Abstract
Functional connectivity (FC) has been widely investigated in many imaging-based neuroscience and clinical studies. Since functional Magnetic Resonance Image (MRI) signal is just an indirect reflection of brain activity, it is difficult to accurately quantify the FC strength only based on signal correlation. To address this limitation, we propose a learning-based tensor model to derive high sensitivity and specificity connectome biomarkers at the individual level from resting-state fMRI images. First, we propose a learning-based approach to estimate the intrinsic functional connectivity. In addition to the low level region-to-region signal correlation, latent module-to-module connection is also estimated and used to provide high level heuristics for measuring connectivity strength. Furthermore, sparsity constraint is employed to automatically remove the spurious connections, thus alleviating the issue of searching for optimal threshold. Second, we integrate our learning-based approach with the sliding-window technique to further reveal the dynamics of functional connectivity. Specifically, we stack the functional connectivity matrix within each sliding window and form a 3D tensor where the third dimension denotes for time. Then we obtain dynamic functional connectivity (dFC) for each individual subject by simultaneously estimating the within-sliding-window functional connectivity and characterizing the across-sliding-window temporal dynamics. Third, in order to enhance the robustness of the connectome patterns extracted from dFC, we extend the individual-based 3D tensors to a population-based 4D tensor (with the fourth dimension stands for the training subjects) and learn the statistics of connectome patterns via 4D tensor analysis. Since our 4D tensor model jointly (1) optimizes dFC for each training subject and (2) captures the principle connectome patterns, our statistical model gains more statistical power of representing new subject than current state-of-the-art methods which in contrast perform above two steps separately. We have applied our tensor statistical model to identify ASD (Autism Spectrum Disorder) by using the learned dFC patterns. Promising classification results have been achieved demonstrating high discrimination power and great potentials in computer assisted diagnosis of neuro-disorders.
1 Introduction
Functional Magnetic Resonance Imaging (fMRI) provides a non-invasive way to study how human brain works. This imaging technique assumes that the change of cerebral blood flow is closely related with brain activity. When an area of the brain is in use, blood flow to that region correspondingly increases [1]. There are various fMRI studies in neuroscience research to understand how the healthy brain works [2], and how that normal function is disrupted in disease [3]. Resting-state fMRI (rs-fMRI) is one of the functional brain imaging techniques that is widely used to measure occurrence of regional interactions that a subject is not performing an explicit task [4]. In resting state, fluctuations in spontaneous neural activity are pre-assumed to underlie the Blood-Oxygen-Level Dependent (BOLD) signal fluctuations, which form inter-regional functional connectivity in human brain network [5]. Since the spatial patterns of resting-state functional brain network are stable and often are overlapped with known anatomical pathways, rs-fMRI has been widely implemented to explore the brain’s functional organization and examine the altered connectivity in neurological or psychiatric diseases [6].
In current functional brain network studies, Pearson’s correlation on BOLD signals is used to measure the strength of FC between two brain regions [5]. It is worth noting that such correlation based connectivity measure is exclusively calculated based on the observed BOLD signals and fixed for the subsequent data analysis. However, the BOLD signal usually has very poor signal-to-noise ratio and is mixed with substantial non-neural noise and artifacts. Therefore, it is hard for current state-of-the-art methods to determine a good threshold of FC strength which can effectively distinguish real and spurious connections [7].
