Abstract
Allene oxide synthase (AOS; hydroperoxide dehydratase; EC 4.2.1.92) catalyzes the first step in the biosynthesis of jasmonic acid from lipoxygenase-derived hydroperoxides of free fatty acids. Using the AOS cDNA from tomato (Lycopersicon esculentum), in which the role of jasmonic acid in wound-induced defense gene activation has been best described, we examined the kinetics of AOS induction in response to wounding and elicitors, in parallel with that of the wound-inducible PIN II (proteinase inhibitor II) gene. AOS was induced in leaves by wounding, systemin, 12-oxophytodienoic acid, and methyl jasmonate. The levels of AOS mRNA started declining by 4 h after induction, whereas the levels of PIN II mRNA continued to increase up to 20 h after induction. Salicylic acid inhibited AOS and PIN II expression, and the addition of 12-oxophytodienoic acid or methyl jasmonate did not prevent the inhibition of PIN II expression in the presence of salicylic acid. Ethylene induced the expression of AOS, but the presence of ethylene alone did not produce an optimal induction of PIN II. The addition of silver thiosulfate, an ethylene action inhibitor, prevented the wound-induced expression of both AOS and PIN II. Products of hydroperoxide lyase affected neither AOS nor PIN II, but induced expression of prosystemin. Based on these results, we propose an updated model for defense gene activation in tomato.
Jasmonic acid (JA), its methyl ester, certain amino acid conjugates, Glc esters, and hydroxylated forms, which are collectively termed jasmonates, occur ubiquitously in all plant species and constitute a major signal in stress-induced gene expression. Stress in the form of mechanical wounding, herbivore damage, desiccation, or pathogen attack triggers the elevation of endogenous JA, which in turn induces the expression of specific jasmonate-responsive genes to combat the stress (for review, see Wasternak and Parthier, 1997). The role of jasmonates in stress-related signaling has been best characterized with respect to the wound-induced expression of proteinase inhibitor genes, which protect the plant against digestive Ser proteinases of herbivorous insects (Farmer and Ryan, 1992; Koiwa et al., 1997). Octadecanoid-deficient mutants of Arabidopsis and tomato (Lycopersicon esculentum) have been reported to be more susceptible to herbivore damage than wild-type plants (Howe et al., 1996; McConn et al., 1997).
Allene oxide synthase (AOS) is the first enzyme in the branch pathway leading to the biosynthesis of JA, and catalyzes the production of unstable allene epoxides that cyclize to form cyclopentenone acids, the precursors for JA (Mueller, 1997). Lipoxygenase-derived hydroperoxides of linolenic and linoleic acids are utilized as substrates in this reaction. With 13S-hydroperoxy-9(Z),11(E),15(Z)-octa-decatrienoic acid (HPOT) derived from α-linolenic acid as a substrate, the jasmonate precursor 12-oxophytodienoic acid (PDA) is synthesized in the reaction initiated by AOS. In addition to octadecanoids, hexadecanoids can also lead to the synthesis of jasmonates (Farmer et al., 1998). The AOS gene has been cloned from flaxseed, guayule rubber particles, and Arabidopsis (Song et al., 1993; Pan et al., 1995; Laudert et al., 1996). Wounding, PDA, and JA induce the expression of both Arabidopsis and flax AOS (Harms et al., 1998; Laudert and Weiler, 1998). Arabidopsis AOS is also induced by ethylene, and this phytohormone is proposed to act together with jasmonates to regulate proteinase inhibitor genes in the wound response of tomato (O'Donnell et al., 1996; Laudert and Weiler, 1998). Salicylic acid (SA), an elicitor of pathogenesis-related gene expression, induces Arabidopsis AOS but represses flax AOS, indicating the existence of possible anomalies among plant species with regard to jasmonate-mediated signal transduction (Harms et al., 1998; Laudert and Weiler, 1998). SA has previously been suggested to be a negative regulator of proteinase inhibitor genes, thus providing a “cross-talk” between the signaling systems involved in pathogen defense and predator defense (Peña-Cortés et al., 1993; Doares et al., 1995).
We have cloned the AOS from tomato and characterized its response to mechanical damage and elicitors, in parallel with that of the wound-induced PIN II (proteinase inhibitor II) gene from tomato. The expression of PIN II has been extensively characterized as a terminal event in the wound- and jasmonate-induced signal transduction cascade in tomato. As such, cloning of tomato AOS provides an excellent opportunity for the concomitant analysis of the potential to synthesize the jasmonate messenger and the development of a response. Our results present additional evidence for better defining the roles of ethylene and SA in defense gene activation in tomato, and also implicate a role for products of hydroperoxide lyase (HPL) in this process.
