Abstract
Fructophily is a rare trait that consists of the preference for fructose over other carbon sources. Here, we show that in a yeast lineage (the Wickerhamiella/Starmerella, W/S clade) comprised of fructophilic species thriving in the high-sugar floral niche, the acquisition of fructophily is concurrent with a wider remodeling of central carbon metabolism. Coupling comparative genomics with biochemical and genetic approaches, we gathered ample evidence for the loss of alcoholic fermentation in an ancestor of the W/S clade and subsequent reinstatement through either horizontal acquisition of homologous bacterial genes or modification of a pre-existing yeast gene. An enzyme required for sucrose assimilation was also acquired from bacteria, suggesting that the genetic novelties identified in the W/S clade may be related to adaptation to the high-sugar environment. This work shows how even central carbon metabolism can be remodeled by a surge of HGT events.
Research organism: Other
eLife digest
Cells build their components, such as the molecular machinery that helps them obtain energy from their environment, by following the instructions contained in genes. This genetic information is usually transferred from parents to offspring. Over the course of several generations, genes can accumulate small changes and the molecules they code for can acquire new roles: yet, this process is normally slow. However, certain organisms can also obtain completely new genes by ‘stealing’ them from other species. For example, yeasts, such as the ones used to make bread and beer, can take genes from nearby bacteria. This ‘horizontal gene transfer’ helps organisms to rapidly gain new characteristics, which is particularly useful if the environment changes quickly.
One way that yeasts get the energy they need is by breaking down sugars through a process called alcoholic fermentation. To do this, most yeast species prefer to use a sugar called glucose, but a small group of ‘fructophilic’ species instead favors a type of sugar known as fructose. Scientists do not know exactly how fructophilic yeasts came to be, but there is some evidence horizontal gene transfers may have been involved in the process.
Now, Gonçalves et al. have compared the genetic material of fructophilic yeasts with that of other groups of yeasts . Comparing genetic material helps scientists identify similarities and differences between species, and gives clues about why specific genetic features first evolved.
The experiments show that, early in their history, fructophilic yeasts lost the genes that allowed them to do alcoholic fermentation, probably since they could obtain energy in a different way. However, at a later point in time, these yeasts had to adapt to survive in flower nectar, an environment rich in sugar. They then favored fructose as their source of energy, possibly because this sugar can compensate more effectively for the absence of alcoholic fermentation. Later, the yeasts acquired a gene from nearby bacteria, which allowed them to do alcoholic fermentation again: this improved their ability to use the other sugars present in flower nectars.
When obtaining energy, yeasts and other organisms produce substances that are relevant to industry. Studying natural processes of evolution can help scientists understand how organisms can change the way they get their energy and adapt to new challenges. In turn, this helps to engineer yeasts into ‘cell factories’ that produce valuable chemicals in environmentally friendly and cost-effective ways.
Introduction
Comparative genomics is a powerful tool for discovering links between phenotypes and genotypes within an evolutionary framework. While extraordinary progress in this respect has been observed in all domains of life, analyses of the rapidly increasing number of fungal genomes available has been particularly useful to highlight important aspects of eukaryotic genomes, including a broader scope of evolutionary mechanisms than was thus far deemed likely. For example, horizontal gene transfers (HGT) are thought to have played a very important role in domestication (Gibbons et al., 2012; Marsit et al., 2015; Ropars et al., 2015) and in the evolution of metabolism in fungi (Alexander et al., 2016; Wisecaver and Rokas, 2015). Instances of the latter are best showcased by the high frequency of HGT events involving gene clusters related to fungal primary and secondary metabolism (Campbell et al., 2012; Khaldi and Wolfe, 2011; Slot and Rokas, 2010; 2011; Wisecaver and Rokas, 2015). When considering the horizontal transfer of single genes, those encoding nutrient transporters seem to be among the most frequently transferred (Coelho et al., 2013; Gonçalves et al., 2016; Richards, 2011). While the identification of HGT events can be straightforward given sufficient sampling of the lineages under study, inferences concerning the evolutionary driving forces behind HGT are often difficult and uncertain, because most HGT events identified are ancient. However, available evidence suggests that HGTs are often associated with rapid adaptation to new environments (Cheeseman et al., 2014; Gojković et al., 2004; Qiu et al., 2013; Richards et al., 2011; Richards and Talbot, 2013).
In line with these findings, we recently reported on the evolutionary history of a unique, high-capacity, specific fructose transporter, Ffz1, which is intimately associated with fructophilic metabolism in ascomycetous budding yeasts (subphylum Saccharomycotina) (Gonçalves et al., 2016). Fructophily is a relatively rare trait that consists in the preference for fructose over other carbon sources, including glucose (Cabral et al., 2015; Gonçalves et al., 2016; Sousa-Dias et al., 1996). The evolution of FFZ1 involved the likely horizontal acquisition of the gene from filamentous fungi (subphylum Pezizomycotina) by the most recent common ancestor (MRCA) of a lineage in the Saccharomycotina, composed so far entirely of fructophilic yeasts (Gonçalves et al., 2016). Most of the approximately one hundred species forming this clade (Wickerhamiella and Starmerella genera, as well as closely related Candida species), are associated with the floral niche and are often isolated from fructose-rich nectar (Canto et al., 2017; de Vega et al., 2017; Lachance et al., 2001). Interestingly, fructophilic lactic acid bacteria, whose metabolism has been dissected in detail, also populate the floral niche (Endo et al., 2009; Endo and Salminen, 2013). These bacteria have been shown to grow poorly on glucose, which can be at least partly explained by their lack of respiratory chain enzymes and alcohol dehydrogenase activity, deficiencies that hinder NAD+ regeneration during growth on this sugar, as shown for Lactobacillus kunkei (Maeno et al., 2016). In contrast to glucose, fructose can be used both as a carbon source and as an electron acceptor for the re-oxidation of NAD(P)H (Zaunmüller et al., 2006), providing an explanation of why it is favored over glucose. Hence, fructophily in lactic acid bacteria seems to be linked to redox homeostasis (Endo et al., 2014). In yeasts, it is still unclear how preferential consumption of fructose may be beneficial, partly because unlike fructophilic bacteria, fructophilic yeasts grow vigorously on glucose when it is the only carbon and energy source available (Sousa-Dias et al., 1996; Tofalo et al., 2012). Our previous work suggested that, although a strict correlation was found so far between the presence of Ffz1 and fructophily in all species investigated (Cabral et al., 2015; Gonçalves et al., 2016; Leandro et al., 2014) and the requirement for FFZ1 was genetically confirmed in the fructophilic species Zygosaccharomyces rouxii (Leandro et al., 2014), it is very likely that there are additional requirements for fructophily. Thus, the FFZ1 gene does not seem to be sufficient to impart a fructophilic character to a previously glucophilic species.
To gain insight into the genetic underpinnings of fructophily in budding yeasts and how it may have become evolutionarily advantageous, here we used comparative genomics to identify traits, in addition to the presence of the FFZ1 gene, that might differentiate yeasts in the fructophilic Wickerhamiella/Starmerella (W/S) clade, focusing on central carbon metabolism. Our results suggest that the evolution of fructophily may have been part of a process of adaptation to sugar-rich environments, which included a profound remodeling of alcoholic fermentation involving the acquisition of bacterial alcohol dehydrogenases, which turned out to be particularly important for glucose metabolism, and an invertase, which is essential for sucrose assimilation. In general, we found a surge of bacterial-derived HGT events in the W/S clade when compared with other lineages in the Saccharomycotina (Marcet-Houben and Gabaldón, 2010), many of which seem to impact redox homeostasis.