For simplicity, many FC characterization methods assume that connectivity patterns in the brain do not change over the course of a resting-state fMRI scan. However, there is a growing consensus in the neuroimaging field, that the spontaneous fluctuations and correlations of signals between two distinct brain regions change with correspondence to cognitive states, even in a task-free environment [8]. Thus, dynamic FC patterns have been investigated recently mainly by using sliding window techniques [8,9]. Due to the large dimension of sliding window numbers and large number of brain regions, it is very difficult to represent the dynamic brain network compactly directly. Therefore, many different machine learning techniques has been proposed to reduce the dimension of dynamic brain FC patterns. The common strategy of those works is to reduce the dimension on sliding windows using clustering [10] or Principle Component Analysis (PCA) [11] along time. And the clustering coefficients or PCA coefficients are used as the compact representation for dynamic FC. However, these methods reduce the dynamic FC dimension at the cost of losing the local temporal changes along time. Furthermore, in all existing dynamic FC methods, the procedures of estimating functional connectivity and extracting connectome features are completely independent. Although both steps are very challenging, we would like to argue that these two steps can help each other in a collaborative manner. Specifically, accurate functional connectivity of individual subjects can derive more reasonable statistical model to better represent the whole population. On the other hand, the learned statistical model can provide additional population-based heuristics to address the uncertainty in measuring functional connectivity for individual subject. To that end, we propose a tensor-based statistical model which jointly optimizes the dynamic functional connectivity at the individual basis and learns the intrinsic connectome patterns for the whole population. Our proposed method is illustrated in Fig. 1, which consists of three parts:
Fig. 1.
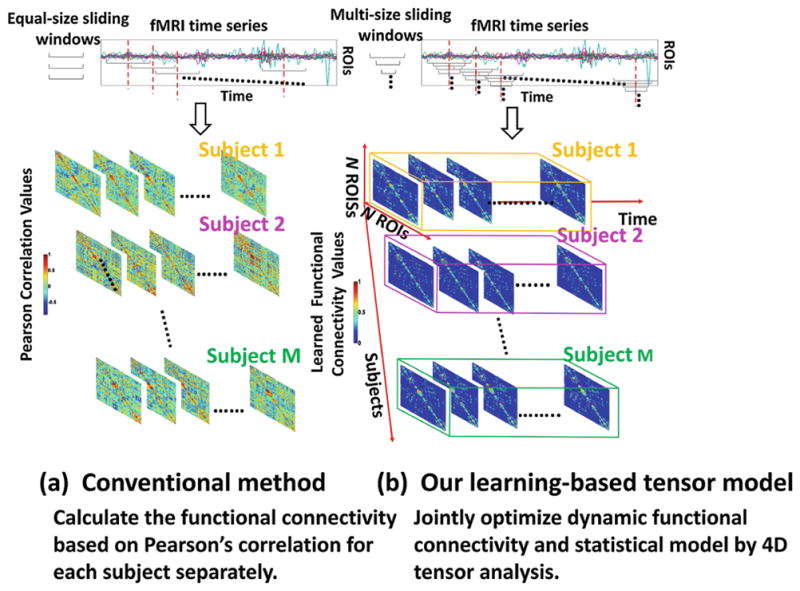
The advantage of our 4D tensor method (b) over the conventional method (a).
First, we present a robust learning-based method to optimize FC from the BOLD signals in a fixed sliding window. In order to avoid the unreliable calculation of FC based on signal correlations, high level feature representation is of necessity to guide the optimization of FC. Specifically, our method seeks for module-wise network structure during the optimization of node-to-node functional connectivity, where the brain region within the same module should have similar connection patterns. Thus, we can optimize functional connections for each brain region based on not only the observed region-to-region signal correlations but also the similarity between high level module-to-module connection patterns. In turn, the refined FC can lead to more reasonable estimation of module-wise connections. Since brain network is intrinsically economic and sparse, sparsity constraint is used to control the number of connections during the joint estimation of principal connection patterns and the optimization of FC.
Second, we further extend the above FC optimization framework from one sliding window (capturing the static FC patterns) to a set of overlapped sliding windows (capturing the dynamic FC patterns). The leverage is that we arrange the FCs along time into a 3D tensor (cubic in Fig. 1) and we employ additional low rank constraint to penalize the oscillatory changes of FC in the temporal domain.
