arose by co-option of ancient genetic patterning modules. Importantly, the third hypothesis makes specific predictions about the function and phylogenetic history of cis-regulatory elements (CREs) involved in appendage patterning.
Agnathans lack paired appendages whereas jawed vertebrates primitively have both paired and unpaired fins, such as the dorsal fin. Structurally, paired fins are typically composed of proximal endoskeletal elements and a distal dermal skeleton, the fin rays. Unpaired fins in most gnathostomes and agnathans exhibit the basic skeletal configuration seen in paired fins, with an endoskeleton and associated fin rays 1. Gene expression studies in paired and median fins reveal similar expression domains for Hox, Tbx, Shh and Fgf during fin bud initiation, including nested expression of Hox genes 2–6. Based on these similarities, one prominent theory of paired fin origins holds that paired fins arose by co-option of ancestral genetic modules first present in the median fin 1,7. As co-option often results from recruitment of existing CREs that evolve to elicit novel temporal or spatial patterns of gene expression, this theory makes specific predictions about the phylogeny, function and structure of enhancers in diverse appendages 8,9. Unfortunately, tests of the co-option hypothesis have been lacking—while a great deal is known regarding the cis-regulatory circuitry controlling limbs and paired fins development 10, virtually no corresponding data exists for median fins.
Sonic hedgehog-mediated control of growth and patterning is a deeply conserved feature of gnathostome paired appendages. In both developing fins and limbs, Shh expression originates from a posteriorly restricted domain called the zone of polarizing activity (ZPA) 11,12. Studies in mouse have shown that Shh expression in the ZPA is controlled by a single long-range cis-regulatory element, termed the “ZPA regulatory sequence” (ZRS) 13,14. The ZRS element is one of the multiple long-range enhancers that control tissue-specific Shh expression and is located approximately at 1 Mb from the Shh transcriptional start site in the 5th intron of the Lmbr1 gene 13. Interestingly, in several vertebrates polydactylous limb mutants have been linked to point mutations within the ZRS, resulting in anterior ectopic expression of Shh 13–15. Whereas point mutations are linked to gain of Shh expression, deletion of the mouse ZRS enhancer results in complete loss of Shh expression and severely truncated distal limb skeleton, a phenotype matching that observed in limbs of the Shh knockout mutant 14,16. The genomes of bony and cartilaginous fish also harbor an orthologous ZRS sequence, which can elicit ZPA-like expression of reporter gene in mouse transgenic assays 13,17. These findings suggest an ancient and conserved regulatory activity for this CRE. In skates and sharks, Shh expression has been detected in both paired and dorsal fin buds 5, yet the regulatory mechanisms driving Shh expression in unpaired fins are unknown.
To gain insight into the functional role of the ZRS in appendage development across vertebrates, we first investigated the evolutionary origins of the ZRS using phylogenetic footprinting (Supplementary Fig. 1a, b). To identify putative agnathan and cephalochordate ZRS sequences, we examined the orthologous intron 5 sequences from the two lmbr1 homologs of the sea lamprey (Petromyzon marinus) and the single lmbr1 gene from amphioxus (Branchiostoma lanceolatum). We found no sequence conservation between these intronic sequences and other vertebrate ZRS enhancers.
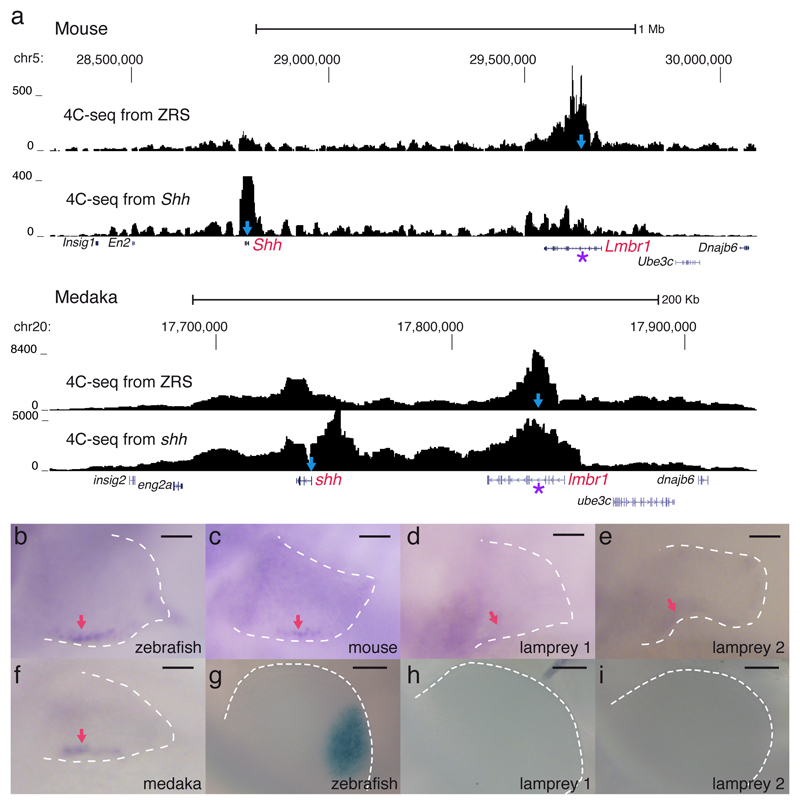
Consistent with the sequence conservation of ZRS across gnathostomes, their corresponding Lmbr1 loci are located in the vicinity of the Shh coding region; by contrast, in amphioxus, these two genes occupy different genomic positions (Irimia et al 2012). Therefore, we used 4C-seq experiments to examine whether the chromatin topology of the Shh locus is conserved among gnathostomes. We found very similar Shh gene regulatory landscapes in medaka and mouse, with strong chromatin contacts observed between the promoter of Shh and the ZRS enhancer (Fig. 1a).