MATERIALS AND METHODS
Reagents and Plant Material
Methyl jasmonate (MeJA; 97% purity) and PDA were obtained, respectively, from Firmenich (Geneva), and Cayman Chemical (Ann Arbor, MI). Systemin was synthesized by Bio-Synthesis (Lewisville, TX). Traumatin and cis-3-hexenal were produced by reacting a HPL/glutathione S-transferase (GST) fusion protein, purified from Escherichia coli, with HPOT (Bate et al., 1998). HPOT was synthesized by mixing soybean lipoxygenase with linolenic acid, as described by Surrey (1964). Trans-2-hexenal, traumatic acid, and all other chemicals were purchased from Sigma-Aldrich (St. Louis).
Tomato (Lycopersicon esculentum var Bonnie Best) plants were grown on soil in growth chambers maintained at 23°C throughout a 16-h/8-h day/night regime under a light intensity of 225 μE m−2 s−1 at canopy level during the daytime. They were harvested 16 to 21 d following germination for the induction experiments, and after fruiting for the collection of plant parts.
Isolation of Tomato AOS
Initially, a partial cDNA clone of 950 bp was isolated by PCR from reverse-transcribed tomato leaf RNA using degenerate primers designed on the basis of published AOS sequences from flaxseed, guayule rubber, and Arabidopsis. The primer used for reverse transcription was TCCGGT/CCCG/ATTA/CGACCAC, which was also the reverse primer in PCR. The forward primer was TTCACT/CGGA/TACTTACATGCC. The 3′ fragment was isolated by 3′ RACE using a dT17-adapter primer and a gene-specific rimer, TCGTCGCCGATCGGTTCAAAGGAG, as described by Innis et al. (1990). To isolate the 5′ fragment, a uni-directional adaptor was first ligated to the 5′ end of double-stranded AOS cDNA prepared from reverse-tran-scribed RNA by second-strand synthesis using RNase H, DNA polymerase, and DNA ligase. The primer used for reverse transcription was TAGAACTCGATAACCGCCTGTGAG. Later, the sense strand of the uni-directional adapter, GCGGTGACCCGGGAGATCTGAATTC, and the gene-specific primer, ACCGCCTGTGAGATCAGTGGA-TGG were used in touch-down PCR to amplify the 5′ fragment. The full-length AOS was then isolated from reverse-transcribed RNA using, respectively, the forward and reverse primers ATGGCATCAACTTCTCTTTCTCTTC-and CGGCTGGTCGACATGCTCTGTTC. The latter was also used in the reverse-transcription reaction.
Expression and Functional Analysis of an AOS Fusion Protein
The tomato AOS cDNA was amplified by PCR using the forward and reverse primers, AGGCTTCGGTGTCTGGGATCCCAC and CGGCTGGTCGACATGCTCTGTTCT, respectively. The amplified fragment was then restricted with BamHI and SalI, ligated in-frame with pGEX 5X-3 (Pharmacia Biotech, Piscataway, NJ), and transformed into E. coli. The AOS protein, which was expressed as a fusion with GST, was purified from E. coli using the protocol specified by the manufacturer. Protein quantity was measured by the method of Bradford (1976).
AOS activity of the AOS-GST fusion protein was measured by following the decrease in A234, which indicated the degradation of the substrate due to the loss of the conjugated diene (Pan et al., 1995). The substrates HPOT and HPOD were prepared as described by Surrey (1964). For functional assay, 1 mL of 50 mm KPO4 (pH 7.0) was modified to contain either 60 μm HPOT or 30 μm HPOD. The reaction was started by addition of the protein. Any possible HPL activity associated with the fusion protein was measured using the coupled enzyme assay described by Vick (1991).
Treatment and Harvest of Plants
Induction and inhibition experiments were performed on 16- to 21-d-old tomato plants. Plants were wounded by slicing all but the youngest leaves twice across the mid-vein with a razor blade. Systemin (5 pmol per plant), PDA (10 pmol per plant), and SA (0.5 μmol per plant), prepared in 15 mm potassium phosphate buffer (pH 6.5) or silver thiosulfate (STS), were fed through cut stems for a period of 1 to 2 h for induction purposes, and the plants were later transferred to water or the appropriate treatment. MeJA and trans-2-hexenal (10 μL of 0.1 m solutions in methanol) were applied on cotton buds placed inside 1-L sealed glass jars in which the plants were placed for exposure. Traumatic acid (1 mm), aminoethoxyvinyl Gly (AVG; 1 mm), and ethephon (7 mm) were sprayed to run-off on plants. All three were prepared in 15 mm KPO4 (pH 6.5). Traumatic acid was initially prepared as a 10 mm stock in methanol, and then diluted in the buffer. Plants were sprayed with traumatin or exposed to cis-3-hexenal, both of which were prepared immediately before use, by reacting a hydroperoxide-lyase/GST fusion protein with HPOT. The exact concentrations used are unknown.