Third, in order to learn the statistical model of intrinsic feature representations derived from the dynamic functional connectivity (dFC) in the population, we arrange the estimated functional dynamics of all subjects into a 4D tensor (right of Fig. 1) with the subjects denoted by fourth dimension. Then we go one step further to alternatively (1) optimize the dFC within each subject-specific 3D tensor under the guidance of learned intrinsic feature representations, and (2) learn the statistical model which can represent majority variations of dynamic functional connectivity patterns in the population. In doing so, we can derive robust statistical model of dynamic functional connectivity and encode the characteristics of dFC for the new unseen subjects. It is worth noting that the output of our statistical model can be regarded as the imaging markers in identifying individuals having or at risk of neuro-disease.
2 Method
2.1 Optimize Functional Connectivity Within Sliding Window
To start our tensor-based statistical model of dFC, we first propose a robust FC optimization method given a sliding window. Let xi ∈ ℝW×1 denote the mean BOLD signal calculated in brain region Oi, (i = 1, ···, N), where W is the length of time course within the sliding window and N is the total number of brain regions under consideration. Conventionally, a N × N connectivity matrix S is used to measure the FC in the whole brain, where each element sij quantitatively measures the strength of FC between region Oi and Oj(i ≠ j). Particularly, the strength of functional connectivity sij is assumed to be measurable based on Pearson’s correlation c(xi, xj) between observed BOLD signals xi and xj, where big value of Pearson’s correlation indicates strong functional connectivity. Thresholding on Pearson’s correlation values is commonly used to remove the spurious connection. However, it is not easy to find a good threshold that works for all subjects.
Since fMRI is just an indirect reflection of brain activity, it is difficult to accurately quantify the FC strength only based on signal correlation. To alleviate this issue, we propose to optimize the reasonable functional connectivities, which should (1) be in consensus with the Pearson’s correlation of low level signals between xi and xj; (2) use the high level information such as module-to-module connection [12] to guide the measurement of low level region-to-region connectivity strength; and (3) represent sparsity since the brain network is intrinsically efficient to have sparse connections [13]. For convenience, we use si ∈ ℝN×1 denote i-th column in connectivity matrix S, which characters the connections of region Oi with respect to other brain regions. Also, we arrange all Pearson’s correlation values into a N × N matrix C = {cij |i, j = 1, ···, N}. Instead of calculating the connectivity sij just based on Pearson’s correlation c(xi, xj) between observed BOLD signals xi and xj, we optimize the connectivity matrix S by integrating the above three criteria:
| (1) |
where α and γ are the scalars which balance the strength of the low rank constraint [14] on S (the second term) and the l1 sparsity constraint [15] on S (the third term).
Discussion
Specifically, the first term requires the optimized functional connectivity matrix S should be close to the observed Pearson’s correlation matrix C. Sparse representation technique has been used in [16] to establish the functional connection of one brain region to all other regions. As the toy examples shown in Fig. 2(a), there might be a lot of spurious connections (gray blocks) in the FC matrix calculated based on Pearson’s correlation. After applying signal sparse representation at each brain regions independently, only a small number of connections remain such that the sparse constraint can make the estimation of FC more robust, as shown in Fig. 2(b).
Fig. 2.
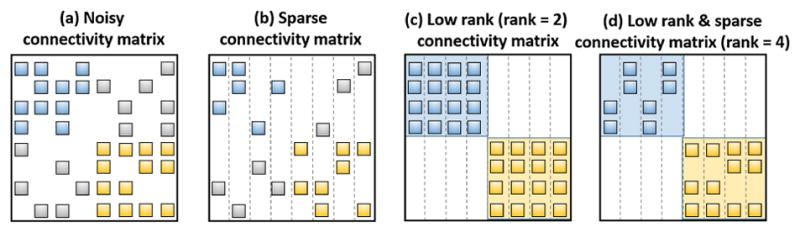
The intuition behind low rank and sparsity constraint in optimizing functional connectivity.
