Cdc48/p97 and the ubiquilin family of UBA-UBL proteins are known for their role in the retrotranslocation of damaged proteins from the endoplasmic reticulum. We demonstrate that Cdc48 and the ubiquilin-like proteins in yeast also play a role in the anterograde trafficking of proteins, in this case the vacuolar protease, Cps1.
Abstract
Cdc48/p97 is known primarily for the retrotranslocation of misfolded proteins in endoplasmic reticulum (ER)-associated protein degradation (ERAD). Here we uncover a novel function for both Cdc48 and the conserved ubiquitin-associated/ubiquitin-like ubiquitin receptor (ubiquilin) proteins in yeast (e.g., Ddi1, Dsk2, and Rad23), which deliver ubiquitinated proteins to the proteasome for degradation. We show that Cdc48, its core adaptors Npl4 and Ufd1, and the ubiquilins confer the constitutive anterograde delivery of carboxypeptidase S (Cps1), a membranal hydrolase, to the multivesicular body (MVB) and vacuolar lumen. Cdc48 and Ddi1 act downstream of Rsp5-dependent Cps1 ubiquitination to facilitate the disassembly of insoluble Cps1 oligomers and upstream of ESCRT-0 to facilitate the entry of soluble protein into the MVB. Consequentially, detergent-insoluble Cps1 accumulates in cells bearing mutations in CDC48, DDI1, and all three ubiquilins (ddi1Δ, dsk2Δ, rad23Δ). Thus, Cdc48 and the ubiquilins have ERAD- and proteasome-independent functions in the anterograde delivery of specific proteins to the yeast vacuole for proteolytic activation. As Cdc48/p97 and the ubiquilins are major linkage groups associated with the onset of human neurodegenerative disease (e.g., amytrophic lateral sclerosis, Alzheimer’s, and Paget’s disease of the bone), there may be a connection between their involvement in anterograde protein sorting and disease pathogenesis.
INTRODUCTION
Cdc48 in yeast and p97/VCP in higher eukaryotes are AAA-ATPases known for their involvement in the ubiquitin/proteasome system (UPS) and elimination of endoplasmic reticulum (ER)-associated degradation (ERAD) substrates (Jentsch and Rumpf, 2007; Wolf and Stolz, 2012). Via its heterodimeric core cofactors, Ufd1 and Npl4, Cdc48/p97 binds to polyubiquitinated misfolded substrates and allows for their extraction from the ER and delivery to the proteasome (Wolf and Stolz, 2012). Alternative cofactors include ubiquitin regulatory X (UBX) and UBX-like ubiquitin-fold proteins (Schuberth and Buchberger, 2008), as well as ubiquitin receptors known as ubiquilins, that may play specific roles in facilitating Cdc48/p97-mediated ERAD (Raasi and Wolf, 2007) and perhaps other proteolytic events, like endolysosomal protein sorting, autophagy, and the proteasome-dependent degradation of mitochondrial outer membrane proteins (Rothenberg and Monteiro, 2010; Karbowski and Youle, 2011; Bug and Meyer, 2012; Lee and Brown, 2012). Thus, Cdc48/p97 is an ubiquitin-selective chaperone having proteasome-dependent and potentiallyindependent roles that lead to proteolysis.
Ubiquilins contain a ubiquitin-like (UBL) domain located at the amino terminus that is involved in binding to proteasome subunits and a carboxy terminal ubiquitin-associated (UBA) domain involved in binding mono- and poly-ubiquitinated cargo proteins destined for degradation (Raasi and Wolf, 2007; Finley, 2009; Lee and Brown, 2012). During ERAD, ubiquilins may undergo recruitment to the ER where they bind polyubiquitinated substrates and deliver them to Cdc48-Npl4-Ufd1 for proteasome-mediated degradation. Based on their organization and structurally conserved UBL and UBA domains there are three ubiquilin-like proteins in yeast: Dsk2 (most similar to the ubiquilins), Rad23, and Ddi1; and all three mediate the degradation of polyubiquitinated cargo proteins via the proteasome (Dantuma et al., 2009; Finley, 2009). Rad23 and Ddi1 were identified in viability screens for genes involved in DNA damage repair and surviving genotoxic stress, while Dsk2 was identified as a suppressor for defects in spindle pole duplication (Dantuma et al., 2009). More recent work shows a role for Dsk2 in the formation of inclusion bodies and clearance of a misfolded substrate (Htt1Q103) via autophagy-dependent delivery to the yeast vacuole (Chuang et al., 2016). Thus, some ubiquilins may have additional functions unrelated to ERAD.
Of the three yeast UBL-UBA/ubiquilin proteins, only Dsk2 and Rad23 have been shown to act on Cdc48-mediated ERAD (Raasi and Wolf, 2007), while Ddi1 clearly appears to have different functions. In a nonbiased screen for SNARE-interacting proteins in yeast, we isolated Ddi1 (Vsm1; Lustgarten and Gerst, 1999) and demonstrated that it interacts directly with the autoregulatory domain of Sso exocytic Q/t-SNAREs in their closed (inactive) form and in a phosphorylation-dependent manner (Marash and Gerst, 2003). The Sso-binding site is located proximal to the carboxy-terminal UBA domain (Gabriely et al., 2008) and falls into a variable central region that differs between the three homologues. In addition to its SNARE-binding site, Ddi1, but not Dsk2 or Rad23, also possesses an evolutionarily conserved retroviral aspartyl protease domain whose catalytic function and substrates remain unknown (Sirkis et al., 2006; Finley, 2009; Trempe et al., 2016). Although the repertoire of yeast ubiquilin-interacting substrates is unclear, studies have shown that their UBA domains interact with ubiquitinated substrates, such as polyubiquitin, the Pds1 checkpoint factor, and the Ufo1 E3 ligase (Raasi and Wolf, 2007; Dantuma et al., 2009; Finley, 2009; Lee and Brown, 2012). In contrast, the UBL domains all bind proteasome subunits, although those of Rad23 and Dsk2 also interact with Ufd2, an E4 ubiquitin chain-elongating ligase associated with Cdc48 (Raasi and Wolf, 2007; Dantuma et al., 2009; Finley, 2009). Moreover, recent structural studies have also implicated the UBL domain of Ddi1 in ubiquitin binding, suggesting that it may have two independent sites for interacting with ubiquitin or ubiquitinylated substrates (Nowicka et al., 2015; Trempe et al., 2016). Thus, all three yeast ubiquilins interact directly with ubiquitin, ubiquitin ligases, and with proteasome subunits, although they are not essential for growth either when deleted either individually or together (Kim et al., 2004).
In this article, we describe a novel function for Cdc48 and the yeast ubiquilins in the delivery of a specific membrane-bound hydrolase, Cps1, to the vacuole. We find that Ddi1 interacts physically and genetically with Cdc48 and the Npl4-Ufd1 core adapters to regulate Cps1 sorting to the multivesicular body (MVB) and, subsequently, to the vacuolar lumen. Cps1 is mislocalized to the vacuolar limiting membrane in cells bearing mutations in these genes (i.e., in cdc48, npl4, ufd1, ddi1∆, and dsk2∆ cells) and both Ddi1 and Rad23 colocalize to an endosomal compartment adjacent to the vacuole when MVB formation is blocked. We demonstrate that the Rsp5 E3 ligase is required for the ubiquitination of Cps1 and its interaction with Ddi1, although the manipulation of free ubiquitin levels does not rescue the defects in Cps1 sorting observed here. Finally, neither Cdc48 nor Ddi1 are involved in regulating the ubiquitination or deubiquitination of Cps1 but disperse insoluble Cps1 oligomers and facilitate monomer entry into the MVB compartment. Thus, we propose a new cellular function for Cdc48 and the yeast ubiquilins, which constitute prominent gene products associated with amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD), in MVB-mediated endosome-vacuole anterograde protein transport.
RESULTS
Ddi1 associates with Cdc48 and rescues defects associated with the Npl4 adaptor
The full involvement of the UBL-UBA ubiquitin receptors in proteolytic degradation or protein trafficking is not known. To better understand the role of Ddi1 in yeast, we performed pull downs of HA-tagged Ddi1 and examined the precipitates for coprecipitating proteins using SDS–PAGE, Coomassie labeling, and mass-spectometry (Figure 1A). A band of ∼120 kDa coprecipitated with HA-tagged native Ddi1 and was more prominent when using a presumed catalytically inactive form of the protein, Ddi1D220A, which bears a substitution in the conserved aspartyl residue necessary for putative proteolytic activity (n = 3 experiments). Mass spectometry revealed the protein to be Cdc48, based on ∼40% coverage over multiple nonoverlapping peptides (Supplemental Figure S1A). Thus, Cdc48 associates with Ddi1, which parallels interactions observed between p97/VCP and the ubiquilins (Raasi and Wolf, 2007; Finley, 2009).
FIGURE 1:
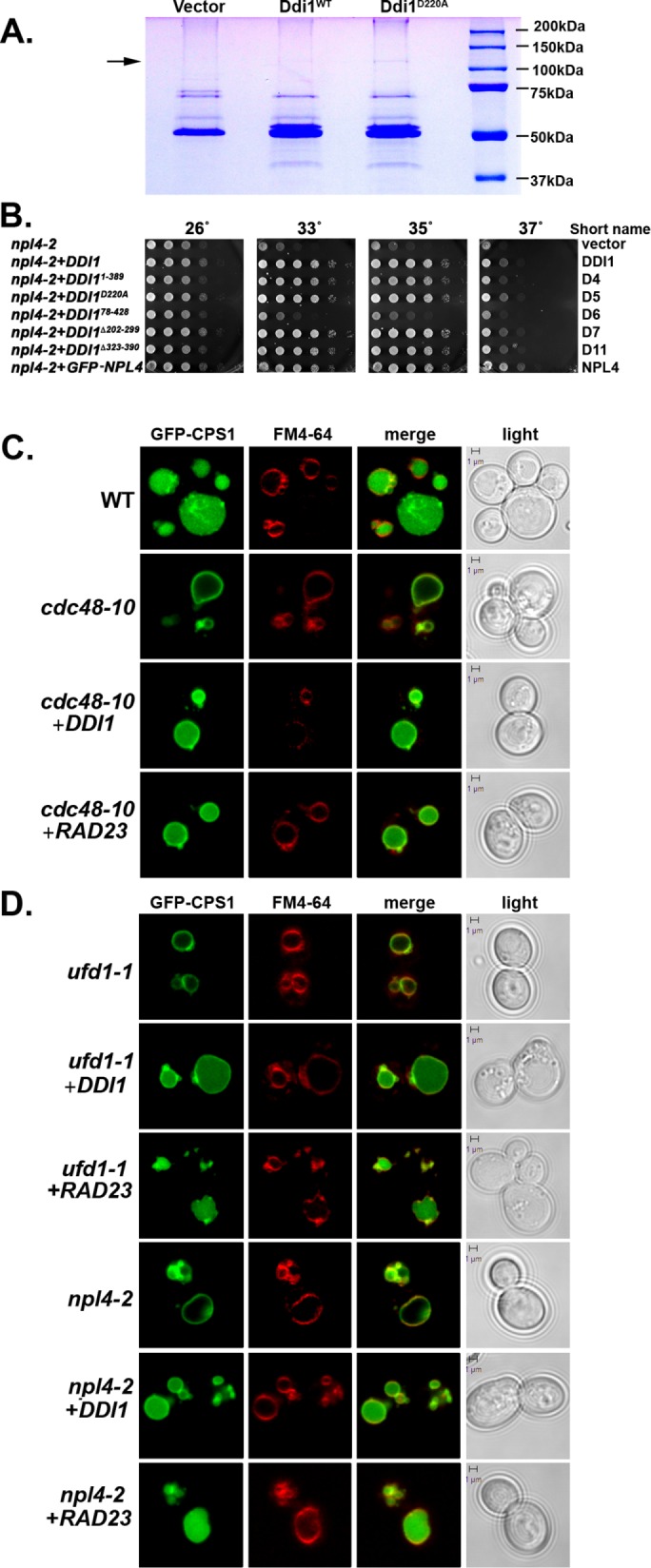
Ddi1 interacts physically with Cdc48, and both are required for Cps1 sorting to the vacuolar lumen. (A) Cdc48 is a Ddi1-binding protein. ddi1Δ cells (W303 background) were transformed with control plasmid (Vector; pAD54) or the same vector expressing either HA-tagged native Ddi1 (Ddi1WT) or the inactive protease mutant (Ddi1D220A). Cells were grown to mid–log phase at 26°C and subjected to co-IP with anti-HA antibodies. Precipitated proteins were resolved by SDS–PAGE and stained with Coomassie, and the bands were excised and analyzed by mass spectrometry. Molecular mass is indicated in kilodaltons (kDa). The arrow indicates Cdc48. The doublet migrating at >50 kDa in the noncontrol lanes is Ddi1; its lower nonphosphorylated form comigrates with a nonspecific band present in the control lane. (B) The UBL of Ddi1 is required to rescue npl4-2. npl4-2 cells were transformed with vector alone (pAD54; Vector) or plasmids expressing either HA-tagged DDI1 (Ddi1) or a truncation mutant (e.g., Ddi11-389, Ddi1D220A, Ddi178-428, Ddi1∆202-299, and Ddi1∆323-390) or GFP-tagged Npl4. Cells were grown to mid–log phase at 26°C before serial dilution and plating onto solid medium. Plates were grown for 2–3 d at the indicated temperatures before documentation. (C) Cdc48 and Ddi1 are required for Cps1 sorting to the vacuolar lumen. WT cells from the cdc48-10 background (WT) and cdc48-10 cells (cdc48-10) expressing GFP-Cps1 were transformed with a control vector or a multi-copy plasmid (2μm) expressing HA-tagged Ddi1 (+Ddi1) or Rad23 (+Rad23), as indicated. Cells were grown at 26°C to mid–log phase, pulse-chase labeled with FM4-64, and examined by confocal microscopy. Merge indicates merger of the GFP and FM4-64 windows. Light indicates the DIC window. Bar = 1 μm. (D) Ufd1 and Npl4 are required for Cps1 sorting to the vacuolar lumen. ufd1-1 and npl4-2 ts mutants expressing GFP-Cps1 from a 2μm plasmid were transformed with a control vector or a plasmid expressing HA-tagged Ddi1 or Rad23. Cells were grown, labeled, and visualized as in C.
