Abstract
Unlike adult mammals, adult frogs regrow their optic nerve following a crush injury, making Xenopus laevis a compelling model for studying the molecular mechanisms that underlie neuronal regeneration. Using Translational Ribosome Affinity Purification (TRAP), a method to isolate ribosome-associated mRNAs from a target cell population, we have generated a transcriptional profile by RNA-Seq for retinal ganglion cells (RGC) during the period of recovery following an optic nerve injury. Based on bioinformatic analysis using the Xenopus laevis 9.1 genome assembly, our results reveal a profound shift in the composition of ribosome-associated mRNAs during the early stages of RGC regeneration. As factors involved in cell signaling are rapidly down-regulated, those involved in protein biosynthesis are up-regulated alongside key initiators of axon development. Using the new genome assembly, we were also able to analyze gene expression profiles of homeologous gene pairs arising from a whole-genome duplication in the Xenopus lineage. Here we see evidence of divergence in regulatory control among a significant proportion of pairs. Our data should provide a valuable resource for identifying genes involved in the regeneration process to target for future functional studies, in both naturally regenerative and non-regenerative vertebrates.
Keywords: Optic Nerve Crush Injury, TRAP, Expression profile
Introduction
Optic neuropathies such as glaucoma typically lead to progressive and irreversible loss of vision due to injury to the retinal ganglion cell (RGC) axons (Fischer and Leibinger, 2012). While very different from glaucoma in the mode of axon damage, and severity and timing of RGC loss, animal models of optic nerve trauma have led to important insights into how RGCs respond to injury. Experiments in which rodent RGCs were co-cultured with the optic tectum, have shown that RGC neurons lose the potential to regenerate at embryonic day 18, two days prior to birth (Chen et al., 1997). In mammals, studies using the optic nerve crush injury and nerve trauma models have also shown most RGCs quickly die; those that attempt regeneration fail (Bernhardt, 1999). In mice, over 80% of RGCs die within the first week and by week 50 virtually all RGCs are dead (Leung et al., 2008). Injury to these axons triggers a series of molecular events leading to Wallerian degeneration of axons and the death of these cells (Wang et al., 2012). Once a CNS axon is severed, to regenerate it must repair damage to the membrane and then initiate assembly of a new growth cone. This process includes regulation of calcium signaling, cytoskeletal restructuring, axonal mRNA and protein transport, and up-regulation of mRNA translation (Bradke, 2012; Wang et al., 2012). While extrinsic factors such as accumulation of inhibitory myelin debris at the injury site (Fawcett, 2006; Fischer and Leibinger, 2012; Yin et al., 2009) and inflammatory responses (Benowitz and Popovich, 2011) can influence axonal regeneration, mammalian RGCs show intrinsic differences in their regenerative capacity from those of regeneration-capable animals (Fischer and Leibinger, 2012). Non-mammalian vertebrates such as fish and frogs retain the ability to regenerate their optic nerves and restore vision into adulthood (Sperry 1944; 1948; Gaze, 1959). Because of the high level of conservation among vertebrate genomes, elucidating the molecular mechanisms used to recover from injury in regeneration-capable model organisms helps us to identify key gaps in regeneration in mammals.
In order to regenerate vision after injury to the optic nerve, RGCs must either 1) die and be replaced by de novo RGCs or 2) be protected from cell death and then repair injury to the membrane and regrow their axons. Both strategies require the assembly of a growth cone, axon growth and navigation, and finally establishment of synapses with the appropriate targets in the brain. Many cold-blooded vertebrates such as frogs and fish follow this second strategy and retain the ability to regenerate the optic nerve into adulthood, making them ideal systems for exploring the dynamics of neuronal regeneration (Gaze, 1959; Sperry, 1944, 1948). In both frogs and fish, a number of regeneration-associated factors, such as gap43 and neurofilaments, are used to assess the progress of axonal regrowth (Diekmann et al., 2015; Zhao and Szaro, 1994). In Xenopus laevis, axons regrow across the site of injury as early as 5 days post crush, extend into lateral margins of the optic tectum by the 12th to 15th day, but can take several months to fully regain their vision (Zhao and Szaro, 1994). In frog tadpoles and zebrafish, RGC axons begin regrowth as early as 2-4 days following injury, have reached the optic chiasm by 5 days, and have recovered their vision 2-4 weeks post injury (Diekmann et al., 2015; Wilson et al., 1992). In both frogs (R S Beaver, 2001) and fish (Meyer et al., 1985), the majority of the axons that regenerate come from pre-existing RGCs that have re-grown from the site of injury. In Xenopus, up to 20% of RGCs die by two weeks after being crushed (Liu et al., 2012), while about 25% of RGCs in zebrafish have died by seven weeks (Zou et al., 2013). Recent profiling studies have identified key roles for the kruppel -like family of transcription factors in regeneration: the factors klf6 and klf7 are positive regulators in zebrafish while klf4 and klf9 are negative regulators in mammals (Moore et al., 2009; Veldman et al., 2007). Other cell-autonomous molecules shown to play a role in optic nerve regeneration include members of the Jak/Stat signaling pathways (Elsaeidi et al., 2014; Smith et al., 2009) and the mTor pathway (Kurimoto et al., 2010; Park et al., 2008), the nitric oxide (NO)-cGMP signaling pathways (Koriyama et al., 2011) and Rho-GTPase pathway (Lehmann et al., 1999).
The heterogeneous nature of the CNS, which contains hundreds of different cell types, physically intertwined and morphologically similar, has posed a formidable technical and scientific challenge (Emery and Barres, 2008). Moreover, post-transcriptional regulation of mRNAs, which includes transport to axons or dendrites for localized translation and RNA storage, means that the total mRNA population of a neuron may not be representative of the pool of transcripts being actively translated (Holt and Schuman, 2013). Solutions for isolating a single cell type include laser capture single cell microdissections, fluorescence activated cell sorting, immunopanning, and more recently translational ribosomal affinity purification (TRAP) (Heiman et al., 2008; Okaty et al., 2011). In TRAP, a transgenic animal line is designed that expresses an epitope tagged variant of a ribosomal subunit. For example, strategies have included eGFP-tagged rpl10a or HA-tagged rpl22 (Heiman et al., 2008, 2014; Sanz et al., 2009). This allows rapid isolation of ribosome-associated mRNAs from only those cells expressing the epitope tag. In this study, eGFP tagged rpl10a is expressed under an RGC-specific promoter, allowing us to isolate mRNAs from RGC cell-bodies and associated dendrites. As others have reported for different cells in various animal models, we have previously found that this technique provides a rapid and robust method for quantifying gene expression in the RGCs of Xenopus laevis (Jiao and Meyerowitz, 2010; Mustroph et al., 2009; Thomas et al., 2012; Yoon et al. 2012; Watson et al., 2012).
Here, we have coupled TRAP with RNA-Seq analysis to generate time-resolved expression profiles for adult Xenopus laevis RGCs recovering from optic nerve crush injury. These profiles reveal clear patterns of temporal-regulation in the days following nerve crush. Because our analysis is specific to RGCs, the transcript profiling is of sufficiently high resolution to reveal clear functional specificity in the groups of up- and down-regulated genes. These data significantly enhance our understanding of the molecular mechanisms that underlie the largely uncharacterized optic nerve regenerative response in Xenopus laevis. We find key transition points in the recovery and regrowth programs between the 1st and 3rd day post-injury and then again between the 3rd and 7th. By mapping our RNA-Seq results to the new Xenopus laevis 9.1 genome assembly, we also find strong evidence of divergent regulation of homeolog-pairs across this time course. Finally, the pattern of global changes in gene expression at these key transition points suggests that RGC regrowth requires a return to an earlier developmental stage, with cell-type specific factors down-regulated in favor of increased biosynthetic capacity and the expression of developmental initiators. In addition to providing access to the raw and processed data for this study through a public repository (GEO), we have also provided interactive access to the data through a custom web application that makes it easy to filter and download subsets. We hope these resources will facilitate exploration of our results.
Material and Methods
Frogs
Restriction enzyme mediated integration (Kroll and Amaya, 1996) was used to create lines of Xenopus laevis that stably express TRAP transgenes in RGCs (Watson et al., 2012). Biologically independent experiments were each carried out using F1 progeny from single Tg(Islet2b:EGFP-RPL10a) female founder lines (Watson et al., 2012). These progeny were screened for presence of the eGFP-rpl10a transgene using a fluorescent dissecting microscope. eGFP positive progeny were then grown to post-metamorphic stage (> 6 months) under 12L:12D photoperiod at 22° C. All animal experiments were carried out using procedures approved by the Washington and Lee University and Johns Hopkins University School of Medicine’s IACUCs.
Optic Nerve Crush
The retinas from each of 10 post-metamorphic transgenic Tg(Islet2b:EGFP-RPL10a) Xenopus laevis frogs, 3.5 - 5.0 cm in length, were either left untreated (naïve) or underwent a monocular surgery (operated; Fig. 1C). Operated individuals were anesthetized with 0.05% ethyl 3-aminobenzoate methanesulfonate (Sigma, USA) and received either a sham surgery (sham) or a crush injury (crush) to the right optic nerve, and no treatment the left optic nerve (control; Fig. 1C). To avoid the surrounding vasculature, an initial surgical incision was made in the right medial roof of the buccal cavity, the muscles were separated, and the optic nerve was exposed at a 45° angle. The exposed nerve was gently separated from the adjacent ophthalmic artery and #55 forceps were used to perform a five second crush located approximately 5 mm from the optic nerve head. Crush injuries were verified by visual inspection, observing a region of clear sheath flanked by opaque areas. Frogs with extensive bleeding were excluded from further study. Following the surgery, frogs were allowed to fully recover from anesthesia in a shallow bath of 0.1x MMR (10 mM NaCl, 0.2 mM KCl, 0.1 mM MgCl2, 0.2 mM CaCl2, 0.5 mM HEPES; pH 7.5) before being immersed in 0.1x MMR. The ten untreated (naïve) frogs were also moved to individual tanks for a day. At 24 hours post-surgery, frogs were transferred to 18.25″ × 12″ × 6.25″ communal tanks with a maximum of ten frogs per tank. Surgically manipulated Tg(islet2b:GFP) frogs used to asses RGC axonal regrowth were allowed to recover for 1, 3, 7, 11, 21, 35, 70, 120, and 210 days while Tg(islet2b:eGFP-L10a) frogs used for TRAP were allowed to recover for 1, 3, 7, or 11 days post optic nerve crush. Following the crush surgery at these time points, the frogs were euthanized by 0.5% ethyl 3-aminobenzoate methanesulfonate followed by decapitation.
Figure 1. Profiling retinal ganglion cell regeneration.