It is clear that the l1 norm is effective to remove redundant connections in functional connectivity [17]. However, one limitation of l1 norm is that the modular relationship are not jointly considered [12]. Figure 2(b) shows the connection patterns obtained using l1 norm, which shows no modular structure. Since brain network exhibit modular organization, we further introduce low rank constraint ||S||* to achieve strong modular organization. As shown in Fig. 2(c), the connection patterns are highly consistent within the same module but the intra modular regions are over connected. To achieve a reasonable sparse and modularized brain connections, we combine low rank and sparsity constraint on S to achieve connection patterns as shown in Fig. 2.
2.2 Characterize Dynamic Functional Connectivity by 3D Tensor Analysis
Here, we extend the learning-based FC optimization method to the temporal domain, in order to capture dynamics of functional connectivity. First, we follow the sliding window technique to obtain T overlapped multiple scale sliding windows which cover the whole time course for one subject. Let St denote for the FC matrix in sliding window t. Then we stack all St along time and form a tensor 𝒮 = {St|t = 1, ···, T} ∈ ℝN×N×T which represents the complete information of dynamic connectivity for each subject. Similarly, we can also construct another tensor 𝒞 = {Ct|t = 1, ···, T} ∈ ℝN×N×T, where each is the N × N matrices in t – th sliding window. Next, we propose a learning-based optimization method to characterize dFC using tensor analysis by:
| (2) |
Compared to the objective function in Eq. 1, the objective function here also encourages low rank on the brain connectivity patterns. Since brain in resting state generally transverses a small number of discrete stages during a short period of time [8], it is reasonable to apply low rank constraint on 𝒮 (by minimizing nuclear norm ||𝒮||*) to penalize too rapid FC change in the temporal domain and also find the optimal connectivity patterns in each sliding window.
2.3 Conventional Linear PCA Statistical Model
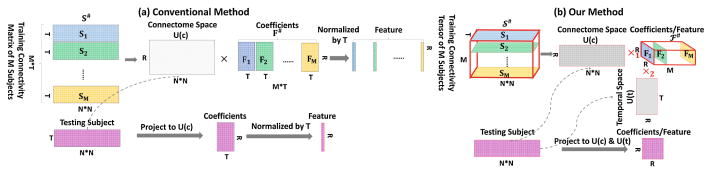
The learned 3D tensor 𝒮m for subject m is N ×N ×T which can not be used as the dynamic FC feature representation due to the high dimension. Conventional work proposed to reshape 𝒮m to a matrix Sm ∈ ℝN2×T, where each column stands for a connection pattern, then M subjects are stacked together to formulate a matrix S# ∈ ℝN2×MT as shown in Fig. 3(a). A low dimensional principle connectome space U(c) ∈ ℝN2×R can be learned by decomposing S# ∈ ℝN2×MT into a orthonormal space U(c) and coefficients matrix using linear Principle Component Analysis (PCA),
Fig. 3.
The comparison of conventional linear principle component analysis brain connectome model and our proposed tensor analysis connectome model.
| (3) |
where U(c) is the brain connectome space, F# = [F1, ···, FM] ∈ ℝN2×MT is the projected coefficients for S# as shown in Fig. 3. The high dimensional dynamic FC Sm ∈ ℝN2×T for subject m can be represented as a coefficients matrix . Since the sliding window number is large, this coefficients matrix is further normalized by the sliding window number T as the compact feature representation for dynamic FC patterns.
Conventional Method Issues
Conventional PCA based brain connectome space based method suffers from several issues: (1) How to set up optimal connection matrix threshold. (2) Temporal dynamics space is not modelled. Using the matrix techniques, the conventional method can only model the connectome space on the population, but the temporal dynamics space are not modelled (shown in Fig. 3(a)).