Figure 1.
The regulatory landscape of Shh and the enhancer activity of ZRS in fin/limb ZPAs are conserved across gnathostomes. a, Regulatory landscapes of mouse and medaka Shh/shh gene determined by 4C-seq from the promoter of the gene and the ZRS enhancer. In each track the viewpoint for the 4C-seq is depicted by a light-blue arrow and the ZRS location with a purple asterisk. Genome coordinates are shown in the x axis and normalized interacting counts in the y axis. b-f, stable transgenic zebrafish larvae showing the activity in 72 hpf pectoral fins of the ZRS region from different species and the two orthologous Lmbr1 introns from lamprey. Red arrows point to the ZPA domain. Scale bars, 50µm. g-i, transgenic mouse embryos showing the activity of the ZRS region from different species in E10.5 developing limbs. Scale bars, 200µm. Three or more transgenic lines were generated for each construct in b-i.
To determine whether the trans-acting environment of developing fins and limbs can recognize ZRS sequences from fish and tetrapods, we performed heterologous transgenic assays in zebrafish and mouse. We found that ZRS elements from zebrafish, mouse and medaka drove reporter gene expression in zebrafish fins within the presumptive ZPA domain (Fig. 1b, c, f). In contrast, lamprey or amphioxus constructs failed to elicit reporter gene expression in zebrafish (Fig. 1d, e and Supplementary Fig. 1k). Next, we used mouse transgenic assays to test the cis-regulatory potential of ZRS elements to drive appendage expression in a tetrapod host. We found that zebrafish, but not lamprey constructs, can drive reporter gene expression in mouse limbs within the presumptive ZPA domain (Fig. 1g-i). To expand our heterologous analysis of ZRS function, we performed transgenic assays with ZRS from other gnathostomes and found that ZRS from skate, gar, coelacanth and anole elicit reporter gene expression in the posteriorly localized ZPA domain of developing zebrafish fins and mouse limbs (Supplementary Fig. 1). In sum, reciprocal transgenic reporters indicate that Shh gene regulation via ZRS enhancer arose in gnathostomes and is conserved in cis and trans, since both mouse and zebrafish host transcription factors can properly decode the information from fish and tetrapod donor cis elements and elicit a ZPA expression pattern. The hypothesis that ZRS emerged after the splitting of agnathans and gnathostomes is further supported by the lack of Hh expression in lamprey dorsal fins (Supplementary Fig. 2).
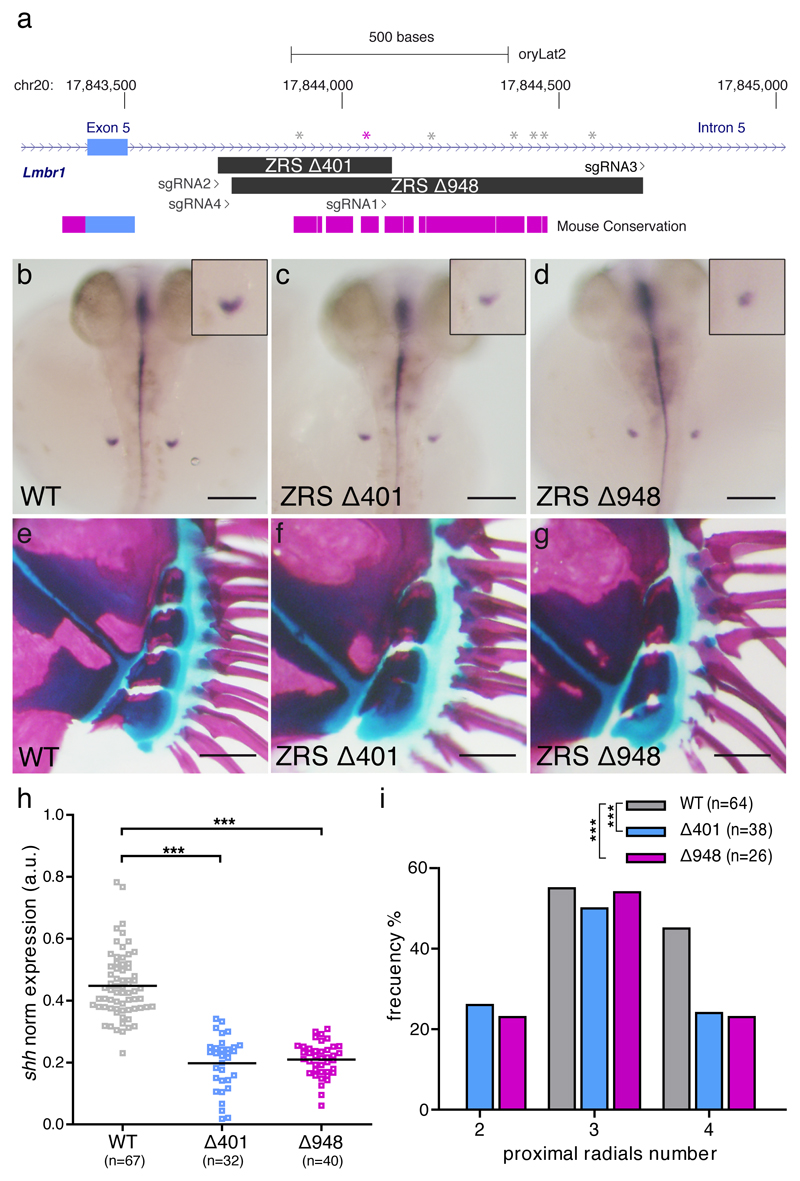
Next, we examined the role of ZRS regulation of shh expression during pectoral fin development. To this end, we chose medaka (O. latipes) as a model organism because, unlike zebrafish, which have duplicated copies of the shh gene after the teleost whole genome duplication 18,19, medaka as other euteleostei retains only one copy of Shh, making deletion analyses less confounding. Thus, we used CRISPR/Cas9 technology to delete the core of the ZRS enhancer in