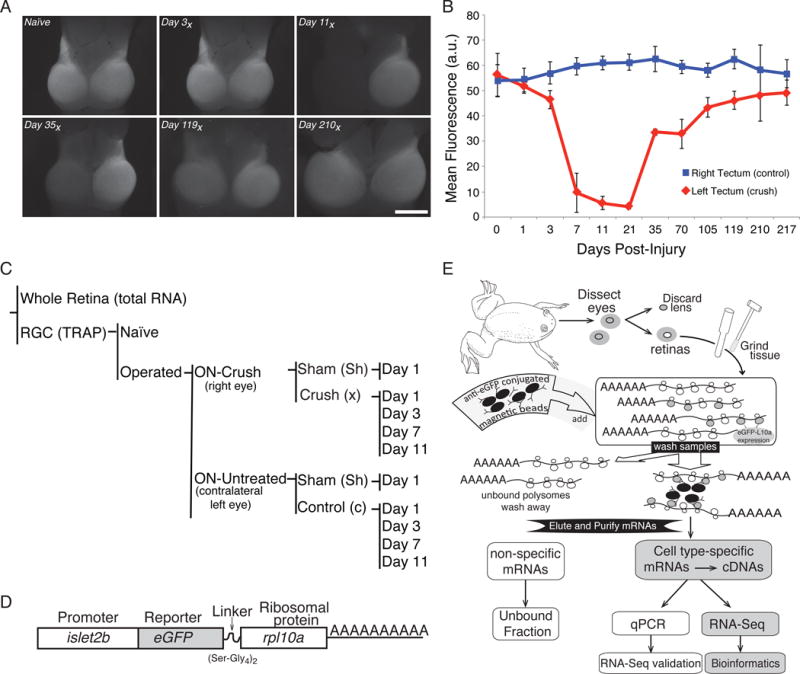
(A) The effects of axonal injury on retinal ganglion cells (RGCs) in the optic tectum can be visualized using frog lines expressing GFP under the control of an RGC-specific promoter (islet2b). An example time series shows the key transition point falls between 3 and 7 days post-injury, with full recovery occurring by 210 days (210x) post-injury. (B) Quantification of mean GFP fluorescence intensity in the tectum, as seen in panel (A). Data were averaged from at least 5 biological replicates per day and error bars represent the standard deviation from the mean. (C) In this study, gene expression in RGCs is directly compared between a right eye in which the optic nerve has undergone a surgical crush (Crush) to the untreated left eye of the same animal (Control) for various days after surgery (1, 3, 7, 11). Additional controls include a sham surgery (Sham), non-surgical animals (Naive), and RNA from whole retina (Total RNA). (D) To allow for tissue specific isolation of ribosome-associated mRNAs from RGCs, a transgenic line of Xenopus laevis is used that expresses an eGFP tagged variant of rpl10a under the control of an RGC-specific promoter (islet2b). (E) Following retina dissection, ribosome-associated RNAs in RGCs are purified using eGFP coated magnetic beads; subsequent poly(A) selection enriches for mRNA species. This mRNA fraction is then used for RT-qPCR validation and RNA-Seq library construction.
Tectal Fluorescence Recovery Following Injury
Progeny from a single maternal line of frogs expressing the GFP gene under a RCC specific promoter, Tg(islet2b:GFP), were used to characterize the regrowth of axon following injury and confirm the crush injury. At the various time points, frogs were terminally anesthetized (0.5% ethyl 3-aminobenzoate methanesulfonate) and the optic nerve and tectum were dissected and imaged immediately using a fluorescence stereomicroscope with a black and white camera cooled for fluorescence (DS-Qi1Mc; Nikon, USA). To compare the images from different animals, all images were taken using the same exposure, magnification, and gain settings. Quantitative analysis was performed using ImageJ (Abramoff and Magelhaes, 2003). A 300-pixel-radius rolling-ball subtraction algorithm was used to remove background noise (Sternberg, 1983). For each time point, the mean fluorescence level over the area (µm2) was compared between the left tecta (corresponding with the right optic nerve crush) and the right tecta (corresponding with the left unoperated optic nerve). The same measurements were made for the sham animals where the left tecta corresponds with the right optic nerve sham and the right tecta with the unoperated left optic nerve. Significance between the two treatment groups was measured for each time point using two-factor ANOVA and Tukey’s post-hoc test.
In Situ Hybridization
Freshly dissected eyes were fixed in 4% MEMPFA (0.1 MOPS (pH 7.4), 2 mM EGTA, 1 mM MgSO4, 4% paraformaldehyde) overnight at 4°C, infiltrated with 30% sucrose in PBS for 24 hours. For each of the time points, both right (crush) and the left (control) eyes from each of two or three WT individuals were cryo-embedded into the same mold using Shandon M-1 embedding matrix (Thermo Scientific Inc.), stored at −80°C and cryosectioned at 14 μm thickness. Hybridization was carried out with hydrolyzed digoxigenin labeled RNA probes transcribed from cDNAs as described previously (Zhang et al., 2008) (see Table S1), most of which were commercially available (Open Biosystems and ThermoFisher). Detection was carried out using a fluorescent peroxidase substrate, cy3-tyramides (Perkin Elmer). Quantification of the uchl1 in situ hybridization signal in retina sections was carried out by manually segmenting the retinal ganglion cell layer and half of the inner plexiform layer based on the DAPI signal in an average of two sections per retina. This was followed by multiple segmentation based quantifications as previously described (Mills et al., 2015), reporting values for the most representative fold-change at the given statistical significance value, as determined by a 2-tailed Student’s t-test.
Immunofluorescence
Tissue sections stored at −80° C, were dried at room temperature for 20 minutes, rinsed in 1x PBS (10 mM PO43−, 137 mM NaCl, and 2.7 mM KCl) for 10 minutes, permeabilized by two sequential 10 minute washes in 1x PBT (1x PBS, 0.1% Triton, 0.1%BSA), and blocked for 30 minutes by immersion in 10% normal goat serum in 1x PBT (1x PBS, 0.1% Triton, 0.1% bovine serum albumin) at room temperature. Antibody labeling was carried out as previously described (Zhang et al., 2008). Briefly, we used an anti-cleaved caspase-3 (ASP175) (Cell Signaling; at 1:300 dilution) and an affinity purified rabbit polyclonal antibody generated against the 14 terminal amino acids (EATESTEQVGDGEN) of X. tropicalis γ-synuclein (Covance; at 1:20,000 dilution; Watson et al., 2012) and incubated overnight at 4° C. A secondary antibody conjugated to Alexa-546 (Life Technologies; at 1:1,000 dilution) was incubated at room temperature for 60 minutes. Sections were rinsed in three sequential 5-minute 1x PBT washes followed by three 5-minute 1× PBS washes, mounted with ProLong mounting media with DAPI (Life Technologies) and covered using No. 1.5 coverslips. Sub-cellular and cellular localization of the protein products was determined using epifluorescence microscopy (Nikon Eclipse 80i) and images were captured using a digital camera (Hamamatsu C4742-80 with imaging software Volocity 6.3). Fluorescent images of retinal cross sections from different treatments were acquired using the same gain and exposure settings. Quantitative analysis of fluorescence in RGCs from retinal cross sections from different treatments was carried out using ImageJ (Abramoff and Magelhaes, 2003) by line integration across the retinal ganglion cell layer based on the DAPI signal in an average of two sections per retina and four retinas per sample group.
Quantitative PCR
Quantitative PCR for 11 genes was carried out on the original RNA-Seq samples and from two additional biologically independent experiments using the same time points. Real time quantitative PCR (qPCR) was performed as previously described (Watson et al., 2012) with the exception of using an IQ5 Real-Time PCR Detection System (Bio-Rad). Briefly, RNA samples were reverse transcribed using oligo-d(T)20 and the SuperScript III kit (Life Technologies) according to the manufacturer’s instructions, using no more than 50 ng of TRAP isolated mRNA per sample. Real time quantitative PCR (qPCR) using 1 ng RNA per sample was carried out using SYBR-green compatible primers to amplify 90-200 bp amplicons using IQ SYBR green Supermix (Bio-Rad). Primers were designed using Primer 3 (Untergasser et al., 2012) and were selected based on their amplification efficiency and their ability to amplify a single dominant peak from a template specific plasmid cDNA (Table S2). To obtain absolute transcript levels, 3- to 8-point standard curves ranging from 101 to 107 plasmid copies were generated for each of the 10 amplicons. Technical replicates were pooled. To determine average expression levels, either two or three independent biological replicates were averaged.
TRAP
Left and right eyes were pooled separately. Retinas were isolated by removing the lens, then peeling the retina away from the retinal pigment epithelial layer (Fig. 1C). Freshly dissected retinas were immersed in ice-cold lysis buffer (20 mM HEPES KOH, pH 7.4, 5 mM MgCl2, 150 mM KCl) to which was added freshly prepared 100 μg/ml cycloheximide, 0.5 mM DTT, protease inhibitors (Roche Mini Complete, EDTA-Free) and 40 U/ml recombinant RNAsin (Promega). The translation ribosomal affinity purification (TRAP) protocol used to isolate and purify the RNAs specific to the RGCs has been described previously (Watson et al., 2012). To immunoprecipitate ribosomes and associated RNAs, equal amounts of anti-eGFP antibodies (19C8 and 19F7; Memorial Sloan-Kettering Monoclonal Antibody Facility) at a concentration of 100 µg of total anti-eGFP antibody per 375 µl of Dynal Protein G-magnetic beads (Life Technologies) were used. TRAP RNA samples were purified using Qiagen’s Micro RNeasy kit (Qiagen) as per the manufacturer’s instructions. Due to the small recovery of RNA, the quantity of RNA was assessed using a Quant-iT Ribogreen (Thermo Fisher Scientific) and the quality was assessed using microfluidic analysis (Agilent Technologies’ Bioanalyzer 1200 picochip). The unbound fractions containing the non-specific mRNAs from other retinal cell types were also collected but not sequenced. Total RNAs from whole naïve retinas were purified using an RNeasy kit as per the manufacturer’s instructions (Qiagen). Purified RNA samples were stored at −80° C. Three independent biological replicate experiments were collected over three days (36 samples).
RNA-Seq
To obtain a minimum of 120 ng/sample of purified TRAP-isolated mRNAs, equal quantities from each of the three independent biological replicates were combined to generate a single pooled sample. RNA samples were then sent to the Johns Hopkins Deep Sequencing Core for RNA-Seq cDNA library construction with poly(A) selection. These cDNA libraries were then sequenced using the Illumina Hi-Seq 2000 platform with 50 bp single-end reads at the Johns Hopkins Sequencing facility in March 2013. Twelve samples were run in six lanes with 2x multiplexing. Control and Crush samples for a given time point were run in the same lane.