To establish relevance of the Ddi1-Cdc48 interaction in vivo, we examined whether Ddi1 is important for the function of Cdc48 and its Ufd1-Npl4 cofactors. We overexpressed native Ddi1 or its various deletion mutants in cells bearing temperature-sensitive (ts) alleles of CDC48, NPL4, and UFD1 and examined them for growth at different temperatures. We employed the cdc48-3 allele, which bears two mutations in the D1 domain (Gallagher et al., 2014)] and cdc48-10, which has two mutations in the D1 and one in the D2 domain (see Materials and Methods). Neither the overproduction of native Ddi1 nor that of its mutant forms, including Ddi1D220A, ameliorated growth defects of the cdc48-3 or cdc48-10 alleles at semirestrictive or restrictive temperatures (Supplemental Figure S1B). In contrast, the overproduction of full-length Ddi1, as well as mutants bearing the UBL domain (e.g., Ddi11-389, Ddi1D220A, Ddi1∆202-296, and Ddi1∆323-390), but not a mutant that lacks the UBL (e.g., Ddi178-428), strongly ameliorated the growth of npl4-2 cells at the different temperatures (Figure 1B). Similar results were observed for npl4-1 cells (unpublished data), but ufd1-1 cells could not be examined since they are not temperature sensitive (Supplemental Figure S1C). Thus, Ddi1 and its UBL-related functions restore functionality to a mutant Cdc48 cofactor but not to Cdc48 itself.
Next, we examined whether the deletion of DDI1 has an effect on the growth of the cdc48 and npl4 mutants. We examined ddi1∆ cdc48-10 and ddi1∆ npl4∆ double mutants at different temperatures but found that their temperature sensitivity was not enhanced relative to control cdc48-10 and npl4∆ cells, respectively (Supplemental Figure S1C).
Cdc48, Npl4, Ufd1, and Ddi1 are required for Cps1 sorting to the vacuolar lumen
Since Ddi1 does not play a role in ERAD and as another Cdc48 cofactor, Ufd3/Doa1, was shown to regulate ubiquitinated carboxypeptidase S (CPS; Cps1) protease sorting to the MVB (Ren et al., 2008), we examined the possible involvement of Ddi1 and Cdc48 in protease delivery to this compartment. We first determined the localization of green fluorescent protein (GFP)-tagged Cps1 expressed in cells bearing attenuated alleles of CDC48, UFD1, and NPL4. In wild-type (WT) cells, Cps1 localized to the vacuolar lumen (Figure 1C and Table 1), while in cdc48-10 cells it localized to the vacuolar limiting membrane (colabeled with FM4-64 [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide]) in 88% of cells even at temperatures permissive for growth (26°C). This is indicative of an inability to access the MVB compartment. Interestingly, overexpression of DDI1 or RAD23 restored Cps1 sorting to the vacuolar lumen in 84 and 70% of cells, respectively (Figure 1C and Table 1). Similar results were observed in ufd1-1 and npl4-2 cells, wherein Cps1 localization to the lumen was inhibited by 74 and 75%, respectively, but could be largely restored upon DDI1 or RAD23 overexpression (Figure 1D and Table 1). We did not use the Euroscarf BY4741 strain background for these experiments, as some Cps1 mislocalizes to the vacuolar limiting membrane even in the WT control cells (unpublished data).
TABLE 1:
Localization of GFP-Cps1.
| GFP-Cps1 localization (% of cells)a | ||
|---|---|---|
| Strain | Vacuole lumen | Vacuole membrane |
| WT | 97.0 ± 0.8 | 3.0 ± 0.8 |
| cdc48-10 | 11.7 ± 2.5 | 88.3 ± 2.5 |
| cdc48-10+DDI1b | 84.0 ± 3.7 | 16.0 ± 3.7 |
| cdc48-10+RAD23c | 70.3 ± 3.3 | 29.7 ± 3.3 |
| npl4-2 | 24.7 ± 3.9 | 75.3 ± 3.9 |
| npl4-2+DDI1b | 73.7 ± 4.5 | 26.3 ± 4.5 |
| npl4-2+RAD23c | 75.7 ± 1.2 | 24.3 ± 1.2 |
| ufd1-1 | 26.0 ± 4.5 | 74.0 ± 4.5 |
| ufd1-1+DDI1b | 63.3 ± 3.7 | 36.7 ± 3.7 |
| ufd1-1+RAD23c | 69.0 ± 7.9 | 31.0 ± 7.9 |
| ddi1Δ | 31.3 ± 6.6 | 68.7 ± 6.6 |
| ddi1Δ+DDI1b | 98.3 ± 0.9 | 1.7 ± 0.9 |
| ddi1Δ+RAD23c | 69.3 ± 3.7 | 30.7 ± 3.7 |
| dsk2Δ | 35.7 ± 2.9 | 64.3 ± 2.9 |
| rad23Δ | 92.0 ± 2.1 | 8.0 ± 2.1 |
| ddi1Δ dsk2Δ rad23Δ | 14.3 ± 1.7 | 85.7 ± 1.7 |
aLocalization is based on scoring 100 cells; n = 3 experiments. Average ± SD is given.
bComplemented with DDI1 expressed from a 2μm plasmid.
cComplemented with RAD23 expressed from a 2μm plasmid.
Next, we examined Cps1 sorting in cells lacking DDI1. GFP-Cps1 was restricted to the vacuolar limiting membrane in 69% of ddi1∆ cells (Figure 2A, Supplemental Figure S1D, and Table 1), similarly to that seen in the cdc48, npl4, and ufd1 mutants (Figure 1C and Table 1). We also observed perivacuolar-localized puncta reminiscent of the class E compartment (Raymond et al., 1992) in ddi1∆ cells. In contrast, GFP-Cps1 localized to the vacuolar lumen in 98 and 69% of ddi1∆ cells expressing either DDI1 or RAD23 from a multicopy plasmid, respectively (Supplemental Figure S1D and Table 1). Thus, the deletion of DDI1 parallels the inactivation of CDC48, with respect to Cps1 sorting. GFP-Cps1 was also mistrafficked in dsk2∆, but not rad23∆, cells (i.e., vacuole membrane labeling in 64 and 8% of cells, respectively; Supplemental Figure S1D and Table 1). Therefore, Ddi1 and Dsk2 regulate Cps1 sorting under native conditions of expression, whereas Rad23 overexpression can compensate, in part, for loss-of-function mutations in DDI1 (or CDC48, NPL4, UFD1) (Figure 1,C and D, and Table 1). Correspondingly, a ddi1∆ dsk2∆ rad23∆ triple mutant showed maximal levels of vacuolar membrane labeling, similarly to that of cdc48-10 cells (Table 1). Thus, all three yeast ubiquilins bear overlapping functions to some degree with respect to Cps1 sorting, although Ddi1 and Dsk2 are most similar.
FIGURE 2:
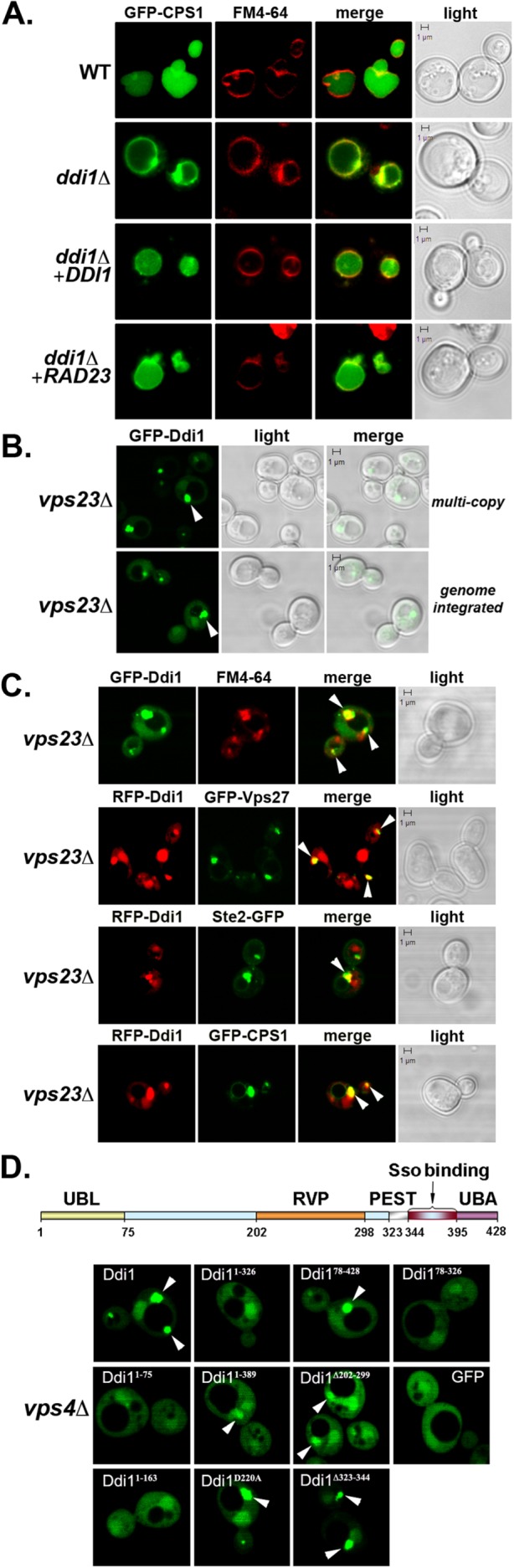
Ddi1 is required for Cps1 sorting to the vacuolar lumen and localizes to the class E compartment in cells defective for MVB formation. (A) Ddi1 is required for Cps1 sorting to the vacuolar lumen. WT (W303) and DDI1 deletion (ddi1Δ; W303) cells expressing GFP-Cps1 from a 2μm plasmid were transformed with either a control vector or a 2μm plasmid expressing HA-Ddi1 (+DDI1) or HA-Rad23 (+RAD23), as indicated. Cells were grown to mid–log at 26°C, pulse-chase labeled with FM4-64, and examined by confocal microscopy. Merge indicates merger of the GFP and FM4-64 windows. Light indicates the DIC window. Bar = 1 μm. (B) GFP-Ddi1 localizes to a perivacuolar compartment in vps23Δ cells. Class E vps mutant yeast cells (vps23Δ) were transformed with a 2μm plasmid expressing GFP-Ddi1 (multi-copy) and examined by fluorescence microscopy. Alternatively, vps23Δ cells carrying a genome-integrated copy of GFP-DDI1 under the control of the GAL promoter (genome-integrated) were examined in parallel. Cells were grown on glucose- or galactose-containing medium at 26°C, respectively, and examined by confocal microscopy. (C) Ddi1 localizes to the class E compartment and colocalizes with Vps27, Ste2, and Cps1. A class E vps mutant (vps23Δ) expressing either GFP- or RFP-tagged Ddi1 (GFP-Ddi1, RFP-Ddi1, respectively) was transformed either with a control vector or 2μm plasmids expressing GFP-Vps27, Ste2-GFP, or GFP-Cps1, as indicated. The cells expressing GFP-Ddi1 or RFP-Ddi1 were grown, pulse-labeled with FM4-64, and visualized as described in A. Arrowheads indicate the class E compartment wherein GFP- and RFP-Ddi1 colocalize with LE substrates. (D) The Ddi1 UBA domain is necessary for MVB localization. Top panel: Schematic shows the general domain structure of Ddi1 (Gabriely et al., 2008). The UBL domain corresponds to residues 1–75, while the UBA domain corresponds to 395–428, although the entire UBA-like region stretches from residues 363 to 428. The retroviral protease (RVP) domain is shown from residues 202 to 297, the PEST domain from 325 to 343, and the Sso SNARE-binding domain from 344 to 395. Bottom panel: A class E mutant (vps4Δ) was transformed with a 2μm plasmid expressing GFP (as control), GFP-tagged full-length Ddi1 or Ddi1 truncation mutants (e.g., Ddi11-75, Ddi11-163, Ddi11-326, Ddi11-389, Ddi178-428, Ddi178-326, Ddi1Δ202-299, Ddi1Δ323-344), or the protease-inactive mutant (e.g., Ddi1D220A). Cells were grown at 26°C and examined by confocal microscopy. White arrows designate the class E compartment.
To determine whether the MVB sorting defects seen in the cdc48, npl4, and ufd1 mutants and ddi1∆ cells are specific to proteins of the biosynthetic pathway, we examined the localization of other ubiquitinated and MVB-sorted cargo proteins, such as Fur4-GFP and Ste2-GFP. As documented previously by others, we found that these proteins localized mainly to the plasma membrane and somewhat to the vacuolar lumen both in WT cells and in cells bearing the cdc48-10 allele but not in control cells bearing a deletion in VPS27, which encodes an ESCRT-0 protein necessary for protein entry into the MVB (Supplemental Figure S2, A and B). Likewise, neither Fur4-GFP nor Ste2-GFP localization was significantly affected in cells lacking DDI1 or all three ubiquilins (e.g., ddi1∆ rad23∆ dsk2∆ cells; Supplemental Figure S2, A and B). Thus, ubiquitinated and endocytosed membrane proteins enter the MVB independently of Cdc48 or Ddi1, Rad23, and Dsk2. Similarly, we found that cells lacking DDI1, bearing the cdc48-10 allele, or both, were no more sensitive to exposure 5-fluoro-orotic acid than WT cells, indicating that there are no obvious general defects in the sorting of Fur4 (unpublished data).
