Abstract
Introduction
Sleep plays an important role in cardiometabolic health. The sleep-wake cycle is partially driven by the endogenous circadian clock, which governs a range of metabolic pathways. The association between sleep and cardiometabolic health may be mediated by alterations of the human metabolome
Objectives
To better understand the biological mechanism underlying the association between sleep and health, we examined human plasma metabolites in relation to sleep duration and sleep timing.
Methods
Using an untargeted approach, 329 fasting plasma metabolites were measured in 277 Chinese participants. We measured sleep timing (midpoint between bedtime and wake up time) using repeated time-use surveys (4 weeks during one year) and previous night sleep duration from questionnaires completed before sample donation.
Results
We found 64 metabolites that were associated with sleep timing with a false discovery rate of 0.2 or lower, after adjusting for potential confounders. Notably, we found that later sleep timing was associated with higher levels of multiple metabolites in amino acid metabolism, including branched chain amino acids and their gamma-glutamyl dipeptides. We also found widespread associations between sleep timing and numerous metabolites in lipid metabolism, including bile acids, carnitines and fatty acids. In contrast, previous night sleep duration was not associated with plasma metabolites in our study.
Conclusion
Sleep timing was associated with a large number of metabolites across a variety of biochemical pathways. Some metabolite associations are consistent with a relationship between late chronotype and adverse effects on cardiometabolic health.
Keywords: Sleep duration, sleep timing, metabolomics
Introduction
Growing evidence has suggested that sleep plays an important role in multiple cardiometabolic conditions: sleep deficiency has been repeatedly associated with higher risks of obesity (Wu et al., 2014), type 2 diabetes (Shan et al., 2015, Cappuccio et al., 2010), and cardiovascular diseases (CVD) (Cappuccio et al., 2011). Night-shift workers who commonly suffer circadian disruption experience larger weight gain (van Drongelen et al., 2011) and are more likely to develop metabolic syndrome (Wang et al., 2014), diabetes (Gan et al., 2015), and CVD (Vyas et al., 2012). Moreover, several recent studies also reported that a preference of later sleep schedules (late chronotype) and larger differences in weekday/weekend schedules (social jetlag) are associated with worse metabolic health (Wittmann et al., 2006, Wong et al., 2015).
The sleep-wake cycle is an important behavioral manifestation of the endogenous circadian clock, which governs a range of metabolic pathways. Therefore, the association between sleep and cardiometabolic health may be mediated by alterations of the human metabolome (Bass and Takahashi, 2010). To date, three studies examined the acute effects of total and partial sleep deprivation in controlled laboratory conditions on human metabolome (Bell et al., 2013, Davies et al., 2014, Weljie et al., 2015). All three studies reported that sleep restriction resulted in widespread changes in circulating metabolites, including reduction of carbohydrates and increased levels of certain lipids and amino acids. However, such effects may not reflect those caused by habitual sleep conditions, such as chronic sleep deprivation or general sleep timing. Moreover, previous studies suggested that late sleep timing may be associated with more severe circadian disruption and may be a risk factor for multiple cardiometabolic conditions independent of sleep duration (Wong et al., 2015). However, no study has examined sleep timing in relation to human metabolome.
In a group of Chinese men and women who completed daily time-use log for four separate weeks in a 1-year period, we used an untargeted approach to measure over 300 metabolites from fasting plasma samples. We examined metabolite levels in relation to multiple measures of sleep timing and sleep duration. The aim of our study is to identify metabolite markers that are associated with natural variations in sleep behavior in the general population, which may help elucidate the biological mechanisms driving the health effects of sleep and circadian rhythm.
Methods
Study population
Our study included subjects from the Shanghai Physical Activity Study. Details of this study have been previously reported (Peters et al., 2010). Briefly, a total of 619 participants were randomly selected from two population-based prospective studies, the Shanghai Women’s Health Study (Zheng et al., 2005) and Shanghai Men’s Health Study (Shu et al., 2015). Participants were asked to complete a daily activity log for seven consecutive days on four separate occasions during a one-year study period (roughly one administration in each season). On average, each participant provided 27.3 days of log data. They also donated blood samples at the beginning and at the end of the one-year period, but only the samples donated at the end of the study period were used for metabolomics assay. A previous study selected 339 men and women for metabolomics assay using the blood samples donated at the end of the study (Xiao et al., 2016), and among them we excluded subjects with non-fasting blood samples. The final analytic sample included 277 men and women. A diagram depicting the study design and sleep variables are presented in Supplementary figure 1.
Measurement of sleep variables
In the activity log, the participants reported the time they went to bed at night and got up in the morning. Additionally, before each blood donation, they completed a short questionnaire and reported sleep duration in the previous night. From these we calculated two main sleep variables: 1) yearly average of the midpoint between bedtime and wake-up time (midpoint of time in bed), a common measure of chronotype (Kantermann et al., 2015, Roenneberg et al., 2003); and 2) sleep duration during the night prior to end-of-study blood donation.
We also calculated several other sleep variables for additional analysis (Supplementary figure 1). These include yearly average of total time spent in bed as a proxy of habitual sleep duration; average midpoint on nights before weekends, difference between average midpoint in weekdays and weekends, standard deviation of repeated measures of midpoint over the 1-year period, and average previous night sleep duration between the two sample donations at the beginning and the end of the study. Moreover, to address the potential U-shaped association between sleep duration and metabolites, we also created two categorical variables to indicate short, normal, and long sleep for average total time in bed and previous-night sleep. For previous-night sleep, we defined <6 hr as short sleep and >9 hr as long sleep. For total time in bed, we designated the bottom 10% (<7.3 hr) to the short and the upper 10% (≥10 hr) to the long category.
Measurement of metabolites
We used EDTA treated fasting plasma samples donated at the end of study year. Metabolite levels were measured by Metabolon, Inc. whose platform and procedures have been described previously (Evans et al., 2009, DeHaven et al., 2010). Briefly, samples were analyzed using ultra high performance liquid-phase chromatography coupled with mass spectrometry and tandem mass spectrometry (LC/MS and LC/MS2) and gas chromatography coupled with mass spectrometry (GC/MS). For each sample, the batch and position within a batch of 32 samples were randomly assigned, and the value for each metabolite was e normalized metabolite to the median for that batch. Individual metabolites were identified by comparing the mass spectra peaks to a chemical reference library. Of the 445 metabolites that were detected, 329 were of known identity. Identified metabolites were grouped into 8 chemical classes (amino acids, carbohydrates, cofactors and vitamins, energy metabolites, lipids, nucleotide metabolites, peptides, and xenobiotics) and 55 sub-pathways. Previous studies have reported a high level of reliability for the metabolomics platform used in this study (Sampson et al., 2013). Detailed methods for LC/MS, LC/MS2, and /GC-MS, as well as compound identification and curation are reported in Supplementary materials. Reference spectral data including retention time and m/z for the 329 metabolites detected in our study are presented in Supplementary table 1.