Read Alignment and Expression Profiling
Raw sequencing reads were filtered for quality and adapter sequences using Trimmomatic (v. 0.33) with the following parameters: a 4-base sliding window with average quality cutoff of 15, removal of 3 leading and trailing low quality bases and a minimum read length of 36 bases (Bolger et al., 2014). For most of the analysis, except where noted, surviving reads were aligned to the Xenopus laevis genome using the JGI gene models version 9.1, retrieved from xenbase.org on 11/21/2015 (Session et al., 2016). Alternatively, to allow for comparison of alignment strategies, reads were also mapped onto a reference database of 30,592 Xenopus laevis and 41,042 Xenopus tropicalis transcripts from the NCBI EST database that had previously been mapped to genomic loci by the Xenbase project (Karpinka et al., 2015). For a comparison of alignment rates obtained with these two strategies, see Figure S1.
In both cases, read alignments were performed using Bowtie2 (v. 2.2.5) with ‘sensitive’ alignment presets augmented with a minimum acceptable alignment score of −0.1 (Langmead and Salzberg, 2012). Following alignment, gene expression estimation was performed using RSEM (v. 1.2.21) with default options (Li and Dewey, 2011). All subsequent analysis was performed in R using custom scripts (R Core Team, 2015).
Distance Analysis and Clustering
Distance matrices for pairwise distance analysis were calculated with the ‘’ClassDiscovery” package (Coombes, 2013), using Euclidean distance. K-means clustering and pairwise correlation analyses were performed using the default implementations in the base R ‘stats’ package (R Core Team, 2015). For K-means clustering, clusters were generated for k ranging from 2 to 16 (Fig. S2.a), with k = 5 being used for the visualizations (Fig. S3A, B). Hierarchical clusters were calculated using the ‘amap’ package (Lucas, 2014), with Euclidean distance and the ‘centroid2’ agglomeration method based on the gene expression matrix containing samples taken 3, 7, and 11-days post optic nerve crush.
Gene Ontology Annotation and Enrichment Analysis
Gene Ontology (GO) terms were associated with gene symbols from the JGI 9.1 gene models using two sources. First, the set of GO terms associated with earlier builds of the Xenopus laevis genome by Xenbase (Karpinka et al., 2015) were mapped to the new JGI 9.1 gene symbols by matching gene symbols in the former with the root names for assigned homeologs in the latter. Mismatches due to non-standard syntax were corrected. Using this approach, only 131 of the 13,771 genes mapped in our RNA-Seq dataset with assigned gene symbols failed to match to Xenbase symbols, resulting in GO annotations for 6,605 genes. Second, GO annotations from the human genome were used where gene symbols could be mapped unambiguously between the two genomes (ftp.geneontology.org). A combination of forward partial matching on homeolog symbols, and gene alias searching on failures, resulted in the annotation of 9,897 genes in the 9.1 set, of which 3,354 were genes not annotated by the Xenbase tables. This gave GO annotations for a total of 9,982 JGI 9.1 gene symbols. GO term enrichment analysis was performed on the set union of terms with the ‘goseq’ package (Young et al., 2010) using standard parameters and custom gene symbol to term mapping (Tables S3 and S4). See Data Access and Source Code below for access to the source code used to assemble our custom GO annotation reference.
Data Access and Source Code
All raw read files (FASTQ) and gene expression count results (FPKM) will be available as of the date of publication through GEO (GSE77724). Additionally, this dataset will be made available through an interactive data viewer that supports downloads of custom subsets of the data, hosted on the companion site: http://rna.wlu.edu/papers. To support emerging reproducible research standards, all source code used in the data analysis pipeline and to generate the figures shown here will also be hosted on this site and licensed under GNU GPLv3.
Results
To focus our study of gene expression in RGCs recovering from injury, we first characterized the time-line of axonal regrowth. After injury, axons regrow past the site of the initial nerve crush to reconnect to the contra lateral optic tectum, the principle visual target in frogs. To characterize the injury and regrowth periods, we used Xenopus laevis frogs expressing green fluorescence (GFP) under regulatory control of an RGC-specific promoter (Tg(islet2b:GFP)), which allows us to visualize the RGC cell bodies, dendrites and axons that make up the optic nerve. In this analysis, we see a modest effect of injury on fluorescence in the tectum from 1 to 3 days post- crush, followed by a significant loss of fluorescence between days 3 and 7 (p<10−4) (Fig 1.A, B). Fluorescence in the left tectum (crush) begins to recover by day 35 and is complete by day 210. While full recovery in the tectum occurs between 119 and 210 days post-injury, we would predict that changes in gene expression that occur during the initial injury response may be the most illuminating to compare between regenerative and non-regenerative species. Therefore, for this study we have focused on two time-points preceding (day 1 and 3) and following (day 7 and 11) loss of RGCs in the optic tectum.
To investigate the changes in gene expression that occur as RGCs recover from an optic nerve crush and begin to regrow their axons, we have used the TRAP method (Fig. 1C). With TRAP we are able to isolate ribosome-associated mRNAs, which are more representative of the actively translating pool than total mRNA, from a specific cell type. To do this, we created lines of transgenic frogs which express an eGFP-tagged variant of the ribosomal protein rpl10a under the control of an RGC-specific promoter from the islet2b locus (Fig. 1D) (Pittman et al., 2008; Watson et al., 2012). This subunit is an attractive target to tag because it is localized to the surface of the large ribosomal subunit. Moreover, our previous work has demonstrated that, in the eye, this construct is expressed exclusively in retinal ganglion cells and can be used to isolate and enrich for RGC-specific mRNAs (Watson et al., 2012).
In our experimental framework (Fig. 1C), we quantify gene expression changes in RGCs recovering from optic nerve crush by comparing mRNA levels in samples collected from the eye undergoing the crush (right) to the contralateral eye (left). At discrete time points following optic nerve crush in the right eye, both eyes were rapidly dissected and the ribosome-associated RNAs were purified from tissue extracts using eGFP antibodies conjugated to magnetic beads (Fig. 1E). To control for the effects of surgery on RGCs, gene expression was also quantified in animals that underwent sham surgeries with no optic nerve crush (sham). To control for the systemic effects of the surgical procedure per se, gene expression was also quantified in animals that did not undergo any surgery (naïve). These mRNA pools were used to construct libraries for RNA-Seq and RT-qPCR validation.
To explore global changes in gene expression after optic nerve injury, we quantified transcript levels in our samples using RNA-Seq with 50-base pair single-end reads. Raw sequencing reads were preprocessed with standard quality filters and adapter trimming and mapped to genes in the Xenopus laevis genome using the new version 9.1 assembly, with Bowtie2 (Langmead and Salzberg, 2012; Session et al., 2016). Across our samples, an average of 70% of reads were aligned to reference genes (Fig. 2A, S1C). Following mapping, read counts for reference transcripts were quantified using RSEM and expressed as fragments sequenced per kilobase of reference transcript per million reads (FPKM) (Li and Dewey, 2011). By directly comparing raw expression levels for this set of genes in crush and control samples we see a shift in global patterns of gene expression across our experimental time course (Fig. 2B). Notably, genes across a wide range of absolute expression levels were both up- and down-regulated in the days following the nerve crush with the magnitude of change increasing across our experimental time course.
Figure 2. RNA-Seq read alignment and analysis.
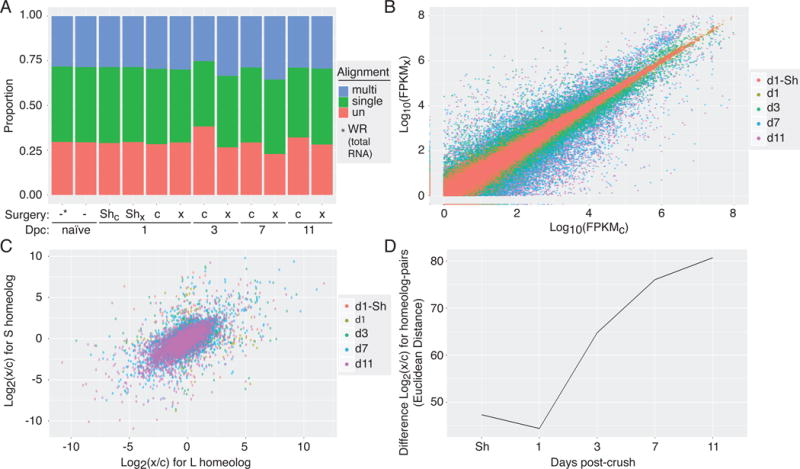
(A) Proportion of RNA-Seq reads that were mapped to the Xenopus laevis 9.1 genome assembly across experimental naïve (-), contralateral control (c) and crush (x) samples demonstrates good mapping rates across the various days post-crush (Dpc). (B) Read counts, expressed as FPKM, for each gene sequenced in the crushed (FPKMC; x-axis) versus control eye (FPKMX; y-axis) showing both up- and down-regulation in the days following injury. (C) Comparison of changes in gene expression following optic nerve crush between homeolog-pairs (L x-axis, S y-axis) across the post-injury time course, shows overall correlation between pairs with clear outliers. (D) The magnitude of difference between changes in gene express among homeolog-pairs, quantified by Euclidean distance, increases over time after optic nerve injury.
The key innovation in the new Xenopus laevis 9.1 genome assembly is to systematically account for an ancestral genome duplication event in the Xenopus lineage that resulted in pseudo-tetraploidy in the modern laevis genome (Session et al., 2016). This new genome annotation explicitly annotates gene homeologs, denoting paralogs on ancestral orthologous chromosomes, with unique gene symbols. Our results support the consensus that this new reference genome represents a major step forward in improving the resolution of gene expression analysis by RNA-Seq in Xenopus laevis (Fig. S1A versus C). For comparison, reads were also mapped to a set of reference transcripts containing 30,592 Xenopus laevis and 41,042 Xenopus tropicalis sequences from the NCBI EST database with assignments to genomic loci by the Xenbase project (Karpinka et al., 2015). Mapping reads against this set of reference transcripts gave far fewer results than our alignments to the 9.1 genome assembly, with an average of only 50% of reads mapping to reference transcripts (Fig. S1A, B); aligning against earlier versions of the gene models fared worse (data not shown).
More importantly, with this new assembly Xenopus laevis serves as a compelling model for genome evolution, if homeolog gene-pairs demonstrate comparable levels of functional specificity seen in simpler organisms (Komili et al., 2007). To this end, we analyzed the expression of homeolog pairs across our experimental time course: although there is a strong correlation in homeolog pair expression levels globally, many pairs diverge, particularly at later time points (Fig. 2C). The increase in the magnitude of divergence at later time points can easily be seen if we compare the effects of optic nerve crush on gene expression of homeolog pairs in the days following injury (Fig. 2D, S7). This finding strongly suggests that a significant portion of homeologs expressed in RGC are under independent transcriptional control. Even though L-copy constitutive expression levels are generally higher than the S-copy across the genome as a whole (Session et al., 2016), there is no clear bias in the change in relative expression levels in our injury model.