Results
The horizontally transferred Ffz1 transporter is essential for fructophily in St. bombicola
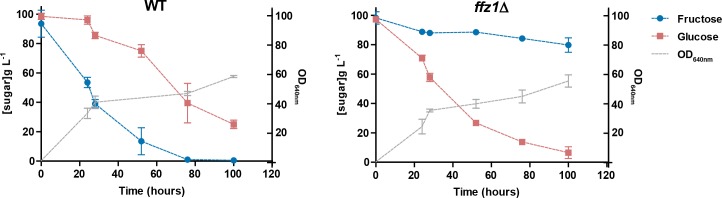
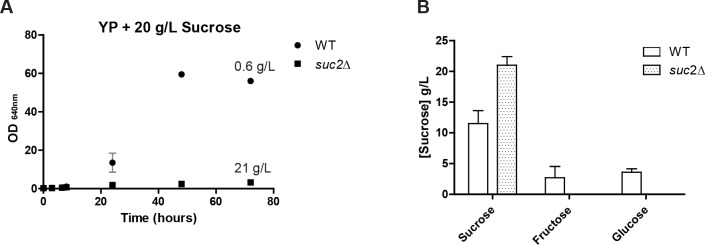
We previously reported the acquisition of a high-capacity fructose transporter (Ffz1) through HGT by the MRCA of W/S-clade species. This transporter was lost in the MRCA of the Saccharomycotina and was later transferred from a Pezizomycotina-related species to the MRCA of the W/S clade, and then from the W/S clade to the MRCA of the Zygosaccharomyces genus (Gonçalves et al., 2016). A putative role for Ffz1 in fructophily in the W/S clade was hypothesized based on its kinetic properties (Pina et al., 2004) and the evidence that it is indispensable for fructophily in the phylogenetically distant species Z. rouxii (Leandro et al., 2014). To test this hypothesis, a FFZ1 deletion mutant was constructed in the genetically tractable W/S-clade species Starmerella bombicola. The sugar-consumption profile in YP medium supplemented with 10% (w/v) fructose and 10% (w/v) glucose (conditions where fructophily is apparent, hereafter referred to as 20FG medium), showed that fructophilic behavior was completely abolished in the ffz1∆ mutant (Figure 1), similarly to what was found in Z. rouxii (Leandro et al., 2014). A slight increase in the glucose consumption rate was also observed for the ffz1∆ mutant compared to the wild type when cultures were grown in 20FG medium (Figure 1).
Figure 1. Sugar consumption profiles of St. bombicola wild type (wt) and ffz1Δ.
Strains were grown in YP supplemented with 10% (w/v) fructose and 10% (w/v) glucose for 100 hr at 30°C with aeration. Error bars represent standard deviation of assays performed in duplicate in two biological replicates.
Alcoholic fermentation genes in W/S-clade yeasts
One of the most distinctive metabolic characteristics found in fructophilic bacteria that distinguished them from closely related non-fructophilic species was the lack of the enzymatic activity required for ethanol production and concomitant NAD(P)+ regeneration. In one well-studied species, the gene encoding the bifunctional alcohol dehydrogenase (ADH)/aldehyde dehydrogenase (ALDH) normally responsible for ethanol production was absent (Endo et al., 2014), whereas in another species it was present but the encoded protein lacked the domain responsible for ethanol production (ADH), while maintaining the domain that conducts the preceding reaction (Maeno et al., 2016). On the other hand, some W/S-clade yeasts were previously known to be efficient producers of sugar alcohols or lipids to the detriment of ethanol (Magyar and Tóth, 2011;Lee et al., 2003a; Kurtzman et al., 2010). These observations prompted us to investigate whether alcohol dehydrogenase genes in fructophilic yeasts might also provide clues pertaining to a relation between fructophily and cofactor recycling in yeasts, as a first step toward unraveling other metabolic determinants of fructophily.
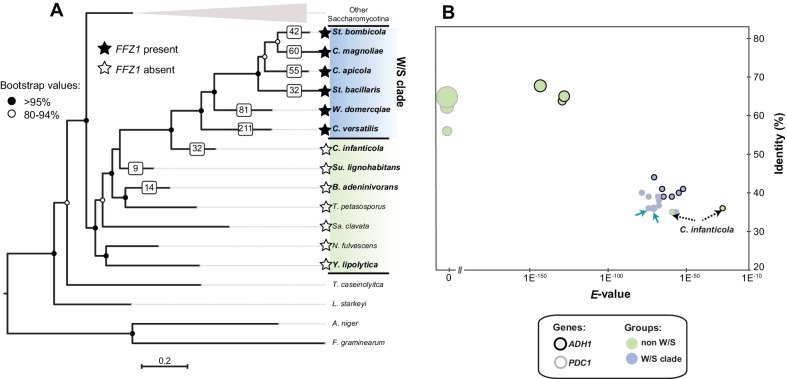
In S. cerevisiae, the ADH1 gene encodes the enzyme mainly responsible for the conversion of acetaldehyde into ethanol (de Smidt et al., 2008). Hence, we started by retrieving homologs of S. cerevisiae ADH1 from the genomes of six W/S-clade species as well as from four of their closest relatives (Figure 2A) using tBLASTx. Among the non-W/S-clade species considered, Candida infanticola (Kurtzman, 2007) occupies a particularly informative position, since it was phylogenetically placed as an outgroup of the W/S clade in our species phylogeny (Figure 2A), being its closest relative among the species included in this analysis. It has presently not been considered part of the W/S clade because it lacks the Ffz1 transporter (Figure 2A) and has not been isolated so far from sugar-rich habitats (Kurtzman, 2007). Notably, while the phylogenetic distance between all the species surveyed and S. cerevisiae was similar, protein sequence identity, E-value, and bitscore values denoted that predicted Adh1 proteins retrieved from W/S-clade species and C. infanticola as top hits of the tBLASTx search were much less similar to the S. cerevisiae ADH1 query than the genes recovered from their non-fructophilic counterparts Sugiyamaella lignohabitans, Blastobotrys adeninivorans, and Yarrowia lipolytica (Figure 2B). Moreover, when Adh1 protein sequences from W/S-clade species and C. infanticola were used as queries in BLASTp searches in the NCBI non-redundant (nr) database, the top 1000 hits consisted entirely of bacterial proteins, while when Adh1 sequences of Su. lignohabitans, B. adeninivorans, and Y. lipolytica yeasts were similarly employed as queries, the top 1000 hits recovered were fungal proteins. Taken together, these results suggest that W/S-clade species and C. infanticola have Adh1 homologs of bacterial origin, in contrast to the remaining three species.
Figure 2. Maximum Likelihood phylogeny of Saccharomycotina (A) and tBLASTx results for alcoholic fermentation genes (B).