2.4 Statistical Model of Dynamic Functional Connectivity by Tensor Analysis on Population
Motivated by those above issues in conventional method, we extend the linear PCA model to a non-linear tensor analysis model in order to learn the optimal connectivity matrices for each subject at different sliding windows from the data without setting thresholds and a better brain connectome representation space and temporal dynamic space based on population jointly. Suppose we have M training subjects in total. As demonstrated in the right of Fig. 1, we stack the individual-based 3D tensors to a population-based 4D tensor (with the fourth dimension stands for M training subjects). Specifically, let the subscript m denote for the subject index. We construct a 4D tensor 𝕊 = {𝒮m|m = 1, ···, M} ∈ ℝN×N×T×M to include the dFC of all M training subjects. It is worth noting that different subject may have different number of sliding windows, here, T = max(Tm), m = 1, ···, M. We also construct another 4D tensor of Pearson’s correlation values ℂ = {𝒞m|m = 1, ···, M, t = 1, ···, T} ∈ ℝN×N×T×M. Then, we learn the dynamic FC pattern using tensor analysis:
| (4) |
In order to learn the principle connectome space as conventional methods, we reshape the 4D tensor 𝕊 to a 3D tensor 𝒮# ∈ ℝN2×T×M, where 1st dimension stands for the vectorized connectivity matrix, 2nd dimension stands for the sliding windows and 3rd model stands for M subjects as shown in Fig. 3(b). We learn a low rank and sparse 4D tensor 𝒮 and simultaneously decompose the reshaped tensor 𝒮# into a brain connetome space U(c) ∈ ℝR×N2, a temporal dynamic space U(t) ∈ ℝR×T and a coefficients tensor ℱ# ∈ ℝR×R×M using the tensor high-order singular value decomposition as,
| (5) |
where ×1,×2 represent the tensor mode multiplication. When the brain connectome space U(c) and U(t) are fixed, we can use the coefficients tensor ℱ# as the low dimension feature representation for all training subjects. Equation 5 can be solved using Alternative Lagrange Multipliers method iteratively [14,15].
The Novelties of Our Method
Compared with the conventional linear PCA connectome space method, our model has several new contributions: (1) Our method is able to learn the optimized sparse and low rank dynamic FC patterns based on data without setting optimal thresholds for brain connectome matrices. (2) Tensor analysis enables our method to learn a low dimensional connectome space U(c) along with a low dimensional temporal dynamic space U(t) which models FC temporal changes. Therefore, our method is more suitable for modeling dynamic FC patterns as shown in Fig. 3(b).
Encode Dynamic FC Feature and Apply Classifier for New Subjects
In the testing stage, we assume that each testing subject is independent to others. Thus, we obtain the compact connectome representations for each testing subject separately. Specifically, We first construct the 3D tensor of Pearson’s correlation 𝒞* ∈ ℝN×N×T. The goal here has two folds: (i) estimate the dynamic functional connectivity 𝒮* (a 3D tensor with the same dimension as 𝒞*) for the underlying subject; and (ii) obtain the compact connectome feature representation F* ∈ ℝR×R by projecting to the subspace spanned by learned connectivity space U(c), U(t) on the training dataset. 𝒮* can be learn using Eq. 2. After 𝒮* is learned, we reshape it to a matrix S*# ∈ ℝN2×T and compute the compact feature representations by projecting S onto the learned connectome space U(c) and temporal dynamic space U(t) from training data,
| (6) |
where F* ∈ ℝR×R is the projected low dimensional feature representation on the trained connectome space and temporal dynamic space. We apply our trained classifier to the learned dynamic FC feature representation to determine the class label for each testing subject.
3 Experiment Results
In this section, we evaluate our proposed tensor connectome model of dynamic functional connectivity by comparing the discriminative power in identifying ASD subjects with conventional state-of-the-art methods.