To substantiate our findings using RNA-Seq, we independently validated changes in gene expression for select up- and down-regulated factors. Semi-quantitative analyses of one such newly discovered RGC-specific factor, uchl1, shows that uchl1 mRNA levels detected in retina sections dramatically increased in the retinal ganglion cell layer compared to that in other retinal cell layers and only in the operated eye (Fig. 3A, B). We also performed RT-qPCR to quantify levels of uchl1, comparing these results to our RNA-Seq transcript alignments; in both we see strong up-regulation in crush versus control samples by day 11 (Fig. 3C). By contrast the synucleins, including snca and sncg show strong down regulation in the RNA-Seq results, which we have confirmed by RT-qPCR and immunofluorescence, respectively (Fig. 3C-E). Additional time points sampled by qPCR at 21 days post-crush reveal the uchl1 and snca mRNA expressions return to levels comparable to those of the control (Fig. 3C).
Figure 3. Independent biological validation of RNA-Seq.
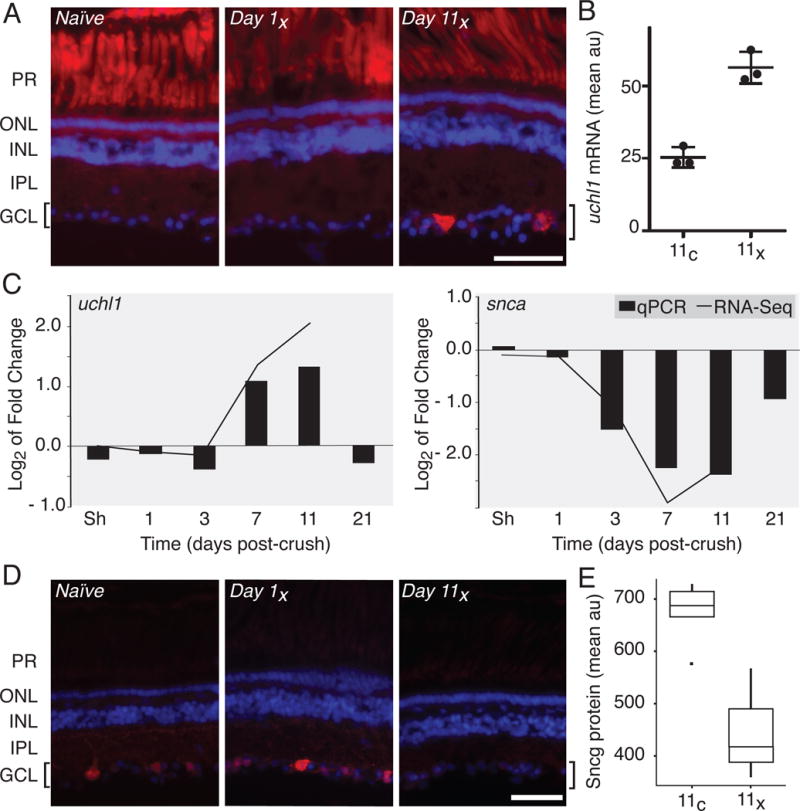
The up- and down-regulation of select factors in response to optic nerve injury described in the TRAP RNA-Seq analysis was independently verified by in situ hybridization, RT-qPCR and immunofluorescence. (A) In situ hybridization demonstrates that uchl1 transcript is up-regulated unilaterally in RGCs by 11 days post-injury (Day 11x). Autofluorescence of photoreceptor cells (PR) shows no injury-dependent variability. (B) Semi-quantitative analysis of fluorescence intensity from uchl1 in situ, as shown in panel A, shows a significant up-regulation of uchl1 mRNA in the crushed eye relative to control eye in the RGC layer by day 11 (p < 0.01). Results were averaged for three retinal sections from each of three different individuals and data represent arbitrary fluorescence units (au). (C) Gene expression levels assayed using RT-qPCR for uchl1 and snca (bars) are highly correlated with changes in fold gene expression quantified by RNA-Seq (line graph). (D) As predicted by RNA-Seq, protein levels for sncg decrease over the experimental time course in injured RGCs, as seen by immunofluorescence. (E) Semi-quantitative analysis of fluorescence intensity in the RGC layer immunostained for sncg protein, as in panel D, shows strong down-regulation of sncg in crush relative to control eyes by day 11. This boxplot shows averaged results for two retinal sections from each of four different individuals and values represent arbitrary fluorescence units (au). In A and D, location of the RGCs observed in the ganglion cell layer (GCL) and highlighted by brackets, photoreceptor cell (PR) outer (ONL) and inner (INL) nuclear and inner plexiform (IPL) layers was visualized using DAPI. The scale bar is 50 μm.
We can compare global patterns of gene expression between our samples using pair-wise distance analysis (Fig. 4A). In the visualization of this matrix, the boxes at the intersection of two samples with very similar gene expression patterns (low distance) are colored with light blue; darker colors represent samples that are more divergent. We can see that there is very little variability between the naïve sample, surgical shams and all of the controls. All of these samples are also strongly correlated with the crush sample taken just one day after treatment. However, there is a large shift in the gene expression pattern between the 1st and 3rd day post-crush. There is a second even larger shift in global gene expression between days 3 and 7, and again between days 7 and 11 post-crush.
Figure 4. Global changes in RGC gene expression following optic nerve injury.
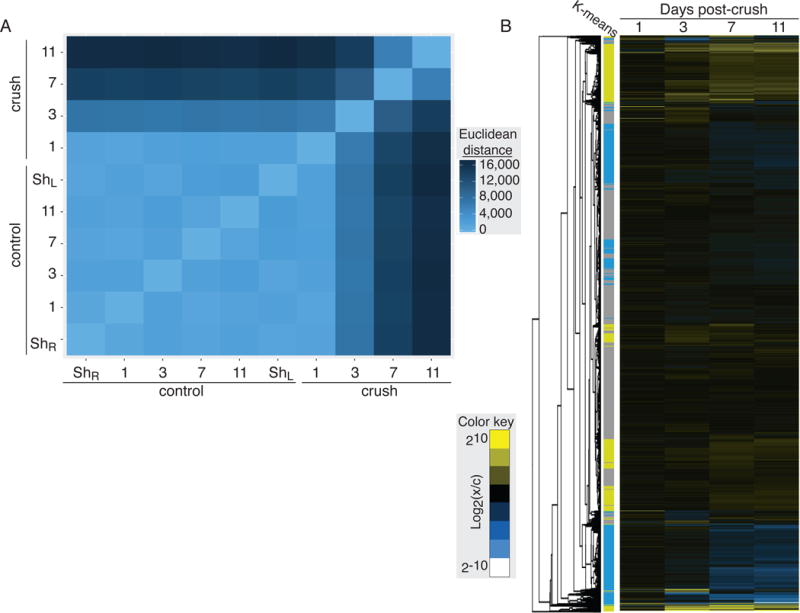
(A) The differences in transcript abundance in each sample was compared pair-wise using the Euclidean distance between FPKM values. In this heat map, the brightest blue represents the most closely related FPKM profiles. Control samples are highly similar, while samples from the optic-nerve crush show large shifts in gene expression pattern in the days following injury. (B) Hierarchical clustering of genes sequenced in RGCs across the experimental time course. Clusters of up- and down-regulated genes shows strong correspondence to groups identified by k-means clustering (groups 1 & 2 yellow, up-regulated; groups 3 & 4 blue, down-regulated; group 5 grey, unchanged).
To identify groups of genes showing similar patterns of changes in gene expression across our experimental time course we performed hierarchical clustering (Fig. 4B). This heat map and associated dendrogram show clear clusters of up- and down-regulated genes. Independent k-means clustering analysis converged on a similar set of groupings (Fig. 4B colored bar, Fig. S4). Both of these clustering methods reveal that expression changes at days 7 and 11 drive the grouping of up- and down-regulated factors. We can also see that the majority of genes expressed at detectable levels in RGCs, however, fall into neither of these categories showing that only discrete sets of genes change expression following optic nerve injury.
To systematically characterize the groups of up- and down-regulated genes, a custom Gene Ontology (GO) database was created to map terms onto the Xenopus laevis 9.1 gene models used to map RNA-Seq reads (Fig. 5). This reference set incorporated assignments from pre-existing Xenbase terms and supplemented these by term associations from the human genome where gene symbols could be unambiguously matched between the human genome and frog. When comparing biological process annotations enriched in the sets of up- and down-regulated genes, there was little functional overlap. This global analysis reveals that genes up-regulated following optic nerve crush are highly enriched in factors necessary to increase the biosynthetic capacity of cells (Fig. 5B). We also see that regenerating RGCs also undergo a robust endoplasmic reticulum stress response, along with an oxidative stress and inflammatory response, as previously reported in mouse (Sharma et al., 2014; Yasuda et al., 2014) and zebrafish models (McCurley and Callard, 2010; Veldman et al., 2007). By contrast, the groups of genes down-regulated following optic nerve crush are highly enriched for biological processes that are relevant in functional, connected RGCs, like synaptic communication factors.
Figure 5. Functional specificity in the set of transcripts that are up- and down-regulated following retinal ganglion crush.
