Arrhythmogenic cardiomyopathy (AC) is a genetic disorder characterized by high risk of life-threatening ventricular arrhythmias, sudden cardiac death (SCD) and progressive heart failure. Currently, there is evidence that AC includes a spectrum of cardiomyopathy phenotypes, ranging from the classical form of arrhythmogenic right ventricular cardiomyopathy (ARVC) to more recently identified forms of arrhythmogenic left ventricular cardiomyopathy.1, 2
ARVC is considered a ‘disease of the desmosome’, since in the majority of cases, it is caused by mutations in genes encoding proteins of the cardiac desmosomes.3 However, mutations in non-desmosomal genes have been also found in ARVC, such as the genes encoding the cardiac ryanodine receptor 2 (RYR2), the transforming growth factor β-3 (TGFB3), the nuclear transmembrane protein 43 (TMEM43) and desmin (DES). Mutations in LMNA, encoding the nuclear envelope protein lamin A/C, known to cause dilated cardiomyopathy (DCM) with an arrhythmic phenotype, was reported to cause also an ARVC phenotype,3 even though the association between LMNA and ARVC is still questioned.
In the left dominant form, which clinically presents as an “arrhythmogenic” DCM, the same desmosomal genes causing ARVC can be found,1 as well as LMNA and the more recently identified filamin C gene (FLNC), encoding an actin cross-linker, Z-line and integrin-binding protein.4–6 These findings clearly show that the AC phenotype may involve both the right and left ventricle with variable expressivity, with mechanisms that are still not well understood and that extend beyond the desmosome to include several components of the cellular mechanotransduction machinery.
In the current issue, Bermúdez-Jimnez et al. describe a Spanish family in which approximately 30 affected family members with AC phenotype were found to carry an rare missense variant of DES gene (c.1203G>C; p.Glu401Asp).7 DES mutations have previously been reported in myopathies, conduction disease and cardiomyopathies, in particular in cases of DCM8 and ARVC.9 While truncation variants have a deleterious effect and therefore, in general, are considered to be pathogenic, missense variants are much more difficult to classify. In DES, missense mutations are frequent in controls:10 however, this study provides convincing evidence that the unique DES Glu401Asp variant causes the disease in this family, with 100% penetrance and variable expressivity. The mutation carriers presented an arrhythmogenic phenotype with high risk of sudden cardiac death and progressive heart failure. It should be noted that in this case of AC caused by DES mutation, as in the cases of other non-desmosomal AC genes, such as FLNC and LMNA, the phenotype is not typical of ARVC: it is unclear if any of the subjects fulfilled the ARVC 2010 Task Force Criteria, although four out of 31 had right ventricular involvement and two of them epsilon waves. In the cardiac tissue, the investigators found fibro-fatty infiltration predominantly in the left ventricle. Cardiomyocytes showed reduced cellular adhesion, reminiscent of the defect found in ARVC, and reduced expression of DES and cell-cell junction proteins.
Indeed, one of the aspects that deserves more investigations in AC is the role of altered mechanotransduction. An essential necessity for cells is the capacity to adapt to their mechanical environment (mechanosensing and mechanotrasduction). To accomplish this task, cardiomyocytes can use members of the G-protein family, integrins or strain-activated ion channels.11 Moreover, the cytoskeleton supplies a network through which the mechanical stresses can be transmitted by actin, microtubules and intermediate filaments (IFs) to the nucleus, inducing protein conformational changes and nuclear deformation that can alter biological activity.