We next attempted to localize GFP-tagged ubiquitin, which was suggested to access the vacuolar lumen in WT cells but be retained in the cytosol in doa1∆ cells (Ren et al., 2008). However, we were unable to observe significant localization of this reporter to the vacuole in the different WT backgrounds examined (e.g., BY4741, SP1, and W303; unpublished data). Thus, we examined the localization of other endomembrane markers, such as GFP-Yif1, which localizes to the trans Golgi in WT cells but undergoes relocalization to the late endosome (LE) and vacuolar lumen in cells defective in selective LE-Golgi retrograde transport (e.g., btn1∆, btn2∆ cells) (Kama et al., 2007, 2011). We examined GFP-Yif1 localization in cdc48-3, cdc48-10, npl4∆, npl4-1, npl4-2, and ufd1-1 cells but did not see any effect (Supplemental Figure S3A). Similar results were observed for Yif1 in ddi1∆ cells (Supplemental Figure S3B) and other markers, such as GFP-tagged carboxypeptidase Y (CPY)1-50 and the CPY receptor, Vps10, were not affected in these mutants (Supplemental Figure S3, C and D). Finally, sorting of a vacuolar integral membrane protein, Vph1-GFP, to the vacuolar limiting membrane was also unaffected (unpublished data).
Since Cps1 sorting was strongly altered in the cdc48, npl4, ufd1, and ddi1∆ mutants, we hypothesized that it might relate solely to vacuolar proteases, although we did not observe defects in GFP-CPY1-50 and Vps10 sorting. We examined ddi1∆, rad23∆, dsk2∆, ddi1∆ rad23∆, and ddi1∆ rad23∆ dsk2∆ cells for defects in full-length CPY sorting by examining secretion onto nitrocellulose filters (Supplemental Figure S3E). We found that only the triple mutant (e.g., ddi1∆ rad23∆ dsk2∆ cells) secreted CPY, although this was not at the level seen with a vps23∆ control strain. Nevertheless, it suggests that there are defects in CPY sorting when all three ubiquilins are absent. Next, we examined cdc48 (e.g., cdc48-3, cdc48-10), npl4 (e.g., npl4-1, npl4-2, npl4∆), and ufd1-1 cells for CPY secretion and found that none secreted more CPY than their WT controls at 26°C (Supplemental Figure S3E). Likewise, the deletion of DDI1 in the npl4∆ strain did not increase CPY secretion, although a slight increase was observed in cdc48-10 ddi1∆ cells but was less than that seen with an ESCRT-0 mutant (e.g., vps27∆). Thus, defects in Ddi1 or Cdc48 alone do not result in the missorting of CPY, although there is an accentuated mislocalization phenotype when combined with mutations in the other ubiquilins (Supplemental Figure S3, E and F).
Ddi1 and Npl4, but not Cdc48, localize to a perivacuolar compartment in cells defective in MVB formation.
Because Cdc48 and ubiquilins play a role in Cps1 sorting to the MVB, it suggests that they may localize to the LE/MVB to function. Previously, we demonstrated that GFP-tagged Ddi1 localizes to the cytoplasm and nucleus in WT cells, nuclear enrichment being dependent on both the UBL and UBA domains (Gabriely et al., 2008). Here we examined the localization of both plasmid- and genomic locus-expressed GFP-Ddi1 in cells defective for MVB formation (e.g., vps4∆, vps20∆, vps23∆, vps25∆, vps27∆, vps37∆ cells; Figure 2B and Supplemental Figure S4, A and B). We found that GFP-Ddi1 expressed either from a multicopy plasmid or from its genomic locus showed labeling of puncta proximal to the vacuole in vps23∆ cells, in addition to fainter cytosolic and nuclear labeling (Figure 2B). This perivacuolar labeling was nonnuclear, as demonstrated using Hoechst dye to label the nucleus (Supplemental Figure S4A). Correspondingly, we observed that either GFP- or red fluorescent protein (RFP)-labeled Ddi1 colocalized with endomembrane markers (e.g., FM4-64, GFP-Vps27, Ste2-GFP, and GFP-Cps1) that accumulate at a perivacuolar (class E) endosomal compartment in cells unable to confer MVB sorting (e.g., vps23∆ cells; Figure 2C) but not in cells that lacked CPS1 alone (e.g., cps1∆; unpublished data). RFP-Ddi1 labeling of the class E compartment was observed in other MVB mutants (e.g., vps4∆, vps20∆, vps25∆, vps27∆, and vps37∆ cells) and, interestingly, GFP-Rad23 also colabeled these structures (Supplemental Figure S4B). In contrast, Rad23 and Ddi1 colabeled only the nucleus in WT cells (Supplemental Figure S4B). Thus ubiquilins, such as Ddi1 and Rad23, localize to the site of MVB formation when MVB protein sorting is inhibited. Mechanistically, this necessitates only the UBA domain and not the UBL or RVP domains as GFP-tagged Ddi178-428, Ddi1∆202-299, Ddi1D220A, and Ddi1∆323-344 but not Ddi11-75, Ddi11-163, Ddi11-326, or Ddi178-326 localized to the perivacuolar compartment in an MVB mutant (e.g., vps4∆ cells; Figure 2D). We note that MVB labeling was observed in some cells with Ddi11-389, which lacks most, but not all, of the UBA and UBA-like domain (residues 363–428; Saccharomyces Genome Database).
Since mutants in CDC48, NPL4, and UFD1 show defects in Cps1 sorting (Figure 1, C and D), we examined GFP-Ddi1 localization therein. In addition to cytoplasmic and nuclear labeling, we observed labeling of a perivacuolar compartment in cdc48-10, npl4∆, and ufd1-1 cells (Supplemental Figure S4C) identical to that observed in the MVB mutants (Figure 2, B and C) but not in WT cells. This compartment also colabeled strongly with FM4-64 (Supplemental Figure S4C). Thus, mutants in the Cdc48-Npl4-Ufd1 pathway show phenotypes (e.g., Cps1 missorting and Ddi1 localization to perivacuolar/class E-like compartments) that are strongly reminiscent of those observed in MVB mutants.
Next, we examined the localization of Cdc48 in WT cells and cells defective in MVB protein sorting. Cdc48-GFP labeled the cytosol along with small puncta that were often located adjacent to vacuoles in WT cells (Figure 3A). Strikingly, when Cdc48-GFP was coexpressed with an early-late endosome marker, Snx4-RFP (Hettema et al., 2003), we observed colocalization to numerous cytoplasmic and perivacuolar puncta (∼90% colocalization, n = 3 experiments; Figure 3A). Moreover, we noticed that there are more labeled puncta in cells coexpressing Cdc48-GFP and Snx4-RFP than in cells expressing Cdc48 alone. Although the reason for this remains unclear at this point, it may indicate that Snx4 contributes to the recruitment of Cdc48 to endosomes. In contrast to Cdc48-Snx4 endosomal colabeling, however, we did not observe Cdc48 colocalization with Vps10-RFP (Deloche and Schekman, 2002), which labels a distinct LE population (Kama et al., 2007), or with RFP-Snf7 (an ESCRT-III component) or RFP-Vps27 (an ESCRT-0 component), which both label the MVB. Thus, Cdc48 associates with a specific endosomal population in WT cells.
FIGURE 3:
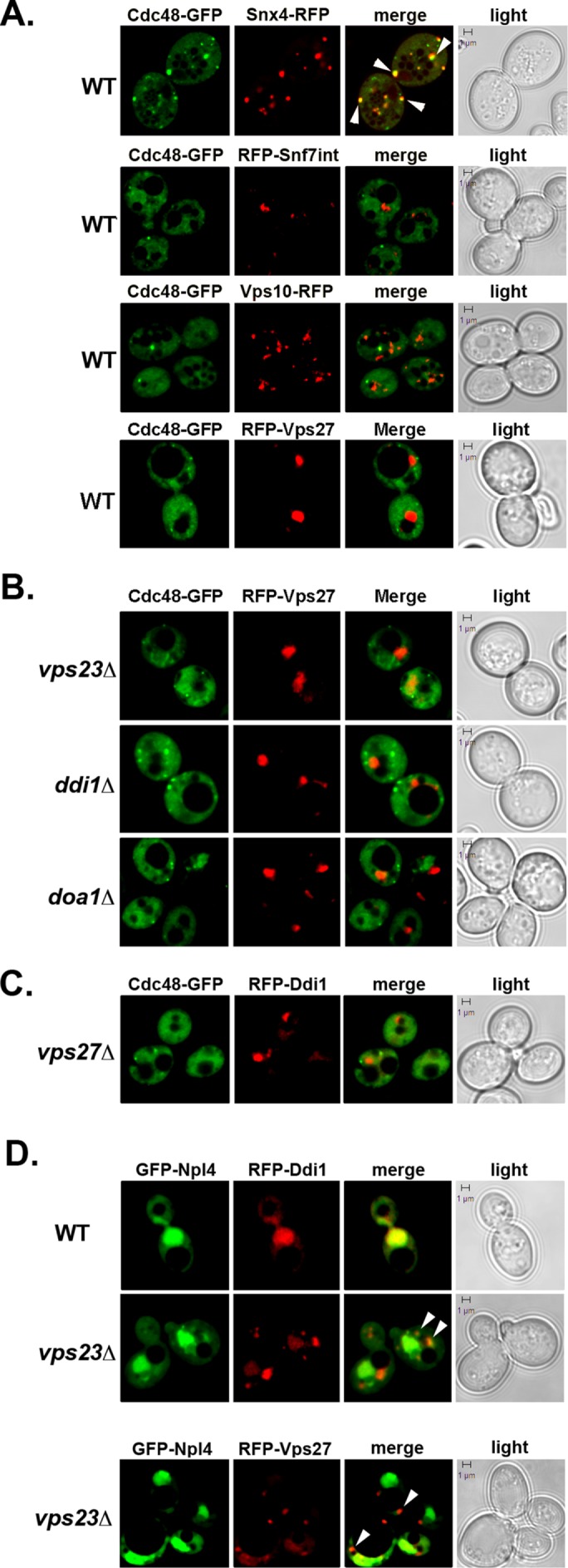
Npl4, but not Cdc48, colocalizes with Ddi1 to the class E compartment in an MVB mutant. (A) Localization of Cdc48 in WT cells. WT (W303) cells expressing Cdc48-GFP from a single-copy plasmid were transformed with plasmids expressing either Snx4-GFP, RFP-Vps27, or Vps10-RFP, as indicated. Alternatively, WT cells expressing RFP-Snf7 from the genome were transformed with the same plasmid expressing Cdc48-GFP. Cells were grown to mid–log phase at 26°C and examined by confocal microscopy. White arrows indicate colocalization. Merge indicates merger of the GFP and RFP windows. Light indicates the DIC window. Bar = 1 μm. (B) Cdc48 does not colocalize with the LE/MVB in MVB sorting mutants. vps23Δ, ddi1Δ, and doa1Δ cells expressing Cdc48-GFP from a single-copy plasmid were transformed with a 2μm plasmid expressing RFP-Vps27. Cells were grown and visualized as in A. (C) Cdc48 does not colocalize with Ddi1 in an MVB sorting mutant. vps27Δ cells expressing Cdc48-GFP from a single-copy plasmid were transformed with a 2μm plasmid expressing RFP-Ddi1. Cells were grown and visualized as in A. (D) Npl4 and Ddi1 colocalize to the nucleus and perivacuolar structures in an MVB mutant. WT (BY4741) and vps23Δ cells expressing GFP-Npl4 from a 2μm plasmid were transformed with plasmids expressing RFP-Ddi1 or RFP-Vps27, as indicated. Cells were grown and visualized as in A. White arrowheads designate colocalization at the class E compartment.
We next examined Cdc48 localization in MVB protein sorting and formation mutants (e.g., ddi1∆ and doa1∆ cells, which are blocked in Cps1 sorting to the MVB, and vps23∆ cells, which are blocked in MVB formation). We found that Cdc48 localization was similar to that seen in WT cells and did not colocalize with RFP-Vps27 (Figure 3B). We also examined for colocalization with Ddi1 under conditions where RFP-Ddi1 accumulates at the perivacuolar compartment (e.g., in vps27∆ cells). However, we did not observe significant colocalization (Figure 3C). Thus, despite the fact that Cdc48 is involved along with Ddi1 in Cps1 sorting, it does not stably colocalize with Ddi1, the LE/MVB compartment, or the class E compartment observed in MVB mutants. This finding, however, may not be entirely surprising given that Cdc48 does not readily colocalize with its other known cofactors, Npl4 and Ufd1, except for general cytoplasmic and nuclear staining (Breker et al., 2013).
Since Ddi1, but not Cdc48, localizes to a class E-like compartment in MVB mutants, we examined whether it colocalizes instead with a Cdc48 adaptor. We examined the localization of GFP-tagged Npl4 in WT, cdc48-10, ufd1-1, and vps23∆ cells, along with RFP-Ddi1. Interestingly, Npl4 and Ddi1 colocalized to the nucleus in the different cell types but colocalized only to perivacuolar structures in the vps23∆ MVB mutant (Figure 3D and unpublished data). GFP-Npl4 localization to the MVB was verified in part using RFP-Vps27 (Figure 3D). Thus, Npl4, but not Cdc48, colocalizes with Ddi1 at the LE/MVB and only when protein entry into the MVB is abrogated.
Ddi1 binds Cps1 but is not required for ubiquitination or deubiquitination
If Ddi1 is required for Cdc48-mediated entry of Cps1 into the MVB, then it is likely to interact with Cps1 (Figure 1C), probably via the UBA domain necessary for Ddi1 localization to the perivacuolar compartment (Figure 2D). To verify this, we expressed myc-tagged Ddi1 in WT cells along with various GFP-tagged proteins that access or enter into the MVB (e.g., Vps27, Ste2, Cps1, and Fur4) and performed coimmunoprecipitation (co-IP) using anti-GFP antibodies (Figure 4A). GFP-Cps1 coprecipitated Ddi1 and both the lower molecular mass (nonphosphorylated) and higher molecular mass (phosphorylated) forms (Gabriely et al., 2008) were detected. In contrast, GFP-Vps27, GFP-Ste2, or GFP-Fur4 could not precipitate myc-Ddi1 (Figure 4A), although we note that GFP-Ste2 appeared to have undergone cleavage/degradation in this experiment. Overall, we propose that Ddi1 interacts with specific MVB-trafficked proteins, as predicted by experiments showing that only Cps1 localization is altered in ddi1∆ cells (Figure 2A and Supplemental Figures S1D and S3, B–D).