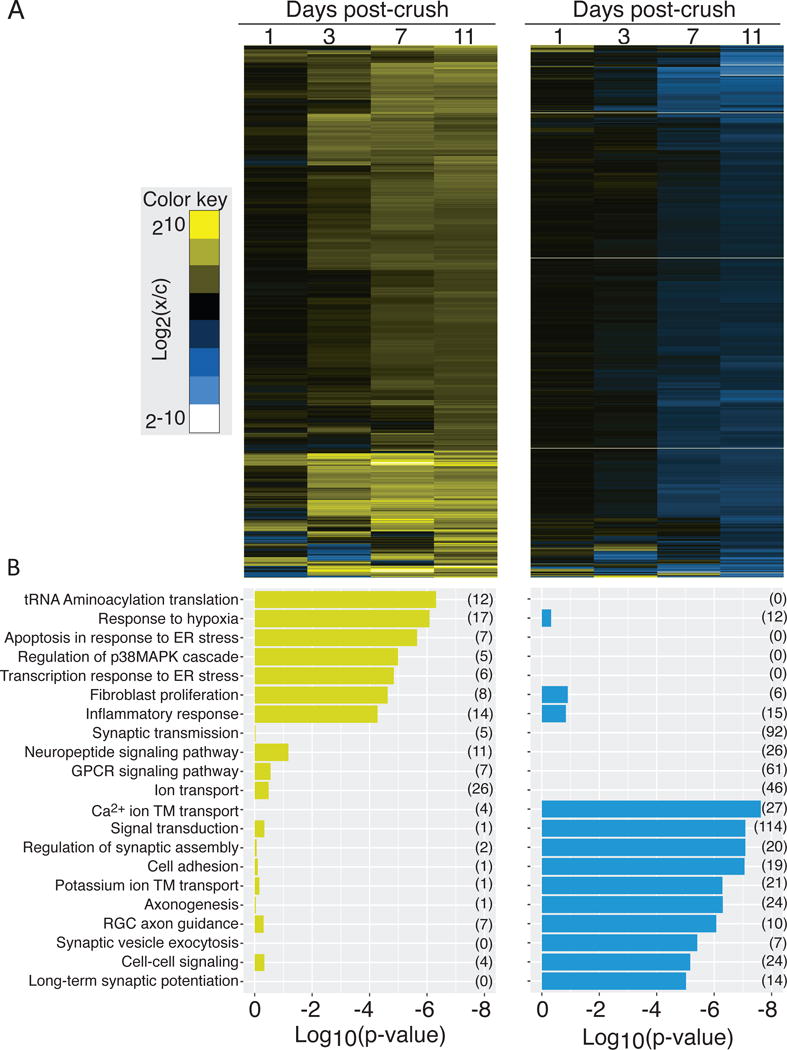
(A) Heat maps showing hierarchical clustering of transcripts that are up- (left) and down-regulated (right), identified by k-means clustering, following retinal ganglion crush. (B) The set of Gene Ontology biological process terms that are significantly over represented in each group of transcripts. The evidence for enrichment of each term (log10(p-value)) is plotted for the group of up- (left) and down-regulated genes (right); there is little overlap in significantly enriched terms.
In addition to using GO analysis to identify broad functional categories in the sets of up- and down-regulated factors, we can also examine the response of specific genes of interest. For example, we find strong enrichment in our TRAP samples, relative to total retina, for known RGC-specific genes including the pou/brn family, the synucleins, and rbpms2 (Fig. 6A). Conversely, the gene most highly enriched in RGCs, gng8, has not been previously characterized as an RGC-specific factor. Two other members of this gene family, encoding G-protein gamma subunits, are also highly enriched.
Figure 6. RGC enriched factors.
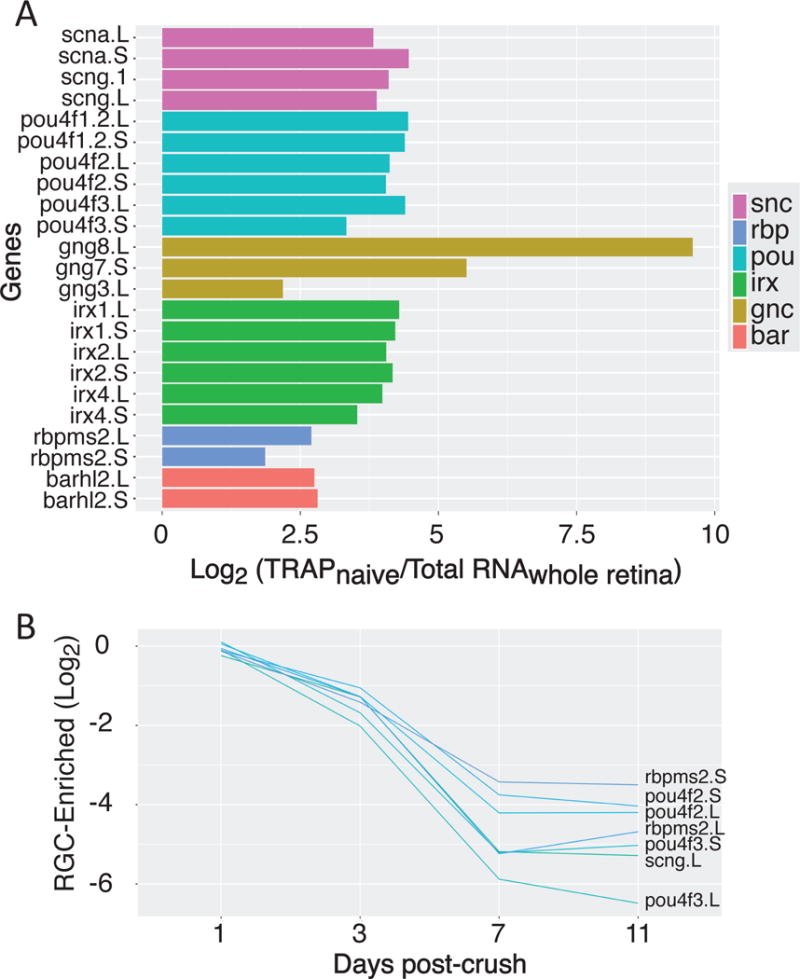
(A) Gene families that are highly enriched in naïve TRAP samples compared to total retina include key RGC-specific factors along-side novel groups. (B) When we compare gene expression in crush versus control samples, we see that optic nerve injury leads to strong down-regulation of many of these RGC-enriched genes.
Interestingly, many of the canonical RGC-specific factors are down-regulated following optic nerve injury, most strongly between days 7 and 11, including rpbms2 (Fig. 6B, S8). By contrast, other RGC-enriched factors like tubulin and gap43 are up-regulated (Fig. 7A, S8). The kruppel-like family of transcription factors, which have been implicated in both positive and negative regulation of axon growth, show highly divergent patterns in expression: both klf6 homeologs are up-regulated 3-days after injury, while klf4 is strongly down-regulated across the time-course (Fig. 7A). Interestingly, the expression pattern of klf6 in retinal cross sections show this mRNA is upregulated in the inner nuclear cells relative to its expression pattern in RGCs in the naïve and at post-injury day 1, an injury-dependent result not observed in the RGC-specific RNA-Seq screen (Fig. S8). As expected from the GO analysis above and previous work in both the mouse and zebrafish, we also see up-regulation of the ER (atfs) (Fig. 7B, S8) and oxidative stress-response (hmox1) systems (Fig. 7C) (Sharma et al., 2014; Veldman et al., 2007; Yasuda et al., 2014). Finally, we also see dramatic up-regulation in some, but not all, members of the Jak/Stat pathway, for example socs3 and IL-10 (Fig. 7D, S8).
Figure 7. Gene groups differentially regulated following optic-nerve injury.
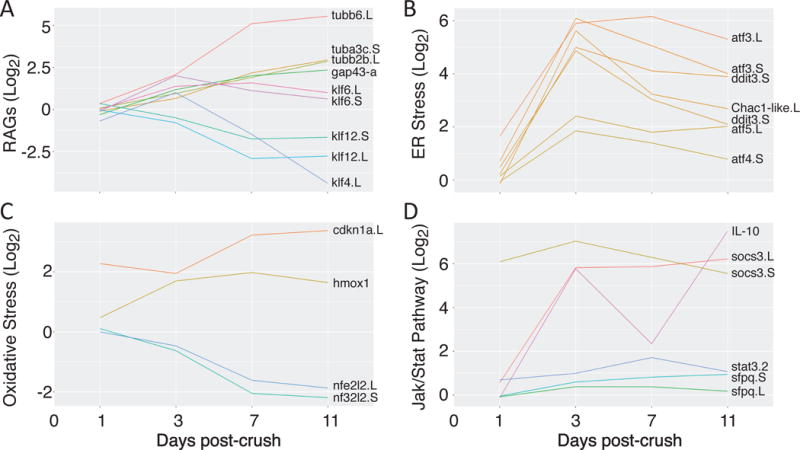
(A) Many axonal regeneration-associated genes (RAGs) and development associated factors are up-regulated following crush (tubulin, gap43, klf6), while some factors shown to inhibit axonal growth are down-regulated (klf4). (B & C) Stress response pathways shown to be up-regulated by optic nerve injury in other vertebrate species are up-regulated in Xenopus laevis RGCs as well. (D) Some members of the Jak/Stat signaling pathway are dramatically up-regulated. (A – D) In all panels, we compare the log2 ratio of expression levels for genes in the crush versus control samples. Genes with names not appended “.L” or “.S” (hmox1, IL-10), represent symbols not yet assigned in the Xenopus laevis 9.1 genome assembly. In these cases, data are shown from read alignments to a transcript reference (detailed in Materials and Methods).
Discussion
Here we have presented time-resolved gene expression profiles describing the response of retinal ganglion cells to optic nerve crush as they enter a period of recovery and regrowth. The first of its kind in adult Xenopus laevis, our analysis benefits from the use of TRAP which allows us to focus our investigation on gene expression in just RGCs. Because we have profiled only the RGCs, which represent a small percentage of all retinal cells, this study represents the highest resolution analysis of the response of RGCs to optic nerve crush injury to date. Also in contrast to previous studies, the use of TRAP ensures that our measurement of gene expression is a closer reflection of the pool of actively translating mRNAs than could be gained by profiling total cellular mRNA. In the days following optic nerve injury, we see large, global, changes in transcript levels across the genome. This highlights the importance of identifying the ribosome-associated mRNA population, as these messages encode factors that are directly involved in the immediate biological response to acute injury and neuronal regrowth in the frog.
Taken broadly, our findings point to two key transitions in the timeline following optic nerve injury: an initial response between the 1st and 3rd day post-crush and a larger magnitude shift between the 3rd and 7th. Overall, across the time course, a larger number of genes enriched in our samples showed decreased (~872) rather than increased (~107) mRNA expression levels in response to optic nerve injury. We also find evidence of similarities between Xenopus laevis and mammalian models, in which the injured optic nerve do not regrow, along with some striking differences; among these differences may lie the keys to understanding the capacity for regeneration in non-mammalian vertebrates.
The Xenopus laevis genome
A recent analysis of a new assembly of the Xenopus laevis genome has extensively characterized the ancient genome duplication event that resulted in allotetraploidy in the modern African Clawed frog (Session et al., 2016). The new Xenopus laevis 9.1 genome assembly accounts for this history: eight of the nine pairs of homeologous chromosomes were matched to orthologous chromosomes in the diploid Xenopus tropicalis genome. Symbols for these genes are appended with “L” or “S” to denote which sequence was found for the “longer” and “shorter” chromosomes in the pair. We found this new genome assembly to be a far better alignment target for our RNA-Seq reads than earlier drafts of the genome or a custom set of reference transcripts.