medaka, specifically aiming at disrupting a conserved ETS1 binding site important for regulation of Shh expression in the ZPA in mouse limbs 17,20 (Fig. 2a). Surprisingly, when compared to wild type (WT) (Fig. 2b, e, h), we found that stable germ-line medaka mutants carrying a 401 bp deletion in ZRS (∆401) showed a statistically significant, yet modest reduction of shh expression at 3dpf, and mild phenotypic outcomes in adults (Fig. 2c, f, h). WT medaka display a normal range of three to four small, square-shaped proximal radials which support the fin rays (Fig. 2e, i), whereas homozygous ZRS mutants displayed two to four proximal radials (Fig. 2f, i). Since the ∆401 deletion still left intact conserved ZRS flanking genomic sequences, we produced a larger, 948 bp deletion (∆948) that encompassed the entire medaka ZRS (Fig. 2a). Once again, we found that, as with the ∆401 deletion, ∆948 mutants displayed only modest reduction of shh expression (Fig. 2d, h). As observed for the ∆401 deletion, ∆948 mutants displayed two to four proximal radials (Fig. 2g, i). µCT-derived reconstructions of the pectoral fins from three-month-old WT and ZRS mutant medaka did not show a significant difference in the total volume of the radials (Supplementary Fig. 3). Our results suggest that, contrary to the mouse ZRS, the medaka ZRS is not solely responsible for ZPA expression of shh in developing pectoral fins.
Figure 2.
Medaka ZRS ablation results in mild pectoral fin defects. a, Stable lines harboring ∆401 and ∆948 ZRS deletions were generated by CRISPR/Cas9. The scheme shows the position of the deletions relative to the sgRNAs used and the box of conservation with the mouse genome. ETS1 binding sites are indicated with gray asterisks and a conserved ETS1 site important for regulation of Shh expression in the ZPA in mouse limbs is depicted in purple. b-d, h, shh expression in pectoral fin buds (insets) appears significantly reduced in ZRS deleted embryos at 3dpf. Scale bars, 200µm. e-g, i alcian blue/alizarin red staining of pectoral fin skeleton reveals a significantly reduced number of proximal radials in adult (5-month-old) mutants. Scale bars, 250µm. h, quantification of shh expression in medaka pectoral fin buds at 3dpf. Each point in the graph represent the measurement of shh expression in a single embryo. A one-way ANOVA test was used for the statistical analysis of shh expression. p-value=4.27x10-20 (***) for the comparison between WT (mean=0.448) and ∆401 (mean =0.198) and p-value=4.42x10-24 (***) for the comparison between WT and ∆948 (mean =0.209). i, Distribution of pectoral fin proximal radials number in adult medaka fish. Difference in the number of proximal radial bones was analysed using a chi-square test. N=number of adult pectoral fins analyzed. p-value=8.21x10-8 (***) for WT versus ∆401 and p-value=2.87x10-7 (***) for the WT versus ∆948 mutant comparison. Both shh expression at 3dpf and bone staining procedures in adults were performed in three independent experiments.