FIGURE 4:
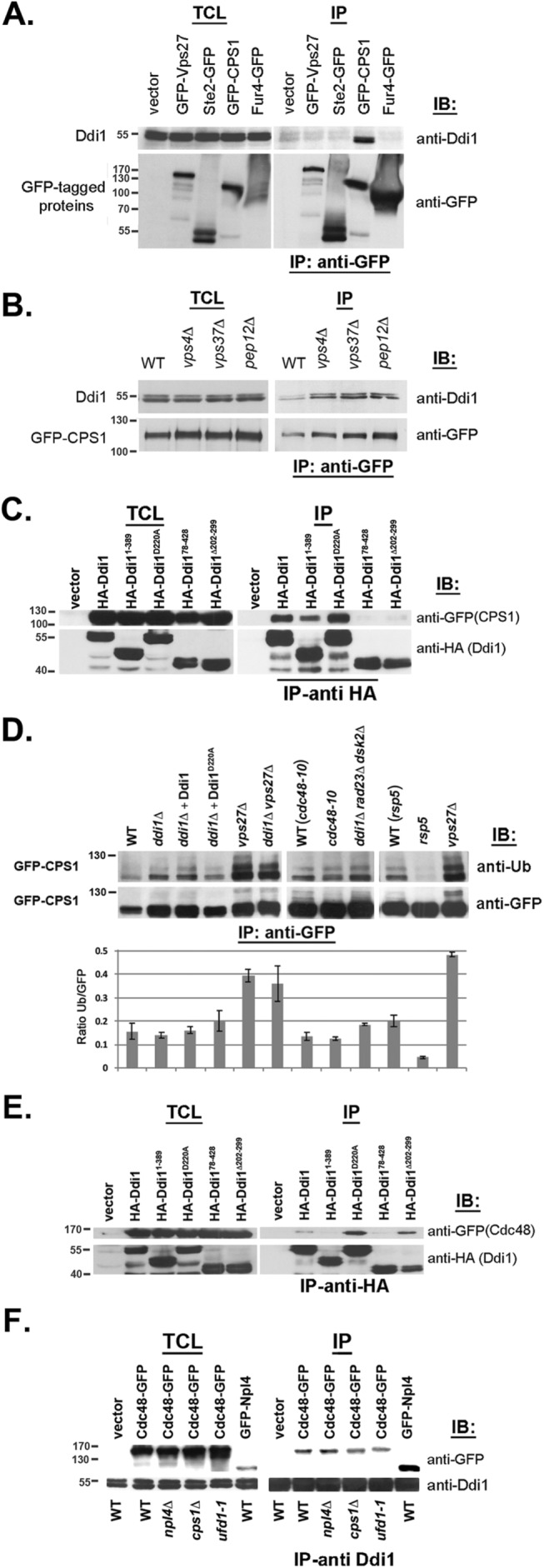
Ddi1 and Cdc48 are not involved in Cps1 ubiquitination/deubiquitination and act upstream of ESCRT-0. (A) Ddi1 binds Cps1 but not endocytosed cargo or ESCRT-0. WT cells (W303) expressing myc-tagged Ddi1 were transformed with plasmids expressing GFP-tagged Vps27, -Ste2, -Fur4, or -Cps1 or a control plasmid (vector). Cells were subjected to IP using monoclonal anti-GFP antibodies (Abs). Proteins in the total cell lysate (TCL) and IP lanes were detected with either anti-GFP (1:1000) or -Ddi1 (1:5000) Abs in immunoblots (IB). (B) Ddi1 binds GFP-Cps1 in class D and E vps mutants. WT cells (BY4741) and vps mutant cells (e.g., vps4Δ, vps37Δ, and pep12Δ) were transformed with 2μm plasmid expressing GFP-Cps1 and subjected to IP with anti-GFP Abs. Proteins were detected as in A. (C) The UBL, UBA, and RVP domains of Ddi1 are required for Cps1 binding. ddi1Δ cells expressing GFP-Cps1 from a 2μm plasmid were transformed with plasmids expressing HA-tagged full-length Ddi1, Ddi1 mutants (e.g., Ddi11-389, Ddi1D220A, Ddi178-428, or Ddi1Δ202-299), or a control vector pAD54 (vector). Cells were subjected to IP with monoclonal anti-HA Abs. Proteins in the TCL and IP lanes were detected with either anti-GFP (1:1000) to detect Cps1 or anti-HA Abs (1:1000) to detect Ddi1 proteins. (D) Ddi1 and Cdc48 are not required for Cps1 ubiquitination or deubiquitination. WT control cells (e.g., W303, cdc48-10, and rsp5-326 backgrounds), ddi1∆ deletion cells (e.g., ddi1∆, ddi1∆ rad23∆ dsk2∆, and ddi1∆ vps27∆ cells), ddi1∆ deletion cells expressing full-length Ddi1 or Ddi1D220A (ddi1∆+Ddi1 or ddi1∆+Ddi1D220A), vps27∆ cells, or cells bearing ts mutations in CDC48 or RSP5 (e.g., cdc48-10, rsp5-326) were transformed with a 2μm plasmid expressing GFP-Cps1. Cells were subjected to IP with monoclonal anti-GFP Abs. Only the IP information is shown and proteins in the IP lanes were detected with anti-ubiquitin (1:1000) or -GFP (1:1000) Abs. Mass (on left) is in kilodaltons. The corresponding histogram below shows the ratio of the ubiquitination signal relative to the GFP signal after normalization (for the GFP signals). The results of two identical experiments are shown. Error bars indicate the SD. (E) The UBL and UBA domains, but not the RVP or catalytic aspartate, are required to bind Cdc48. ddi1Δ cells expressing Cdc48-GFP from a single-copy plasmid were transformed with plasmids expressing HA-tagged Ddi1, Ddi1 truncation mutants (e.g., Ddi11-389, Ddi178-428, and Ddi1∆202-298) or the D220A substitution mutant (e.g., Ddi1D220A). Cells were subjected to IP with anti-HA Abs. Proteins in the TCL and IP lanes were detected with either anti-GFP (1:1000) or -HA (1:1000) Abs. (F) Ddi1 binds Npl4 but interacts with Cdc48 in the absence of Npl4 or native Ufd1. WT control cells (BY4741), npl4Δ, ufd1-1, and cps1Δ cells expressing Cdc48-GFP from a single-copy plasmid or WT cells expressing GFP-Npl4 from a 2μm plasmid were subjected to IP with anti-Ddi1 Abs. Proteins in the TCL and IP lanes were detected with monoclonal anti-GFP (1:1000) or polyclonal anti-Ddi1 Abs.
Next we examined whether the association of Ddi1 with Cps1 is altered in cells defective in MVB and vacuolar protein sorting (e.g., vps4∆, vps37∆, and pep12∆, the latter affected in Golgi-endosome sorting). We precipitated GFP-Cps1 from WT and mutant cells and probed for native Ddi1 but did not observe any significant differences (Figure 4B). Thus, the Ddi1-Cps1 interaction occurs independently of active MVB and vacuolar protein sorting. We note that the IP of GFP-Cps1 from cells led to the appearance of a tight doublet of ∼110 kDa, which is more strongly visible in WT cells and is more apparent in Figure 4, C and D. While we do not definitively know the cause for doublet formation, native Cps1 can be differentially glycosylated and its maturation (and activation as a soluble enzyme) involves cleavage from an immature ∼77-kDa form to yield a mature ∼74 kDa form (Spormann et al., 1992). While only the mature differentially glycosylated forms should be apparent in lysates derived from WT cells, both smaller and larger immature forms of GFP-Cps1 should be present in lysates derived from cells where protein sorting into the MVB or vacuole lumen is compromised (e.g., vps27∆, ddi1∆, and cdc48-10 cells). We predict that only the immature form(s) of Cps1 should interact with Ddi1, which is present in the cytosol. It is important to note, however, that we cannot readily resolve mature GFP-Cps1 from the smallest immature form in these SDS–PAGE gels, due to the small relative size differences (Figure 4, B–D). Thus, while it may appear that mature GFP-Cps1 coprecipitates with Ddi1 (especially in lysates derived from WT cells, where the mature form is present), it is far more likely that it is an immature form (which maintains its cytoplasmic tail) that coprecipitates. We also note that no free GFP was observed in these blots, indicating that both mature and immature forms of GFP-Cps1 are relatively stable (unpublished data).
With this in mind, we examined the molecular requirements for the interaction between Ddi1 and Cps1 by expressing Ddi1 deletion mutants in ddi1∆ cells and using co-IP (Figure 4C). HA-tagged full-length Ddi1 and mutants bearing the UBL domain (e.g., Ddi11-389 and Ddi1D220A) bound a GFP-tagged Cps1 doublet, whereas mutants lacking the UBL or RVP domain (e.g., Ddi178-428 and Ddi1Δ202-298) could not. Again, this doublet is likely to represent the different immature glycosylated forms of Cps1 (and that the lower band of the doublet is not the mature form). We also note that Ddi11-389 bound Cps1, though less than that of full-length Ddi1 or the D220A mutant. This is probably because some (e.g., residues 363–389) of the UBA-like region is still present in this mutant. Thus, we predict that the UBL, RVP, and, perhaps, the UBA domain are all required for the interaction with Cps1.
Since the mechanism by which Cdc48-Npl4-Ufd1 and Ddi1 confer Cps1 sorting is not known (i.e., do they act prior to, or during, MVB formation to regulate Cps1 entry?), we examined whether they control Cps1 ubiquitination (Figure 4D). By immunoprecipitating GFP-Cps1 and detecting with anti-ubiquitin antibodies, we examined the amount of ubiquitinated Cps1 in WT cells (i.e., in which Cps1 reaches the vacuole lumen and, therefore, is deubiquitinated) or in cells either lacking DDI1 or having attenuated Cdc48 function (i.e., in which Cps1 remains on the limiting membrane and may be monoubiquitinated on residue K8 and/or perhaps K12). In parallel, we assessed Cps1 ubiquitination in vps27∆ cells, in which Cps1 should remain in its fully ubiquitinated state, as well as in ddi1∆ vps27∆ double mutants to determine whether Ddi1 is necessary for the ubiquitination of Cps1. We observed only minor differences in the level of GFP-Cps1 precipitated from the different WT controls (e.g., W303 and the mutant backgrounds) and cells lacking DDI1 or VPS27 (e.g., ddi1∆, vps27∆, and ddi1∆ vps27∆ cells) or bearing ts alleles of CDC48 or RSP5 (the E3 ligase for Cps1; Morvan et al., 2004). In contrast, we observed differing levels of ubiquitinated Cps1, whereby no ubiquitination was observed in the rsp5-326 ts mutant, while elevated levels of ubiquitination were seen in cells lacking VPS27 (e.g., vps27∆ and ddi1∆ vps27∆ cells). Importantly, no differences in the level of ubiquitination were observed in cells lacking DDI1 (or all three ubiquitin receptors, e.g., ddi1∆ rad23∆ dsk2∆ cells) or in the cdc48-10 cells (Figure 4D). Thus, it is unlikely that either Ddi1 (and the other ubiquilins) or Cdc48 regulate the ubiquitination or deubiquitination of Cps1. We note that multiple molecular mass forms are detected using anti-ubiquitin antibodies (Figure 4D, upper panel), indicating that in addition to possible differential glycosylation, there are size shifts fully consistent with perhaps di- and polyubiquitinated forms of Cps1 being present in the mutant cells. Although we do not know the full nature of Cps1 modification, the results clearly show however that the level of ubiquitination itself is not significantly changed between immature forms in WT cells and mutants in DDI1 or CDC48 (Figure 4D, histogram).
Our results also suggest that the RVP domain does not confer deubiquitinase activity, since expression of the inactive catalytic form, Ddi1D220A, in ddi1∆ cells did not increase the level of Cps1 ubiquitination relative to native Ddi1. Thus, Ddi1 and Cdc48 act downstream of ubiquitination machinery and, based on the Cps1 localization data (Figures 1C and 2A), upstream to ESCRT-0 and the VPS pathway. It is also important to note that neither the deletion of DDI1 nor its overexpression affected the levels of native Cdc48, as examined in Westerns using anti-Cdc48 antibodies (unpublished data).
The UBL and UBA domains of Ddi1 interact with Cdc48
Since Cdc48 binds Ddi1 and together are necessary for Cps1 sorting to the MVB (Figures 1, A and C, and 2A), we examined their requirements for interaction. We first verified the interaction between native Cdc48 and Ddi1 by performing immunoprecipitation with anti-Ddi1 antibodies using extracts from WT and ddi1∆ cells and examining the precipitates with anti-Cdc48 antibodies. We found that a faint band corresponding to Cdc48 could be precipitated from WT but not ddi1∆ cells (Supplemental Figure S5A). Thus, there is an interaction between Ddi1 and Cdc48 at native levels of expression, as suggested from the initial pull-down experiment (Figure 1A).
Next, we expressed the HA-tagged Ddi1 mutants (Gabriely et al., 2008) in ddi1∆ cells and examined their ability to coprecipitate GFP-Cdc48 (Figure 4E). Full-length Ddi1 and mutants having both the UBL and UBA domains (e.g., Ddi1D220A, Ddi1Δ202-299) interacted with Cdc48. In contrast, removal of either the UBL or most of the UBA domain (e.g., Ddi178-428 and Ddi11-389, respectively; see schematic in Figure 2D) led to the loss of Cdc48 binding. As observed initially (Figure 1A), the putative catalytically inactive RVP form of Ddi1 (Ddi1D220A) was better able to bind Cdc48 (Figure 4E). Moreover, deletion of the RVP domain, which mediates dimerization (Gabriely et al., 2008), also bound Cdc48 more strongly (2.1 ± 0.1-fold more, as normalized for the amount of precipitated Ddi1Δ202-299; n = 2 experiments). Thus, both the UBL and UBA domains are required for Ddi1 to interact with Cdc48, and removal of the RVP/dimerization domain enhances the interaction.