With this new assembly, Xenopus laevis can be used as a compelling model for genome evolution, particularly if homeolog gene-pairs demonstrate the levels of functional divergence seen following whole-genome duplication events in simpler eukaryotes (Komili et al., 2007). Indeed, genomic analysis has revealed evidence of nucleotide sequence divergence at the level of single nucleotide polymorphism between homeolog pairs, functional specificity in the groups of genes maintained over time as homeolog pairs, and differences in gene expression between pairs across developmental stage (Session et al., 2016). Our findings add recovery from acute injury to this list: in RGCs gene expression widely diverged amongst homeolog pairs in the days following optic nerve crush. Interestingly, the magnitude of divergence in expression level increases as well, particularly between the 3rd and 7th and then again between the 7th and 11th days post-crush. In RGCs, therefore, we do find evidence for divergence in regulatory control of many homeolog pairs.
Shared responses: oxidative and endoplasmic reticulum stress
Among the earliest and most robust responses that occur after optic nerve injury in RGCs is the up-regulation of factors involved in responding to endoplasmic reticulum (ER) stress. ER stress results from the accumulation of unfolded and misfolded proteins in the ER lumen and triggers the unfolded protein response, which is critical for cellular survival (Oslowski and Urano, 2011; Walter and Ron, 2011). The large increase seen in ER stress-associated mRNAs expressed 3 days after injury and its subsequent gradual decrease observed as early as 7 days, indicates that regeneration-capable animals must also activate these critical signaling pathways involved in promoting neuronal survival. This response to early optic nerve injury in our results in frogs is highly consistent with results in both the mouse (Yasuda et al., 2014) and zebrafish (Veldman et al., 2007) (Fig. S6). For example, genes that are strongly up-regulated in both mouse total retina and frog RGCs include chac1, ddit3, atf3, atf4 and atf5 (Yasuda et al., 2014). Interestingly, in Xenopus laevis, atf5 has also been shown to be up-regulated during tail regeneration in larvae (Tazaki et al., 2005).
Previous work in other models also suggested a strong activation of the oxidative stress response following injury. It is reasonable to anticipate that RGCs would experience attack by free radicals released during injury. In our results, we do see up-regulation of some key oxidative stress response factors, such as hmox1. However, others like the antioxidant factor nfe2l2 are actually down-regulated in frog RGCs. Moreover, we do not find evidence that oxidative stress response factors as a gene category are strongly enriched in the group of genes that are up-regulated following crush (p > 10−1). This finding may be indicative of a difference in the timing or magnitude of the oxidative stress response experienced in the injured frog RGC population as it initiates repair and regrowth programs, as compared to the mouse retina (Yasuda et al., 2014).
Both oxidative and ER stresses can induce apoptotic cell death. Indeed, strongly up-regulated ER stress factors such as atf4 and atf5 are also directly implicated in the control of apoptosis induced by ER stress (Tabas and Ron, 2011). However, we do not see evidence of a general activation of apoptosis in our RGCs after injury, with little to no change in the expression levels of canonical pro-apototic factors (such as aifm1) and no increase in apoptosis in RGCs as measured by activated caspase3 (Fig. S9) or TUNEL (data not shown). This is consistent with a population of cells recovering from injury and initiating a set of growth programs rather than cell death pathways.
The retinal ganglion cell: identity and adaptation to injury
Our injury model shows an up-regulation of many of the factors typically used as hallmarks for nerve regeneration. For example, gap43 and tubulin mRNA appear as early as 3-days post-crush and continue to increase in magnitude the 7th - 11th day, consistent with regeneration studies in Xenopus and zebrafish (Diekmann et al., 2015; Veldman et al., 2007; Zhao and Szaro, 1994). In Xenopus, gap43 levels have been shown to increase at the site of axon injury through the 9th day following surgery (Zhao and Szaro, 1994). In zebrafish, Diekman et al. (2015) used a transgenic line expressing gap43 in the optic nerve to visually assess and define the period of re-growth following injury. Here they found gap43 mRNA expression was up-regulated during the initial 2nd - 8th day post-crush, during the growth extension stage, but was subsequently down-regulated on days 8 – 21, after contact and functional recovery had occurred (Diekmann et al., 2015). Using gap43 expression and the peak change in global expression-levels as a guide, our data define a recovery period between the 3rd and 7th day post injury, a time-course consistent with loss of fluorescence in RGC axons within the optic tectum.
Some of the temporal difference between regrowth in the zebrafish and Xenopus might be attributable to differences between species and distance of the crush injury from the optic nerve head (McCurley and Callard, 2010). However, our optic nerve crush injuries are located 5 mm from the eye orbit, whereas most crush injuries in zebrafish and other Xenopus studies are located adjacent or very close to the eye optic nerve head (<0.5 mm) (McCurley and Callard, 2010; Veldman et al., 2007; Zhao and Szaro, 1994). Axonal regrowth across the crush site and into the optic tecta as measured by the recovery of fluorescence in the tecta provides a timeline consistent with our injury model. Temporal differences between our results and previous studies may also be attributed to the more sensitive TRAP method which isolates only the ribosome-associated mRNAs that may be masked in expression data isolated from total RNA pools. For instance differences between total cellular RNA levels and actively translating mRNAs in juvenile Xenopus show that neurofilament (nef-l, - m, -h) expression is tightly controlled not only at the transcriptional level but also through post-transcriptional regulation of mRNA localization and translational efficiency, underscoring the need for more sensitive screens that focus on the pools of translationally active mRNAs (Szaro and Strong, 2010).
Comparison between our total retinal RNA and TRAP samples demonstrate a strong enrichment for RGC-specific genes with the TRAP isolation, including hallmark factors like the 4pouf (brn3) and synucleins (alpha, beta and gamma) and uchl1. Some RGC-enriched genes show up-regulation in response to nerve injury, including factors like cytoskeletal components (actb, tubb2b, tubb3, and tuba1b). Up-regulated factors also include RGC-specific genes such as uchl1. uchl1 is a neuron-specific ubiquitin C-terminal hydrolase involved in proteasome mediated protein degradation (Day and Thompson, 2010; Esteve-Rudd et al., 2010). In mammals, uchl1 is also expressed in RGCs and human mutations in this gene are associated with late-onset Parkinson’s disease. In our data, it is both highly enriched in RGCs and dramatically up-regulated in response to optic nerve crush. Finally, the most highly-enriched RGC specific gene in our dataset, gng8, was also up-regulated following nerve injury. This gene encodes a G-protein gamma subunit and presents a compelling opportunity for future investigation because it has not previously been characterized as an RGC-specific factor or implicated in neuronal regeneration.
Provocatively, many canonical RGC-enriched genes were strongly down-regulated following optic nerve crush. This included comparatively general neuronal factors like cell adhesion (a host of cadherins, amigo1), axonogenesis (dcc), and ion channels (kcnf1, kcnp3, kcns1, scn2b). Axon guidance receptors (robo3, robo2-like) also appeared down-regulated, however this may be due to localized translation in the axon. We also observed more than a two-fold decrease in expression levels in many canonical RGC-specific genes including: the pou homeodomain proteins (pou4f1/brn3a, pou4f2/brn3b, pouf4f3/brn3c), an RNA-binding protein (rbpms2), and the synucleins (snca, scnb, scng). This intriguing finding strongly suggests that the initiation of RGC recovery and regrowth requires a concomitant down-regulation of factors that define cell-type specificity in this population of neurons.
Does regeneration recapitulate development?
During development neurons form growth cones, initiate axon elongation, navigate to, and connect with their appropriate target. Following axonal injury, similar genetic programs are re-activated during the growth phase following membrane repair and growth cone assembly. As during development, this active growth mode is associated with specific axonal growth associated genes and signaling pathways (Fischer and Leibinger, 2012). Based on developmental studies (Paranjpe et al., 2013; Peshkin et al., 2015; Shigeoka, 2016) and both mouse and zebrafish regenerative studies (Fischer and Leibinger, 2012; Sharma et al., 2014; Veldman et al., 2007; Yasuda et al., 2014), we predicted that we would find many of the same axonal growth associated genes and signaling. The kruppel-like family (klf) of transcription factors provides one example of a gene family that has been shown to be active during the period of axonal growth that occurs during normal development. In fish and mouse, two klfs (klf6 and klf7) increase axon growth in response to injury (Moore et al., 2009; Veldman et al., 2007). In Xenopus laevis RGCs, klf6 and klf7 show increased expression following the optic nerve crush. Interestingly, eight Klf family members including klf4 and klf9 were shown to suppress axon growth in the optic nerve of mouse (Moore et al., 2009). Consistent with this, we have found that both klf4 and klf12 are strongly down regulated following injury in the regeneration-capable frog RGC population.
More broadly, we also observe a dramatic up-regulation in the cellular biosynthetic machinery as RGCs recover from injury and initiate regrowth, to levels that would normally be unusual in a population of mature, differentiated neurons. For example, every cellular aminoacyl-tRNA synthetase is dramatically up-regulated by the 7th day following nerve crush. Indeed, tRNA aminoacylation is the most highly enriched category of factors in the set of up-regulated genes. In addition, we also see strong up-regulation in the downstream protein biosynthetic machinery, including translational elongation factors (eef1b2, eef1a1) and at least one isoform of every mapped ribosomal protein gene.
Many cell-autonomous molecules have been shown to play a role in optic nerve regeneration including: the mTor pathway, the negative regulator phosphatase and tensin homolog (pten) (Kurimoto et al., 2010; Park et al., 2008), the Jak/Stat signaling system, sfpq and socs3. Previous work has demonstrated a compelling relationship between the latter two. In mammals, socs3-dependant inhibition of the Jak/Stat pathway is linked to failure to regenerate optic axons; in zebrafish this effect may be overcome through cytokine signaling (Elsaeidi et al., 2014; Smith et al., 2009). Interestingly, activation of both these pathways leads to some of the most extensive optic nerve regrowth observed in mice (Sun et al., 2011). In our results, we find that socs3 is strongly upregulated following injury, along with some cytokines (interleukin IL-10 and IL-34).
The timing of the up-regulation of these factors in response to optic nerve crush coincides with a massive down-regulation in genes involved in cell-cell signaling, ion transport, synaptic maintenance, and synaptic vesicle exocytosis. Therefore, the recovering RGC demonstrates a highly unique gene expression program. On the one hand, cell-type specific factors decrease while the biosynthetic machinery is dramatically up-regulated alongside certain initiators of axon development. However, we do not see a broad upregulation of axonogenesis or neurogenesis factors. So the global pattern of changes in gene expression described here suggests that regenerating RGCs partially regress to a gene expression program more similar to that of an earlier stage in development than a mature RGC. It is likely that retaining this level of cellular plasticity in the global gene expression program is key to the ability of RGCs to fully regenerate following injury in the frog.