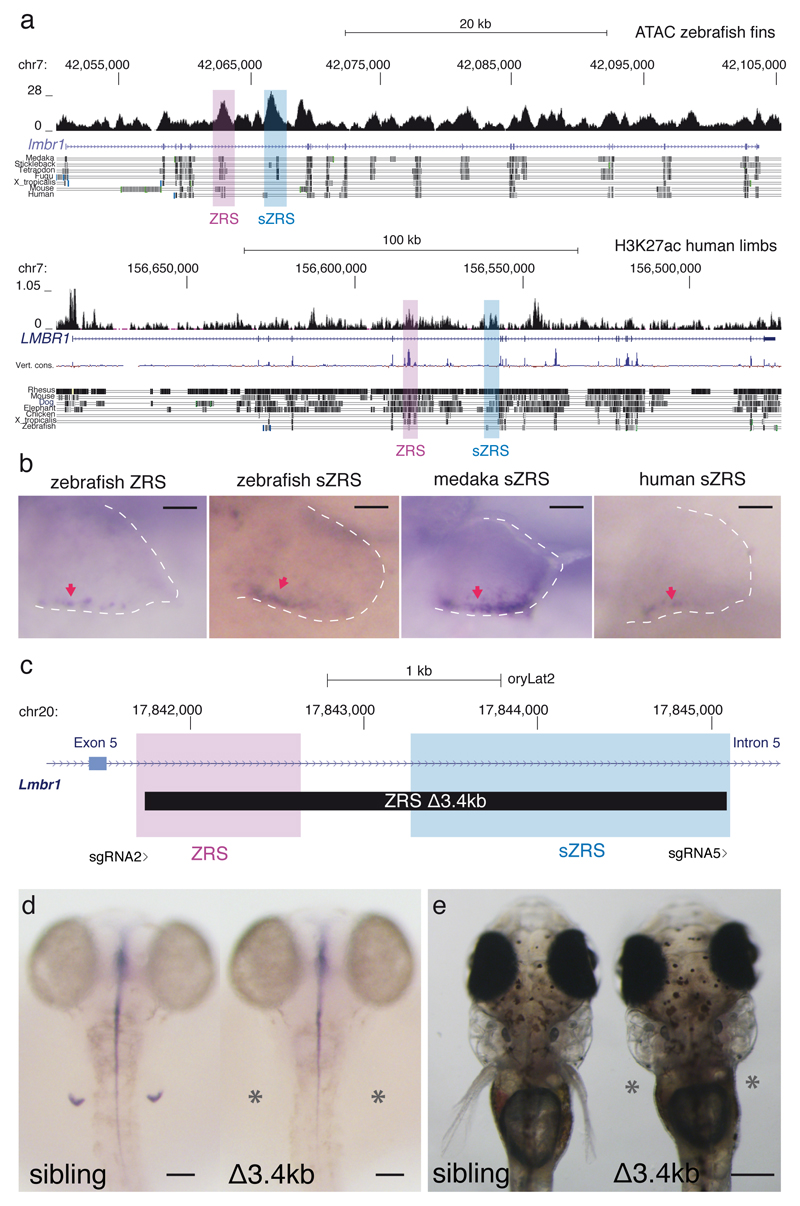
In order to find putative ZRS shadow enhancers, we searched the zebrafish lmbr1 genomic landscape for candidate sequences showing epigenetic marks and sequence conservation evidence consistent with an active CRE. Following this criterion, we identified a candidate sequence located within the same lmbr1 intron as the ZRS, conserved among teleosts and displaying a strong ATAC-seq peak derived from zebrafish developing fins 21 (Fig. 3a).
Figure 3.
The Lmbr1 intron containing ZRS harbors shadow enhancers for ZRS (sZRS) in zebrafish, medaka and humans. Deleting both ZRS and sZRS completely truncate pectoral fin development. a, ATAC-seq signal in zebrafish fins and H3K27ac distribution in human limbs along the lmbr1/LMBR1 genes. Genome coordinates are shown in the x axis and reads counts in the y axis. b, stable transgenic zebrafish larvae showing the activity in the ZPA (red arrows) of the zebrafish ZRS, zebrafish sZRS, medaka sZRS and human sZRS regions in 72 hpf pectoral fins. For each construct, three independent transgenic lines were generated. Scale bars, 50µm. c, Scheme summarizing the ∆3.4kb mutation deleting ZRS and sZRS from medaka genome and the position of the sgRNAs used. d, at 3dpf, shh is absent in the pectoral fins (asterisks) of ∆3.4kb homozygous mutants (n=6). Scale bars, 100µm. e, in contrast to their siblings, no pectoral fins are observed (asterisks) in ∆3.4kb mutant larvae at 9dpf (n=12). Scale bar, 400µm.
To functionally test this potential ZRS shadow enhancer (sZRS), we generated stable zebrafish transgenic lines carrying this element and the orthologous sequence from medaka. We found that at 60 hpf, developing larvae showed sZRS-driven GFP expression in the presumptive ZPA domain for both elements, mirroring very closely the expression pattern elicited by the zebrafish ZRS (Fig. 3b). To determine whether additional ZRS-like elements might exist in the genome of mammals, we searched for potential shadow ZRS sequences in the human genome. We discovered several regions within the human LMBR1 introns that showed H3K27ac epigenetic mark in limbs 22, which is associated with active enhancers (Supplementary Fig. 4). One of these sequences shares partial conservation to the zebrafish sZRS and is located in an equivalent position within the intron 5 of the human LMBR1 gene (Fig. 3a). Transgenic zebrafish lines carrying this human sZRS drive GFP expression in the presumptive ZPA domain in developing fins, as seen for the zebrafish and medaka elements (Fig. 3b). These results suggest that additional ZRS-like enhancers exist in both teleost and human genomes and may explain the modest shh downregulation and phenotypic outcome after ZRS deletion in medaka. To functionally test if sZRS is necessary for the expression of shh in pectoral fin buds, accounting for the mild phenotypes observed upon ∆948 ZRS deletion, we generated a mutant line deleting both ZRS and sZRS (∆3.4kb deletion) in the medaka genome (Fig. 3c). Interestingly, we found that in contrast to control animals, medaka mutants carrying the ∆3.4kb deletion showed a complete loss of shh expression in pectoral fin buds at 3dpf (Fig. 3d). In addition, these mutant fish completely fail to develop pectoral fins (Fig. 3e; Supplementary video 1). Detailed analysis of pectoral fin formation in WT and ∆3.4kb ZRS mutants confirmed the requirement of shh for their early development (Supplementary Fig. 5).