As shown, the overexpression of Ddi1 constructs that possess the UBL domain (e.g., DDI1, DDI11-389, DDI1D220A, DDI1∆202-299, and DDI1∆323-390; see schematic in Figure 2D) rescued the temperature sensitivity of npl4-2, but not cdc48-3 or cdc48-10, cells (Figure 1B and Supplemental Figure S1B). While we did not expect that DDI1 overexpression would alleviate all cdc48 phenotypes, the result suggested that Ddi1 might not bind directly to Cdc48 but perhaps via Npl4 (or Ufd1). To test for an interaction between Npl4 and Ddi1, we examined whether GFP- or HA-tagged Npl4 coprecipitates with Ddi1. Indeed, endogenously expressed Ddi1 interacted with both GFP-Npl4 (Figure 4F) and HA-Npl4 (Supplemental Figure S5B) in WT cells, and this interaction was lessened greatly in cdc48-10 cells at elevated temperatures (Supplemental Figure S5B). Thus, Ddi1 might precipitate Npl4 via Cdc48.
To determine whether Npl4 or Ufd1 are required for the Cdc48-Ddi1 interaction, we expressed GFP-tagged Cdc48 in the npl4∆ or ufd1-1 cells and performed co-IP with anti-Ddi1 antibodies (Figure 4F). Endogenously expressed Ddi1 interacted with Cdc48 even in the absence of Npl4 or under conditions when Ufd1 function is attenuated (i.e., in ufd1-1 cells). Interestingly, we observed an increase in Ddi1 binding to Cdc48 in the absence of Npl4 (+45.8% ± 10.3% after normalization for precipitated Ddi1; n = 3 experiments, p < 0.015) and a decrease in Ddi1 binding in the ufd1-1 strain (–51.4% ± 13.5%; n = 3 experiments, p < 0.01). Thus, neither Npl4 nor fully active Ufd1 are absolutely required for the interaction of Ddi1 with Cdc48, although we cannot exclude the possibility that an additional factor (e.g., Ufd1) is necessary. Of consequence, we also observed that the Ddi1-Cdc48 interaction is reduced in the absence of substrate, since Ddi1 binding to Cdc48 was reduced by >20% (22.1% ± 1.0% after normalization for precipitated Ddi1; n = 3 experiments) in cells lacking CPS1 (Figure 4F). Thus, there may be additional substrates recognized by both Ddi1 and Cdc48-Npl4-Ufd1 and that facilitate their interaction in vivo.
Cdc48 releases Cps1 from detergent-insoluble aggregates
Cdc48-Npl4-Ufd1 plays an essential role in the retrotranslocation of misfolded proteins during ERAD (Jentsch and Rumpf, 2007; Dantuma and Hoppe, 2012; Wolf and Stolz, 2012). Yet Cdc48-Npl4-Ufd1 and the ubiquilins also act upstream of the MVB pathway to control Cps1 sorting to the vacuole, as shown here. Since other studies have also demonstrated an aggregate prevention role for Cdc48 orthologs (Kobayashi et al., 2007; Song et al., 2007; Nishikori et al., 2008; Yamanaka et al., 2012), we hypothesized that perhaps Cps1 forms oligomers or aggregates that necessitate Cdc48-mediated disassembly prior to being recognized by ESCRT-0 and attaining entry into the MVB.
To verify this, we examined the endogenous and plasmid-based expression of myc-Cps1 and HA-Cps1, respectively, in WT control and mutant (e.g., cdc48-10, ddi1∆, cdc48-10 ddi1∆, and ddi1∆ dsk2∆ rad23∆) yeast and examined their distribution to ice-cold soluble and detergent- (e.g., Triton X-100, NP-40; 1%) insoluble (pellet) fractions derived from lysates of cells treated either with or without cycloheximide (see Materials and Methods). This procedure determines the level of insoluble Cps1 before treatment and 15–60 min after cycloheximide treatment, the latter being indicative of clearance of the insoluble form via the MVB pathway. Overall, for both myc- and HA-tagged Cps1, we routinely observed significantly increased levels of detergent-insoluble Cps1 in the pellet fractions derived from all the mutants examined, relative to WT control cells (Figure 5 and Supplemental Figure S6). This increase occurred using either Triton X-100 or NP-40 as the detergent for solubilization and corresponded with a decrease in Cps1 in the soluble/supernatant fractions derived from these same strains (Supplemental Figure S6). We note that the decrease of Cps1 in the pellet fraction derived from WT cells likely results from the proteolytic processing of Cps1 to create the mature cleaved form (i.e., minus the epitope) that reaches the vacuole through the MVB pathway.
FIGURE 5:
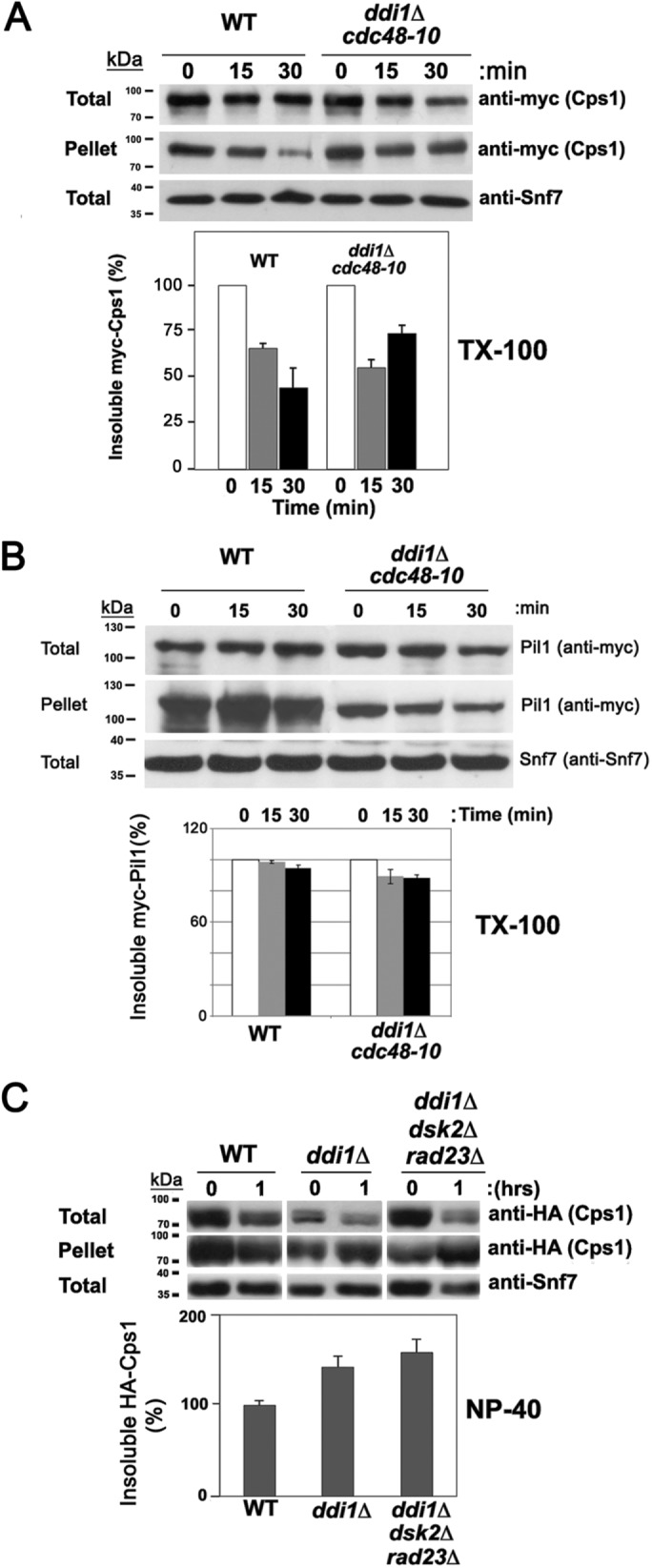
Cdc48 and the ubiquilins reduce the detergent-insoluble fraction of Cps1. (A) The detergent-insoluble fraction of Cps1 decreases substantially in WT but less so cdc48-10 ddi1Δ cells. WT and cdc48-10 ddi1Δ cells expressing endogenous myc-Cps1 from its genomic locus were subjected to a cycloheximide (CHX)-chase degradation/sedimentation assay to resolve the amount of Triton X-100 detergent-insoluble Cps1 in the pellet fraction, as described under Materials and Methods. Samples of were removed after 0, 15, and 30 min of CHX treatment before processing to determine the relative amounts of myc-Cps1 in the total, pellet, and supernatant (see Supplemental Figure S6A) fractions by Western analysis with anti-myc antibodies. Anti-Snf7 antibodies were employed to assess the levels of total protein loaded and in subsequent quantitative analyses used for normalization of the results (unlike actin, Snf7 is not degraded upon CHX treatment). The amount of insoluble Cps1 in the pellet was normalized to the loading control, after which the percentage was calculated relative to the amount at 0 h. A representative experiment is shown in the top panel. A histogram of the quantification of three repetitive experiments is shown beneath. kDa = kilodaltons. (B) The detergent-insoluble fraction of eisosome protein, Pil1, does not change in cdc48-10 ddi1Δ cells. WT and cdc48-10 ddi1Δ yeast expressing myc-Pil1 from its genomic locus were subjected to the cycloheximide-chase degradation/sedimentation assay in A to resolve the amount of Triton X-100 detergent-insoluble Pil1 by Western analysis using anti-myc antibodies as in A. A histogram of showing the results of three repetitive experiments is shown beneath. (C) The level of NP-40 detergent-insoluble Cps1 increases in ddi1Δ and ddi1Δ dsk2Δ rad23Δ cells. WT control cells (W303), ddi1Δ, and ddi1Δ dsk2Δ rad23Δ cells expressing HA-Cps1 were subjected to the same procedure as in A, and the percentage of insoluble Cps1 in the pellet fraction after 1 h was calculated after the normalization for gel loading. In the representative experiment, the level of NP-40-insoluble Cps1 increased in the ddi1Δ and ddi1Δ dsk2Δ rad23Δ pellet fractions by 34 and 78%, respectively. A histogram of three repetitive experiments is shown at the bottom.
For myc-Cps1, we specifically observed ∼60% more insoluble myc-Cps1 in the Triton X-100 pellet fraction derived from cdc48-10 ddi1∆ cells than in WT cells at 30 min after treatment (Figure 5A; average remaining from time 0 = 68.8 ± 5.6% and 43.0 ± 10.9%, respectively, n = 3 experiments; p = 0.04). This was concomitant with a ∼30% decrease of myc-Cps1 in the supernatant of cdc48-10 ddi1∆ cells relative to WT cells after normalization for loading (Supplemental Figure S6A; average remaining from time 0 = 66.7 ± 10.6% and 91.1 ± 7.2%, respectively, n = 3 experiments; p = 0.05). Similarly, we observed that the levels of HA-Cps1 increased by ∼70% and ∼65% in the NP-40–insoluble fraction of both cdc48-10 and cdc48-10 ddi1∆ cells (Supplemental Figure S6B; average remaining from time 0 = 163.4 ± 28.5% and 159.1 ± 22.1%, respectively, p = 0.02 and p = 0.015; n = 3) relative to WT cells, in which the level was essentially unchanged (97.2 ± 22.9%), after normalization for loading. Correspondingly, we observed somewhat lesser amounts of HA-Cps1 in the soluble fraction derived from cdc48-10 and cdc48-10 ddi1∆ cells (Supplemental Figure S6B; average remaining from time 0 = 72.1 ± 2.3%, p = 0.01 and 66.4 ± 1.6%, p = 0.05, respectively; n = 3) than that observed in WT cells (78.5 ± 3.3%, n = 3). Similar results were also observed for both cdc48-10 and cdc48-10 ddi1∆ cells shifted to 37°C (1 h; unpublished data). Thus, we observed an increase in insoluble Cps1 concomitant with a decrease in the soluble protein in cdc48-10 and cdc48-10 ddi1∆ cells, but not WT cells, on cycloheximide treatment.
As described above (Figure 5A and Supplemental Figure S6B), tagged Cps1 accumulates in the insoluble fraction on cycloheximide treatment in cdc48-10 and cdc48-10 ddi1∆ cells. To verify whether Cps1 indeed accesses a bona fide detergent-insoluble fraction, we examined the level of myc-tagged Pil1, an eisosome resident known to associate with detergent-insoluble aggregates, in the Triton X-100–insoluble and -soluble fractions after 30 min of cycloheximide treatment. We found that the levels of myc-Pil1 remained mostly steady in the Triton X-100–insoluble fraction of both WT and cdc48-10 ddi1∆ cells (Figure 5B; average remaining from time 0 = 94.6 ± 2.0 and 88.0 ± 2.3%, respectively, p = 0.04; n = 3). Thus, the loss of DDI1 and attenuation of CDC48 did not alter Pil1 recruitment/localization to detergent-insoluble aggregates. We also noted that the levels of myc-Pil1 in the detergent-soluble fraction (Supplemental Figure S6A) remained largely unchanged in WT cells (average remaining from time 0 = 108.4 ± 15.0%, p = 0.05; n = 3) but did decline in cdc48-10 ddi1∆ cells (66.1 ± 15.2%, n = 3) after treatment. However, unlike myc-Cps1, whose levels declined consistently in the soluble fraction in all experiments, we noted that changes in the levels of soluble myc-Pil1 were not always observed (e.g., see representative experiment in Supplemental Figure S6A). Thus, while Pil1 accesses a detergent-insoluble fraction along with Cps1, its localization and overall stability in the different cellular fractions is mainly independent of Ddi1/Cdc48 functions. We note that both bands of the (immature) Cps1 doublet are plainly visible in the soluble fraction derived from cdc48-10 ddi1∆ cells (Supplemental Figure S6A) and decrease in a similar manner after CHX treatment. Thus, the chase of these forms to the pellet fraction probably occurs using the same mechanism.