Covariates
The baseline questionnaire of the Shanghai Women’s Health Study and Shanghai Men’s Health Study collected demographic information including age and gender. Follow-up in-person interviews were conducted every two years. Height was measured at baseline and weight was measured repeatedly in follow-up interviews. To calculate body mass index (BMI, weight (kg)/height(m)2), we used baseline height and weight from the interview that was the closest to the date of sample collection (<2 years). Smoking status (current smoker or non-smoker) was reported when the plasma samples were taken. We calculated day-time napping duration based on activity log. In the same periods when activity log was completed, the participants were asked to wear an Actigraph accelerometer on the left hip at all times except when sleeping, showering and swimming. Total physical activity was calculated using actigraphy data using previously described methods (Peters et al., 2010). The time of sample donation was recorded in the study. The distribution of these covariates in the overall study and by sex is presented in Supplementary table 2.
Statistical analysis
Metabolite levels were first batch normalized and then log-transformed. Values below the detection threshold were set to the minimum observed value of the metabolite. The median level of “missingness” before imputation was 1%. Metabolites that were observed in ≤90% of the samples were excluded from the analysis. Pairwise correlations among metabolites were determined using the Pearson correlation coefficient, and correlations among sleep variables were determined using the Spearman correlation coefficient. Linear regression was used to estimate the association between each metabolite and sleep variables, adjusted for age (continuous), clock time at sample collection (continuous), sex (male, female), smoking status (yes, no), BMI (continuous), napping time (continuous), and physical activity energy expenditure (continuous). Moreover, because age and time of sample collection are important confounders and may have a nonlinear relationship with metabolites, we explored using spline terms for age and time at sample collection, but found the additional flexibility had little impact on the results. To account for multiple comparisons, we primarily used a false discovery rate (FDR) <0.2 to define statistical significance for purposes of reporting associations, but we also reported statistical significance using more stringent Bonferroni correction method (0.05/329=0.00015). We used the likelihood-ratio test to determine whether is a statistically significant interaction between sex and sleep in relation to metabolites. Because a statistically significant interaction was detected for some metabolites, we also performed stratified analysis in men and women separately.
We also evaluated the association between sleep variables and metabolomic patterns. Specifically, we used sparse principal component analysis (Zou et al., 2006) to create 10 principal components (PCs) with 10 non-zero loadings. We then performed step-wise regression between sleep variables and 10 PCs adjusted for the same set of covariates. We calculated the additional percent of variance in sleep variables explained by each of the PCs. We used Bonferroni correction to evaluate the statistical significance for interaction with sex (p<0.0078 (0.05/64)). All analyses were performed with SAS (version 9.1.3, SAS Institute, Cary, NC) and the R statistical language package (version 3.1.2).
Results
The distribution of the two main sleep variables by study characteristics is presented in Supplementary table 3. Participants with less than elementary school education had earlier average midpoint of time in bed, and time at sample donation was positively associated with midpoint. In contrast, sleep duration in the previous night before sample collection was only associated with clock time at sample collection.
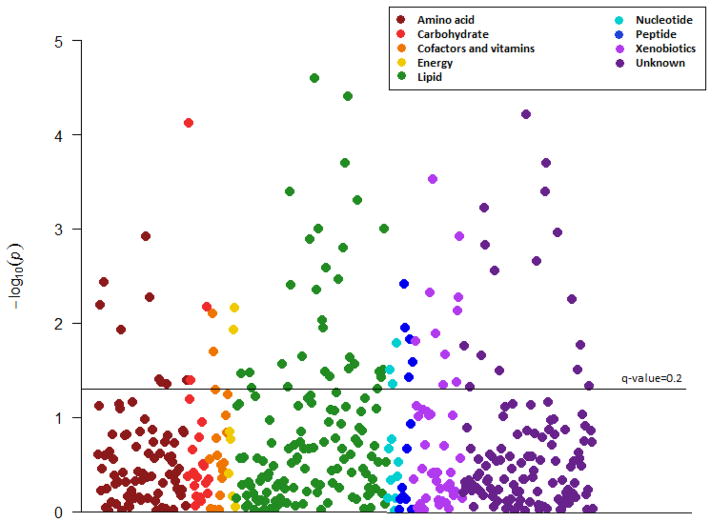
After controlling for all covariates, 64 of the 329 metabolites with known identity were found to be associated with average midpoint of time in bed with an FDR<0.2 (Table 1, Figure 1), and they represented all 8 biochemical classes and 29 out of the 55 sub-pathways. Late midpoint was associated with higher levels of amino acids, carbohydrates, bile acids, steroids, dipeptides and several xenobiotics. In contrast, midpoint point was inversely associated with all the carnitines and fatty acids (except for CMPF), as well as bilirubin and AMP. When we used Bonferroni correction, only three metabolites had a significant association with midpoint of sleep, and these include cis-4-decenoyl carnitine, laurylcarnitine, and 1,6-anhydroglucose. Detailed information on the specific metabolites as well as their effect estimates and p-values are presented in Figure 2, Table 2 (for the 64 metabolites associated with midpoint) and Supplementary table 4 (full list). When we conducted subgroup analysis by sex focusing on the 64 metabolites significantly associated with midpoint, the metabolite associations were the same in direction for both men and women and we did not find any statistically significant interaction with sex (Supplementary table 5). Additional adjustment of previous night sleep duration or average time spent in bed had little impact on the results (data not shown).
Table 1.
Number of metabolites with known identity detected by chemical class, and the number of metabolites that were significantly a associated with midpoint of time in bed
| Chemical class | Total number of known metabolites | Number (%) of known metabolites with significant association |
|---|---|---|
| All | 329 | 64 (19.5) |
| Amino acids | 81 | 9 (11.1) |
| Carbohydrates | 20 | 3 (15.0) |
| Cofactors and vitamins | 17 | 2 (11.8) |
| Energy metabolites | 7 | 2 (28.6) |
| Lipids | 136 | 29 (21.3) |
| Nucleotide metabolites | 11 | 3 (27.3) |
| Peptides | 13 | 5 (38.5) |
| Xenobiotics | 44 | 10 (22.7) |
Statistical significance defined as false discovery rate (FDR) <0.2
Figure 1.