(A) ML phylogeny depicting the phylogenetic relationship between W/S species (highlighted in blue) and closest relatives (highlighted in green); other Saccharomycotina lineages were collapsed as indicated. Names of the species are abbreviated as indicated in Figure 2—source data 1. Presence (black stars) and absence (white stars) of the Ffz1 transporter is indicated for each species. AI results are shown as the number of trees in which phylogenetic clustering with bacteria was confirmed for each species tested in the respective branch (white squares). (B) tBLASTx results for ADH1 and PDC1 searches in W/S-clade species and closest relatives Su. lignohabitans, B. adeninivorans, and Y. lipolytica (highlighted in bold in Figure 2A). For each gene and species, the plot depicts the E-value (x axis), identity (y axis), and bitscore (z axis, diameter of the circle) relative to the S. cerevisiae orthologs. C. versatilis Pdc proteins of bacterial origin are indicated by the blue arrows.
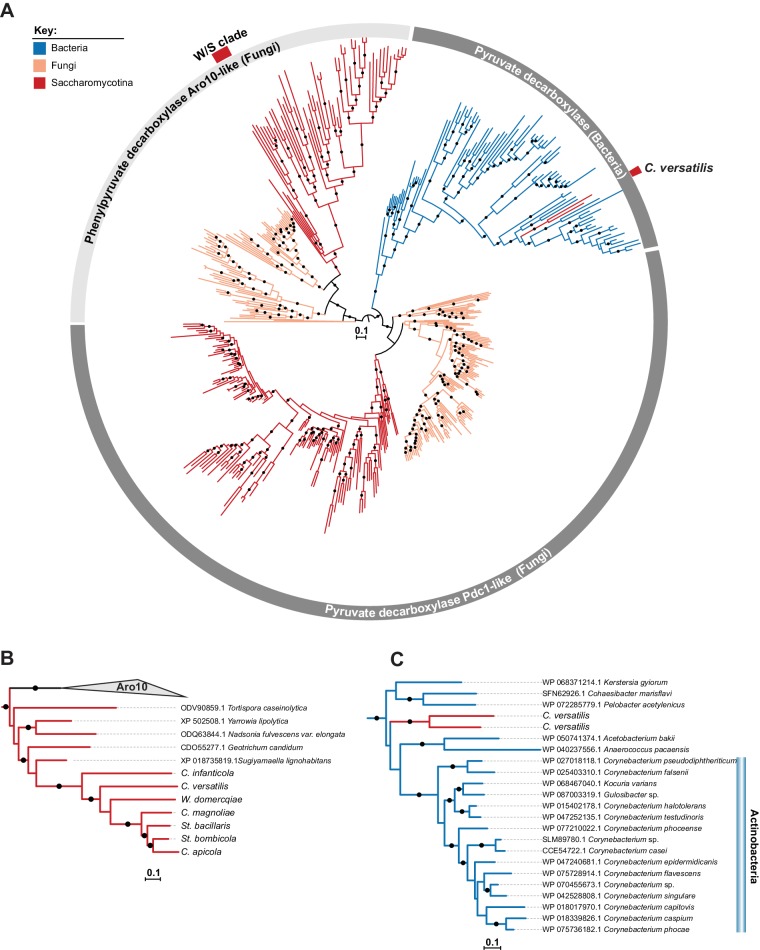
The first step in the alcoholic fermentation pathway consists in the conversion of pyruvate in acetaldehyde, catalyzed by pyruvate decarboxylase (Pdc) (Hohmann and Cederberg, 1990). Since the gene encoding the ‘native’ enzyme catalyzing the second step, ADH1, is missing from W/S-clade genomes and seems to have been ‘replaced’ by a bacterial version, we next examined whether PDC genes mirrored somehow the peculiarities in the evolution of ADH1 observed in the W/S clade. To this end, the sequence of the gene encoding the enzyme mainly responsible for conversion of pyruvate to acetaldehyde in S. cerevisiae, Pdc1, was used to retrieve its homologs in the set of species identified in Figure 2A. Remarkably, the Pdc sequences retrieved in this manner from the genomes of W/S-clade species and C. infanticola were also found to be more dissimilar to S. cerevisiae Pdc1 than those recovered from the three remaining non-fructophilic species, based on sequence identity, E-values, and bitscores (Figure 2B). In line with the observations for Adh1 sequences, two out of the three Pdc sequences identified in C. versatilis, were more closely related to bacterial Pdc proteins than to fungal Pdc enzymes (Figure 2B). However, a BLASTp search using the third Pdc sequence from C. versatilis and the remaining Pdc sequences from W/S-clade species as queries, showed that their closest relatives were fungal proteins. In this case, the lower E-value appears to reflect the fact that the Pdc orthologs found in W/S-clade species and C. infanticola seem to belong to a decarboxylase family that is phylogenetically related to S. cerevisiae Aro10. In S. cerevisiae, Aro10 acts preferentially on substrates other than pyruvate and is not involved in alcoholic fermentation (Kneen et al., 2011; Romagnoli et al., 2012; Vuralhan et al., 2005). To better assess the phylogenetic relation between Aro10 and Pdc1-related sequences, and determine the evolutionary origin of the sequences identified in the W/S clade, a Maximum Likelihood (ML) phylogeny was reconstructed using the top 500 NCBI BLASTp hits using S. cerevisiae Pdc1 (CAA97573.1), St. bombicola putative Pdc ortholog, and C. versatilis Pdc sequences from apparent bacterial origin as queries. Putative Pdc sequences from the other W/S-clade species not available at the NCBI database were also included. This phylogeny (Figure 3A and B) confirmed the clustering of the W/S-clade sequences with Aro10 proteins from fungi, which indicates that PDC1 was lost in the W/S clade. Additionally, as suggested by the BLASTp results, the two Pdc1-like proteins from C. versatilis were clustered with bacterial pyruvate decarboxylases (Figure 3C).
Figure 3. ML phylogeny of Pdc1 and Aro10.
A) ML phylogeny of Pdc1-like proteins. W/S-clade and C. versatilis sequences are indicated by red blocks. Branches with bootstrap support higher than 90% are indicated by black dots. The different lineages are represented with different branch colors (red for Saccharomycotina, blue for bacteria and orange for other Fungi (i.e. non-Saccharomycotina). Clades highlighted in grey (Aro10- and Pdc1-like) were assigned according to the phylogenetic position of functionally characterized S. cerevisiae proteins. (B, C) Pruned ML phylogenies depicting the phylogenetic relationship between W/S-clade Aro10 proteins and their closest relatives in the Saccharomycotina (B) and between C. versatilis Pdc1 xenologs and the closest related bacterial pyruvate decarboxylases (C). For W/S- clade sequences, protein ID is indicated before the abbreviated species name.
All glycolytic genes were examined in the same set of species and were all found to be present and to exhibit the expected level of similarity to S. cerevisiae query proteins (Figure 2—source data 3). This, together with the fact that other inspected publicly available genome assemblies of W/S-clade species are of very high quality (e.g. Wickerhamiella domercqiae JCM 9478, PRJDB3620 or St. bombicola JCM 9596 from RIKEN Center), makes it very unlikely that alcoholic fermentation genes were missed in W/S-clade species because of insufficient coverage or quality of the genome assemblies used.