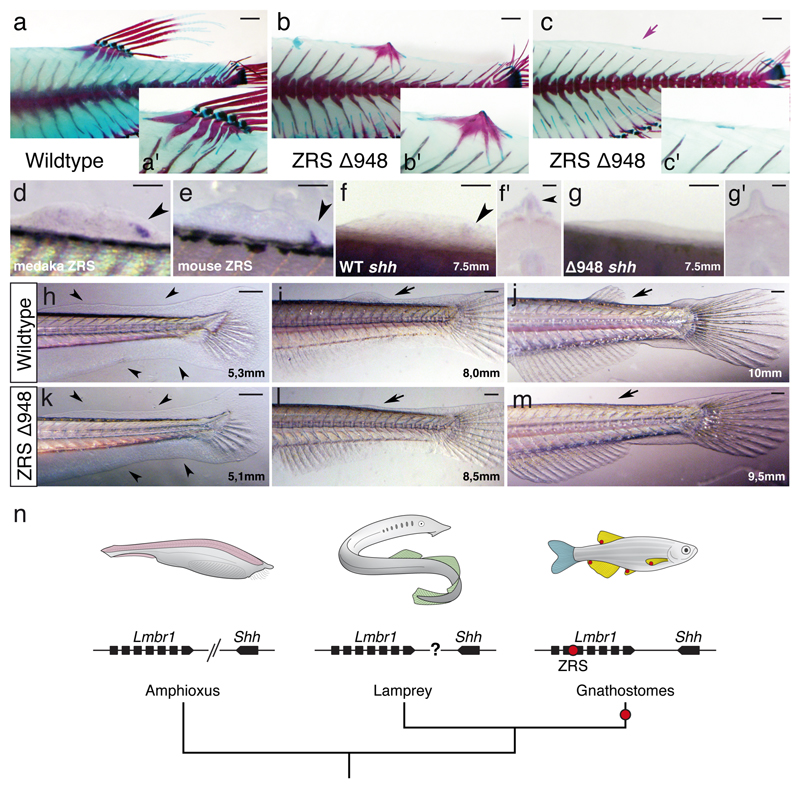
Finally, to establish the role of the ZRS enhancer in median fin development, we examined unpaired fins in our ZRS medaka mutant. Remarkably, we found that the ∆948 ZRS deletion resulted in near complete ablation of the dorsal fin (Fig. 4a-c; Supplementary Fig. 6). Detailed evaluation of the dorsal fin phenotype in this mutant revealed that adult fish either completely lack endoskeletal elements and fin rays 74% (31/42), or they are very reduced 26% (11/42) (Supplementary Fig. 6). Our analysis of transgenic zebrafish lines carrying either the medaka or the mouse ZRS revealed that both enhancers drive expression in developing dorsal fins, in a posteriorly restricted domain (Fig. 4d, e). In line with these findings, we detected a posteriorly restricted, mesodermal expression of shh in the medaka developing dorsal fin as previously reported in chondrichthyans 5 (Fig. 4f, f’). Furthermore, medaka ∆948 ZRS mutants did not show shh expression in the dorsal fin bud (Fig. 4g, g’). These results suggest an early arrest of dorsal fin development in ZRS ∆948 mutants in the absence of shh signaling. Analysis of dorsal fin formation in WT and ZRS mutants confirmed this early requirement (Fig. 4h-m). Interestingly, ZRS activity was also detected in developing pelvic and anal fins, while sZRS only show expression in paired fins but not in median fins (Supplementary Fig. 7). Whereas in pelvic fins ZRS deletion resulted in some endoskeletal and fin ray defects, no phenotype was observed for the anal fin (Supplementary Fig. 7).
Figure 4.
Dorsal fin do not form in ZRS medaka mutants. a-c, Skeletal staining and fin morphology in wild type and mutant fish. Alcian blue/alizarin red staining (insets) reveals an almost complete ablation of the fin rays and very reduced endoskeletal elements (purple arrow). Scale bars, 1mm. d, e, Posteriorly restricted expression of medaka and mouse ZRS in the zebrafish dorsal fin bud (black arrowheads). Scale bars, 100µm. f-g, Whole mount ISH and transverse sections (f’-g’) in medaka showing shh expression in the posterior dorsal fin bud of wild type (black arrowheads) but not in the ∆948 mutant. Bone staining and ZRS or shh expression assessment were performed in three or more independent experiments. For f-g and f’-g’, scale bars 100µm and 50µm, respectively. h-m, Temporal series showing normal development of the fin-fold in mutant embryos (black arrowheads), but arrested dorsal fin bud growth later during development (black arrows). For each stage represented in h-m, five or more fish were analyzed. Scale bars, 250µm. n, summary of the evolution of the Shh locus. Lmbr1 and Shh are linked in gnathostomes genomes but not in amphioxus. The genomic linkage between the 4 Hh and the two Lmbr1 genes in lamprey is unclear. Red dots in fins reflect the expression domain of Shh driven by ZRS in developing fin buds. The detailed mechanisms patterning median fins in amphioxus (pink), lampreys (green), and the caudal fin in gnathostomes (blue) remain to be identified, but appear to be independent of ZRS and, in amphioxus and lamprey, may not involve hedgehog signaling.