Manhattan plot of metabolites associations with midpoint of time spent in bed
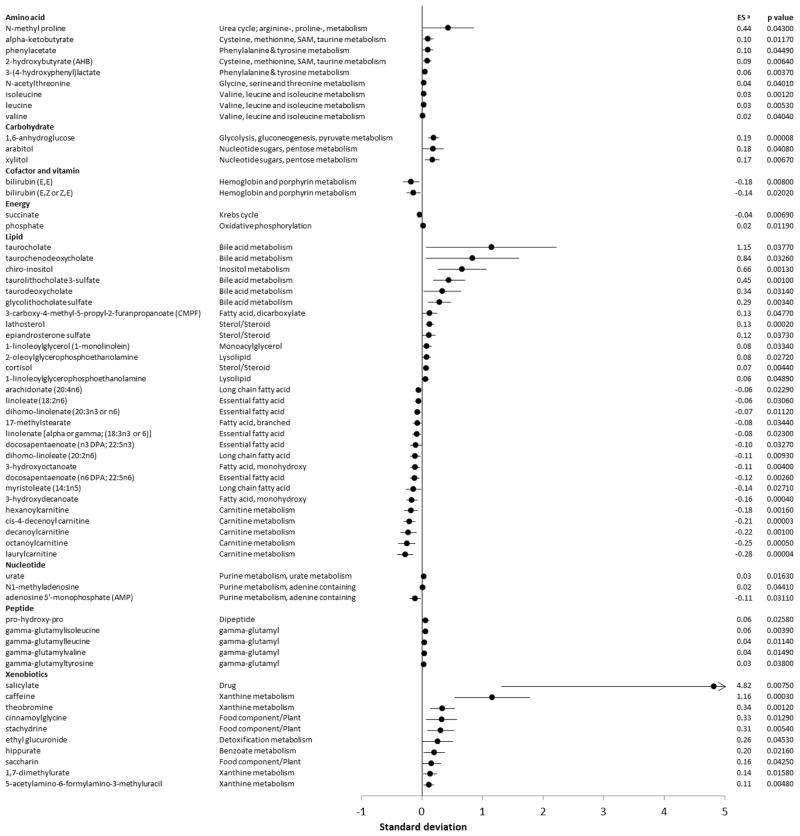
Figure 2.
Forest plot of metabolites associations with midpoint of time spent in bed. Associations shown on the plot are statistically significant, defined as false discovery rate<0.2. a Effect estimate expressed as changes in metabolite level (standard deviation, log scale) per 1 hour delay in midpoint of time. Results adjusted for age (continuous), sex (male, female), body mass index (continuous), smoking status (nonsmoker, smoker), napping (continuous), physical activity (continuous) and sampling time (continuous). Abbreviation: ES, effect size.
Table 2.
Metabolites associated a with midpoint of time in bed in the Shanghai Physical Activity Study
| Metabolite | Pathway | Effect estimate b | Standard error | P value | Q value | HMDB ID | KEGG ID |
|---|---|---|---|---|---|---|---|
| Amino acid | |||||||
| isoleucine | Valine, leucine and isoleucine metabolism | 0.03 | 0.01 | 0.0012 | 0.02 | HMDB00172 | C00407 |
| 3-(4-hydroxyphenyl)lactate | Phenylalanine & tyrosine metabolism | 0.06 | 0.02 | 0.0037 | 0.05 | HMDB00755 | C03672 |
| leucine | Valine, leucine and isoleucine metabolism | 0.03 | 0.01 | 0.0053 | 0.06 | HMDB00687 | C00123 |
| 2-hydroxybutyrate (AHB) | Cysteine, methionine, SAM, taurine metabolism | 0.09 | 0.03 | 0.0064 | 0.06 | HMDB00008 | C05984 |
| alpha-ketobutyrate | Cysteine, methionine, SAM, taurine metabolism | 0.1 | 0.04 | 0.0117 | 0.09 | HMDB00005 | C00109 |
| N-acetylthreonine | Glycine, serine and threonine metabolism | 0.04 | 0.02 | 0.0401 | 0.19 | C01118 | |
| valine | Valine, leucine and isoleucine metabolism | 0.02 | 0.01 | 0.0404 | 0.19 | HMDB00883 | C00183 |
| N-methyl proline | Urea cycle; arginine-, proline-, metabolism | 0.44 | 0.21 | 0.043 | 0.19 | ||
| phenylacetate | Phenylalanine & tyrosine metabolism | 0.1 | 0.05 | 0.0449 | 0.19 | HMDB00209 | C07086 |
| Carbohydrate | |||||||
| 1,6-anhydroglucose | Glycolysis, gluconeogenesis, pyruvate metabolism | 0.19 | 0.05 | 0.00008 | 0.01 | HMDB00640 | |
| xylitol | Nucleotide sugars, pentose metabolism | 0.17 | 0.06 | 0.0067 | 0.06 | HMDB00568 | C00379 |
| arabitol | Nucleotide sugars, pentose metabolism | 0.18 | 0.09 | 0.0408 | 0.19 | HMDB01851 | C00474 |
| Cofactor and vitamin | |||||||
| bilirubin (E,E) | Hemoglobin and porphyrin metabolism | −0.18 | 0.07 | 0.008 | 0.07 | ||
| bilirubin (E,Z or Z,E) | Hemoglobin and porphyrin metabolism | −0.14 | 0.06 | 0.0202 | 0.13 | ||
| Energy | |||||||
| succinate | Krebs cycle | −0.04 | 0.01 | 0.0069 | 0.06 | HMDB00254 | C00042 |
| phosphate | Oxidative phosphorylation | 0.02 | 0.01 | 0.0119 | 0.09 | HMDB01429 | C00009 |
| Lipid | |||||||
| cis-4-decenoyl carnitine | Carnitine metabolism | −0.21 | 0.05 | 0.00003 | 0.01 | ||
| laurylcarnitine | Carnitine metabolism | −0.28 | 0.07 | 0.00004 | 0.01 | HMDB02250 | |
| lathosterol | Sterol/Steroid | 0.13 | 0.03 | 0.0002 | 0.01 | HMDB01170 | C01189 |
| 3-hydroxydecanoate | Fatty acid, monohydroxy | −0.16 | 0.05 | 0.0004 | 0.01 | HMDB02203 | |
| octanoylcarnitine | Carnitine metabolism | −0.25 | 0.07 | 0.0005 | 0.02 | ||
| decanoylcarnitine | Carnitine metabolism | −0.22 | 0.07 | 0.001 | 0.02 | HMDB00651 | |
| taurolithocholate 3-sulfate | Bile acid metabolism | 0.45 | 0.13 | 0.001 | 0.02 | HMDB02580 | C03642 |
| chiro-inositol | Inositol metabolism | 0.66 | 0.2 | 0.0013 | 0.02 | ||
| hexanoylcarnitine | Carnitine metabolism | −0.18 | 0.06 | 0.0016 | 0.03 | HMDB00705 | C01585 |
| docosapentaenoate (n6 DPA; 22:5n6) | Essential fatty acid | −0.12 | 0.04 | 0.0026 | 0.04 | HMDB13123 | C06429 |
| glycolithocholate sulfate | Bile acid metabolism | 0.29 | 0.1 | 0.0034 | 0.05 | ||
| 3-hydroxyoctanoate | Fatty acid, monohydroxy | −0.11 | 0.04 | 0.004 | 0.05 | HMDB01954 | |
| cortisol | Sterol/Steroid | 0.07 | 0.03 | 0.0044 | 0.05 | HMDB00063 | C00735 |
| dihomo-linoleate (20:2n6) | Long chain fatty acid | −0.11 | 0.04 | 0.0093 | 0.08 | C16525 | |
| dihomo-linolenate (20:3n3 or n6) | Essential fatty acid | −0.07 | 0.03 | 0.0112 | 0.09 | HMDB02925 | C03242 |
| arachidonate (20:4n6) | Long chain fatty acid | −0.06 | 0.02 | 0.0229 | 0.14 | HMDB01043 | C00219 |