Identification of other genes of bacterial origin in the W/S clade
The tBLASTx analyses revealed that extant ADH1 genes in W/S-clade species and also in C. infanticola were likely horizontally transferred from bacteria, and the same was observed for PDC1 in C. versatilis. To identify other genes of bacterial origin in the W/S clade that might be related to central carbon metabolism and therefore to fructophily, a systematic high-throughput analytical pipeline based on the Alien Index (AI) score (Alexander et al., 2016; Gladyshev et al., 2008) was employed for HGT-detection in the previously defined group of species comprising W/S-clade representatives and close relatives outside the clade.
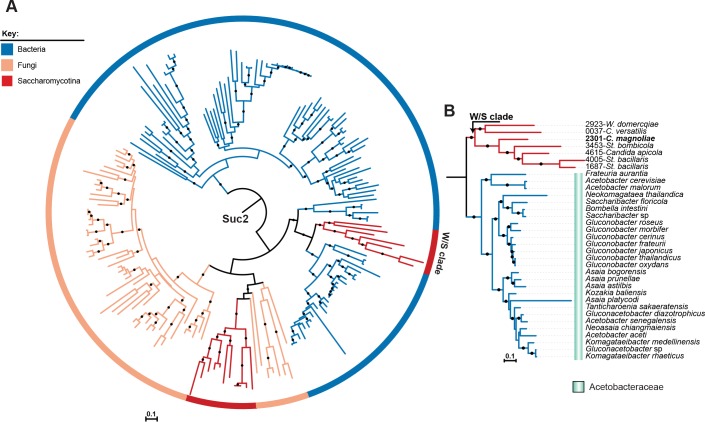
We found a considerably larger number of genes of bacterial origin in W/S-clade species and also in C. infanticola when compared with the two species more distantly related to the W/S clade (Su. lignohabitans and B. adeninivorans, Figure 2A, Figure 2—source data 2). Notably, C. versatilis displayed the highest number of putative HGT-derived genes from bacteria (211), followed by W. domercqiae with 80 genes for which phylogenetic clustering with bacteria could be confirmed (Figure 2A). Given that all W/S-clade species possess a higher number of HGT-derived genes when compared to their closest relatives, it is possible that a surge of HGT events occurred in the MRCA of the W/S clade. In line with this hypothesis, we predicted that it would be possible to identify a meaningful number of genes that were retained in more than one W/S-clade species. Indeed, after implementation of alignment thresholds (protein sequences > 150 amino acids) and collapsing the replicate phylogenies and lineage-specific gene duplications, it was possible to define 52 ortholog groups with representatives in two or more W/S-clade genomes. Not excluding the possibility that independent events also occurred in the various species, this suggests that a surge of HGT events took place in the MRCA of the W/S clade and that different species subsequently retained different sets of genes, C. versatilis being the species that retained the most genes of bacterial origin (Figure 2A). As expected, ADH1 was present in this group (Figure 2—source data 2). PDC1 was absent because only ortholog groups of bacterial origin detected in at least two W/S-clade species were selected, and bacterial Pdc1 proteins were only identified in C. versatilis. Another ortholog group relevant to fructose metabolism found among the 52 defined in this manner was SUC2, which encodes an invertase in S. cerevisiae that extracellularly hydrolyzes sucrose into fructose and glucose (Carlson et al., 1981). In S. cerevisiae, the MAL and the IMA genes have also been shown to play a role in sucrose hydrolysis (Deng et al., 2014; Voordeckers et al., 2012), but these genes are absent in the genomes of W/S-clade species, meaning that the horizontally transferred invertase appears to be the only enzyme with sucrose-hydrolyzing capacity encoded in the W/S-clade genomes investigated. In a phylogenetic tree reconstructed using the 200 top phmmer hits to W/S-clade species Candida magnoliae (protein ID 2301, see Figure 2—source data 2), strong support for clustering of W/S-clade sequences to Acetobacteraceae SacC sequences was observed (Figure 4A and Figure 4B). A topology comparison test (Approximately Unbiased, AU) also strongly supported the HGT hypothesis (p-value=4e−07).
Figure 4. ML phylogeny of fungal Suc2 and bacterial SacC proteins.
A) ML phylogeny of Suc2/SacC proteins (top 200 phmmer hits). W/S-clade species are highlighted. The different lineages are represented with different branch colors (red for Saccharomycotina, blue for bacteria, and orange for other Fungi (i.e. non-Saccharomycotina)). Branches with bootstrap support higher than 90% are indicated by black dots. (B) Pruned ML phylogeny of Suc2 depicting the phylogenetic relationship between the W/S clade and Acetobacteraceae. For W/S sequences, protein ID is indicated before the abbreviated species name.
Figure 4—figure supplement 1. Growth and consumption of sucrose in St.
To check for putative enrichment in other protein functions among the remainder of the transferred genes, the 52 ortholog groups were subsequently cross-referenced with the Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and InterPro annotations. Notably, out of 43 proteins to which a GO molecular function was assigned, 16 impacted redox homeostasis and were associated with oxidoreductase activity (GO:0016491 and GO:0016616, 14 genes) and peroxidase activity (GO:004601, two genes), while a BLAST KOALA annotation (Kanehisa et al., 2016) indicated that the biological processes most frequently involved were amino acid metabolism, carbohydrate metabolism, and metabolism of cofactors and vitamins (Figure 2—source data 4). Notably, some of these genes appeared to have undergone several intraspecific duplications, in particular those encoding oxidorreductases participating in various metabolic pathways.
Evolution of alcohol dehydrogenase genes in W/S-clade species
We noted that, in addition to ADH1, other alcohol dehydrogenase genes seem to have been horizontally acquired by W/S-clade species (Figure 2—source data 4), including putative ADH6 and SFA1 orthologs, which can also participate in alcoholic fermentation in S. cerevisiae (Drewke et al., 1990; Ida et al., 2012). In all cases, except for SFA1, the ‘native’ yeast orthologs appear to have been lost in W/S-clade genomes. This could imply that ethanol production is conducted by alcohol dehydrogenases of bacterial origin in W/S-clade species. To learn more about the evolutionary history of these genes, detailed phylogenetic analyses were conducted for Adh1 and Adh6, for which maximum likelihood phylogenies were reconstructed using the top phmmer hits obtained using St. bombicola Adh1 and Adh6 proteins as queries.