Conclusions
Here we generated a temporal expression profile for Xenopus laevis RGCs that provides an initial view of the genes active in the RGC cell bodies during the early period of repair and regrowth. Among the most highly up-regulated injury-induced genes are two newly identified genes, gng8 and uchl1, that are likely to play a role in the regeneration process. Use of ribosomal profiling such as TRAP provides a sensitive and rapid technique to generate expression profiles for specific cell types not masked by RNA expression from multiple cell types and total RNA pools. Given the regrowth of axons from the site of injury, combined with the large amount of mRNA translation occurring locally within the axon (Holt and Schuman, 2013), we plan to use this approach to generate additional expression profiles for RGC axons, oligodendrocytes, astrocytes and monocytes at the site of injury. Coupled with data from this study, such profiles will provide a comprehensive view of injury-related genetic programs involved in both the intrinsic and extrinsic factors that permit regeneration to occur in regeneration-capable animals such as the frog.
Supplementary Material
In (A) and (C) the total number of reads for each sample, across days post-crush (dpc), is shown for alignment to a custom transcript reference (A) and the Xenopus laevis 9.1 gene models (C), using Bowtie. Total bar height indicates the number of reads aligned for each sample. The salmon proportion represents reads that were not aligned to the reference, the green portion the number of reads aligned to a single reference, and blue the number of reads aligned to more than one valid reference (in which case the best match is used). We see that using the Xenopus laevis 9.1 assembly not only improves overall alignment percentage but results in a much larger percentage of unique hits. (B) Number of reads aligned to the Xenopus laevis versus tropicalis reference transcripts for read alignments shown in (A).
Comparison of the log2-fold change in gene expression in crush versus control samples using read mapping results from the Xenopus laevis 9.1 genome assembly versus our custom transcript reference. Although there is a strong correlation, the gene model alignments feature better dynamic range.
(A) The distribution of normalized read count values, expressed as FPKM, is relatively uniform across samples, indicating no sources of technical bias in sequencing or alignment. (B) The distribution of the log−-fold change in gene expression in crush versus control samples shifts across the experimental time course, indicating global changes in gene expression.
(A) Total, within- and between-cluster sums of squares for different numbers of clusters. (B) Comparison of log2-fold changes in gene expression for crush versus control samples and days 1 and days 11, colored by k-means group (with k = 5) demonstrates the k-clusters effectively identify groups of up- and down-regulated genes.
Known RGC-specific genes (black) are all strongly enriched in the naïve TRAP samples compared to RNA isolated from the total retina.
Correlation matrix comparing read counts for genes matched between the Xenopus laevis and Mus musculus genomes in control and crush experiments (Yasuda et al., 2014). All values represent Pearson correlation.
Comparison of log2-fold changes in gene expression for crush versus control samples across the experimental time course using on (A) L-homeologs or (B) S-homeologs. (C) Comparison of log2-fold changes in gene expression for crush versus control samples across the experimental time course in L- versus S-homeologs. All values represent Euclidean distance.
In all figures, signal from in situ hybridization probes are shown alone (grey scale) and pseudo-colored red in merged images with DAPI signal (blue), to show the location of the RGCs observed in the ganglion cell layer (GCL) and highlighted by brackets, photoreceptor cell (PR) outer (ONL) and inner (INL) nuclear and inner plexiform (IPL) layers. Scale bar is 50 μm. (A) In RNA-Seq analysis, atf3 is strongly up-regulated in injured RGCs by day 11 (log2(x/c) = 5.3) (Fig. 7B). (B) In RNA-Seq analysis, using the transcript assembly, prph is strongly up-regulated in injured RGCs by day 11 (log2(x/c) = 3.3). (C) In RNA-Seq analysis, using the transcript assembly, gap43 is highly up-regulated in injured RGCs by day 11 (log2(x/c) = 2.3). (D) In RNA-Seq analysis socs3 is strongly up-regulated in injured RGCs by day 11 (log2(x/c) = 6.2). (E) In RNA-Seq analysis, klf6 is transiently up-regulated in injured RGCs at day 3 but returns to normal levels by day 11. (F) In RNA-Seq analysis, rpbms2 is strongly down-regulated in injured RGCs by day 11 (log2(x/c) = −4.7).
(A) The upper panel shows retinal flatmounts from naïve (unoperated) retinas immunostained with anti-cleaved caspase-3 and the lower panel is merged with DAPI. We see high levels of activated caspase3 indicating that some cells in the retinal are undergoing apoptosis. (B) Representative images from retinal cross sections from naïve eyes and crush eyes 1 or 11 days post-injury (Day 1x and Day 11x). Here we see that the RGCs of the ganglion cell layer (GCL) are not expressing activated caspase3. However other retinal cells are, in a non-injury dependent manner, likely the horizontal cells located in the inner nuclear layer (INL). Signal from immunostain with anti-cleaved caspase-3 are shown alone (grey scale) and pseudo-colored red in merged images with DAPI signal, to show the location of the RGCs in the ganglion cell layer (GCL) and in indicated by brackets, photoreceptor cell (PR) outer (ONL) and inner plexiform (IPL) layers. Scale bar is 50 µm.
Acknowledgments
Authors thank Daniel Rhoades for help with the initial bioinformatics analyses and Luke Dreary and Ferdinand Kaya for technical assistance and Christine Holt for use of microscopy equipment. Authors also thank Jose Sotelo-Silveira and Jean-Michel Cioni for helpful comments on the manuscript along with John Bowman, Jennifer Lee, Ron Perets and Virginia McGhee for help proofreading. This work was supported in part by grants to the Washington and Lee University from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program (DH, AW, BM, GBW, FLW), the Dr. G. Ashley Allen ’65 Student Research Grant (DR, SA), the Levy Neuroscience Undergraduate Research Fellowships (DR), the H. F. Lenfest Undergraduate Research Fellowship (CI, AW), the H. F. Lenfest Endowment for Faculty Support (GBW, FLW), the Foundation Fighting Blindness and the Glaucoma Research Foundation (NMA) and the Thomas F. and Kate Miller Jeffress Memorial Trust J-1026, NSF DBI-1126118 (FLW).
List of abbreviations
- TRAP
translational ribosome affinity purification
- RGC
retinal ganglion cell
- FPKM
fragments sequenced per kilobase per million reads
- ONC
optic nerve crush
Footnotes
Competing interests:
The authors declare they have no competing interests
Literature Cited
- Abramoff M, Magelhaes P. Image Processing with ImageJ. Biophotonics Int. 2003;11:36–42. [Google Scholar]
- Benowitz LI, Popovich PG. Inflammation and axon regeneration. Curr Opin Neurol. 2011;24:577–583. doi: 10.1097/WCO.0b013e32834c208d. [DOI] [PubMed] [Google Scholar]
- Bernhardt RR. Cellular and molecular bases of axonal regeneration in the fish central nervous system. Exp Neurol. 1999;157:223–240. doi: 10.1006/exnr.1999.7059. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradke FJWSME. Assembly of a new growth cone after axotomy: the precursor to axon regeneration. Nat Rev Neurosci. 2012;13:183–193. doi: 10.1038/nrn3176. [DOI] [PubMed] [Google Scholar]
- Chen DF, Schneider GE, Martinou JC, Tonegawa S. Bcl-2 promotes regeneration of severed axons in mammalian CNS. Nature. 1997;385:434–439. doi: 10.1038/385434a0. [DOI] [PubMed] [Google Scholar]
- Coombes KR. ClassDiscovery: Classes and methods for “class discovery” with microarrays or proteomics 2013 [Google Scholar]
- Day INM, Thompson RJ. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog Neurobiol. 2010;90:327–362. doi: 10.1016/j.pneurobio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Diekmann H, Kalbhen P, Fischer D. Active mechanistic target of rapamycin plays an ancillary rather than essential role in zebrafish CNS axon regeneration. Front Cell Neurosci. 2015;9:251. doi: 10.3389/fncel.2015.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaeidi F, Bemben MA, Zhao XF, Goldman D. Jak/Stat signaling stimulates zebrafish optic nerve regeneration and overcomes the inhibitory actions of Socs3 and Sfpq. J Neurosci Off J Soc Neurosci. 2014;34:2632–2644. doi: 10.1523/JNEUROSCI.3898-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Barres BA. Unlocking CNS Cell Type Heterogeneity. Cell. 2008;135:596–598. doi: 10.1016/j.cell.2008.10.031. [DOI] [PubMed] [Google Scholar]
- Esteve-Rudd J, Campello L, Herrero MT, Cuenca N, Martin-Nieto J. Expression in the mammalian retina of parkin and UCH-L1, two components of the ubiquitin-proteasome system. Brain Res. 2010;1352:70–82. doi: 10.1016/j.brainres.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Fawcett JW. Overcoming inhibition in the damaged spinal cord. J Neurotrauma. 2006;23:371–383. doi: 10.1089/neu.2006.23.371. [DOI] [PubMed] [Google Scholar]
- Fischer D, Leibinger M. Promoting optic nerve regeneration. Prog Retin Eye Res. 2012;31:688–701. doi: 10.1016/j.preteyeres.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Gaze RM. Regeneration of the optic nerve in Xenopus laevis. Q J Exp Physiol Cogn Med Sci. 1959;44:290–308. doi: 10.1113/expphysiol.1959.sp001402. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type–specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. The Central Dogma Decentralized: New Perspectives on RNA Function and Local Translation in Neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinka JB, Fortriede JD, Burns KA, James-Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 2015;43:D756–763. doi: 10.1093/nar/gku956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koriyama Y, Takagi Y, Chiba K, Yamazaki M, Arai K, Matsukawa T, Suzuki H, Sugitani K, Kagechika H, Kato S. Neuritogenic activity of a genipin derivative in retinal ganglion cells is mediated by retinoic acid receptor β expression through nitric oxide/S-nitrosylation signaling. J Neurochem. 2011;119:1232–1242. doi: 10.1111/j.1471-4159.2011.07533.x. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Kurimoto T, Yin Y, Omura K, Gilbert H, Kim D, Cen LP, Moko L, Kügler S, Benowitz LI. Long-Distance Axon Regeneration in the Mature Optic Nerve: Contributions of Oncomodulin, cAMP, and pten Gene Deletion. J Neurosci. 2010;30:15654–15663. doi: 10.1523/JNEUROSCI.4340-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Fournier A, Selles-Navarro I, Dergham P, Sebok A, Leclerc N, Tigyi G, McKerracher L. Inactivation of Rho Signaling Pathway Promotes CNS Axon Regeneration. J Neurosci. 1999;19:7537–7547. doi: 10.1523/JNEUROSCI.19-17-07537.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CK, Lindsey JD, Crowston JG, Lijia C, Chiang S, Weinreb RN. Longitudinal Profile of Retinal Ganglion Cell Damage after Optic Nerve Crush with Blue-Light Confocal Scanning Laser Ophthalmoscopy. Investig Opthalmology Vis Sci. 2008;49:4898. doi: 10.1167/iovs.07-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu H, Deaton SK, Szaro BG. Heterogeneous Nuclear Ribonucleoprotein K, an RNA-Binding Protein, Is Required for Optic Axon Regeneration in Xenopus laevis. J Neurosci. 2012;32:3563–3574. doi: 10.1523/JNEUROSCI.5197-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas A. amap: Another Multidimensional Analysis Package 2014 [Google Scholar]