| linolenate [alpha or gamma; (18:3n3 or 6)] | Essential fatty acid | −0.08 | 0.04 | 0.023 | 0.14 | HMDB01388 | C06427 |
| myristoleate (14:1n5) | Long chain fatty acid | −0.14 | 0.06 | 0.0271 | 0.16 | HMDB02000 | C08322 |
| 2-oleoylglycerophosphoethanolamine | Lysolipid | 0.08 | 0.04 | 0.0272 | 0.16 | ||
| linoleate (18:2n6) | Essential fatty acid | −0.06 | 0.03 | 0.0306 | 0.17 | HMDB00673 | C01595 |
| taurodeoxycholate | Bile acid metabolism | 0.34 | 0.16 | 0.0314 | 0.17 | HMDB00896 | C05463 |
| taurochenodeoxycholate | Bile acid metabolism | 0.84 | 0.39 | 0.0326 | 0.17 | HMDB00951 | C05465 |
| docosapentaenoate (n3 DPA; 22:5n3) | Essential fatty acid | −0.1 | 0.05 | 0.0327 | 0.17 | HMDB01976 | C16513 |
| 1-linoleoylglycerol (1-monolinolein) | Monoacylglycerol | 0.08 | 0.04 | 0.0334 | 0.17 | ||
| 17-methylstearate | Fatty acid, branched | −0.08 | 0.04 | 0.0344 | 0.17 | ||
| epiandrosterone sulfate | Sterol/Steroid | 0.12 | 0.06 | 0.0373 | 0.18 | HMDB00365 | C07635 |
| taurocholate | Bile acid metabolism | 1.15 | 0.55 | 0.0377 | 0.18 | HMDB00036 | C05122 |
| 3-carboxy-4-methyl-5-propyl-2-furanpropanoate (CMPF) | Fatty acid, dicarboxylate | 0.13 | 0.06 | 0.0477 | 0.2 | ||
| 1-linoleoylglycerophosphoethanolamine | Lysolipid | 0.06 | 0.03 | 0.0489 | 0.2 | ||
| Nucleotide | |||||||
| urate | Purine metabolism, urate metabolism | 0.03 | 0.01 | 0.0163 | 0.12 | HMDB00289 | C00366 |
| adenosine 5′-monophosphate (AMP) | Purine metabolism, adenine containing | −0.11 | 0.05 | 0.0311 | 0.17 | HMDB00045 | C00020 |
| N1-methyladenosine | Purine metabolism, adenine containing | 0.02 | 0.01 | 0.0441 | 0.19 | HMDB03331 | C02494 |
| Peptide | |||||||
| gamma-glutamylisoleucine | gamma-glutamyl | 0.06 | 0.02 | 0.0039 | 0.05 | ||
| gamma-glutamylleucine | gamma-glutamyl | 0.04 | 0.02 | 0.0114 | 0.09 | HMDB11171 | |
| gamma-glutamylvaline | gamma-glutamyl | 0.04 | 0.02 | 0.0149 | 0.11 | HMDB11172 | |
| pro-hydroxy-pro | Dipeptide | 0.06 | 0.03 | 0.0258 | 0.16 | HMDB06695 | |
| gamma-glutamyltyrosine | gamma-glutamyl | 0.03 | 0.02 | 0.038 | 0.18 | ||
| Xenobiotics | |||||||
| caffeine | Xanthine metabolism | 1.16 | 0.32 | 0.0003 | 0.01 | HMDB01847 | C07481 |
| theobromine | Xanthine metabolism | 0.34 | 0.1 | 0.0012 | 0.02 | HMDB02825 | C07480 |
| 5-acetylamino-6-formylamino-3-methyluracil | Xanthine metabolism | 0.11 | 0.04 | 0.0048 | 0.05 | HMDB11105 | C16365 |
| stachydrine | Food component/Plant | 0.31 | 0.11 | 0.0054 | 0.06 | HMDB04827 | C10172 |
| salicylate | Drug | 4.82 | 1.79 | 0.0075 | 0.07 | HMDB01895 | C00805 |
| cinnamoylglycine | Food component/Plant | 0.33 | 0.13 | 0.0129 | 0.1 | ||
| 1,7-dimethylurate | Xanthine metabolism | 0.14 | 0.06 | 0.0158 | 0.11 | HMDB11103 | C16356 |
| hippurate | Benzoate metabolism | 0.2 | 0.09 | 0.0216 | 0.14 | HMDB00714 | C01586 |
| saccharin | Food component/Plant | 0.16 | 0.08 | 0.0425 | 0.19 | D01085 | |
| ethyl glucuronide | Detoxification metabolism | 0.26 | 0.13 | 0.0453 | 0.19 | ||
Associations presented in this table are statistically significant using a false discovery rate threshold of 0.2
Expressed as changes in metabolite level (standard deviation, log scale) per 1 hour delay in midpoint of time. Results adjusted for age (continuous), gender (male, female), body mass index (continuous), smoking status (nonsmoker, smoker), napping (continuous), physical activity (continuous) and sampling time (continuous).
An individual’s sleep schedule often differs between weekdays and weekends (Wong et al., 2015). However, in our population the average differences between midpoints on weekdays and weekends were small (<30 minutes for over 90% subjects and <10 minutes for over 50%). When we conducted a sensitivity analysis focusing on average midpoint on weekend evenings (Friday and Saturday nights) alone, we found that the results were largely similar to those for average of all nights (Supplementary table 6). Moreover, we did not find any metabolite that was associated with standard deviation of repeated measures of midpoint over the one-year period or the difference between averaged midpoints in weekdays and weekends (data not shown).