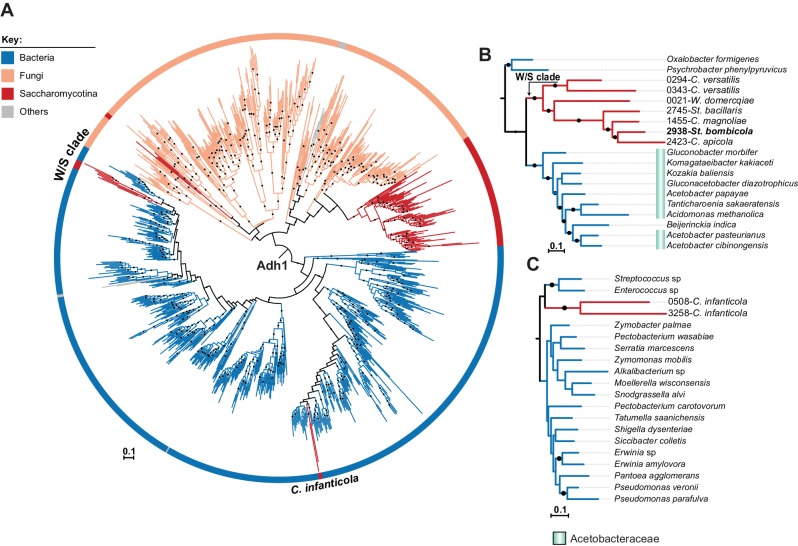
The resulting Adh1 tree (Figure 5A) included protein sequences from both bacteria and fungi. All W/S-clade species clustered with strong support with the Acetobacteraceae (Proteobacteria) Adh1 proteins (Figure 5A and Figure 5B). Within the Adh1 W/S-clade cluster, the overall phylogenetic relationships were in line with the expected relationships between the species (Figure 2A), suggesting that a single HGT event occurred in the MRCA of this clade. Topology comparison tests (AU) strongly supported the Adh1 HGT event to the W/S clade (p-value=8e−03), adding to the strong evidence of HGT provided by the robustly supported branch that clusters the W/S-clade xenologs with bacteria and the AI results. Adh1 sequences from C. infanticola also clustered with those of proteins of bacterial origin. However, the two Adh1 sequences from C. infanticola are not similar to those of the W/S clade (Figure 5C), as might be expected if a single HGT event were responsible for the acquisition of the ADH1 gene in both lineages. These sequences instead grouped, albeit with weak support, with Adh1 sequences from the distantly related Lactobacillales and Enterobacteriales (Figure 5C), implying that an independent HGT event may have occurred in the C. infanticola lineage. Nonetheless, topology tests did not strongly support an independent origin for W/S and C. infanticola Adh1 proteins (p-value=0.073).
Figure 5. ML phylogeny of fungal and bacterial Adh1 proteins.
(A) ML phylogeny of Adh1 proteins (top 4000 phmmer hits). W/S-clade species and C. infanticola are highlighted. The different lineages are represented with different branch colors (red for Saccharomycotina, blue for bacteria, and orange for other Fungi (i.e. non-Saccharomycotina)). Branches with bootstrap support higher than 90% are indicated by black dots. Poorly represented lineages (<10 sequences) are shown in grey. (B, C) Pruned ML phylogenies of Adh1 depicting the phylogenetic relationship between the W/S clade and Acetobacteraceae (B) and between C. infanticola and other groups of bacteria (C). For W/S-clade sequences, the protein ID is indicated before the abbreviated species name.
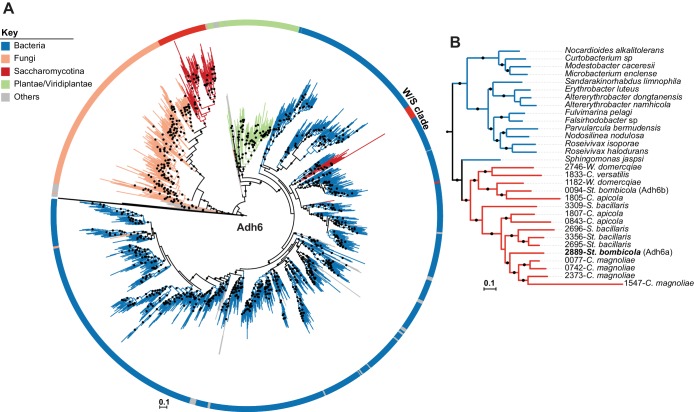
An extended Adh6 ML phylogeny (Figure 6A) was reconstructed with the top 10,000 phmmer hits to show that the W/S-clade sequences are indeed Adh6 orthologs. In this phylogeny, W/S-clade sequences clustered with strong support (>95%) with Proteobacteria Adh6 sequences, within a large cluster that also encompasses known fungal Adh6 proteins. Remarkably, while Adh1 W/S-clade sequences grouped with those of the Acetobacteraceae (Figure 5A and Figure 5B), the Adh6 sequences are more closely related to those of other bacterial families, as highlighted in Figure 6B. The phmmer search failed to uncover Adh6 sequences in the C. infanticola proteome. The absence of an ADH6 ortholog was further confirmed by a tBLASTx search against the C. infanticola genome using Adh6 sequences from both S. cerevisiae (KZV09178.1) and St. bombicola as queries. This result suggests that both ADH1 and ADH6 were lost in an ancestor of C. infanticola and the W/S clade. Interestingly, the ADH6 xenologs were apparently duplicated several times within each W/S-clade species (Figure 6A, Figure 6B and Figure 2—source data 4), and Starmerella bacillaris and C. magnoliae harbor the most paralogs (four in total).
Figure 6. ML Phylogeny of Adh6 proteins.
(A) The phylogeny was constructed with the top 10,000 phmmer hits using St. bombicola Adh6 as a query (in bold, Panel B). Sequences with more than 80% similarity were eliminated. Branches with bootstrap support higher than 90% are indicated by black dots. Poorly represented lineages (<10 sequences) are shown in grey. The different lineages are represented with different branch colors (red for Saccharomycotina, blue for bacteria, and orange for other Fungi (i.e. non-Saccharomycotina)). Adh1-like sequences were collapsed as indicated. (B) Pruned ML phylogeny depicting the phylogenetic relationship between Adh6 sequences from the W/S clade and their closest bacterial relatives. For W/S-clade sequences, the protein ID is indicated before the abbreviated species name.
Kinetic properties of alcohol dehydrogenases of bacterial origin in W/S-clade species
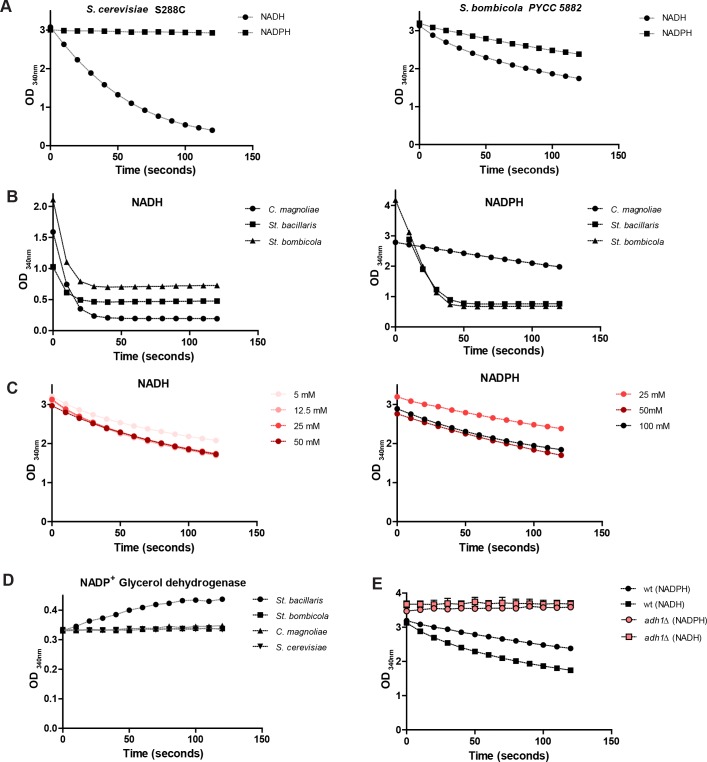
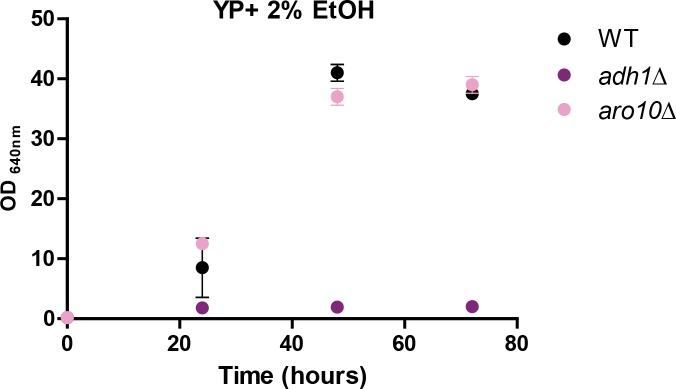
Acquisition of bacterial alcohol dehydrogenases by W/S-clade yeasts could have been driven by putative benefits afforded by an enzyme with kinetic characteristics that provide some advantage that the ‘native’ enzyme presumably lacked or, alternatively, by the need to restore alcoholic fermentation after an ancestral loss event, possibly in connection to adaptation to a new environment. To help elucidate this and also taking into account the link found between Adh activity and fructophily in bacteria, we set out to expound putative functional differences between Adh xenologs found in the W/S clade and ‘native’ Adh enzymes. Specifically, we compared alcohol dehydrogenase (Adh) activity in three W/S-clade species, in the closely related yeast B. adeninivorans, and in the model species S. cerevisiae, as well as in a distantly related fructophilic yeast species Zygosaccharomyces kombuchaensis. All W/S-clade species tested were capable of using ethanol as carbon and energy source and have a Crabtree-negative behavior when growing on sugars, meaning that ethanol production in aerated batch cultures starts only when cell densities are very high,limiting oxygen availability (typically OD640 nm 30,~5–10 g/L ethanol in St. bombicola).