Experiments performed in parallel using ddi1∆ and ddi1∆ dsk2∆ rad23∆ cells (Figure 5C) demonstrated fairly similar results with tagged Cps1, as those seen with cdc48-10 and cdc48-10 ddi1∆ cells. Increases of ∼40% and ∼80% of NP-40–insoluble HA-Cps1 were observed (average remaining from time 0 = 133.6 ± 18.0% and 171.4 ± 26.7%, respectively, p = 0.02 and p = 0.003; n = 3) in ddi1∆ and ddi1∆ dsk2∆ rad23∆ cells after treatment for 1 h relative to WT cells, in which the level was largely unchanged (average remaining from time 0 = 96.4 ± 7.9%, respectively). Correspondingly, the levels of detergent-soluble HA-Cps1 derived from ddi1∆ and ddi1∆ dsk2∆ rad23∆ cells (Supplemental Figure S6C) were observed to decline by ∼25% and ∼50% (average remaining from time 0 = 73.9 ± 8.1% and 51.2 ± 11.2%, respectively, p = 0.01; n = 3) relative to WT cells, in which the level was essentially unchanged (98.9 ± 9.2%, p = 0.04; n = 3).
Overall, these results suggest that Cdc48 and the yeast ubiquilins act to disassemble insoluble Cps1 oligomers/aggregates for them to enter the MVB pathway and reach the vacuole. This appears to be independent of the endosomal accumulation of Cps1, as no increase in the levels of NP-40–soluble or -insoluble HA-Cps1 were observed in cells lacking VPS27 (vps27∆ cells) relative to WT (VPS27) cells (Supplemental Figure S6D; average remaining from time 0 = 68.2 ± 4.2% and 50.6 ± 2.2%, respectively, in vps27∆ cells [n = 3] and 76.2 ± 3.4% and 58.0 ± 2.8%, respectively, in WT cells [n = 3]). Moreover, as combined CDC48 and DDI1 mutations (e.g., cdc48-10 ddi1∆) did not necessarily result in additive increases in the levels of insoluble Cps1, it would seem likely that these proteins function together on the same process.
Rsp5 inactivation reduces the interaction between Ddi1 and Cps1
Since Cps1 is ubiquitinated by Rsp5 (Katzmann et al., 2004 and Figure 4D), we examined whether inactivation of the rsp5ts allele would influence the recognition of Cps1 by Ddi1 (Figure 6). Indeed, on shifting rsp5-326 cells to 37°C for 1 h, we observed a significant decline in the level of native Ddi1 that coprecipitated with HA-tagged Cps1 (e.g., –37.0 ± 0.1% of the level observed at 26°C; n = 2). In contrast, the amount of Ddi1 that coprecipitated with HA-tagged Cps1 in WT cells actually increased 40.0 ± 9.0% (n = 2) under the same conditions. Thus, Rsp5 inactivation lessens the ability of Ddi1 to bind to Cps1, suggesting that Rsp5-mediated ubiquitination of Cps1 mediates this interaction. That said, we also examined the binding of Ddi1 to GFP-Cps1K8R and GFP-Cps1K12R, which bear arginine substitutions in the cytosolic domain of Cps1, as well as a double lysine substitution mutant, GFP-Cps1K8,12R. However, these substitutions did not abolish the interaction with native Ddi1 in coimmunoprecipitation experiments (unpublished data). Thus, while Rsp5 function facilitates the Ddi1-Cps1 interaction, the precise residues involved were not revealed.
FIGURE 6:
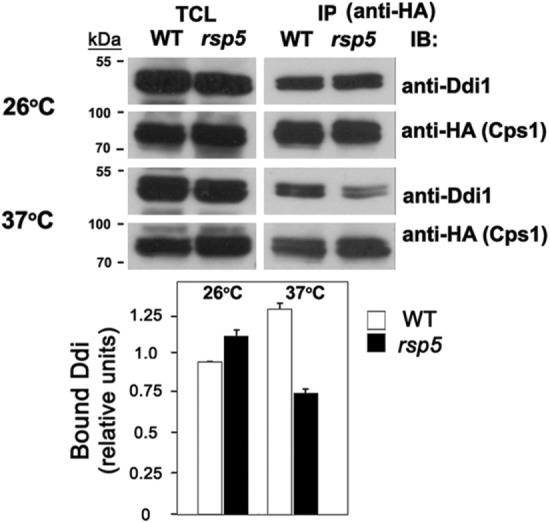
Cps1 binding to Ddi1 is Rsp5 dependent. WT cells (SEY 6210) and rsp5ts cells (SSY22) expressing HA-tagged Cps1 were grown and maintained at 26° or shifted to 37° for 1 h and were subjected to IP using monoclonal anti-HA antibodies. Proteins in the total cell lysate (TCL) and IP lanes were detected with anti-Ddi1 (1:5000) or anti-HA (1:1000) antibodies in immunoblots (IB).
Overexpression of ubiquitin does not restore Cps1 sorting in ddi1∆, cdc48-10, or cdc48-10 ddi1∆ cells
Since free ubiquitin levels may become depleted when ubiquitin-dependent processes are altered, we examined whether the overexpression of ubiquitin (UBL4) could alleviate defects in Cps1 sorting in ddi1∆, cdc48-10, or cdc48-10 ddi1∆ cells (Supplemental Figure S7). However, GFP-Cps1 remained limited to the vacuolar membrane or class E-like compartment even on the overexpression of ubiquitin in these mutants. Thus, it is unlikely that ubiquitin is limiting in the ddi1∆, cdc48-10, or cdc48-10 ddi1∆ mutants and would compromise the level of Cps1 ubiquitination and result in its missorting.
DISCUSSION
The Cdc48-Npl4-Ufd1 complex and Ddi1/Dsk2/Rad23 ubiquilins are proposed to mediate selective protein targeting to the proteasome for proteolytic degradation (Jentsch and Rumpf, 2007; Raasi and Wolf, 2007; Dantuma and Hoppe, 2012; Lee and Brown, 2012; Wolf and Stolz, 2012). We now show that these factors also mediate selective anterograde protein sorting to the vacuole via the MVB. We identified Ddi1 as an interacting protein and potential cofactor of Cdc48 (Figures 1A and 4, E and F, and Supplemental Figure S1A) and show that Cdc48, its core adaptors (Npl4, Ufd1), and both Ddi1 and Dsk2 are necessary for Cps1 hydrolase localization to the vacuolar lumen (Figures 1, C and D, and 2A, Supplemental Figure S1D, and Table 1). In contrast, this enzyme is not internalized and localizes to the vacuolar membrane on their diminution (i.e., mutation or deletion). This phenotype parallels that seen for mutants in MVB formation (Henne et al., 2011); hence, Cdc48 and its adaptors act effectively as MVB protein sorting elements. This is highly specific, as other ubiquitinated and subsequently MVB-targeted proteins, such as internalized Ste2 or Fur4 were not mislocalized (Supplemental Figure S2) and do not bind to Ddi1, unlike Cps1 (Figure 4A). The vacuolar localization of a soluble hydrolase, CPY, was also not affected by mutations in either DDI1 or CDC48, although combined ddi1Δ and cdc48-10 mutations led to a partial secretion phenotype (Supplemental Figure S3, C and F). This was also observed on the deletion of all three ubiquilins (Supplemental Figure S3E), indicating that their loss has an overall deleterious effect on vacuolar protein sorting.
In support of a role in protein entry into the MVB, both Ddi1 and Rad23 accumulate (and colocalize) on the LE compartment present in MVB mutants (Figure 2, B–D, and Supplemental S4, A and B). Similar results were observed in cdc48, npl4, and ufd1 mutants wherein GFP-Ddi1 localized to class E-like puncta adjacent to the vacuole (Supplemental Figure S4C). Thus, aside from its cytosolic and nuclear localization in WT cells (Gabriely et al., 2008; Supplemental Figure S4C), Ddi1 accumulates on a LE/class E-like compartment when MVB formation is blocked. On the other hand, Cdc48 labels the cytoplasm and small puncta that colocalize with specific endosome markers (i.e., Snx4; Figure 3A) but does not accumulate on the class E-like compartment (Figure 3B). Thus, we predict that Ddi1 associates with ubiquitinated Cps1 that accumulates on this compartment when the MVB pathway is blocked. Indeed, Rsp5 E3 ligase function is required for Cps1 ubiquitination (Figure 4D; Katzmann et al., 2004) and appears necessary for the interaction of Ddi1 with Cps1 (Figure 6). The interaction with Cps1 was verified by co-IP and appeared specific, as Ddi1 could not bind other MVB substrates (Figure 4A), and this was unchanged in the MVB mutants (Figure 4B). More importantly, the deletion of DDI1 (or the other ubiquilins) or a mutation in CDC48 did not affect the level of Cps1 ubiquitination relative to WT cells (Figure 4D). This again is in contrast to a mutation in RSP5, which blocked Cps1 ubiquitination, or a deletion in VPS27, which resulted in the accumulation of ubiquitinated Cps1 (Figure 4D). Thus, neither Ddi1 nor Cdc48 affects Cps1 ubiquitination, indicating that they act downstream thereof. Moreover, Cps1 missorting as a consequence of mutations in DDI1 or CDC48 does not appear to result from changes in the level of free ubiquitin, since ubiquitin overexpression does not rescue the phenotype of ddi1Δ, cdc48-10, or ddi1Δ cdc48-10 cells (Supplemental Figure S7).
Overall, our results show that Ddi1 and Cdc48 facilitate the dispersal of insoluble Cps1 oligomers (see the model in Figure 7). This is because immature (uncleaved) Cps1 accumulates in a detergent-insoluble fraction when DDI1, CDC48 or both are mutated (Figure 5, A and C, and Supplemental Figure S6B), concurrent with its localization on the MVB/vacuole membrane (Figures 1C and 2A and Supplemental Figure S1D; Table 1). The latter finding precludes the possibility that unfolded Cps1 (a potential ERAD substrate) might accumulate in the ER on CDC48 or DDI1 inactivation. Moreover, the deletion of all three yeast ubiquilins led to the same accumulation of insoluble Cps1 as in cdc48-10 cells (Figure 5C). Thus, their recognition of ubiquitinated Cps1 probably recruits Cdc48-Npl4-Ufd1 to the endosome membrane to create monomers that interact better with ESCRT-0 and, thereby, enter intralumenal vesicles (ILVs). This entry into ILVs might also promote the proteolytic processing of Cps1, since the apparent bypass of MVB entry (i.e., observed in ddi1Δ, cdc48-10, or ddi1Δ cdc48-10 cells) results in the accumulation of immature (and insoluble) Cps1 on the limiting membrane of the vacuole (Figures 1C, 2A, 4D, 5, A and C, and Supplemental Figure S6, A–C), likely indicating deficient cleavage. Importantly, this function of Cdc48 and the yeast ubiquilins is independent of conditions whereby Cps1 accumulates on the MVB endosomes due to defects in the MVB pathway alone (e.g., in vps27Δ cells; Supplemental Figure S6D). Thus, we conclude that in addition to their role in ERAD and proteasome-mediated proteolysis, Cdc48 and the ubiquilins play a biosynthetic role in the anterograde delivery of a hydrolase to the vacuole/lysosome for its proteolytic activation. This result might have been predicted since another adaptor, Ufd3/Doa1, was implicated in Cps1 sorting to the MVB (Ren et al., 2008). However, the necessity for either Cdc48 or ubiquilin function in facilitating Cps1 entry into the MVB in that work was not demonstrated. While we did observe an accumulation of multiple forms of ubiquitinated Cps1 in vps27Δ cells (Figure 4D) concomitantly with its perivacuolar accumulation (e.g., in vps23Δ cells; Figure 2C), this was not the case for cdc48-10 and ddi1∆ cells, in which Cps1 preferentially accumulates on the vacuolar membrane instead (Figures 1C and 2A) and appears no more ubiquitinated in the WT control cells (Figure 4D). Although not entirely clear at this point, it may be that Cps1 oligomers become deubiquitinated after their arrival to the vacuolar membrane, unlike in MVB mutants whereby Cps1 primarily accumulates on the perivacuolar compartment and not the vacuolar membrane.
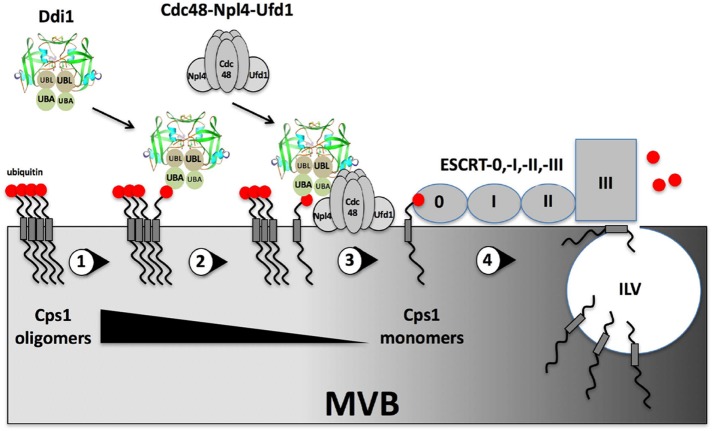
FIGURE 7:
A possible model for Ddi1-Cdc48 function in dispersing Cps1 oligomers and conferring MVB entry. (1) Ubiquitinated-Cps1 oligomers reside on the surface of the late endosome/MVB compartment. (2) The oligomers recruit Ddi1 via the UBA domain (note: the UBL and UBA domains of each monomer are arrayed anti-parallel, based on the RVP crystal structure [ Sirkis et al., 2006]). (3) Ddi1 recruits Cdc48-Npl4-Ufd1 via the UBL domain to dissolve oligomers. (4) Cps1 in its monomeric state is recognized by ESCRT-0 and handed off to the downstream ESCRT machinery (e.g., ESCRT-I, -II, and -III). Following de-ubiquitination, Cps1 enters intralumenal vesicles (ILVs) and accesses the vacuolar lumen on MVB fusion with the vacuole (not shown). Note: under conditions whereby Cdc48 and/or Ddi1 are dysfunctional (i.e., cdc48 and/or ddi1Δ cells; not shown), Cps1 may remain in an oligomeric state but reaches the vacuolar membrane, unlike in class E MVB mutants where it is primarily perivacuolar localized and ubiquitinated.