For induction experiments, the leaves of plants were harvested at 0, 1, 4, and 20 h following induction. For all other experiments, the period of exposure to treatments and the time of harvest are given in the figure legends.
Isolation and Blotting of DNA and RNA
DNA and RNA were isolated from plant tissue as described by Sambrook et al. (1989) and Chang et al. (1993), respectively. The nucleic acids were blotted and probed as described by Sambrook et al. (1989). Hybridization and washes were performed at 65°C. An AOS partial fragment from 522 to 1040 bp, lacking the C-terminal cytochrome P450 (Cyt P450) domain, was used in northern blots for AOS. The probes for PIN II, prosystemin, and PR1b1 were amplified by PCR as partial fragments from reverse-transcribed tomato leaf RNA. For northern blots, 20 μg of total RNA was loaded per lane. The ethidium bromide stain of rRNA was used to affirm uniformity in loading for RNA blots.
RESULTS AND DISCUSSION
Isolation, Sequence Analysis, and Functional Assay of Tomato AOS
A partial cDNA for the tomato AOS was isolated using degenerate primers designed on the basis of published sequences of the Arabidopsis, flax, and guayule rubber AOS sequences. The 5′ and 3′ ends of the gene were later isolated by RACE, and the complete cDNA was sequenced in its entirety (Fig. 1a). The presence of a stop codon 42 bp upstream of an AUG codon suggests that this AUG is the start codon. A transit peptide for chloroplast-targeting proteins was identified at the N-terminal end of the AOS protein, based on established criteria (von Heijne et al., 1989). This peptide is 39 residues long, has a preponderance of Ser, an absence of Asp, Glu, and Tyr (except for Tyr-30), an Ala residue within −3 to −1 of the proposed cleavage site (Fig. 1a, marked by asterisk), and a β-sheet immediately prior to the proposed cleavage site. The mature AOS polypeptide from tomato is presumed to start at Ser-30, whereas that from Arabidopsis and flax starts at Leu-22 and Ser-46, respectively. Significant homology between the tomato AOS and the other AOS proteins begins at residue −70 of the former. The tomato AOS shares maximum sequence identity with the flax AOS, which is 61% at the nucleotide level and 62% identical at the protein level. The Arabidopsis and rubber AOS proteins are, respectively, 60% and 56% identical with the tomato AOS. The four characteristic Cyt P450 domains are present at the C terminus of the tomato protein, as they are in the other AOS proteins. The Cyt P450 domains of the tomato AOS are 77% identical to those of the flax AOS and 74% identical to those of the Arabidopsis and rubber AOS proteins (Fig. 1b).
Figure 1.
a, Nucleotide and deduced amino acid sequence of the tomato AOS cDNA. The large arrow indicates the transcription start site and the small arrow indicates the point at which homology with other published AOS sequences start. The asterisk indicates the proposed cleavage site of the mature leader sequence. The amino acids constituting the four Cyt P450 domains at the C terminus of the sequence are underlined, and the primers used for amplifying the coding region for the fusion protein are double-underlined. b, Alignment of amino acids constituting the Cyt P450 domains A through D in flax, rubber, Arabidopsis, and tomato AOS proteins.