In all non-W/S-clade species tested, the characteristic NADH-dependent Adh activity was readily observed, but no NADPH-dependent Adh activity was detected (Figure 5—figure supplement 2A), in line with available information concerning yeast enzymes (Cho and Jeffries, 1998; Dashko et al., 2014; Ganzhorn et al., 1987; Leskovac et al., 2002). Conversely, all W/S-clade species tested (St. bombicola, C. magnoliae, and St. bacillaris) exhibited Adh activity when either NADH or NADPH was added to the reaction mixture (Figure 5—figure supplement 2B). In fact, although both cofactors could be used for conversion of acetaldehyde into ethanol, there was a lower affinity for the substrate (higher Km) for NADPH-dependent activity in St. bombicola (Figure 5—figure supplement 2C). Interestingly, in Acetobacter pasteurianus a bacterial species in the Acetobacteraceae, the same family as the likely donor of W/S-clade Adh1, alcohol dehydrogenase activity was found to be NADH-dependent, although it is unclear whether NADPH was also tested as a cofactor (Masud et al., 2011).
Genetic dissection of alcoholic fermentation in Starmerella bombicola
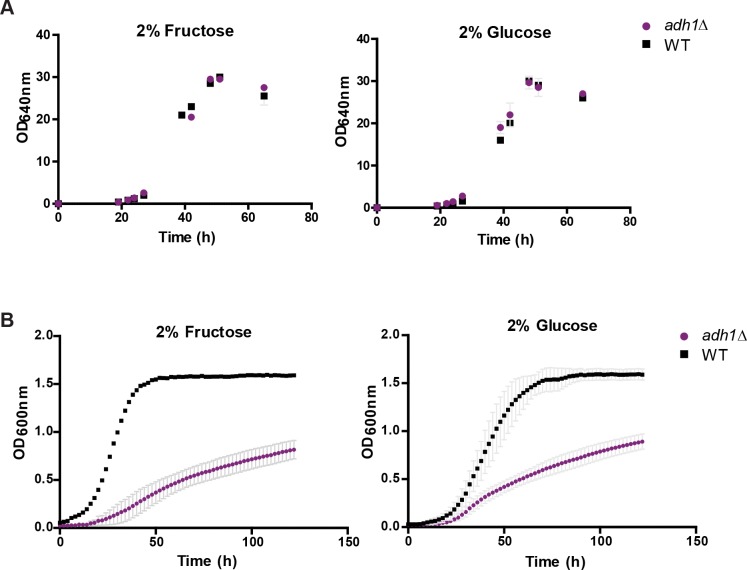
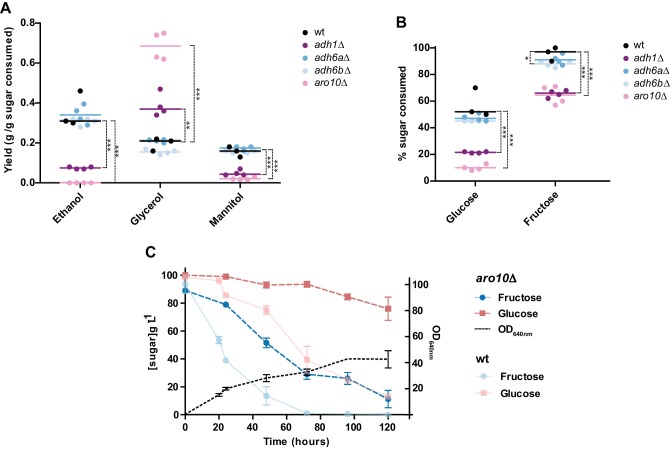
In S. cerevisiae, the paralogous enzymes Adh1, Adh2, Adh3, and Adh5 were all shown to contribute to different degrees to the inter-conversion of ethanol and acetaldehyde in a NADH-dependent manner. Although their participation in alcoholic fermentation is not substantial, as may be inferred from the lack of detectable (NADP+ dependent) activity in S. cerevisiae crude cell extracts (Figure 5—figure supplement 2A), Adh6 and Adh7 can, in principle, also catalyze this type of reaction, using NADPH instead of NADH (de Smidt et al., 2008). Since both ADH1- and ADH6-like genes are present in the genomes of all W/S-clade species studied, it was not clear which enzyme (Adh1-type or Adh6-type) was responsible for the NADPH-dependent inter-conversion of ethanol and acetaldehyde observed in W/S-clade species. To elucidate this, and to evaluate the impact of alcohol dehydrogenases on metabolism, three deletion mutants were constructed in St. bombicola (adh1∆, adh6a∆, and adh6b∆). During aerated growth, deletion of ADH1 (adh1∆) did not seem to significantly affect specific growth rates in glucose or fructose when compared to the wild type (Figure 5—figure supplement 3A). However, we noted a five-fold decrease in ethanol production (Figure 7A) and the absence of growth on ethanol as sole carbon and energy source in the adh1∆ mutant (Figure 7—figure supplement 1). Although some ethanol was produced, no Adh activity was detected in cell-free extracts of the adh1∆ mutant when either NADH or NADPH was used (Figure 5—figure supplement 2E). These results suggest that Adh1 is the main enzyme used in alcoholic fermentation in St. bombicola and that it therefore likely accepts both NADH and NADPH as cofactors. We predicted that, if W/S-clade Adh1 enzymes were mainly used in the recycling of NADPH, which does not normally occur in yeasts, in its absence, compensation would be expected to occur in other NADP+ regenerating reactions. If on the contrary, W/S-clade Adh1 enzymes were mainly used in the recycling of NADH, the compensatory increase of a NAD+ producing reaction would be observed. In line with the cofactor preference measured in cell extracts, the significant decrease in ethanol yield seems to be counterbalanced by a concomitant increase in glycerol production (Figure 7A), similarly to what has been observed in the S. cerevisiae adh1∆ mutant (de Smidt et al., 2012). Moreover, growth of the adh1∆ mutant cultivated on identical growth medium but under limited aeration was severely affected (Figure 5—figure supplement 3B). Taken together, these observations strongly suggest that Adh1 plays an important role in redox homeostasis, namely in NAD+ regeneration in the absence of oxygen because, similarly to S. cerevisiae, glycerol formation in St. bombicola is probably a NAD+ regenerating reaction. Consistent with this hypothesis, we did not detect NADP+ dependent glycerol dehydrogenase activity in cell-free extracts (Figure 5—figure supplement 2D), and in at least one W/S-clade species, this reaction was shown to be NADH-dependent (Van Bogaert et al., 2008). In contrast, mannitol production, which is a NADP+ regenerating reaction (Lee et al., 2003b), was significantly decreased in the adh1∆ mutant (Figure 7A).