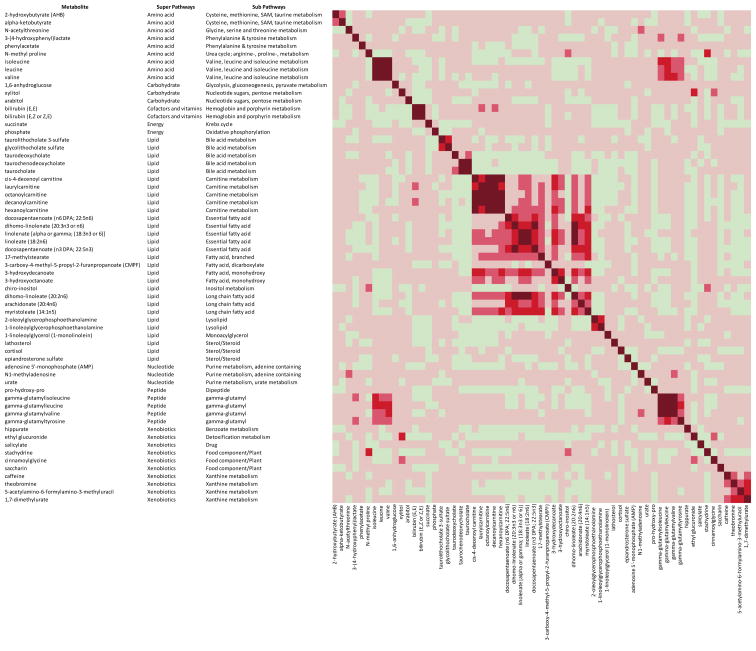
We investigated the clustering of the 64 metabolites that were significantly associated with midpoint of time in bed. The overall correlations were moderate to low (Pearson correlation coefficient<0.4). However, we observed very high correlations (Pearson correlation coefficient>0.8) among carnitines, branched chain amino acids and dipeptides. We also found moderate to high correlations (Pearson correlation coefficient, 0.4–0.8) among various fatty acids, between fatty acids and carnitines, and between amino acids and their dipeptide derivatives (Figure 3).
Figure 3.
This heat map shows the correlation among the known metabolites significantly a associated with midpoint of time in bed. The colors represent Pearson correlation coefficient: very dark red, ≥0.8; dark red, 0.6-<0.8; medium red, 0.4-<0.6; light red, 0-<0.4; light green, −0.4 - <0. a Statistical significance was determined using a false discovery rate threshold of 0.2.
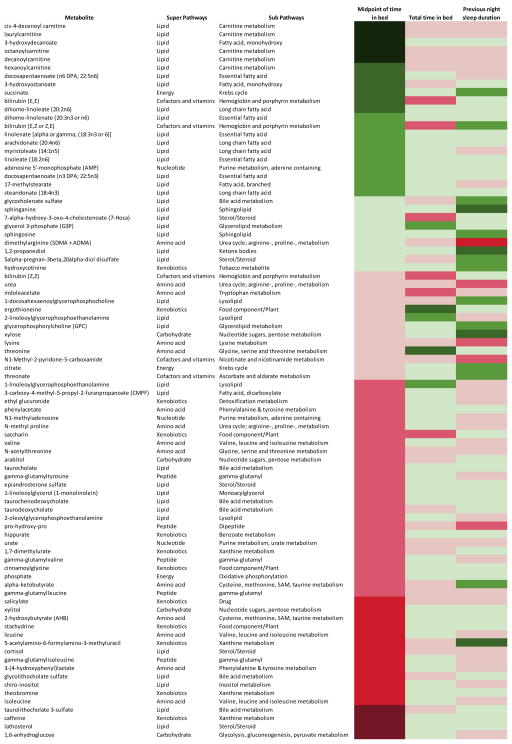
We examined metabolites in relation to previous night sleep duration, and we did not find any metabolite with a significantly (FDR<0.2) association (full list of metabolites associations are shown in Supplementary table 7). We also did not find any metabolite associations with yearly average of total time spent in bed as a proxy of habitual sleep duration (Supplementary table 8). Further analysis showed no metabolite associations with the short or long categories of total time in bed or previous night sleep duration (data not shown). Finally, we further compared the metabolite associations for sleep timing, previous night sleep duration and average time in bed (Figure 4). For a more complete comparison, we present all the metabolites that were associated with at least one of the sleep variables at p<0.05. Overall we found the pattern of metabolite associations for the three variables was quite distinct. Metabolites that showed stronger association with total time in bed and previous night sleep duration tended to be only weakly associated with midpoint of time in bed.
Figure 4.
This heat map shows the associations between metabolites and three sleep variables, average sleep timing, average time in bed and previous night sleep duration. Metabolites that are included in this figure have at least one association with a p-value<0.05. The 8 color codes represent different z-scores: very dark green, <−3.29 (p-value <0.001); dark green, −3.29 – <−2.58 (0.001-<0.01); medium green, −2.58 – <−1.96 (0.01-<0.05); light green, −1.96 – <0 (0.05–1); light red, >0 – 1.96 (0.05–1); medium red, >1.96 – 2.58 (0.01–<0.05); dark red, >2.58 – 3.29 (0.001–<0.01); very dark red >3.29 (<0.001). Positive z-scores suggest positive association (higher levels of metabolites with later midpoint) while negative z-scores suggest inverse association (higher levels of metabolites with earlier midpoint). All models were adjusted for age (continuous), sex (male, female), body mass index (continuous), smoking status (nonsmoker, smoker), napping (continuous), physical activity (continuous) and sampling time (continuous).
Discussion
Our study is the first to investigate the human metabolome in relation to habitual sleep. The sleep-wake cycle is driven by the internal circadian clock, which orchestrates a wide range of metabolic pathways, including carbohydrate, protein and amino acid, and lipid metabolism (Bailey et al., 2014). We found that sleep timing was associated with the fasting levels of a large number of metabolites across multiple biochemical pathways, highlighting the central role of circadian rhythms and sleep in human metabolism.
Amino acids and peptides
Both protein degradation and synthesis are regulated by the circadian system, with increased protein degradation during sleep and higher protein synthesis during awake time. (Bailey et al., 2014). Microarray studies have found diurnal variations in genes involved in protein turnover (Duffield et al., 2002), and previous studies have reported daily fluctuations in circulating amino acid levels (Dallmann et al., 2012, Weljie et al., 2015, Ang et al., 2012). We found sleep timing was associated with multiple amino acids. Notably, later sleep timing was associated higher circulating levels of valine, leucine, isoleucine (branched chain amino acids (BCAA)) and their gamma-glutamyl dipeptides, as well as several metabolites from the sulfur amino acids metabolism, and tyrosine and phenylalanine metabolism (alpha-hydroxybutyrate, alpha-ketobutyrate, Phenylacetate and 3-(4-hydroxyphenyl)lactate).
A growing body of research has linked BCAA metabolism, sulfur amino acids metabolism, and tyrosine and phenylalanine metabolism pathways to impaired cardiometabolic health (Adams, 2011, Lynch and Adams, 2014). For example, it has been suggested that higher concentrations of circulating BCAAs, alpha-hydroxybutyrate and alpha-ketobutyrate may serve as markers of insulin resistance (Batch et al., 2014, Tom and Nair, 2006, Ferrannini et al., 2013, Gall et al., 2010). Moreover, circulating levels of BCAAs and 3-(4-hydroxyphenyl)lactate were found to be associated with higher BMI (Moore et al., 2014). Interestingly, a recent study found fasting isoleucine level was elevated after eight nights of sleep restriction (5.5 hour) in 12 healthy adults (Bell et al., 2013), and another study found higher levels of leucine and valine following one night sleep restriction (4 hour) in rats (Weljie et al., 2015). It is worth noting that the magnitude of the associations between sleep timing and amino acids observed in our study was relatively small – one hour of delay in midpoint was associated with only <0.1 SD increase in many amino acids in this population, which may not lead to clinically meaningful difference. However, on average, our study subjects had an early sleep timing: the average midpoint of sleep of our study population was 2:30 am, 1 hour earlier than what was reported in previous literature for the same age group on work-free days (Roenneberg et al., 2007). We cannot exclude the possibility that the effect of sleep timing on metabolites may be nonlinear, and the potentially adverse effect of sleep timing may become more pronounced when sleep timing becomes more extremely late.