ZRS phylogeny and function confirms a specific prediction of the co-option hypothesis for the origin of paired appendages: the ZRS is deeply conserved in cis and trans across gnathostome fins, it appears to be lacking from the genome of agnathans, and deletion of the ZRS enhancer in fish results in loss of the dorsal fin (Fig. 4n). This study reveals the necessity of comparative functional genomic studies that integrate analyses across diverse taxa and organs in a phylogenetic context. The cis-regulatory code in ZRS clearly integrates complex information with shadow enhancers and diverse effects on Shh expression across different appendages. Indeed, the striking dorsal fin phenotype in ZRS mutants, coupled with the knockdown in expression observed suggest that shh expression in the dorsal fin is chiefly driven by the ZRS enhancer. In contrast, in pectoral fin development, Shh signaling gained additional robustness with the recruitment of shadow enhancers. Both phenomena can now be seen as evolutionary novelties of jawed vertebrates.
Online Methods
Animal experimentation
All experiments involving animals conform national and European Community standards for the use of animals in experimentation and were approved by the Ethical committees from the University Pablo de Olavide, CSIC and the Andalucian Government.
Fish stocks
Wild type strains for zebrafish and medaka, respectively AB/Tübingen (AB/Tu) and iCab, were maintained and bred under standard conditions 23,24. Embryos were staged in hours post-fertilization (hpf) as described 25,26.
Phylogenetic analyses and isolation of ZRS orthologues
Orthologous ZRS sequences from various species where retrieved from the UCSC genome database (http://genome.ucsc.edu), the gar genome from the ENSEMBL database. and the skate genome from Skatebase. Sequence alignments and conservation peaks were visualized using the mVista program Shuffle-LAGAN. Putative ZRS elements were isolated from: mouse (Mus musculus), anole (Anolis carolinensis), coelacanth (Latimeria menadoensis), zebrafish (Danio rerio), gar (Lepisosteus oculatus), skate (Leucoraja erinacea), and sea lamprey (Petromyzon marinus). Using mouse or zebrafish as the reference genome, two Lmbr1-like genes were identified in the sea lamprey genome and the intronic region spanning exons five and six was subsequently tested for regulatory activity in transgenic assays. Supplementary Table 1 lists the oligonucleotide sequences used to amplify the genomic fragments from their corresponding genomes. Genomic DNA fragments were isolated using the Platinum® Taq DNA polymerase High Fidelity Kit (Life Technologies). Fragments were cloned into an entry vector (PCR8/GW/TOPO; except for D. rerio, which was cloned into pENTR/D-TOPO) and transferred to the appropriate destination vectors using the Gateway® LR recombination reaction (Invitrogen).
Mouse transgenesis
After subcloning into the entry vector, the DNA inserts were transferred to a destination vector containing the human minimal β-globin promoter upstream of the LacZ/SV40polyA reporter gene (for coelacanth fragment), or a vector containing the mouse hsp68 minimal promoter (all other organisms) and LacZ/SV40polyA (a kind gift from Marcelo Nobrega). Final destination vectors were confirmed by restriction digest and sequencing. Cyagen Biosciences (Cyagen.com) performed injections and LacZ staining for all DNA elements. Mouse embryos were harvested, stained and fixed as per 27. Embryos were analyzed and imaged using a Leica M205FA microscope.
Skeletal staining
Skeletal staining was performed as previously described 28. Briefly, fish were fixed by 10% neutral-buffered formalin overnight. After washing with distilled and deionized water (ddH2O), specimens were placed in a graded series of 70% EtOH followed by 30% acetic acid/70% EtOH. Cartilage was stained overnight using a 0.02% Alcian blue solution in 30% acetic acid /70% EtOH. Specimens were then briefly rinsed using ddH2O and the solution was changed to a 30% saturated sodium borate solution and incubated for an hour. After, specimens were immersed in a 1% trypsin/30% saturated sodium borate and incubated at room temperature for eight hours. Following another ddH2O rinse, specimens were transferred into 1% KOH solution with 0.005% Alizarin Red S. On the following day, specimens were rinsed in ddH2O and subjected to a graded glycerol series for photographing using a Leica M205FA microscope followed by storage.
PMA staining and µCT scanning
After skeletal staining, fins were separated from the body. Fins were stained using a 0.5% (weight/volume) PMA (Phosphomolybdic acid) stain for 17 hours followed by two washes using ddH2O. Specimens were placed into 1.5mL microcentrifuge tubes with ddH2O and kept overnight to settle. On the following day, specimens were scanned using the UChicago PaleoCT (GE Phoenix v/tome/x 240kv/180kv scanner), at 50 kVp, 160 µA, no filtration, 5x-averaging, exposure timing of 1000 ms per image, and a resolution of 6.000 µm per slice (216 µm3 per voxel). Scanned images were analyzed and segmented using Amira 3D Software 6.0 (FEI).
Zebrafish transgenesis
After subcloning in the PCR8/GW/TOPO entry vector, the DNA inserts were transferred to an enhancer detection vector composed of a gata2 minimal promoter, an enhanced GFP reporter gene and a strong midbrain enhancer (z48) that works as an internal control for transgenesis in zebrafish 29. Zebrafish transgenic embryos were generated using the Tol2 method 30. One-cell stage embryos were injected with 2 nl of 25 ng/µl of transposase mRNA, 20 ng/µl of phenol:chloroform-purified constructs and 0.05% phenol red solution. Three or more independent stable transgenic lines were generated for each construct.
Zebrafish and medaka in situ hybridization
Antisense digoxigenin-labeled (Boehringer-Mannheim) RNA probes were prepared from cDNA. Specimens were prepared, hybridized, and stained as previously described for zebrafish 31 and medaka 32. Specimens were visualized with and Olympus SZX16 binocular microscope and photographed with an Olympus DP71 camera.