Figure 7. Metabolite production and sugar consumption in St. bombicola wild type (wt) and deletion mutants (adh1Δ, adh6aΔ, adh6bΔ, and aro10Δ).
(A) Ethanol, glycerol, and mannitol yields determined after 72 hr of growth. (B) Percentage of sugar (fructose and glucose) consumed after 72 hr of growth. Mean values are represented by the colored horizontal lines. Assays were performed in duplicate in two biological replicates. All strains were grown in 20FG medium at 30°C with aeration. Statistically significant differences (one way ANOVA, using the Bonferroni’s correction) between wt and deletion mutants for sugar consumption and metabolite production are shown (* p-value<0.05; ** p-value<0.01; *** p-value<0.001). Additional p-values for other pairwise comparisons are shown in Figure 7—source data 1. (C) Sugar consumption profile of aro10∆ mutant grown in 20FG medium. Sugar consumption profile is also shown for the wt strain as indicated in the key and was previously reported in Figure 1. Error bars represent standard deviation of assays performed in duplicate in two biological replicates.
The deletion of each of the two ADH6 paralogous genes (adh6a∆ and adh6b∆ mutants) did not significantly affect ethanol production (Figure 7A). This means that, although some of the enzymes encoded by these genes might be involved in the production of ethanol in the absence of Adh1, when Adh1 is functional, they are not essential for ethanol metabolism and are probably mainly involved in other metabolic reactions. Finally, to ascertain how perturbations in alcoholic fermentation might affect the relative preference of St. bombicola for fructose over glucose, we monitored the consumption of both sugars in aerated cultures over time in the adh1∆, adh6a∆, and adh6b∆ mutants. There was a significant decrease in sugar consumption rates in the adh1∆ mutant (Figure 7B), but fructophily was still observed in all mutants and became even more pronounced as the lack of Adh1 affected glucose consumption more than it did fructose consumption.
Orthologs of S. cerevisiae Pdc1, which catalyzes the first step in the fermentative pathway, appeared to be absent in W/S-clade genomes. Since all W/S-clade species investigated produce ethanol, which requires prior decarboxylation of pyruvate to acetaldehyde, it follows that the role normally fulfilled by Pdc1 in S. cerevisiae must have been taken over by a different enzyme in W/S-clade yeasts. According to our phylogenetic analysis, the only candidate likely to assume this function would be the product of the ARO10 gene (Figure 3A). However, and although it displays some amino acid sequence similarity with Pdc1, S. cerevisiae Aro10 displayed extremely low affinity for pyruvate as a substrate (Kneen et al., 2011). Therefore, to ascertain whether Aro10 is fulfilling a role in alcoholic fermentation in W/S-clade yeasts, an ARO10 deletion mutant (aro10Δ) was constructed in St. bombicola. This mutant failed to produce ethanol, as might be expected in the complete absence of pyruvate decarboxylase activity (Figure 7A). Similarly to the adh1Δ mutant, it exhibited increased glycerol production, but unlike the former, it grew considerably slower than the wild type strain in aerated conditions (Figure 7C). This decrease was probably mainly due to the fact that the absence of pyruvate decarboxylase activity affects other metabolic routes in addition to alcoholic fermentation, such as the production of acetate and acetoin (Flikweert et al., 1996). The phenotype observed in this mutant confirmed that Aro10 is the only decarboxylase involved in alcoholic fermentation in St. bombicola and that, therefore, the modification of enzymatic specificities was also involved in remodeling alcoholic fermentation in W/S-clade yeasts. Importantly, in the aro10∆ mutant, glucose consumption seems to be even more seriously affected than in the adh1Δ mutant (Figure 7B) and, in fact, glucose was left all but untouched even after 120 hr of growth (Figure 7C), while fructose was almost totally consumed.
Identification of a yeast lineage devoid of an ADH1 gene
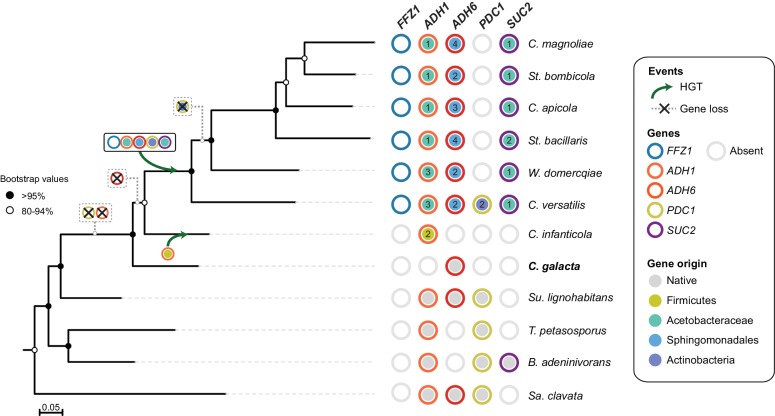
The present study describes a series of evolutionary events affecting the genes involved in alcoholic fermentation in all the species studied so far in the W/S clade, a lineage of more than 100 species that is very distantly related to S. cerevisiae. Two alternative hypotheses can be put forward concerning the order of the events underlying the observed remodeling of the fermentative pathway. Both our comparative genomics data and our experimental results suggest that loss of ‘native’ ADH1 and PDC1 orthologs preceded acquisition of bacterial counterparts, whose extant functions seem to be similar to the roles normally fulfilled by alcoholic fermentation enzymes in yeasts. However, while we found one species currently placed outside the W/S clade (C. infanticola) lacking PDC1, which is consistent with loss of this gene having preceded acquisition of the bacterial versions of this gene by C. versatilis, all species in our analysis possessed either a ‘native’ or the bacterial version of the ADH1 gene. In fact, all publicly available genomes across the entire subphylum Saccharomycotina possess at least one ADH1 ortholog. Remarkably, sequencing of the genome of Candida galacta (Figure 8) in the context of a distinct project (the Y1000+ Project sequencing the genomes of all known species of Saccharomycotina; http://y1000plus.org)(Hittinger et al., 2015) showed that it lacks both PDC1 and ADH1 (Figure 8, Figure 2—source data 3). The phylogenetic position of this species strongly supports the hypothesis that loss of native ADH1 preceded the (likely independent) acquisition of the bacterial versions of the gene by the C. infanticola lineage and by the MRCA of the W/S clade.