Initial structure–function studies suggest that the UBA/UBA-like domain (residues 363–428) is required for Ddi1 localization to a class E-like compartment/MVB (Figure 2D), the interaction with Cps1 (Figure 4C), and to some degree with Cdc48 (Figure 4E). Thus, the interaction of the Ddi1 UBA with ubiquitinated cargo appears to confer protein localization and an ability to engage the Cdc48 complex. In contrast, the UBL and RVP domains are not necessary for localization, though the UBL is necessary to bind to Cdc48 (Figure 4E). This result is interesting in light of recent structural and biochemical studies showing that the Ddi1 UBL is unique among those of the yeast ubiquilin-like proteins, as it is also capable of binding directly to ubiquitin (Nowicka et al., 2015; Trempe et al., 2016), as well as to the proteasome via Rpn1. This dual ubiquitin/proteasome binding function is facilitated by basic residues that surround the hydrophobic contacts on the surface of the UBL β-sheet and it was postulated (Nowicka et al., 2015) that these properties might allow Ddi1 to interact with either monoubiquitinated or polyubiquitinated substrates (and the proteasome) when dimerized in its head-to-tail conformation (Sirkis et al., 2006). Our results suggest that although the full role of the RVP/dimerization domain (unique to Ddi1) remains unclear, it is required for binding to Cps1 though not for the interaction with Cdc48. In fact, removal of the RVP domain or mutation of the catalytic aspartate improved the interaction of Ddi1 with Cdc48 (Figures 1A and 4E). Although dimerization is necessary for interactions with ubiquitinated substrate (e.g., Cps1; Figure 4C), apparently the monomer better recognizes Cdc48 through both the UBL and UBA domains. Although the interaction between Ddi1 and Cdc48 does not appear to be necessarily strong, it is consistent with both overexpression and endogenous expression studies (Figures 1A and 4, E and F, and Supplemental Figure S5A). The reason behind this is unclear and may relate to a low level of affinity or colocalization, for example. More work is required to elucidate the mechanism of substrate recognition and Cdc48 complex recruitment.
This is not the first prediction of ERAD- or proteasome-independent functions for Cdc48/p97 and the ubiquilins. ERAD-independent Cdc48/p97 functions in the nuclear environment include the clearance of San1-ubiquitinated misfolded proteins (Gallagher et al., 2014) and protein extraction from chromosomes (Dantuma and Hoppe, 2012) to facilitate proteasome-mediated degradation. Cdc48 was shown to act as a ubiquitin-selective chaperone that regulates nuclear entry of a membrane-bound transcription factor to control transcription (Rape et al., 2001) and plays a role in the proteasome-mediated degradation of mitochondrial-associated membrane proteins (MAD) (Karbowski and Youle, 2011). In contrast to these proteasome-dependent roles, Cdc48/p97 also acts on endolysosomal routes. For instance, the targeting of stress granules and P-bodies for degradation via autophagy necessitates Cdc48/p97 (Buchan et al., 2013). Likewise, the sorting of internalized EEA1 and monoubiquitinated caveolin to endolysosomes is p97 dependent (Bug and Meyer, 2012), and Cdc48/p97 attenuation results in defects in autophagosome biogenesis (Ju et al., 2009; Krick et al., 2010; Bug and Meyer, 2012), while even earlier studies suggested nondegradative roles for Cdc48/p97 in homotypic ER, Golgi, and nuclear envelope membrane fusion (Meyer et al., 2012). Importantly, Cdc48/p97/VCP has been shown to play a preventative role in protein aggregation connected to inclusion body myopathies (Yamanaka et al., 2012) and to prevent the aggregation of polyQ-expanded huntingtin exon 1 (Nishikori et al., 2008) and TAR DNA-binding protein 43 (TDP-43) (Ritson et al., 2010), for example.
Ubiquilins also have non-ERAD or -UPS roles. For example, endosomal proteins containing ubiquitin-interacting motifs (e.g., Eps15, Hrs) interact with the UBL domain of ubiquilin (Regan-Klapisz et al., 2005), and UBXD1 (along with p97) is required for the endolysosomal sorting of caveolin (Bug and Meyer, 2012). Mammalian Rad23 does not bind to ERAD-related erasin, which suggests that it confers alternative functions (Lim et al., 2009) and, like Cdc48/p97, ubiquilins facilitate autophagy by promoting autophagosome maturation (Rothenberg and Monteiro, 2010; Lee and Brown, 2012).
Importantly, Cdc48/p97 and the mammalian ubiquilins are associated with neurodegenerative disease. Defects in the p97 and UPS system correlate with inclusion body myopathies associated with systemic degenerative diseases, such as Paget’s disease of bone and frontotemporal dementia (Ju and Weihl, 2010; Yamanaka et al., 2012), as well as neuropathies, such as Parkinson’s disease (PD), spinocerebellar ataxia, ALS, and AD (Franz et al., 2014; Parakh and Atkin, 2016). Global changes in protein quality control are incurred by mutations in p97 and, together, have cumulative effects leading to deficient proteostasis (Amm et al., 2014). Ubiquilins concentrate in inclusion bodies associated with AD, PD, and ALS, and which result from inefficient ERAD (Haapasalo et al., 2010; Zhang et al., 2014), as well as with huntingtin poly-Q expansion aggregates (Rutherford et al., 2013). In the case of AD and ALS, mutations in UBQLN1 and UBQLN2 are genetically linked to disease susceptibility (Haapasalo et al., 2010; Deng et al., 2011; Zhang et al., 2014; Parakh and Atkin, 2016). In AD, ubiquilin-1 interacts with amyloid precursor protein (APP) and influences both its levels and sorting to the plasma membrane (Haapasalo et al., 2010). Ubiquilin-1 also interacts with presenilin, the catalytically active component involved in APP processing, and to control its levels, functionality, and aggresome formation (Haapasalo et al., 2010; Viswanathan et al., 2011). Finally, ubiquilin potentiates the aggregation of ubiquitinated TDP-43 in cell culture models (Kim et al., 2009; Deng et al., 2011). Thus, p97 and the ubiquilins are both strongly connected to proteostasis disorders.
We show in this study that Cdc48 and the yeast ubiquilin-like proteins have additional functions that relate to selective anterograde endolysosomal protein transport. Although mammalian Ddi1 lost its UBA domain during evolution, its potential to homodimerize (Diaz-Martinez et al., 2006; Gabriely et al., 2008), interact directly with ubiquitin via the UBL (Nowicka et al., 2015; Trempe et al., 2016), and heterodimerize with other UBA-containing ubiquilins (e.g., Rad23) (Bertolaet et al., 2001) may allow for similar transport steps to be affected in Cdc48/p97- and ubiquilin-affiliated neuropathies. Whether these disorders arise due to defects in proteasome-mediated protein degradation (i.e., ERAD, MAD, nuclear quality control), proteasome-independent degradation (i.e., autophagy, endocytic sorting), biosynthetic endolysosomal sorting (as shown here), or all of the above is still unclear. Unraveling the many roles of Cdc48/p97 and its adaptors in cellular proteostasis is essential to understand why neurons are sensitive to mutations therein.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains used in this study are listed in Supplemental Table S1. Yeast were grown in standard growth media containing 2% glucose; synthetic complete (SC), and drop-out media were prepared as described (Haim-Vilmovsky and Gerst, 2009), while rich growth medium (yeast extract, peptone, dextrose; YPD) and an amino acid–enriched SC medium were prepared according to Rose et al. (1990). Standard methods were used for the introduction of DNA into yeast and the preparation of genomic DNA (Rose et al., 1990). Plasmids used in this study are listed in Supplemental Table S2. The tagging of genes at their genomic loci with GFP or the deletion of genes was performed by homologous recombination using PCR products bearing flanking regions complementary to the appropriate genomic sequences that were generated by PCR with specific primers to genome-tagging plasmids, used as templates (Longtine et al., 1998).
Sequencing of the cdc48-10 allele
The cdc48-10 allele was amplified from the cdc48-10 strain (KFY194) and sequenced. Mutations in the D1 domain (P257L [CCT to CTT] and T413R [ACA to AGA]) and D2 domain (H509Y [CAT to TAT]) were identified.
Growth assays
Yeast were grown on synthetic minimal and synthetic rich growth media. For growth tests on plates, yeast were grown to mid–log phase, normalized for optical density (O.D.600), diluted serially (×10), and plated by drops onto solid medium preincubated at different temperatures.
Fluorescence microscopy
GFP and RFP fluorescence in strains expressing the appropriate GFP- and RFP-tagged fusion proteins was visualized by confocal microscopy. Nuclear staining with Hoechst dye (Hoechst 33342; Molecular Probes; 50 μg/ml) was performed for 10 min at room temperature prior to visualization. For vacuole staining, cells were pulsed with 5.4 μM of FM4-64 (Molecular Probes) for 30 min in the dark at 26°C. Following the pulse, a chase of 20 min in medium lacking FM4-64 was performed. Labeled cells were observed by confocal microscopy. In the figures, Merge indicates either merger of the GFP and FM4-64 windows or GFP and RFP windows, where appropriate. Light indicates the differential interference contrast (DIC) window. Size bars = 1 μm. Images were captured with a Zeiss LSM710 confocal imaging system mounted on an AxioObserver inverted microscope and using a Plan-Apochromat 63×/1.40 N.A. objective. Image acquisition was accomplished at 26°C using the detection system and accompanying software provided by the manufacturer. Adobe Photoshop was used to assemble and size the images for figure preparation.
Immunological assays
Antibodies.
Detection in Western blots was performed using monoclonal anti-HA (1:1000; Covance), monoclonal anti-myc (1:1000; 9E10; Santa Cruz Biotechnology), monoclonal anti-GFP (1:100; Roche), monoclonal anti-ubiquitin (P4G7) (Covance), and polyclonal anti-CPY (1:5000; Abcam), anti-Cdc48 (anti-VCP, ab138298; 1:500; Abcam), and anti-Ddi1 (1:5000; Lustgarten and Gerst, 1999) antibodies.
Coimmunoprecipitation and mass spectrometry.
Following transformation, yeast were grown in synthetic media at 26°C and cells were harvested in the mid–logarithmic phase. Yeast were lysed in 1 mM EDTA, 10 mM Tris-HCl, pH 7.5, 150 mM NaCl containing 0.5% Nonidet P-40 (TE/NP-40 buffer), aprotinin (10 μg/ml), leupeptin (10 μg/ml), soybean trypsin inhibitor (10 μg/ml), pepstatin (10 μg/ml), and 100 μM phenylmethylsulfonylfluoride (PMSF). Next, 25 O.D.600 units were taken for coimmunoprecipitation (co-IP) in 1 ml of TE/NP-40 buffer with 10 μl of monoclonal anti-HA antibody (Roche) by incubation on rotator overnight at 4°C. Next, Ddi1 complexes were immunoprecipitated with a 50-μl suspension of washed Protein-G Agarose beads (Santa Cruz Biotechnology) prewashed in TE/NP-40 buffer and incubated with rotation for 2 h at 4°C. Beads were washed ×5 with 1 ml of TE/NP-40 buffer and Ddi1 precipitates were eluted by the addition of 50 μl sample buffer (0.18 M Tris-HCl, pH 6.8, 4.7% SDS, 0.3% Bromophenol Blue, 0.42M β-mercaptoethanol). The eluted proteins were resolved by SDS–PAGE, and gel was stained using Imperial Protein Stain (Pierce). The band observed in the Ddi1D220A lane at ∼120 kDa, and the corresponding regions in the vector and native Ddi1 (Ddi1WT) lanes (marked by arrow [ Figure 1A]) were excised from the gel and subjected to mass spectrometry analysis. Peptides corresponding to Cdc48 are listed in Supplemental Figure S1A.
Immunoprecipitation and Western analysis.
Interactions between native, HA-, GFP-, or myc-tagged proteins and other proteins present in cell lysates were monitored by immunoprecipitation (IP) from cell extracts, as described in Kama et al. (2011). Samples of total cell lysates (TCLs; 30 μg protein per lane) and immunoprecipitates obtained from 500 to 1000 μg protein of lysate (per IP reaction) were resolved by electrophoresis and detected by Western blotting. Detection was performed by chemiluminescence using secondary anti-mouse (1:10,000; GE Healthcare Life Sciences), anti-rabbit (1:10,000; GE Healthcare Life Sciences), and anti-mouse Igκ light chain (1:10,000; BD Pharmingen) peroxidase-conjugated antibodies. Quantification of the bands was done using GelQuant.NET 1.6.6 and Excel.
Immunoblot assay for CPY secretion.
Yeast were either plated or spotted (as 10-fold serial dilutions of cells) on YPD plates at 26°C and grown for 24 h prior to replica plating onto nitrocellulose filters (Schleicher and Schuell, BA-S85). The filters were placed yeast-side-up on a fresh YPD plate and grown for an additional day at 26°C. Immunoblotting assay for CPY secretion was performed using standard procedures. Cells were first removed by three washes of the filters for 5 min each with phosphate-buffered saline (PBS). No cell lysis was observed on either plates or filters using this technique. Filters were then blocked in 5% nonfat dry-milk in 0.1% Tween-20/PBS for 1 h. After blocking, filters were probed with polyclonal anti-CPY antibodies (1:5000; Abcam) and detected by enhanced chemiluminescence (ECL) using anti-rabbit peroxidase-conjugated antibodies (1:10,000; Amersham Biosciences).
Cycloheximide-chase Cps1 degradation and sedimentation assays
HA- or GFP-Cps1 distribution to a detergent-insoluble fraction was assayed using a cycloheximide-chase degradation assay coupled with sedimentation in the presence of detergent, as adapted from Gallagher et al. (2014) for use with NP-40 and as adapted from Kanneganti et al. (2011), for use with Triton X-100.