4C-seq
4C-seq assays were performed as recently reported 33 using as starting material 500 medaka embryos at 48hpf and 25 mouse embryos at E9.5. Supplementary Table 1 lists primers used as viewpoints for mouse and medaka shh promoters and medaka ZRS.
Cas9 target design and mutant generation
To obtain deletions of the ZRS element in medaka, sgRNAs targeting the flanking regions of the enhancer were designed using the CRISPRscan 34 and CCtop 35 online tools. Guided RNAs (sgRNAs) were generated as previously described 36. 3-5nL of a mixture containing sgRNAs (40ng/uL) and Cas9 protein (300ng/uL) 37, addgene vector #47327) were co-injected into one-cell stage medaka embryos to induce genomic deletions within the locus. ∆401 mutant line was established using the following sgRNAs: sgRNA1 and sgRNA2. ∆948 mutant line was generated using sgRNA3 and sgRNA4. ∆3.4kb mutant line was generated using sgRNA2 (same as for ∆401 mutant) and sgRNA5. Primers used for screening of genomic deletions (∆401, ∆948 and ∆3.4kb ZRS) in F1 progeny are listed together with the sgRNA sequences in Supplementary Table 1. These mutations were further analysed at F1 by sequencing to characterize the exact extent of the chromosomal lesions. Mutants for the ∆3.4kb ZRS deletion were obtained by crossing F0 fish in which each founder had 15-20% of their germline carrying the Δ3.4kb mutation. Approximately 1 out of 30 F1 embryos obtained was homozygous mutant for the Δ3.4kb deletion. With this strategy, we have so far obtained 18 double mutants.
Metamorphic ammocoete larva in situ hybridization
An ammocoete larva of the Far Eastern brook lamprey Lethenteron reissneri (syn. Lampetra reissneri) in metamorphosis was acquired from a local supplier in Nagano, Japan, and fixed with 4% paraformaldehyde for 24 h at 4 °C. First and second dorsal fins were dissected, embedded in paraffin wax and sectioned with a thickness of 10 microns. Embryos of the Arctic lamprey Lethenteron camtschaticum (syn. Lethenteron japonicum) were obtained as previously described 38 and staged according to 39. Digoxigenin-labeled riboprobes of the L. camtschaticum HhA, HhB and HhD genes were obtained from Sugahara et al., (2016). 3’ regions of the coding sequence of the L. camtschaticum MyHC1 (myosin heavy chain 1; 40) and ColA (collagen A or col2a1a; 41) genes were amplified by PCR from cDNA prepared from a mix of embryos of L. camtschaticum at different stages. Supplementary Table 1 lists primers used for cloning MyHC1 and ColA. Amplified fragments were cloned into pCRII/TOPO (Life Technologies) and digoxigenin-labeled riboprobes were prepared as described by the manufacturer (DIG RNA Labeling Mix, ROCHE). In situ hybridization experiments on developing dorsal fin sections (10 μm) of L. reissneri and whole-mount embryos of L. camtschaticum were performed according to 38.
Statistical analyses
Expression of shh in medaka pectoral fin buds was measured using image J software and normalized by the expression of shh in the midline. A one-way ANOVA test was used for the statistical analysis of these measurements. The number of pectoral fin proximal radial bones was counted in WT and mutant fish after alizarin red staining protocol. Differences in proximal radial number were tested by using a chi-square test. All fin length measurements were obtained using image J software and were normalized by the standard length of the fish. In all length cases a t-test was used for analysis.
Supplementary Material
Acknowledgements
We thank to Ana Fernández-Miñan from the CABD Aquatic vertebrate platform for providing the medaka 4C-seq samples, to Rafael D. Acemel for helping with the design of the medaka 4C-seq primers and to all members of JLGSK laboratory and Fernando Casares for fruitful discussions. We thank John Westlund for illustration assistance and Fumiaki Sugahara for providing the clone of lamprey HhA and the metamorphic ammocoete larva. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 740041), the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement #658521, the Spanish Ministerio de Economía y Competitividad (grants BFU2016-74961-P, BFU2014-53765-P and BFU2014-55738-REDT), the Andalusian Government (grant BIO-396), CNPq Universal Program Grant 403248/2016-7 and CAPES/Alexander von Humboldt Foundation fellowship (to I.S.).
Footnotes
Code availability
Custom code for 4C-seq mapping is available upon request.
Data availability
Data sets presented in this study are available under Gene Expression Omnibus (GEO) accession GSE97860.
Author Contributions
J.L. generated and analyzed the medaka mutants. E.d.l.C.-M carried out the 4C-seq experiments and the zebrafish transgenic assays with the help of S.N. and J. L. J.P. generated the mouse transgenic data. T.N. performed the µCT experiments. J. P-A. carried out the lamprey in situ experiments. J.L.G.-S., J.R.M.M., I.S. and N.S. conceived, designed and coordinated the Project with the help of J. L. and N.M. I.S. J.L.G.-S., J.R.M.M., N.S, J.L. and N.M. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
URLs. ENSEMBL database, http://www.ensembl.org/index.html, Skatebase, http://skatebase.org, UChicago PaleoCT, http://luo-lab.uchicago.edu/paleoCT.html.