Together with the previous findings, our findings suggest that late sleep timing was associated with systematic changes in several amino acid pathways, some of which were previously indicated in cardiometabolic disease risk. More future studies are needed to directly examine the potential role of amino acid metabolism in mediating the health effects of sleep timing.
Lipids
Internal circadian clocks control multiple aspects of lipid metabolism (Bailey et al., 2014). Several recent metabolomics studies reported 24-hr oscillations of multiple lipid metabolites (Dallmann et al., 2012, Ang et al., 2012, Davies et al., 2014), even when the subjects followed a constant routine that removed external stimuli such as changes in light, physical activities and food intakes (Dallmann et al., 2012). In our study, we found that a large number of compounds in multiple lipid pathways were associated with sleep timing. However, it is worth noting that because the circulating levels of many lipids show circadian variation, the associations found in our study may reflect changes in average metabolite levels, alterations in the amplitude of the fluctuation, or a phase shift in the fluctuation patterns. Unfortunately we only measured plasma metabolites at one time point during the day, and therefore we cannot determine which of the aforementioned mechanisms drove the findings observed in our study.
Most notably, we found that late sleep timing was associated with lower levels of multiple fatty acids. Moreover we found that sleep timing was also inversely associated with several acylcarnitines, which play an essential role in transporting fatty acids into mitochondria, where fatty acid β-oxidation takes place (McCoin et al., 2015). The molecular clock regulate multiple enzymes involved in fatty acid metabolism (Gooley and Chua, 2014), including CPT-1, the enzyme that converts acyl-CoA to acylcarnitine (Panda et al., 2002, Hughes et al., 2009). Interestingly, in a recent study by Weljie et al., human subjects were subject to 5-day sleep restriction (4 hours of sleep per night) and then a night of recovery sleep. The study found reduced levels of acylcarnitines among subjects who did not recover after sleep restriction (Weljie et al., 2015). However, another study reported higher levels of acylcarnitines during total sleep deprivation (Davies et al., 2014). The discrepancy in these two studies indicates that acute effects induced by sleep deprivation may be different from chronic sleep debt. Our findings suggest that in addition to chronic sleep debt, habitually late sleep timing may also be associated with systematic alterations in fatty acid metabolism, and more future studies are needed to investigate the long-term impact of circadian dysfunction in real-world situations on lipid metabolism.
Higher levels of bile acids were associated with late sleep timing in our study. Bile acids are major cholesterol metabolites that are synthesized in the liver. Elevated bile acid levels have been linked with dyslipidemia and hyperglycemia, and recent studies suggested that bile acid sequestrants that aim at reducing circulating bile acid levels can cause markedly improvement in these two conditions (Brinton, 2008). Moreover, we found that higher levels of lathosteral, an important maker for impaired cholesterol metabolism (Farkkila et al., 1996, Matthan et al., 2013), was also associated with late sleep timing. Taken together, our findings are consistent with a disruptive role of late sleep timing on bile acid and cholesterol homeostasis.
Other metabolites and pathways
A number of xenobiotic metabolites were positively associated with late sleep timing in our study. Higher levels of markers of coffee consumption such as caffeine, hippurate, theobromine, and cinnamoylglycine (Guertin et al., 2015) were strongly associated with late midpoint of time in bed, which is consistent with the established association between coffee consumption and late sleep timing (Roehrs and Roth, 2008). Additionally, late sleep timing was also associated with salicylate, a major ingredient in pain medications, suggesting that certain medical conditions may be in play. Finally, several other metabolites associated with sleep timing are also potential markers of certain dietary exposures, including glucurionide (alcohol) (Dinis-Oliveira, 2016), xylitol (sweetner), CMPF (fish intake), 1-linoleoylglycerol (plant lipids), and prolylhydroxyproline (collagen supplement), stachydrine and chiro inositol (citrus)) (Guertin et al., 2014). Taken together, lifestyle factors may be responsible for some of the observed metabolite associations with sleep timing.
Sleep duration
We did not find any metabolite associations with total time in bed or previous night sleep duration, which may be explained by a number of factors. First, total time in bed may include other non-sleep-related activity such as reading and watching television. Previous work showed that total time in bed is a poor measure of sleep behavior and does not distinguish between controls and patients with insomnia (Natale et al., 2009). On the other hand, self-reported previous night sleep duration may be more accurate. However, it does not necessarily reflect habitual sleep duration. Moreover, we had a relatively narrow distribution of sleep duration and were underpowered to detect meaningful associations with short sleep: ~90% participants reported between 6 and 9 hours of sleep and only 22 participants reported less than 6 hours of sleep. We believe future studies with better measurement of sleep duration and larger sample size will be needed to investigate the metabolic profiles associated with quantity of sleep.
Strengths and limitations
A major strength of our study is that we have repeated measures (~28 days) of sleep over a one-year period, which allowed us to more accurately assess habitual sleep. Moreover, using an untargeted approach to examine the metabolome, we were able to measure a large number of metabolites across a broad range of biochemical pathways, many of which have never been examined before in relation to sleep. However, there are also several limitations of our study. First, as mentioned above, we did not have information on habitual sleep duration. Second, our population has a fairly stable sleep routine with small overall variation and little difference between weekdays and weekends, probably due to their older age and the fact that most of our study participants were retired at the time of the study. This limited our ability to examine metabolites in relation to shift sleep schedule between weekday and weekends, which has been previously shown to be an important cardiometabolic risk factor (Wong et al., 2015). However, the stability in sleep routines also has important advantage, because it suggests that in this population, day-time schedule might have little impact on sleep timing. Therefore, our measurement may well reflect the intrinsic preference of sleep timing (chronotype). Third, we have identified a number of metabolites that were markers of diet, coffee drinking and alcohol consumption, which suggest that uncontrolled lifestyle factors may had an impact on our results. Particularly, dietary intake and timing of the last meal may be associated with both sleep timing and plasma levels of certain metabolites. Unfortunately we don’t have information on the last meal before blood donation and could not control for its confounding effects. Moreover, although we adjusted for several health behaviors, including napping, physical activity and smoking, confounding due to other lifestyle factors and environmental factors may also have an impact on our results. Finally, we did not have multiple measurements of metabolites at different times of the day, and therefore we were unable to assess the diurnal fluctuation patterns of metabolite levels.