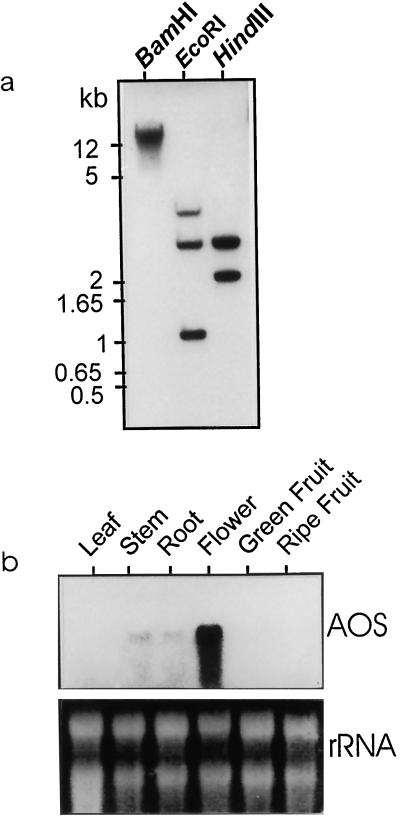
Southern analysis of tomato genomic DNA indicated the existence of a single gene encoding AOS (Fig. 2a). Restriction with BamHI revealed only a single band, while that with either HindIII or EcoRI yielded two or three bands in concurrence with the presence of internal restriction sites for these enzymes within the cDNA sequence. The expression of the gene was highest in flowers, with low amounts of transcript being present in stem and root (Fig. 2b). In leaves and fruits, AOS mRNA was not detectable under non-inducing conditions, such as in the absence of wounding or elicitors. Contrary to this, there was a basal level of constitutive expression of the flax AOS under non-inducing conditions in all plant organs (Harms et al., 1998). A similar constitutive level of the AOS protein has also been observed in Arabidopsis (Laudert and Weiler, 1998). This could be due to age differences between the experimental plants, as the tomato seedlings used in our experiments were 16 to 21 d old, while the flax and Arabidopsis plants were 4 to 6 weeks old. Older leaves have been shown to have higher levels of AOS mRNA than younger leaves (Harms et al., 1998).
Figure 2.
a, Southern blot of tomato genomic DNA restricted with NotI, EcoRI, and HindIII and probed with the tomato AOS cDNA. b, Expression pattern of AOS in different plant parts of tomato.
Functional assay of the tomato AOS protein fused to GST revealed a comparable reactivity toward HPOT and HPOD (Table I). The pure GST protein did not react with HPOT, but did exhibit a capacity to degrade HPOD at a rate that was 7-fold less than that observed with the AOS-GST fusion protein. There was no reactivity toward HPOT or HPOD in the coupled-enzyme assay for HPL, thus conclusively establishing the identity of the cDNA sequence described above as AOS.
Table I.
AOS activity of AOS-GST fusion protein or GST protein purified from E. coli extracts
| Reaction | AOS-GST | GST |
|---|---|---|
| AOS activity | ||
| HPOT | 8.5 | 0 |
| HPOD | 7.4 | 1.1 |
| HPL activity | ||
| HPOT | 0 | ND |
| HPOD | 0 | ND |
Purified protein was added to the assay buffer containing either HPOD or HPOT, and the reduction in A234 was recorded at 15-min intervals over a period of 4 min. AOS activity is expressed as Δ A234 min−1 mg−1 protein. HPL activity is expressed as Δ A340 min−1 mg−1 protein. Data represent the average of two separate experiments. ND, Not determined.
Induction of AOS and PIN II Genes by Wounding and Elicitors
The induction of the AOS transcript in tomato was examined in parallel with that of the PIN-II transcript in the presence of wounding or the elicitors systemin, PDA, and MeJA (Fig. 3). Systemin is a systemic signal processed as an 18-amino acid polypeptide from its precursor, prosystemin, the expression of which increases in response to wounding (Pearce et al., 1991; McGurl et al., 1992). Systemin is upstream of jasmonate in the current model for the signal cascade in wound-induced gene expression. PDA and MeJA are products of the AOS branch pathway. Seedling shoots were harvested 0, 1, 4, and 20 h after wounding or treatment with elicitors. All four of the above treatments were capable of inducing expression of the AOS gene in tomato. Although AOS mRNA was undetectable at 0 h, it accumulated rapidly by 1 h following induction, and either stabilized or started declining by 4 h. At 20 h following induction, it had significantly declined compared with the levels at 1 h. Wound-induced expression of flax and Arabidopsis AOS and accumulation of JA in wounded flax leaves reached a maximum by 6 h and started to decline thereafter, thus following similar kinetics as the tomato AOS (Harms et al., 1998; Laudert and Weiler, 1998). The induction of AOS expression by PDA and MeJA suggests a positive feedback, similar to the one reported for HPL, which initiates the second branch pathway from lipoxygenase (Bate et al., 1998). Regulation of such a feedback loop could occur at the point of release of fatty acid products from the chloroplast, where they are synthesized, or from the peroxisome, a speculative site for the final β-oxidation reactions in the synthesis of JA (Harms et al., 1995).
Figure 3.
Induction of tomato AOS by wounding, systemin (Sys), PDA, and MeJA. Tomato seedlings (16–21-d-old) were wounded, exposed to MeJA, or the cut stems were placed in PDA or systemin solutions. In the case of PDA and systemin, the shoots were transferred to water after the initial 2 h of immersion in the treatment solutions. Seedling shoots were harvested 0, 1, 4, or 20 h after application of treatments, and lanes 1 to 4 represent these time points, respectively.