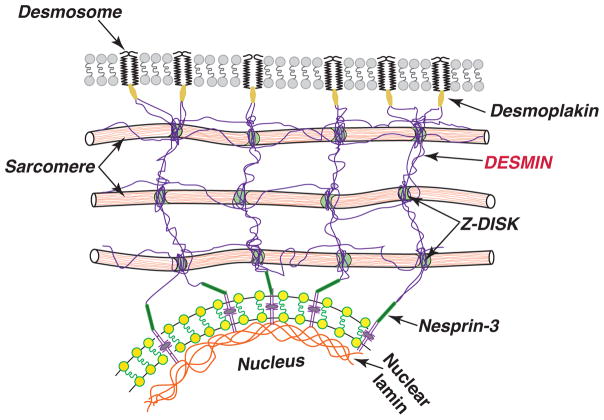
Among the cytoskeleton filaments, IFs are the most flexible and they are also highly stretchable. In cardiomyocytes, the main IFs is DES, which links myofibrils radially along Z-discs to the sarcomeres, longitudinally to the intercalated discs, and attaches directly to the nuclear inner surface interacting with LMNA.12 DES filaments are strategically located to maintain sarcomere alignment and homogeneity, to act as a lateral mechanical link between Z-disks and to transmit force radially. These IFs are crucial for force transmission from the cell membrane to the nuclei and for its mechanical stability. Nuclei in “healthy” cells when stretched increased their aspect ratio in parallel with increased sarcomere length, while nuclei in DES-null cells deform less13. This lack of hard-wiring of the nucleus to the cytoskeleton results in a reduced degree of phosphorylation of the Jun N-terminal kinase (JNK): in fact, DES-null cell showed a near absence of stretch-induced signaling leading to JNK phosphorylation13. Furthermore, DES is connected to the desmosome proteins via DSP (Figure).14 In this way, DES filaments provide a link to desmosomes critical in cardiac mechanotransduction and long-range force transmission across cells. Therefore, it is not surprising that a mutation that affects the DES filaments network will also affect the distribution of DSP at the junctional sites, eventually causing an AC phenotype. Taken together, these data show an essential role for DES being involved in both mechanical stress transmission and stress transduction in cardiac muscle cells.
Figure 1.
Desmosomal proteins, desmin and nuclear lamina provide a continuum structure for mechanotransduction and mechanosensing in cardiomyocytes, which explains their common role in AC.
In conclusion, there are three important insights from the study of Bermúdez-Jimnez et al. that have intriguing scientific and clinical implications: (1) Failure of the proper connection between the cell-cell junction with the intracellular structures (desmosome, nucleus) alters the cardiomyocyte mechanotransduction and mechanosensing, and eventually leads to an arrhythmogenic phenotype. (2) The AC phenotype is a spectrum, even in the same family, and can involve either the right or the left ventricle or both. (3) In this disease, that overlaps with the DCM phenotype, there is a need of appropriate risk stratification as done in ARVC, which takes into account the risk of life-threatening arrhythmias (regardless of the left ventricular dysfunction),2 and should include genetic testing, ECG monitoring, and cardiac magnetic resonance with late gadolinium enhancement.
Acknowledgments
This work is supported in part by the AHA17GRNT33670495 to LM and a Trans-Atlantic Network of Excellence grant from the Fondation Leducq (14-CVD 03) to LM and OS.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Sen-Chowdhry S, Syrris P, Prasad SK, Hughes SE, Merrifield R, Ward D, Pennell DJ, McKenna WJ. Left-dominant arrhythmogenic cardiomyopathy: an under-recognized clinical entity. J Am Coll Cardiol. 2008;52:2175–2187. doi: 10.1016/j.jacc.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Spezzacatene A, Sinagra G, Merlo M, Barbati G, Graw SL, Brun F, Slavov D, Di Lenarda A, Salcedo EE, Towbin JA, Saffitz JE, Marcus FI, Zareba W, Taylor MR, Mestroni L, Familial Cardiomyopathy R. Arrhythmogenic Phenotype in Dilated Cardiomyopathy: Natural History and Predictors of Life-Threatening Arrhythmias. J Am Heart Assoc. 2015;4:e002149. doi: 10.1161/JAHA.115.002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corrado D, Basso C, Judge DP. Arrhythmogenic Cardiomyopathy. Circ Res. 2017;121:784–802. doi: 10.1161/CIRCRESAHA.117.309345. [DOI] [PubMed] [Google Scholar]