Conclusion
In summary, we found late sleep timing was associated with a large number of metabolites across a variety of biochemical pathways. Although we cannot rule out confounding completely, many metabolites associated with sleep timing in our study were also previously linked with cardiometabolic health and warrant further investigation, particularly in lipid and amino acid metabolism. Overall, our study provides new insight into the biological mechanisms underlying the health effects of sleep and points to the need for future studies to better understand the role of circadian rhythms in metabolic regulation.
Supplementary Material
Acknowledgments
Funding: This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Department of Health and Human Services.
Footnotes
Compliance with Ethical Standards:
Conflicts of interest: the authors declare no conflicts of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: Informed consent was obtained from all individual participants included in the study.
References
- ADAMS SH. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv Nutr. 2011;2:445–56. doi: 10.3945/an.111.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANG JE, REVELL V, MANN A, MANTELE S, OTWAY DT, JOHNSTON JD, THUMSER AE, SKENE DJ, RAYNAUD F. Identification of Human Plasma Metabolites Exhibiting Time-of-Day Variation Using an Untargeted Liquid Chromatography-Mass Spectrometry Metabolomic Approach. Chronobiology International. 2012;29:868–881. doi: 10.3109/07420528.2012.699122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAILEY SM, UDOH US, YOUNG ME. Circadian regulation of metabolism. J Endocrinol. 2014;222:R75–96. doi: 10.1530/JOE-14-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASS J, TAKAHASHI JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–54. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BATCH BC, HYLAND K, SVETKEY LP. Branch chain amino acids: biomarkers of health and disease. Curr Opin Clin Nutr Metab Care. 2014;17:86–9. doi: 10.1097/MCO.0000000000000010. [DOI] [PubMed] [Google Scholar]
- BELL LN, KILKUS JM, BOOTH JN, 3RD, BROMLEY LE, IMPERIAL JG, PENEV PD. Effects of sleep restriction on the human plasma metabolome. Physiol Behav. 2013;122:25–31. doi: 10.1016/j.physbeh.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRINTON EA. Novel pathways for glycaemic control in type 2 diabetes: focus on bile acid modulation. Diabetes Obesity & Metabolism. 2008;10:1004–1011. doi: 10.1111/j.1463-1326.2008.00903.x. [DOI] [PubMed] [Google Scholar]
- CAPPUCCIO FP, COOPER D, D’ELIA L, STRAZZULLO P, MILLER MA. Sleep duration predicts cardiovascular outcomes: a systematic review and meta-analysis of prospective studies. Eur Heart J. 2011;32:1484–92. doi: 10.1093/eurheartj/ehr007. [DOI] [PubMed] [Google Scholar]
- CAPPUCCIO FP, D’ELIA L, STRAZZULLO P, MILLER MA. Quantity and quality of sleep and incidence of type 2 diabetes: a systematic review and meta-analysis. Diabetes Care. 2010;33:414–20. doi: 10.2337/dc09-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DALLMANN R, VIOLA AU, TAROKH L, CAJOCHEN C, BROWN SA. The human circadian metabolome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIES SK, ANG JE, REVELL VL, HOLMES B, MANN A, ROBERTSON FP, CUI N, MIDDLETON B, ACKERMANN K, KAYSER M, THUMSER AE, RAYNAUD FI, SKENE DJ. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 2014;111:10761–6. doi: 10.1073/pnas.1402663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEHAVEN CD, EVANS AM, DAI HP, LAWTON KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. Journal of Cheminformatics. 2010:2. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DINIS-OLIVEIRA RJ. Oxidative and Non-Oxidative Metabolomics of Ethanol. Curr Drug Metab. 2016 doi: 10.2174/1389200217666160125113806. [DOI] [PubMed] [Google Scholar]
- DUFFIELD GE, BEST JD, MEURERS BH, BITTNER A, LOROS JJ, DUNLAP JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Current Biology. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- EVANS AM, DEHAVEN CD, BARRETT T, MITCHELL M, MILGRAM E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- FARKKILA MA, KAIREMO KJ, TAAVITSAINEN MJ, STRANDBERG TA, MIETTINEN TA. Plasma lathosterol as a screening test for bile acid malabsorption due to ileal resection: Correlation with (75)SeHCAT test and faecal bile acid excretion. Clinical Science. 1996;90:315–319. doi: 10.1042/cs0900315. [DOI] [PubMed] [Google Scholar]
- FERRANNINI E, NATALI A, CAMASTRA S, NANNIPIERI M, MARI A, ADAM KP, MILBURN MV, KASTENMULLER G, ADAMSKI J, TUOMI T, LYSSENKO V, GROOP L, GALL WE. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62:1730–7. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALL WE, BEEBE K, LAWTON KA, ADAM KP, MITCHELL MW, NAKHLE PJ, RYALS JA, MILBURN MV, NANNIPIERI M, CAMASTRA S, NATALI A, FERRANNINI E, GROUP RS. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAN Y, YANG C, TONG X, SUN H, CONG Y, YIN X, LI L, CAO S, DONG X, GONG Y, SHI O, DENG J, BI H, LU Z. Shift work and diabetes mellitus: a meta-analysis of observational studies. Occup Environ Med. 2015;72:72–8. doi: 10.1136/oemed-2014-102150. [DOI] [PubMed] [Google Scholar]
- GOOLEY JJ, CHUA EC. Diurnal regulation of lipid metabolism and applications of circadian lipidomics. J Genet Genomics. 2014;41:231–50. doi: 10.1016/j.jgg.2014.04.001. [DOI] [PubMed] [Google Scholar]