Wounding and elicitors also induced expression of PIN II, a well-characterized defense gene in tomato, but the kinetics of induction were different from that of AOS. While the AOS mRNA started to decline by 4 h after induction and was reduced considerably by 20 h, PIN II mRNA continued to increase throughout the 20-h duration. Therefore, although the capacity to synthesize the jasmonate messenger is transient, it is sufficient to initiate systems required for the continued induction of defense genes until the time when stress is overcome.
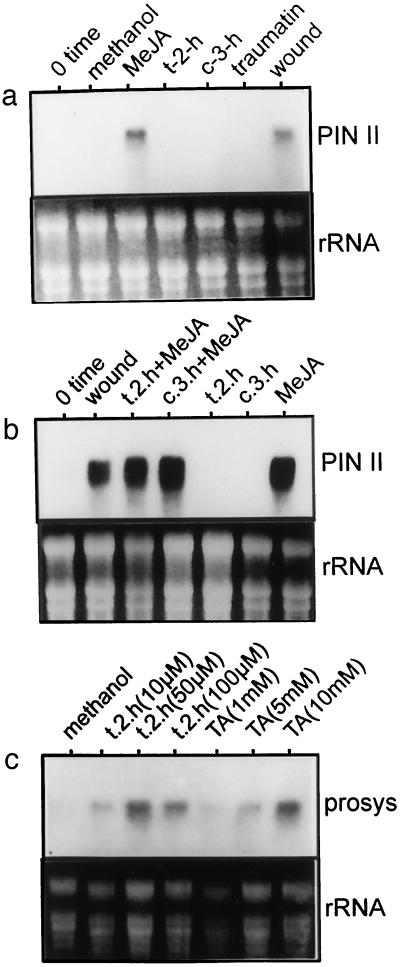
HPL, another enzyme of the lipoxygenase pathway, uses the same substrate as AOS to produce the volatiles trans-2-hexenal and cis-3-hexenal and the wound hormone traumatin. Treatment of tomato plants with products of HPL did not induce the expression of AOS (data not shown) or PIN II (Fig. 4a). When plants were exposed to trans-2-hexenal or cis-3-hexenal in the presence of MeJA, PIN II expression was induced to similar levels as when MeJA was present alone (Fig. 4b). Although products of HPL did not affect the expression of AOS or PIN II, they did induce expression of the prosystemin gene (Fig. 4c). Application of both trans-2-hexenal (10, 50, or 100 μm) and traumatic acid (1, 5, or 10 mm), the commercially available oxidized form of traumatin, produced an induction of prosystemin gene expression.
Figure 4.
Effect of products of HPL on PIN II (a and b) or prosystemin (c) gene expression. Tomato seedlings were exposed to trans-2 hexenal (t-2-h), cis-3-hexenal (c-3-h), traumatin, traumatic acid (TA), or wounded as described in “Materials and Methods.” t-2-h, c-3-h, and MeJA were applied to cotton swabs placed within sealed glass jars containing the potted plants. Traumatin and TA were sprayed on the plant as solutions prepared in potassium phosphate buffer (15 mm, pH 6.5) containing 0.01% (w/v) Triton X-100. Seedlings were harvested 4 h after starting exposure to treatments. Methanol was used as a solvent for MeJA and t-2-h was used as a control, as was the zero-time point.
The HPL gene itself has been shown to be induced by wounding, but not by MeJA (Bate et al., 1998). It is possible that wound-induced expression of HPL constitutes an early step in the signal transduction pathway upstream of systemin. Thus, the initial event in the signaling cascade triggered by wound or herbivore damage could well be the release of the “grassy” or “green-note odor,” a collective signature of volatile products arising from the HPL branch pathway. Parallel events triggered by wounding may, however, be required for the processing of prosystemin and the occurrence of downstream steps leading to defense gene activation.
Effect of SA on the Expression of AOS and PIN II
SA is a secondary signal implicated in pathogen defense, and it induces expression of several pathogenesis-related genes. The effect of SA on AOS and PIN II expression was analyzed in tomato seedlings. Pretreatment with SA for 2 h prior to wounding or treatment with systemin, PDA, or MeJA prevented the optimal induction of AOS that occurred when the inducing treatments were given alone (Fig. 5). The levels of AOS transcript were reduced by feeding SA through the cut stems of seedlings prior to 4 h of exposure to the various inducing conditions. SA also inhibited the induction of PIN II if given as a pretreatment before induction with wounding or elicitors. The application of PDA or MeJA, which are products of AOS, could not prevent the inhibition of PIN II expression produced by SA. Conversely, PR1b1, the pathogenesis-related gene in tomato, was induced by SA, thus affirming that imbibition of SA by the plant did not produce any adverse general effect on gene expression.