- 4.Begay RL, Tharp CA, Martin A, Graw SL, Sinagra G, Miani D, Sweet ME, Slavov DB, Stafford N, Zeller MJ, Alnefaie R, Rowland TJ, Brun F, Jones KL, Gowan K, Mestroni L, Garrity DM, Taylor MR. FLNC Gene Splice Mutations Cause Dilated Cardiomyopathy. JACC Basic Transl Sci. 2016;1:344–359. doi: 10.1016/j.jacbts.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begay RL, Graw SL, Sinagra G, Asimaki A, Rowland TJ, Slavov DB, Gowan K, Jones KL, Brun F, Merlo M, Miani D, Sweet ME, Deveraj K, Wartchow EP, Gigli M, Puggia I, Salcedo EE, Garrity DM, Ambardekar AV, Buttrick P, Rece TB, Bristow MR, Saffitz JE, Mestroni L, Taylor MR. Filamin C Truncation Mutations Are Associated With Arrhythmogenic Dilated Cardiomyopathy and Changes in the Cell–Cell Adhesion Structures. JACC Clin Electrophysiol. 2017 doi: 10.1016/j.jacep.2017.12.003. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortiz-Genga MF, Cuenca S, Dal Ferro M, Zorio E, Salgado-Aranda R, Climent V, Padron-Barthe L, Duro-Aguado I, Jimenez-Jaimez J, Hidalgo-Olivares VM, Garcia-Campo E, Lanzillo C, Suarez-Mier MP, Yonath H, Marcos-Alonso S, Ochoa JP, Santome JL, Garcia-Giustiniani D, Rodriguez-Garrido JL, Dominguez F, Merlo M, Palomino J, Pena ML, Trujillo JP, Martin-Vila A, Stolfo D, Molina P, Lara-Pezzi E, Calvo-Iglesias FE, Nof E, Calo L, Barriales-Villa R, Gimeno-Blanes JR, Arad M, Garcia-Pavia P, Monserrat L. Truncating FLNC Mutations Are Associated With High-Risk Dilated and Arrhythmogenic Cardiomyopathies. J Am Coll Cardiol. 2016;68:2440–2451. doi: 10.1016/j.jacc.2016.09.927. [DOI] [PubMed] [Google Scholar]
- 7.Bermúdez-Jiménez FJ, Carriel V, Brodehl A, Alaminos M, Campos A, Schirmer I, Milting H, BÁ, Álvarez M, López-Fernández S, García-Giustiniani D, Monserrat L, Tercedor L, Jiménez-Jáimez L. The Novel Desmin Mutation p.Glu401Asp Impairs Filament Formation, Disrupts Cell Membrane Integrity and Causes Severe Arrhythmogenic Left Ventricular Cardiomyopathy/Dysplasia. Circulation. 2018 doi: 10.1161/CIRCULATIONAHA.117.028719. XXX:XXX-XXX. [DOI] [PubMed] [Google Scholar]
- 8.McNally EM, Mestroni L. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Tintelen JP, Van Gelder IC, Asimaki A, Suurmeijer AJ, Wiesfeld AC, Jongbloed JD, van den Wijngaard A, Kuks JB, van Spaendonck-Zwarts KY, Notermans N, Boven L, van den Heuvel F, Veenstra-Knol HE, Saffitz JE, Hofstra RM, van den Berg MP. Severe cardiac phenotype with right ventricular predominance in a large cohort of patients with a single missense mutation in the DES gene. Heart Rhythm. 2009;6:1574–1583. doi: 10.1016/j.hrthm.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 10.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, Exome Aggregation C, MacArthur DG, Farrall M, Cook SA, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmisano MG, Bremner SN, Hornberger TA, Meyer GA, Domenighetti AA, Shah SB, Kiss B, Kellermayer M, Ryan AF, Lieber RL. Skeletal muscle intermediate filaments form a stress-transmitting and stress-signaling network. J Cell Sci. 2015;128:219–224. doi: 10.1242/jcs.142463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldfarb LG, Olivé M, Vicart P, Goebel HH. Intermediate filament diseases: desminopathy. Adv Exp Med Biol. 2008;642:131–164. doi: 10.1007/978-0-387-84847-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otten E, Asimaki A, Maass A, van Langen IM, van der Wal A, de Jonge N, van den Berg MP, Saffitz JE, Wilde AA, Jongbloed JD, van Tintelen JP. Desmin mutations as a cause of right ventricular heart failure affect the intercalated disks. Heart Rhythm. 2010;7:1058–1064. doi: 10.1016/j.hrthm.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Moncayo-Arlandi J, Brugada R. Unmasking the molecular link between arrhythmogenic cardiomyopathy and Brugada syndrome. Nat Rev Cardiol. 2017;14:744–756. doi: 10.1038/nrcardio.2017.103. [DOI] [PubMed] [Google Scholar]