References
- 1.Freitas R, Gomez-Skarmeta JL, Rodrigues PN. New frontiers in the evolution of fin development. J Exp Zool B Mol Dev Evol. 2014 doi: 10.1002/jez.b.22563. [DOI] [PubMed] [Google Scholar]
- 2.Sordino P, van der Hoeven F, Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- 3.Neumann CJ, Grandel H, Gaffield W, Schulte-Merker S, Nusslein-Volhard C. Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development. 1999;126:4817–26. doi: 10.1242/dev.126.21.4817. [DOI] [PubMed] [Google Scholar]
- 4.Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417:754–8. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- 5.Dahn RD, Davis MC, Pappano WN, Shubin NH. Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature. 2007;445:311–4. doi: 10.1038/nature05436. [DOI] [PubMed] [Google Scholar]
- 6.Freitas R, Zhang G, Cohn MJ. Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature. 2006;442:1033–7. doi: 10.1038/nature04984. [DOI] [PubMed] [Google Scholar]
- 7.Pieretti J, et al. Organogenesis in deep time: A problem in genomics, development, and paleontology. Proc Natl Acad Sci U S A. 2015;112:4871–6. doi: 10.1073/pnas.1403665112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2011;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 9.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–85. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehrke AR, Shubin NH. Cis-regulatory programs in the development and evolution of vertebrate paired appendages. Semin Cell Dev Biol. 2016;57:31–9. doi: 10.1016/j.semcdb.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogura T, et al. Evidence that Shh cooperates with a retinoic acid inducible co-factor to establish ZPA-like activity. Development. 1996;122:537–42. doi: 10.1242/dev.122.2.537. [DOI] [PubMed] [Google Scholar]
- 12.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 13.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–35. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 14.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 15.Maas SA, Suzuki T, Fallon JF. Identification of spontaneous mutations within the long-range limb-specific Sonic hedgehog enhancer (ZRS) that alter Sonic hedgehog expression in the chicken limb mutants oligozeugodactyly and silkie breed. Dev Dyn. 2011;240:1212–22. doi: 10.1002/dvdy.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 17.Kvon EZ, et al. Progressive Loss of Function in a Limb Enhancer during Snake Evolution. Cell. 2016;167:633–642 e11. doi: 10.1016/j.cell.2016.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–57. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 19.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 20.Lettice LA, et al. Development of five digits is controlled by a bipartite long-range cis-regulator. Development. 2014;141:1715–25. doi: 10.1242/dev.095430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gehrke AR, et al. Deep conservation of wrist and digit enhancers in fish. Proc Natl Acad Sci U S A. 2015;112:803–8. doi: 10.1073/pnas.1420208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotney J, et al. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–96. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene: 1995. [Google Scholar]
- 24.Koster R, Stick R, Loosli F, Wittbrodt J. Medaka spalt acts as a target gene of hedgehog signaling. Development. 1997;124:3147–56. doi: 10.1242/dev.124.16.3147. [DOI] [PubMed] [Google Scholar]
- 25.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 26.Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605–18. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Schneider I, et al. Appendage expression driven by the Hoxd Global Control Region is an ancient gnathostome feature. Proc Natl Acad Sci U S A. 2011;108:12782–6. doi: 10.1073/pnas.1109993108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird NC, Mabee PM. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae) Dev Dyn. 2003;228:337–57. doi: 10.1002/dvdy.10387. [DOI] [PubMed] [Google Scholar]
- 29.Acemel RD, et al. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet. 2016;48:336–41. doi: 10.1038/ng.3497. [DOI] [PubMed] [Google Scholar]
- 30.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–44. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 1994;10:73–4. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Morales JR, et al. Differentiation of the vertebrate retina is coordinated by an FGF signaling center. Dev Cell. 2005;8:565–74. doi: 10.1016/j.devcel.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Minan A, Bessa J, Tena JJ, Gomez-Skarmeta JL. Assay for transposase-accessible chromatin and circularized chromosome conformation capture, two methods to explore the regulatory landscapes of genes in zebrafish. Methods Cell Biol. 2016;135:413–30. doi: 10.1016/bs.mcb.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Moreno-Mateos MA, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–8. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS One. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vejnar CE, Moreno-Mateos MA, Cifuentes D, Bazzini AA, Giraldez AJ. Optimized CRISPR-Cas9 System for Genome Editing in Zebrafish. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.prot086850. pdb prot086850. [DOI] [PubMed] [Google Scholar]
- 37.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugahara F, Murakami Y, Kuratani S. Gene Expression Analysis of Lamprey Embryos. In: Hauptmann G, editor. In Situ Hybridization Methods. Springer New York; New York, NY: 2015. pp. 263–278. [Google Scholar]
- 39.Tahara Y. Normal stages of development in the lamprey, Lampetra reissneri (Dybowski) Zool Sci. 1988;5:109–118. [Google Scholar]
- 40.Kusakabe R, Takechi M, Tochinai S, Kuratani S. Lamprey contractile protein genes mark different populations of skeletal muscles during development. J Exp Zool B Mol Dev Evol. 2004;302:121–33. doi: 10.1002/jez.b.20009. [DOI] [PubMed] [Google Scholar]
- 41.Ohtani K, et al. Expression of Sox and fibrillar collagen genes in lamprey larval chondrogenesis with implications for the evolution of vertebrate cartilage. J Exp Zool B Mol Dev Evol. 2008;310:596–607. doi: 10.1002/jez.b.21231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.