Figure 5.
Effect of SA on the expression of AOS, PIN II, and PR1b1 in tomato. Lane 1, Buffer control; lane 2, SA; lane 3, wounding; lane 4, wounding plus SA; lane 5, PDA; lane 6, PDA plus SA; lane 7, MeJA; lane 8, MeJA plus SA; lane 9, systemin; and lane 10, systemin plus SA. The concentrations used for the various treatments and the method of treatment application are given under “Materials and Methods.” Seedlings were pretreated with SA for a period of 2 h prior to wounding or application of elicitors. Harvesting was done 4 h after treatment application.
The octadecanoid signaling pathway in plants closely resembles that in animals, and is known to be inhibited by non-steroidal anti-inflammatory drugs such as SA and aspirin, which in animals inhibit cyclooxygenase activity, leading to prostaglandin synthesis (van der Ouderaa et al., 1980). In plants, SA and aspirin have been shown to inhibit the synthesis of AOS, thus blocking the formation of PDA and JA (Peña-Cortés et al., 1993; Harms et al., 1998). Aspirin also causes the irreversible inactivation of the AOS protein by acetylation of three Ser residues at the C terminus, a process similar to the inactivation of the animal cyclooxygenase (Pan et al., 1998). Inhibition by SA is also proposed to occur at a step between JA synthesis and the transcription of proteinase-inhibitor genes (Doares et al., 1995). Our experiments confirm that SA inhibits wound- or elicitor-induced expression of PIN II, since supplying SA-treated plants with PDA or JA did not result in the induction of PIN II expression. Contrary to the above results, AOS from Arabidopsis was shown to be induced by SA, resulting in increased mRNA, enzyme activity, and PDA accumulation (Laudert and Weiler, 1998). This could be due to the existence of slight differences between plant species in their metabolic pathways, causing inherent differences in their response to the same stimulus.
Role of Ethylene in Induction of AOS and PIN II Expression
The phytohormone ethylene has been suggested to act concomitantly with jasmonates in inducing the expression of various stress-related genes (Xu et al., 1994; O'Donnell et al., 1996; Penninckx et al., 1998). Treatment of tomato seedlings with ethephon, an ethylene-generating compound, for a period of 4 h induced the expression of AOS to the same extent as that observed with wounding (Fig. 6). Pretreatment with STS, an inhibitor of ethylene action, reduced the wound-induced expression of AOS. Application of ethephon to STS-pretreated plants did not prevent inhibition of AOS expression. The ethylene-induced expression of AOS was completely blocked if seedlings were pretreated with SA. Expression of PIN-II was not induced by ethylene. However, STS reduced the wound-induced expression of PIN II, as in the case of AOS. Pretreatment with AVG, an inhibitor of ethylene biosynthesis, did not affect the wound-induced expression of AOS or PIN II, presumably due to the fact that AVG only reduces ethylene synthesis and does not completely block it.
Figure 6.
Effect of ethylene on the expression of AOS and PIN II in tomato. Lane 1, Buffer control; lane 2, wounding; lane 3, ethephon; lane 4, AVG; lane 5, AVG plus wounding; lane 6, STS; lane 7, STS plus wounding; lane 8, STS plus ethephon; lane 9, SA plus ethephon. Concentrations of the chemicals and the method of application are described in “Materials and Methods.” Pretreatment with AVG, STS, or SA was for 2 h, after which time the treatments were applied. Seedlings were harvested 4 h after treatment application.
Ethylene has previously been shown to induce expression of the AOS gene in Arabidopsis, and wounding increased ethylene production (O'Donnell et al., 1996; Laudert and Weiler, 1998). Our results show that the tomato AOS gene is also induced by ethylene, and STS, an ethylene-action inhibitor, prevents this induction. Wounding or application of ethephon failed to induce the expression of AOS in the presence of STS, indicating that wounding acts through ethylene to induce AOS expression. Although ethylene induced AOS expression, there was no corresponding induction of PIN II expression. Therefore, either activity of the AOS protein or the release of JA to its site of action limits the induction of PIN II expression. The results of O'Donnell et al. (1996) have indicated that ethylene has to act in concert with JA to induce PIN II gene expression. Our results confirm this and also show that JA by itself is insufficient to induce PIN II expression. Ethylene acts together with JA for PIN II induction, since STS prevented the wound-induced expression of PIN II. Also, SA inhibited the ethylene-induced accumulation of AOS mRNA. Thus, the presence of SA completely prevents the induction of AOS, whether this induction is via wounding acting through ethylene, by the intercellular signal, systemin, or by positive feedback exerted through PDA or JA.