Experiment Setup
We randomly partition all subjects into 10 non-overlapping approximately equal size sets. Then, we use one fold for testing and the remaining folds are used for training. The training subjects are further divided into 5 subsets for another 5-fold inner cross validation, where 4 folds are used as training subset and the last fold is used as the validation subset. We found that our method is able to achieve stable performance when α = 1, γ = 1. The tensor connectome space U(c) and temporal dynamic space U(t) are learned from the subjects in the training subset. The low dimension connectome features representations of those training subjects, derived from the learned 4D tensor model, are further used to train the classic SVM (Support Vector Machine) for classification. Here, in order to show the power of our learned dynamic brain connectivity features, we only use a standard linear kernel SVM with l2 norm penalty. The optimal parameters are determined based on validation subset. It is worth noting that any classification model can be applied to our learned dynamic FC feature representations. For each testing subject, we use the approach summarized in Sect. 2.4 to estimate the dynamic FC feature representations, which is considered as the connectome signature to identify the clinical label of the testing subject. For the competing methods, we apply our multiple scale sliding window strategy to calculate the dynamic functional connectivity feature representation (based on Pearson’s correlation and optimal thresholding on validation dataset) for each subject. We first manually set up the sliding window size which ranges from 20 to 100 of the entire time course. It is worthy noting to mention that the brain connection patterns are unstable is the window size is smaller than 20. In optimizing the dynamic FC pattern, we set the shift of sliding window to 1 TR, in order to fully capture the dynamics of FC. In order to reduce feature dimension, we follow the work in to use classic PCA (Principle Component Analysis) model to encode the low dimensional connectome feature representation [11] for comparison.
Data Preprocessing
The subjects in our experiments were scanned for six and ten minutes during resting state, respectively, producing 180 time points and 300 time points at a repetition time (TR) of 2 s. We have corrected the head motion of the data first. It is worthy noting that this work focus on dynamic functional connectivity study and we assume that the dynamics caused by head motion are removed in motion correction. We further processed all these data using Data Processing Assistant for 0 the AAL template with 116 ROIs to the subject image domain and compute the mean BOLD signal in each ROI, where conventional method calculate the 116×116 connectivity matrix S based on the Pearson’s correlation of mean BOLD signals between any pair of two distinct brain regions.
Evaluation Measurements
We use several quantitative measurements including Accuracy (ACC), Specificity (SPEC) and Sensitivity (SEN) to evaluate the classification performance using our learned tensor dynamic FC feature representation. In the following experiments, PCA represents Pearson’s correlation based dynamic FC and feature coded using PCA. OURS represents the learned dynamic FC patterns by our 4D tensor method.
Subject Information
We conducted various experiments on resting-state fMRI images using two Autism data sets in order to demonstrate the generality of our method. We use the Autism Brain Imaging Data Exchange (ABIDE) database including both the data from NewYork University (NYU) and University of Minnesota (UM) site. Specifically, 45 NC and 45 ASD subjects are selected from the NYU site. 74 NC and 57 ASD subjects are selected from UM site.
Evaluation of Learned Dynamic FC Patterns in NC/ASD Classification Using the Same Dataset
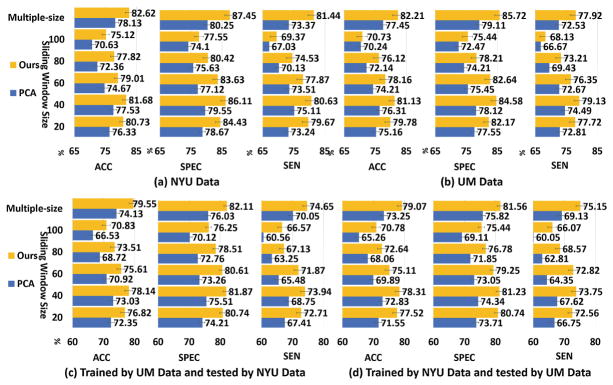
We first evaluate the performance of our model using the same dataset for training and testing. 10-fold cross-validation strategy is employed here on UM and NYU dataset. The NC/ASD classification results using different sliding window setup on UM and NYU dataset are shown in Fig. 4(a) and (b) respectively. It is shown that, first, the optimal sliding window size is 40 for two datasets; secondly, multiple scale sliding window setup achieves the best performance for all methods on two datasets, which improves >1% in terms of Accuracy compared to best performance achieved by fixed sliding window size; our learned 4D tensor feature representation improves the performance at least 4.5% in terms of Accuracy compared with the conventional Pearson’s correlation & PCA method using multiple scale sliding windows.