- GUERTIN KA, LOFTFIELD E, BOCA SM, SAMPSON JN, MOORE SC, XIAO Q, HUANG WY, XIONG X, FREEDMAN ND, CROSS AJ, SINHA R. Serum biomarkers of habitual coffee consumption may provide insight into the mechanism underlying the association between coffee consumption and colorectal cancer. Am J Clin Nutr. 2015;101:1000–11. doi: 10.3945/ajcn.114.096099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUERTIN KA, MOORE SC, SAMPSON JN, HUANG WY, XIAO Q, STOLZENBERG-SOLOMON RZ, SINHA R, CROSS AJ. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr. 2014;100:208–17. doi: 10.3945/ajcn.113.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGHES ME, DITACCHIO L, HAYES KR, VOLLMERS C, PULIVARTHY S, BAGGS JE, PANDA S, HOGENESCH JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANTERMANN T, SUNG H, BURGESS HJ. Comparing the Morningness-Eveningness Questionnaire and Munich ChronoType Questionnaire to the Dim Light Melatonin Onset. J Biol Rhythms. 2015;30:449–53. doi: 10.1177/0748730415597520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LYNCH CJ, ADAMS SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10:723–36. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATTHAN NR, ZHU L, PENCINA M, D’AGOSTINO RB, SCHAEFER EJ, LICHTENSTEIN AH. Sex-Specific Differences in the Predictive Value of Cholesterol Homeostasis Markers and 10-Year Cardiovascular Disease Event Rate in Framingham Offspring Study Participants. Journal of the American Heart Association. 2013:2. doi: 10.1161/JAHA.112.005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCOIN CS, KNOTTS TA, ADAMS SH. Acylcarnitines--old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol. 2015;11:617–25. doi: 10.1038/nrendo.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOORE SC, MATTHEWS CE, SAMPSON JN, STOLZENBERG-SOLOMON RZ, ZHENG W, CAI Q, TAN YT, CHOW WH, JI BT, LIU DK, XIAO Q, BOCA SM, LEITZMANN MF, YANG G, XIANG YB, SINHA R, SHU XO, CROSS AJ. Human metabolic correlates of body mass index. Metabolomics. 2014;10:259–269. doi: 10.1007/s11306-013-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NATALE V, PLAZZI G, MARTONI M. Actigraphy in the assessment of insomnia: a quantitative approach. Sleep. 2009;32:767–71. doi: 10.1093/sleep/32.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PANDA S, ANTOCH MP, MILLER BH, SU AI, SCHOOK AB, STRAUME M, SCHULTZ PG, KAY SA, TAKAHASHI JS, HOGENESCH JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- PETERS TM, MOORE SC, XIANG YB, YANG G, SHU XO, EKELUND U, JI BT, TAN YT, LIU DA K, SCHATZKIN A, ZHENG W, CHOW WH, MATTHEWS CE, LEITZMANN MF. Accelerometer-measured physical activity in Chinese adults. Am J Prev Med. 2010;38:583–91. doi: 10.1016/j.amepre.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROEHRS T, ROTH T. Caffeine: sleep and daytime sleepiness. Sleep Med Rev. 2008;12:153–62. doi: 10.1016/j.smrv.2007.07.004. [DOI] [PubMed] [Google Scholar]
- ROENNEBERG T, KUEHNLE T, JUDA M, KANTERMANN T, ALLEBRANDT K, GORDIJN M, MERROW M. Epidemiology of the human circadian clock. Sleep Med Rev. 2007;11:429–38. doi: 10.1016/j.smrv.2007.07.005. [DOI] [PubMed] [Google Scholar]
- ROENNEBERG T, WIRZ-JUSTICE A, MERROW M. Life between clocks: daily temporal patterns of human chronotypes. J Biol Rhythms. 2003;18:80–90. doi: 10.1177/0748730402239679. [DOI] [PubMed] [Google Scholar]
- SAMPSON JN, BOCA SM, SHU XO, STOLZENBERG-SOLOMON RZ, MATTHEWS CE, HSING AW, TAN YT, JI BT, CHOW WH, CAI Q, LIU DA K, YANG G, XIANG YB, ZHENG W, SINHA R, CROSS AJ, MOORE SC. Metabolomics in epidemiology: sources of variability in metabolite measurements and implications. Cancer Epidemiol Biomarkers Prev. 2013;22:631–40. doi: 10.1158/1055-9965.EPI-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAN Z, MA H, XIE M, YAN P, GUO Y, BAO W, RONG Y, JACKSON CL, HU FB, LIU L. Sleep duration and risk of type 2 diabetes: a meta-analysis of prospective studies. Diabetes Care. 2015;38:529–37. doi: 10.2337/dc14-2073. [DOI] [PubMed] [Google Scholar]
- SHU XO, LI H, YANG G, GAO J, CAI H, TAKATA Y, ZHENG W, XIANG YB. Cohort Profile: The Shanghai Men’s Health Study. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOM A, NAIR KS. Assessment of branched-chain amino Acid status and potential for biomarkers. J Nutr. 2006;136:324S–30S. doi: 10.1093/jn/136.1.324S. [DOI] [PubMed] [Google Scholar]
- VAN DRONGELEN A, BOOT CR, MERKUS SL, SMID T, VAN DER BEEK AJ. The effects of shift work on body weight change - a systematic review of longitudinal studies. Scand J Work Environ Health. 2011;37:263–75. doi: 10.5271/sjweh.3143. [DOI] [PubMed] [Google Scholar]
- VYAS MV, GARG AX, IANSAVICHUS AV, COSTELLA J, DONNER A, LAUGSAND LE, JANSZKY I, MRKOBRADA M, PARRAGA G, HACKAM DG. Shift work and vascular events: systematic review and meta-analysis. BMJ. 2012;345:e4800. doi: 10.1136/bmj.e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG F, ZHANG L, ZHANG Y, ZHANG B, HE Y, XIE S, LI M, MIAO X, CHAN EY, TANG JL, WONG MC, LI Z, YU IT, TSE LA. Meta-analysis on night shift work and risk of metabolic syndrome. Obes Rev. 2014;15:709–20. doi: 10.1111/obr.12194. [DOI] [PubMed] [Google Scholar]
- WELJIE AM, MEERLO P, GOEL N, SENGUPTA A, KAYSER MS, ABEL T, BIRNBAUM MJ, DINGES DF, SEHGAL A. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci U S A. 2015;112:2569–74. doi: 10.1073/pnas.1417432112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WITTMANN M, DINICH J, MERROW M, ROENNEBERG T. Social jetlag: misalignment of biological and social time. Chronobiol Int. 2006;23:497–509. doi: 10.1080/07420520500545979. [DOI] [PubMed] [Google Scholar]
- WONG PM, HASLER BP, KAMARCK TW, MULDOON MF, MANUCK SB. Social Jetlag, Chronotype, and Cardiometabolic Risk. J Clin Endocrinol Metab. 2015 doi: 10.1210/jc.2015-2923. jc20152923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU Y, ZHAI L, ZHANG D. Sleep duration and obesity among adults: a meta-analysis of prospective studies. Sleep Med. 2014;15:1456–62. doi: 10.1016/j.sleep.2014.07.018. [DOI] [PubMed] [Google Scholar]
- XIAO Q, MOORE SC, KEADLE SK, XIANG YB, ZHENG W, PETERS TM, LEITZMANN MF, JI BT, SAMPSON JN, SHU XO, MATTHEWS CE. Objectively measured physical activity and plasma metabolomics in the Shanghai Physical Activity Study. Int J Epidemiol. 2016 doi: 10.1093/ije/dyw033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHENG W, CHOW WH, YANG G, JIN F, ROTHMAN N, BLAIR A, LI HL, WEN W, JI BT, LI Q, SHU XO, GAO YT. The Shanghai Women’s Health Study: rationale, study design, and baseline characteristics. Am J Epidemiol. 2005;162:1123–31. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
- ZOU H, HASTIE T, TIBSHIRANI R. Sparse principal component analysis. Journal of Computational and Graphical Statistics. 2006;15:265–286. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.