Updated Model for Defense Gene Activation in Tomato
The current model for octadecanoid signaling in tomato recognizes wounding, herbivore damage, systemin, oligogalacturonides, chitosan, burning, or UV radiation as the various primary signals that trigger expression of defense genes (Schaller and Ryan, 1995; Koiwa et al., 1997; Wasternack and Parthier, 1997). These signals are transduced to a phospholipase, causing the release of linolenic and linoleic acids from membrane lipids. The free fatty acids serve as substrates for the formation of JA, which acts as second messenger to activate defense genes, specifically the proteinase inhibitor genes in tomato. AOS is the first enzyme in the branch pathway leading to the formation of JA, and as such might represent the rate-limiting step in JA biosynthesis. This assumption is supported by the fact that overexpression of the flax AOS gene in potato led to 8- to 12-fold increases in endogenous JA levels (Harms et al., 1995).
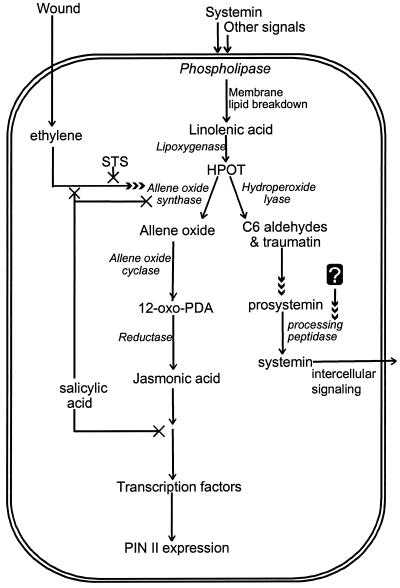
Our new data, together with previous reports, have led us to propose an updated model for octadecanoid signaling in defense gene activation in tomato (Fig. 7). Accordingly, wounding triggers two parallel events: First is the activation of the membrane phospholipase, the production of free fatty acids, and their conversion to corresponding hydroperoxides by lipoxygenase. These hydroperoxides are used by HPL to produce C6 volatiles, which cause induction of prosystemin. An unknown second signal produced in response to wounding is required for the processing of prosystemin to systemin, which then acts as a systemic intercellular signal to induce the production of JA. Ethylene could be involved in this process, although there is as yet no evidence to prove this. The fatty acid hydroperoxides are also used as substrates by AOS to produce JA. HPL and AOS would likely be very tightly regulated at the levels of gene induction and protein activation, because both enzymes use the same substrate and their actions are required consecutively, not simultaneously. The second event in octadecanoid signaling is the action of ethylene, which, in response to wounding, triggers the induction of AOS. In addition, it acts in concert with JA in the downstream activation of PIN II expression. Ethylene and the volatile products of HPL are thus crucial factors in the octadecanoid-signaling cascade. Interdependence of ethylene biosynthesis/action and HPL expression is an interesting prospect for future research.
Figure 7.
An updated model proposed for defense gene activation in tomato. Wounding, systemin, and other signals act through ABA on a phospholipase to release free fatty acids, which form the substrate for lipoxygenase. The fatty acid hydroperoxides produced by phospholipase are used by AOS to form PDA and JA, or by HPL to form six-carbon volatiles and traumatin. PDA and JA activate defense gene expression. The C6 volatiles and traumatin induce expression of prosystemin. Processing of prosystemin to systemin presumably requires an unidentified factor that also responds to wounding. The processed systemin then acts as a systemic intercellular signal. Ethylene, produced in response to wounding, induces the expression of AOS. Ethylene together with JA is required for the optimum expression of PIN II. STS blocks the ethylene- and wound-induced expression of AOS and PIN II. SA inhibits the effect of ethylene on AOS induction, inhibits the expression of AOS, and inhibits the action of PDA and JA in defense gene induction.
In summary, cloning of the tomato AOS has enabled us to examine in parallel the kinetics of gene induction of AOS and PIN II. In addition, we have shown that: (a) the production of C6 volatiles by HPL upon wounding could be the first step in wound-induced defense gene activation, (b) wounding acts through ethylene to induce AOS and PIN II gene expression, and (c) although ethylene induces AOS, it requires the combined presence of ethylene and JA to cause PIN II induction. Our results confirm previous reports that ethylene acts in concert with JA to induce PIN II, and that SA inhibits the octadecanoid-signaling pathway at two steps: the synthesis of the AOS message and the activation of defense genes by JA.