Fig. 4.
Evaluation of learned dynamic FC patterns. (a) and (b) Use the same training and testing dataset. (c) and (d) Use different training and testing dataset.
Evaluation of Learned Dynamic FC Patterns in NC/ASD Classification Using Different Dataset
To evaluate the generalization of the learned dynamic FC pattern representations, we select the training data and testing data from different sites. Two experiments are conducted here: first, we split the NYU and UM data into ten non-overlapped folds and use 9 folds from NYU as the training data and one fold from UM as the testing data; then, we swicth the training and testing dataset. Figure 4(c) and (d) shows the performance of conventional Pearson’s correlation patterns coded by PCA and our learned dynamic FC patterns with respect to different sliding window sizes. The performance is sensitive to different window size setting up. All competing methods achieve the best performance when the sliding window is 40 and multiple scale sliding window setting up improves the performance about >1% compared to sliding window size 40 for all competing methods. Compared with the conventional Pearson’s correlation patterns coded by PCA [11], our learned dynamic FC pattern improves the ASD classification performance >4% on Accuracy using multiple scale sliding windows.
Validation of Learned Dynamic FC Patterns
To evaluate the learned dynamic FC patterns, we checked the learned dynamic connection patterns for vision function regions, such as Lingual gyrus, Cuneus, Parahippocampal gyrus, and for motion function such as Putamen, Globus Pallidus using the NYU ASD dataset. We found that the connection patterns of those regions are inconsistent by the conventional Pearson’s correlation and thresholding method. However, the connection pattern learned by our method is changing smoother along time for those regions, which is consistent with current neuroscience findings.
Figure 5(a) shows the visualization of the positive connected ROIs to the Putamen along time computed by conventional Pearson’s correlation method and our method. Putamen is known to be related to motion function, since the fMRI time series are all collected during the resting and there is no motion task involved, the connection to putamen should be consistent and smooth along time. We found that the connection pattern by our method is stable along time. However, the connection pattern computed by the conventional Pearson’s correlation method is changing randomly along time. We also show the connection pattern variations along time for the Cuneus in Fig. 5(b). Cuneus is known to be related to vision function. The connection patterns to Cuneus are supposed to be consistent along time since no vision tasks are involved in the resting state. Compared to the varying connection patterns along time calculated by conventional Pearson’s correlation method, the connection patterns generated by our method resemble much better at different times.
Fig. 5.
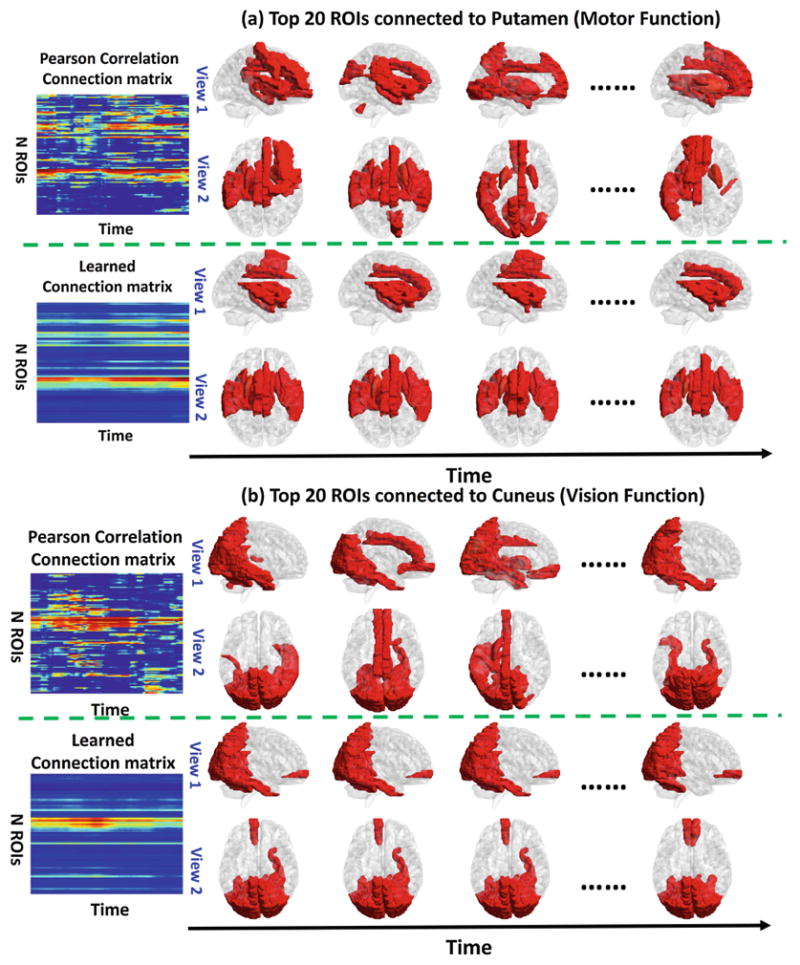
Visualization of the top 20 connection along time of Putamen (motion function related) and Cuneus (relation to vision function) by the Pearson’s correlation and our method.