- McCurley AT, Callard GV. Time Course Analysis of Gene Expression Patterns in Zebrafish Eye During Optic Nerve Regeneration. J Exp Neurosci. 2010;2010:17–33. [PMC free article] [PubMed] [Google Scholar]
- Meyer RL, Sakurai K, Schauwecker E. Topography of regenerating optic fibers in goldfish traced with local wheat germ injections into retina: Evidence for discontinuous microtopography in the retinotectal projection. J Comp Neurol. 1985;239:27–43. doi: 10.1002/cne.902390103. [DOI] [PubMed] [Google Scholar]
- Mills EA, Davis CO, Bushong EA, Boassa D, Kim KY, Ellisman MH, Marsh-Armstrong N. Astrocytes phagocytose focal dystrophies from shortening myelin segments in the optic nerve of Xenopus laevis at metamorphosis. Proc Natl Acad Sci. 2015;112:10509–10514. doi: 10.1073/pnas.1506486112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci. 2009;106:18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Sugino K, Nelson SB. Cell Type-Specific Transcriptomics in the Brain. J Neurosci. 2011;31:6939–6943. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJC. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics. 2013;14:762. doi: 10.1186/1471-2164-14-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L, Wühr M, Pearl E, Haas W, Freeman RM, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW. On the Relationship of Protein and mRNA Dynamics in Vertebrate Embryonic Development. Dev Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Dev Camb Engl. 2008;135:2865–2871. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Beaver RS, D SA. Continued neurogenesis is not a pre-requisite for regeneration of a topographic retino-tectal projection. Vision Res. 2001;41:1765–1770. doi: 10.1016/s0042-6989(01)00053-0. [DOI] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session A, Uno Y, Kwon T, Harland R, Taira M, Rokhsar D. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016 doi: 10.1038/nature19840. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma TP, McDowell CM, Liu Y, Wagner AH, Thole D, Faga BP, Wordinger RJ, Braun TA, Clark AF. Optic nerve crush induces spatial and temporal gene expression patterns in retina and optic nerve of BALB/cJ mice. Mol Neurodegener. 2014;9:14. doi: 10.1186/1750-1326-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung J, Turner-Bridger B, Ohk J, Lin J, Amieux PS, Jung H, Holt CE. The dynamic translatome of retinal ganglion cell axons during assembly and maintenance of the mouse visual system. Cell. 2016 in press. [Google Scholar]
- Smith PD, Sun F, Park KK, Cai B, Wang C, Kuwako K, Martinez-Carrasco I, Connolly L, He Z. SOCS3 Deletion Promotes Optic Nerve Regeneration In Vivo. Neuron. 2009;64:617–623. doi: 10.1016/j.neuron.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry RW. Optic nerve regeneration with return to vision in anurans. J Neurophysiol. 1944;7:57–69. [Google Scholar]
- Sperry RW. Patterning of Central Synapses in Regeneration of the Optic Nerve in Teleosts. Physiol Zool. 1948;21:351–361. doi: 10.1086/physzool.21.4.30152014. [DOI] [PubMed] [Google Scholar]
- Sternberg SR. Biomedical Image Processing. Computer. 1983;16:22–34. [Google Scholar]
- Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaro BG, Strong MJ. Post-transcriptional control of neurofilaments: New roles in development, regeneration and neurodegenerative disease. Trends Neurosci. 2010;33:27–37. doi: 10.1016/j.tins.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazaki A, Kitayama A, Terasaka C, Watanabe K, Ueno N, Mochii M. Macroarray-based analysis of tail regeneration in Xenopus laevis larvae. Dev Dyn Off Publ Am Assoc Anat. 2005;233:1394–1404. doi: 10.1002/dvdy.20472. [DOI] [PubMed] [Google Scholar]
- Thomas A, Lee PJ, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, Dierick HA. A Versatile Method for Cell-Specific Profiling of Translated mRNAs in Drosophila. PLoS ONE. 2012;7:e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang JT, Medress ZA, Barres BA. Axon degeneration: Molecular mechanisms of a self-destruction pathway. J Cell Biol. 2012;196:7–18. doi: 10.1083/jcb.201108111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson FL, Mills EA, Wang X, Guo C, Chen DF, Marsh-Armstrong N. Cell type-specific translational profiling in the Xenopus laevis retina. Dev Dyn Off Publ Am Assoc Anat. 2012;241:1960–1972. doi: 10.1002/dvdy.23880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Gaze RM, Goodbrand IA, Taylor JSH. Regeneration in theXenopus tadpole optic nerve is preceded by a massive macrophage/microglial response. Anat Embryol (Berl) 1992;186:75–89. doi: 10.1007/BF00710404. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Tanaka Y, Ryu M, Tsuda S, Nakazawa T. RNA Sequence Reveals Mouse Retinal Transcriptome Changes Early after Axonal Injury. PLoS ONE. 2014;9:e93258. doi: 10.1371/journal.pone.0093258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Cui Q, Gilbert HY, Yang Y, Yang Z, Berlinicke C, Li Z, Zaverucha-do-Valle C, He H, Petkova V, et al. Oncomodulin links inflammation to optic nerve regeneration. Proc Natl Acad Sci U A. 2009;106:19587–19592. doi: 10.1073/pnas.0907085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Oglesby E, Marsh-Armstrong N. Xenopus laevis P23H rhodopsin transgene causes rod photoreceptor degeneration that is more severe in the ventral retina and is modulated by light. Exp Eye Res. 2008;86:612–621. doi: 10.1016/j.exer.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Szaro BG. The return of phosphorylated and nonphosphorylated epitopes of neurofilament proteins to the regenerating optic nerve of Xenopus laevis. J Comp Neurol. 1994;343:158–172. doi: 10.1002/cne.903430112. [DOI] [PubMed] [Google Scholar]
- Zou S, Tian C, Ge S, Hu B. Neurogenesis of retinal ganglion cells is not essential to visual functional recovery after optic nerve injury in adult zebrafish. PLoS One. 2013;8:e57280. doi: 10.1371/journal.pone.0057280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In (A) and (C) the total number of reads for each sample, across days post-crush (dpc), is shown for alignment to a custom transcript reference (A) and the Xenopus laevis 9.1 gene models (C), using Bowtie. Total bar height indicates the number of reads aligned for each sample. The salmon proportion represents reads that were not aligned to the reference, the green portion the number of reads aligned to a single reference, and blue the number of reads aligned to more than one valid reference (in which case the best match is used). We see that using the Xenopus laevis 9.1 assembly not only improves overall alignment percentage but results in a much larger percentage of unique hits. (B) Number of reads aligned to the Xenopus laevis versus tropicalis reference transcripts for read alignments shown in (A).
Comparison of the log2-fold change in gene expression in crush versus control samples using read mapping results from the Xenopus laevis 9.1 genome assembly versus our custom transcript reference. Although there is a strong correlation, the gene model alignments feature better dynamic range.
(A) The distribution of normalized read count values, expressed as FPKM, is relatively uniform across samples, indicating no sources of technical bias in sequencing or alignment. (B) The distribution of the log−-fold change in gene expression in crush versus control samples shifts across the experimental time course, indicating global changes in gene expression.
(A) Total, within- and between-cluster sums of squares for different numbers of clusters. (B) Comparison of log2-fold changes in gene expression for crush versus control samples and days 1 and days 11, colored by k-means group (with k = 5) demonstrates the k-clusters effectively identify groups of up- and down-regulated genes.
Known RGC-specific genes (black) are all strongly enriched in the naïve TRAP samples compared to RNA isolated from the total retina.
Correlation matrix comparing read counts for genes matched between the Xenopus laevis and Mus musculus genomes in control and crush experiments (Yasuda et al., 2014). All values represent Pearson correlation.
Comparison of log2-fold changes in gene expression for crush versus control samples across the experimental time course using on (A) L-homeologs or (B) S-homeologs. (C) Comparison of log2-fold changes in gene expression for crush versus control samples across the experimental time course in L- versus S-homeologs. All values represent Euclidean distance.
In all figures, signal from in situ hybridization probes are shown alone (grey scale) and pseudo-colored red in merged images with DAPI signal (blue), to show the location of the RGCs observed in the ganglion cell layer (GCL) and highlighted by brackets, photoreceptor cell (PR) outer (ONL) and inner (INL) nuclear and inner plexiform (IPL) layers. Scale bar is 50 μm. (A) In RNA-Seq analysis, atf3 is strongly up-regulated in injured RGCs by day 11 (log2(x/c) = 5.3) (Fig. 7B). (B) In RNA-Seq analysis, using the transcript assembly, prph is strongly up-regulated in injured RGCs by day 11 (log2(x/c) = 3.3). (C) In RNA-Seq analysis, using the transcript assembly, gap43 is highly up-regulated in injured RGCs by day 11 (log2(x/c) = 2.3). (D) In RNA-Seq analysis socs3 is strongly up-regulated in injured RGCs by day 11 (log2(x/c) = 6.2). (E) In RNA-Seq analysis, klf6 is transiently up-regulated in injured RGCs at day 3 but returns to normal levels by day 11. (F) In RNA-Seq analysis, rpbms2 is strongly down-regulated in injured RGCs by day 11 (log2(x/c) = −4.7).
(A) The upper panel shows retinal flatmounts from naïve (unoperated) retinas immunostained with anti-cleaved caspase-3 and the lower panel is merged with DAPI. We see high levels of activated caspase3 indicating that some cells in the retinal are undergoing apoptosis. (B) Representative images from retinal cross sections from naïve eyes and crush eyes 1 or 11 days post-injury (Day 1x and Day 11x). Here we see that the RGCs of the ganglion cell layer (GCL) are not expressing activated caspase3. However other retinal cells are, in a non-injury dependent manner, likely the horizontal cells located in the inner nuclear layer (INL). Signal from immunostain with anti-cleaved caspase-3 are shown alone (grey scale) and pseudo-colored red in merged images with DAPI signal, to show the location of the RGCs in the ganglion cell layer (GCL) and in indicated by brackets, photoreceptor cell (PR) outer (ONL) and inner plexiform (IPL) layers. Scale bar is 50 µm.