ACKNOWLEDGMENTS
We thank Dr. Nicholas Bate (Pioneer Hi-Bred International, Johnston, IA) for critical reading of the manuscript, and Dr. Yuhai Cui (University of Guelph) for help in preparing the figures.
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
LITERATURE CITED
- Bate NJ, Sivasankar S, Moxon M, Riley JCM, Thompson JE, Rothstein SJ. Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound-inducible. Plant Physiol. 1998;117:1393–1400. doi: 10.1104/pp.117.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Doares SH, Narváez-Vásquez J, Conconi A, Ryan CA. Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 1995;108:1741–1746. doi: 10.1104/pp.108.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell. 1992;4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Weber H, Vollenweider S. Fatty acid signaling in Arabidopsis. Planta. 1998;206:167–174. doi: 10.1007/s004250050388. [DOI] [PubMed] [Google Scholar]
- Harms K, Atzorn R, Brash A, Kühn H, Wasternack C, Willmitzer L, Peña-Cortés H. Expression of a flax allene oxide synthase cDNA leads to increased endogenous jasmonic acid (JA) levels in transgenic potato plants but not to a corresponding activation of JA-responding genes. Plant Cell. 1995;7:1645–1654. doi: 10.1105/tpc.7.10.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms K, Ramirez I, Peña-Cortés H. Inhibition of wound-induced accumulation of allene oxide synthase transcripts in flax leaves by aspirin and salicylic acid. Plant Physiol. 1998;118:1057–1065. doi: 10.1104/pp.118.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. An octadecanoid pathway mutant (JL5) of tomato is compromised in signalling for defense against insect attack. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis MA, Gelfand DH, Sninsky JJ, White TJ. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. [Google Scholar]
- Koiwa H, Bressan RA, Hasegawa PM. Regulation of proteinase inhibitors and plant defense. Trends Plant Sci. 1997;2:379–384. [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Holländer-Czytko H, Weiler EW. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- Laudert D, Weiler EW. Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 1998;15:675–684. doi: 10.1046/j.1365-313x.1998.00245.x. [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurl B, Pearce G, Orozco-Cardenas M, Ryan CA. Structure, expression and anti-sense inhibition of the systemin precursor gene. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- Mueller MJ. Enzymes involved in jasmonic acid biosynthesis. Physiol Plant. 1997;100:653–663. [Google Scholar]
- O'Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ. Ethylene as a signal mediating the wound response of tomato plants. Science. 1996;274:1914–1917. doi: 10.1126/science.274.5294.1914. [DOI] [PubMed] [Google Scholar]
- Pan Z, Camara B, Gardner HW, Backhaus RA. Aspirin inhibition and acetylation of the plant cytochrome P450, allene oxide synthase, resembles that of animal prostaglandin endoperoxide H synthase. J Biol Chem. 1998;273:18139–18145. doi: 10.1074/jbc.273.29.18139. [DOI] [PubMed] [Google Scholar]
- Pan Z, Durst F, Werck-Reichhart D, Gardner HW, Camara B, Cornish K, Backhaus RA. The major protein of guayule rubber particles is a cytochrome P450: characterization based on cDNA cloning and spectroscopic analysis of the solubilized enzyme and its reaction products. J Biol Chem. 1995;270:8487–8494. doi: 10.1074/jbc.270.15.8487. [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191:123–128. [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Métraux J-P, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller A, Ryan CA. Systemin: a polypeptide defense signal in plants. Bioessays. 1995;18:27–33. doi: 10.1002/bies.950180108. [DOI] [PubMed] [Google Scholar]
- Song W-C, Funk CD, Brash AR. Molecular cloning of an allene oxide synthase: a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc Natl Acad Sci USA. 1993;90:8519–8523. doi: 10.1073/pnas.90.18.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey K. Spectrophotometric method for determination of lipoxidase activity. Plant Physiol. 1964;39:65–70. doi: 10.1104/pp.39.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ouderaa FJ, Buytenhek M, Nugteren DH, van Dorp DA. Acetylation of prostaglandin endoperoxide synthase with acetylsalicylic acid. Eur J Biochem. 1980;109:1–8. doi: 10.1111/j.1432-1033.1980.tb04760.x. [DOI] [PubMed] [Google Scholar]
- Vick BA. A spectrophotometric assay for hydroperoxide lyase. Lipids. 1991;26:315–320. [Google Scholar]
- Von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Parthier B. Jasmonate-signalled plant gene expression. Trends Plant Sci. 1997;2:302–307. [Google Scholar]
- Xu Y, Chang P-FL, Liu D, Narasimhan ML, Raghothama KG, Hasegawa PM, Bressan RA. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]