4 Conclusion
In this work, we propose a novel learning-based method to discover both static and population based dynamic FC patterns from resting-state fMRI data using multiple linear tensor analysis. We evaluated the learned dynamic FC patterns by apply it as biomarkers for Autism identification and achieved promising results. Future work will explore the application of dynamic FC patterns on diagnosis of more neurological disorders such as, Alzheimer’s diseases and Obesity.
References
- 1.Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412:150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- 2.Mousss MN, Steen MR, Laurienti PJ, Hayasaka S. Consistency of network modules in resting-state fMRI connectome data. PLoS One. 2012;7:44428. doi: 10.1371/journal.pone.0044428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilello M. Correlating cognitive decline with white matter lesion and brain atrophy magnetic resonance imaging measurements in Alzheimer’s disease. J Alzheimers Dis. 2015;48(4):987–994. doi: 10.3233/JAD-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswal BB. Resting state fMRI: a personal history. NeuroImage. 2012;62:938–944. doi: 10.1016/j.neuroimage.2012.01.090. [DOI] [PubMed] [Google Scholar]
- 5.Heuvel M, Pol H. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20:519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Miller DB, O’Callaghan J. Biomarkers of Parkinson’s disease. Metabolism. 2015;64:40–46. doi: 10.1016/j.metabol.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, Zhu X, Zhang H, Gao W, Shen D, Wu G. Reveal consistent spatial-temporal patterns from dynamic functional connectivity for autism spectrum disorder identification. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. MICCAI 2016. LNCS. Vol. 9900. Springer; Cham: 2016. pp. 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wee C, Yap P, Shen D. Diagnosis of autism spectrum disorders using temporally distinct resting-state functional connectivity networks. CNS Neurosci Ther. 2016;22:212–219. doi: 10.1111/cns.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonardi N. Principal components of functional connectivity: a new approach to study dynamic brain connectivity during rest. NeuroImage. 2013;83:937–950. doi: 10.1016/j.neuroimage.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31:75–86. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Huang D, De La Torre F, Lucey S. Complex non-rigid motion 3D reconstruction by union of subspaces. CVPR. 2014:410. [Google Scholar]
- 15.Zhu Y, Lucey S. 3D motion reconstruction for real-world camera motion. CVPR. 2011 [Google Scholar]
- 16.Wee C, Yap P, Zhang D, Wang L, Shen D. Group-constrained sparse fMRI connectivity modeling for mild cognitive impairment identification. Brain Struct Funct. 2014;219:641–656. doi: 10.1007/s00429-013-0524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Lucey S. Convolutional sparse coding for trajectory reconstruction. IEEE Trans Pattern Anal Mach Intell. 2015;37:529–540. doi: 10.1109/TPAMI.2013.2295311. [DOI] [PubMed] [Google Scholar]