For NP-40 insolubility assays, cells were grown to mid–log phase in liquid synthetic medium, and 5 O.D.600 units of cells were taken for each strain and time point examined. Cells were either left untreated with cycloheximide (zero time; 0 h) or were grown for 1 h in the presence of 50 μg/ml cycloheximide at 26°C. Cells were then harvested and lysed in 200 µl Lysis buffer A (0.1% NP-40, 100 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 5% glycerol, 1 mM PMSF, and EDTA-free complete protease inhibitor [Roche]) by vortexing with 100 µl of 0.5 mm acid-washed glass beads for 15 min at 4°C. To remove unlysed cells, lysates were centrifuged at 300 × g for 1 min at 4°C. Protein concentration was determined using the Pierce MicroBCA protein assay kit (Thermo Scientific). Next, 30-µg aliquots of each of the total cell lysates (TCLs) were removed and added to 40 µl Lysis buffer-SUMEB (1:1) buffer (the latter composed of 1% SDS, 8 M urea, 10 mM MOPS, pH 6.8, 10 mM EDTA, 1 mM PMSF, 0.01% bromophenol blue). In parallel, TCL aliquots (150 µg protein each) for each strain/time point were centrifuged at 12,800 × g for 30 min at 4°C. The supernatant (representing the “soluble” fraction) was added to 100 µl SUMEB. The pellet (representing the “insoluble” fraction) was resuspended in 40 µl (Lysis buffer-SUMEB [1:1]). All samples were boiled for 5 min and clarified for 1 min by centrifugation at 13,000 × g.
For Triton X-100 insolubility assays, cells were grown and 10 O.D.600 units each taken for treatment without (0 h) or with cycloheximide (1 h), as described above. Cells were lysed in 200 μl Lysis buffer B (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 0.1 μM MG132, and EDTA-free complete protease inhibitor [Roche]) by vortexing with 100 μl of 0.5-mm acid-washed glass beads for 15 min at 4°C. Lysates were clarified and protein concentration determined, as described above. Ice-cold Triton X-100 (10%) was added to aliquots of the TCLs (350 μg protein each) to reach a final concentration of 1% in a volume of 200 μl. Samples were incubated on ice for 30 min and then centrifuged at ∼21,000 × g for 1 h at 4o to separate the detergent-insoluble pellet fraction from the detergent-soluble supernatant. Pellet fractions were in solubilized in 60 μl of 1× SDS–PAGE sample buffer followed by boiling for 10 min and clarification by centrifugation for 0.5 min at 13,000 × g.
Protein samples from the TCL (30 μg), supernatant (30 μg), and resuspended pellet were resolved on 9% SDS–PAGE gels, transferred to nitrocellulose, and immunoblotted with monoclonal mouse anti-HA (1:1000) and anti-SNF7 (1:2000) rabbit polyclonal antibodies (gift of M. Babst, University of Utah, Salt Lake City, UT). Quantification of the bands was done using GelQuant.NET 1.6.6 and Excel.
To examine the insolubility of endogenous Cps1 and Pil1, we utilized a somewhat different aggregation assay, performed according to Alberti et al. (2010). Units of cells (10 O.D.600 each) were taken for treatment either without (i.e., 0 min) or with (e.g., 15 min and 30 min) added cycloheximide, as described above. Cells were lysed in 300 μl Lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 5% glycerol, 0.1 μM MG132, and EDTA-free complete protease inhibitor [Roche]). The suspension was transferred to a 1.5-ml microfuge tube containing 100 μl of 0.5-mm acid-washed glass beads, and the cells were lysed using a bead beater at 4°C. Afterward, 300 μl of ice-cold buffer B (50 mM Tris, pH 7.0, 150 mM NaCl, and 1% Triton X-100) was added, and the lysate was vortexed for 10 s. Subsequently, the crude lysate was centrifuged for 2 min at 800 × g (at 4°C) to pellet the cell debris, and the protein concentration was determined, as described above. Sedimentation was performed by centrifuging aliquots of the TCLs (e.g., 350 μg protein each) at 14,000 × g for 1 h at 4°C to separate the detergent-insoluble pellet fraction from the detergent-soluble supernatant. The pellet was resuspended in 100 μl of a 1:1 mixture of lysis buffer and buffer B containing protease inhibitors. Protein samples from the TCL (30 μg), supernatant (30 μg), and resuspended pellet (40 μl) were resolved on 9% SDS–PAGE gels, transferred to nitrocellulose membranes, and immunoblotted with mouse monoclonal anti-Myc (1:1000) or anti-Snf7 (1:2000) rabbit polyclonal antibodies (gift of M. Babst). Quantification of the bands was done using GelQuant.NET 1.6.6 and Excel.
Supplementary Material
Acknowledgments
We thank M. Babst, S. Bar-Nun, P. Carvalho, O. Deloche, S. Emr, W. Huh, R. Piper, W. Prinz, M. Sharon, and C. Stefan for generously providing reagents. This work was supported by grants from the Israel Science Foundation (#358/10 and #717/14) to J.E.G. and a fellowship to V.K. from the National Contest for Life Foundation (NCL-Stiftung), Germany. J.E.G. holds the Bessen-Brender Chair in Microbiology and Parasitology.
Abbreviations used:
- AAA-ATPase
ATPases associated with diverse cellular activities
- Abs
antibodies
- AD
Alzheimer’s disease
- ALS
amytrophic lateral sclerosis
- APP
amyloid precursor protein
- CPY
carboxypeptidase Y
- DIC
differential interference contrast
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ESCRT
endosomal sorting complexes required for transport
- FM4-64
N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide
- GFP
green fluorescent protein
- HA
human influenza hemagglutinin
- IB
immunoblot
- ILVs
intralumenal vesicles
- IP
immunoprecipitation
- LE
late endosome
- MVB
multivesicular body
- PD
Parkinson’s disease
- PMSF
phenylmethylsulfonylfluoride
- RFP
red fluorescent protein
- RVP
retroviral protease domain
- SNARE
soluble N-ethylmaleimide sensitive factor activating protein receptor
- TDP-43
TAR DNA-binding protein 43
- UBA
ubiquitin-associated
- UBL
ubiquitin-like
- UBX
ubiquitin regulatory X
- UPS
ubiquitin-proteosome system
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-11-0652) on February 12, 2018.
REFERENCES
- Alberti S, Halfmann R, Lindquist S. (2010). Biochemical, cell biological, and genetic assays to analyze amyloid and prion aggregation in yeast. Methods Enzymol , 709–734. [DOI] [PubMed] [Google Scholar]
- Amm I, Sommer T, Wolf DH. (2014). Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta , 182–196. [DOI] [PubMed] [Google Scholar]
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. (2001). UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol , 417–422. [DOI] [PubMed] [Google Scholar]
- Breker M, Gymrek M, Schuldiner M. (2013). A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol , 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R. (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell , 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug M, Meyer H. (2012). Expanding into new markets–VCP/p97 in endocytosis and autophagy. J Struct Biol , 78–82. [DOI] [PubMed] [Google Scholar]
- Chuang KH, Liang F, Higgins R, Wang Y. (2016). Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol Biol Cell , 2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Heinen C, Hoogstraten D. (2009). The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) , 449–460. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Hoppe T. (2012). Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol , 483–491. [DOI] [PubMed] [Google Scholar]
- Deloche O, Schekman RW. (2002). Vps10p cycles between the TGN and the late endosome via the plasma membrane in clathrin mutants. Mol Biol Cell , 4296–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H. et al (2011). Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature , 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Martinez LA, Kang Y, Walters KJ, Clarke DJ. (2006). Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div , 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. (2009). Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem , 477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A, Ackermann L, Hoppe T. (2014). Create and preserve: proteostasis in development and aging is governed by Cdc48/p97/VCP. Biochim Biophys Acta , 205–215. [DOI] [PubMed] [Google Scholar]
- Gabriely G, Kama R, Gelin-Licht R, Gerst JE. (2008). Different domains of the UBL-UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol Biol Cell , 3625–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher PS, Clowes Candadai SV, Gardner RG. (2014). The requirement for Cdc48/p97 in nuclear protein quality control degradation depends on the substrate and correlates with substrate insolubility. J Cell Sci , 1980–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapasalo A, Viswanathan J, Bertram L, Soininen H, Tanzi RE, Hiltunen M. (2010). Emerging role of Alzheimer’s disease-associated ubiquilin-1 in protein aggregation. Biochem Soc Trans , 150–155. [DOI] [PubMed] [Google Scholar]
- Haim-Vilmovsky L, Gerst JE. (2009). m-TAG: a PCR-based genomic integration method to visualize the localization of specific endogenous mRNAs in vivo in yeast. Nat Protoc , 1274–1284. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. (2011). The ESCRT pathway. Dev Cell , 77–91. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HR. (2003). Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J , 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. (2007). Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway. Trends Biochem Sci , 6–11. [DOI] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. (2009). Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol , 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Weihl CC. (2010). Inclusion body myopathy, Paget’s disease of the bone and fronto-temporal dementia: a disorder of autophagy. Hum Mol Genet , R38–R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R, Kanneganti V, Ungermann C, Gerst JE. (2011). The yeast Batten disease orthologue Btn1 controls endosome-Golgi retrograde transport via SNARE assembly. J Cell Biol , 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R, Robinson M, Gerst JE. (2007). Btn2, a Hook1 ortholog and potential Batten Disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol Cell Biol , 605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti V, Kama R, Gerst JE. (2011). Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol Biol Cell , 1648–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Youle RJ. (2011). Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr Opin Cell Biol , 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. (2004). Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol Biol Cell , 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Mi K, Rao H. (2004). Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell , 3357–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Shi Y, Hanson KA, Williams LM, Sakasai R, Bowler MJ, Tibbetts RS. (2009). Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J Biol Chem , 8083–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Manno A, Kakizuka A. (2007). Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells , 889–901. [DOI] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M. (2010). Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol , 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Brown EJ. (2012). Ubiquilins in the crosstalk among proteolytic pathways. Biol Chem , 441–447. [DOI] [PubMed] [Google Scholar]
- Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, Rothenberg C, Wang H, Ye Y, Fang S, Monteiro MJ. (2009). Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol , 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Lustgarten V, Gerst JE. (1999). Yeast VSM1 encodes a v-SNARE binding protein that may act as a negative regulator of constitutive exocytosis. Mol Cell Biol , 4480–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash M, Gerst JE. (2003). Phosphorylation of the autoinhibitory domain of the Sso t-SNAREs promotes binding of the Vsm1 SNARE regulator in yeast. Mol Biol Cell , 3114–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. (2012). Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol , 117–123. [DOI] [PubMed] [Google Scholar]
- Morvan J, Froissard M, Haguenauer-Tsapis R, Urban-Grimal D. (2004). The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic , 383–392. [DOI] [PubMed] [Google Scholar]
- Nishikori S, Yamanaka K, Sakurai T, Esaki M, Ogura T. (2008). p97 Homologs from Caenorhabditis elegans, CDC-48.1 and CDC-48.2, suppress the aggregate formation of huntingtin exon1 containing expanded polyQ repeat. Genes Cells , 827–838. [DOI] [PubMed] [Google Scholar]
- Nowicka U, Zhang D, Walker O, Krutauz D, Castaneda CA, Chaturvedi A, Chen TY, Reis N, Glickman MH, Fushman D. (2015). DNA-damage-inducible 1 protein (Ddi1) contains an uncharacteristic ubiquitin-like domain that binds ubiquitin. Structure , 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parakh S, Atkin JD. (2016). Protein folding alterations in amyotrophic lateral sclerosis. Brain Res , 633–649. [DOI] [PubMed] [Google Scholar]
- Raasi S, Wolf DH. (2007). Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin Cell Dev Biol , 780–791. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. (2001). Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell , 667–677. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell , 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan-Klapisz E, Sorokina I, Voortman J, de Keizer P, Roovers RC, Verheesen P, Urbe S, Fallon L, Fon EA, Verkleij A. et al (2005). Ubiquilin recruits Eps15 into ubiquitin-rich cytoplasmic aggregates via a UIM-UBL interaction. J Cell Sci , 4437–4450. [DOI] [PubMed] [Google Scholar]
- Ren J, Pashkova N, Winistorfer S, Piper RC. (2008). DOA1/UFD3 plays a role in sorting ubiquitinated membrane proteins into multivesicular bodies. J Biol Chem , 21599–21611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ. et al (2010). TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci , 7729–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. (1990). Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Rothenberg C, Monteiro MJ. (2010). Ubiquilin at a crossroads in protein degradation pathways. Autophagy , 979–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Lewis J, Clippinger AK, Thomas MA, Adamson J, Cruz PE, Cannon A, Xu G, Golde TE, Shaw G. et al (2013). Unbiased screen reveals ubiquilin-1 and -2 highly associated with huntingtin inclusions. Brain Res , 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. (2008). UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci , 2360–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirkis R, Gerst JE, Fass D. (2006). Ddi1, a eukaryotic protein with the retroviral protease fold. J Mol Biol , 376–387. [DOI] [PubMed] [Google Scholar]
- Song C, Wang Q, Li CC. (2007). Characterization of the aggregation-prevention activity of p97/valosin-containing protein. Biochemistry , 14889–14898. [DOI] [PubMed] [Google Scholar]
- Spormann DO, Heim J, Wolf DH. (1992). Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J Biol Chem , 8021–8029. [PubMed] [Google Scholar]
- Trempe JF, Saskova KG, Siva M, Ratcliffe CD, Veverka V, Hoegl A, Menade M, Feng X, Shenker S, Svoboda M. et al (2016). Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family. Sci Rep , 33671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan J, Haapasalo A, Bottcher C, Miettinen R, Kurkinen KM, Lu A, Thomas A, Maynard CJ, Romano D, Hyman BT. et al (2011). Alzheimer’s disease-associated ubiquilin-1 regulates presenilin-1 accumulation and aggresome formation. Traffic , 330–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DH, Stolz A. (2012). The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim Biophys Acta , 117–124. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Sasagawa Y, Ogura T. (2012). Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta , 130–137. [DOI] [PubMed] [Google Scholar]
- Zhang KY, Yang S, Warraich ST, Blair IP. (2014). Ubiquilin 2: a component of the ubiquitin-proteasome system with an emerging role in neurodegeneration. Int J Biochem Cell Biol , 123–126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.