Figure 8. Loss and acquisition of sugar metabolism related genes in the W/S clade and closely related lineages.
Phylogenetic relationship between W/S species and closest relatives are depicted based on the ML phylogeny using the same dataset as in Figure 2A, but with the addition of C. galacta. For each of the four relevant genes: ADH1, ADH6, SUC2, and PDC1, presence, absence, and the native or bacterial origin of the orthologs found in the cognate draft genomes are shown for each species next to the respective branch of the tree. Each gene is represented by circles with different line colors (blue for previously studied FFZ1 (Gonçalves et al., 2016), orange for ADH1, red for ADH6, yellow for PDC1, and purple for SUC2). For xenologs, the different predicted bacterial donor lineages are denoted by different fill colors as indicated in the key. For W/S-clade species, the number of paralogs found in the cognate draft genome is also shown. Inferred gene losses (cross) and HGTs events (arrows) are indicated in the tree using the same color codes.
The invertase of bacterial origin is essential for sucrose assimilation in St. bombicola
Our results so far are consistent with the hypothesis that the surge in HGT observed in the MRCA of the W/S clade is related to its adaptation to the high-sugar environment in the floral niche, as exemplified by the acquisition of the Ffz1 fructose transporter from filamentous fungi and by the likely reacquisition of alcoholic fermentation involving HGT for alcohol dehydrogenase and modification of enzyme specificity (of Aro10). Acquisition of a bacterial invertase (sacC) (Martin et al., 1987) by a lineage lacking this pivotal enzyme for sucrose metabolism is also in line with this hypothesis, since most floral nectars are very rich is sucrose (Mittelbach et al., 2015;Canto et al., 2017).
To assess whether the horizontally acquired invertase gene is indeed responsible for sucrose assimilation in the W/S clade, a sacC deletion mutant (referred henceforth to as suc2∆ to emphasize the functional relation to the well-known S. cerevisiae SUC2 gene) was constructed in St. bombicola. Growth assays in medium supplemented with sucrose as sole carbon and energy source, showed that suc2∆ mutants were unable to grow (Figure 4—figure supplement 1A), while the wild-type strain attained high-cell densities. Furthermore, the suc2∆ mutant failed to consume measurable amounts of sucrose, even after 72 hr (Figure 4—figure supplement 1A). During growth of the wild-type strain on sucrose, the decrease in sucrose concentrations was accompanied by the appearance of fructose and glucose in the growth medium, strongly suggesting that the horizontally transferred SUC2 gene encodes an extracellular invertase (Figure 4—figure supplement 1B). This conclusion is consistent with the apparent absence of genes encoding sucrose transporters in the W/S-clade genomes analyzed, which indicates that sucrose must be first hydrolyzed outside the cell to be used as a carbon and energy source.
Discussion
The yeast lineage here named the Wickerhamiella/Starmerella (W/S) clade comprises several species that have previously attracted attention due to their unusual metabolic features. The most prominent example is St. bombicola, a species used for the production of sophorolipids, which are amphipathic molecules that are employed as biosurfactants (Samad et al., 2015; Takahashi et al., 2011). Starmerella bacillaris is often found in wine fermentations and is known for diverting an important fraction of its carbon flux towards the production of glycerol instead of ethanol (Englezos et al., 2015). Candida magnoliae has been reported to be capable of producing large amounts of erythritol (Ryu et al., 2000). More recently, we reported that fructophily was an important common trait that unified these species and others belonging to the W/S clade, and we also consubstantiated a strict correlation between the presence of the transporter Ffz1 and fructophily. Here, we show that presence of the Ffz1 transporter is a pre-requisite for fructophily in St. bombicola, as previously observed in Z. rouxii (Leandro et al., 2014). The stronger emphasis on the production of sugar alcohols as byproducts of metabolism at the expense of ethanol seemed also to be a common trait between the species examined (Lee et al., 2003b; Ryu et al., 2000), which led us to hypothesize that the preference of these yeasts for fructose was likely to be part of a broader remodeling of metabolism connected to the adaptation to the high-sugar environments where these yeasts thrive. The present work reflects our effort to uncover other aspects of this adaptation using comparative genomics as a starting point.
Our examination of genes acquired from bacteria showed that the number of HGT events from bacteria into the W/S clade and its neighbor lineage (represented by the species C. infanticola), far exceeded the number of events reported for other Saccharomycotina lineages (Marcet-Houben and Gabaldón, 2010) and was also considerably higher than those we identified in two species closely related to, but phylogenetically clearly outside the W/S clade (Su. lignohabitans and B. adeninivorans). The largest number of HGT events was detected in the earliest-diverging species in the W/S clade, C. versatilis, which together with the phylogenetic signal in the genes that were acquired through HGT, suggests that a large surge of acquisitions probably occurred in the MRCA of the clade (Figure 2A). Under this model, most extant lineages subsequently discarded a large portion of the xenologs originally present in the common ancestor. In line with this hypothesis, it was possible to identify 52 xenolog ortholog groups that were present in at least two W/S-clade species. It seems likely that, in addition to HGT events common only to W/S-clade species, additional HGT events took place in the MRCA of C. infanticola and the W/S clade because, from the inspection of the phylogenies constructed for C. infanticola, at least six genes of apparent bacterial origin in this species also have bacterial origin in the W/S clade. While the bacterial donor lineage seems to be quite different between C. infanticola and the W/S clade for one gene (ADH1), strongly suggesting that they were acquired in separate events, the other five genes share a common ancestor, possibly pointing to a single event. As far as can be presently assessed from the output of our AI pipeline, the remaining HGT-derived genes seem to be specific to C. infanticola. In addition, we noted that the set of 52 xenologs present in at least two extant W/S-clade species is enriched for genes encoding proteins that affect redox balance in the cell. In fact, changes in fluxes through main metabolic pathways were previously shown to impact redox balance and oxidative stress in yeasts (González-Siso et al., 2009), which is consistent with our hypothesis that associates the acquisition of bacterial genes with adaptive changes in metabolism.
The most striking finding concerning the function of the transferred genes is the profound remodeling of the ubiquitous alcoholic fermentation pathway used by yeasts to convert pyruvate into ethanol with concomitant regeneration of NAD+. The first step of the pathway, consisting of the conversion of pyruvate to acetaldehyde, is normally catalyzed by the enzyme Pdc1. However, in most W/S-clade yeasts, Pdc1 is absent. Here, we provided genetic evidence for one W/S-clade species, St. bombicola, that the role of Pdc1 was taken over by a related decarboxylase encoded in S. cerevisiae by the ARO10 gene. In S. cerevisiae, phenylpyruvate is the primary substrate of Aro10, which links this enzyme to amino acid catabolism, rather than alcoholic fermentation (Romagnoli et al., 2012; Vuralhan et al., 2005). However, it has been shown that site-directed mutagenesis of a few selected sites was capable of considerably increasing the affinity of S. cerevisiae Aro10 for pyruvate (Kneen et al., 2011), which supports the notion that this shift in substrate specificity may have occurred naturally in the course of evolution of W/S-clade yeasts. Intriguingly, C. versatilis, also lacks a native PDC1 gene but possesses two genes of bacterial origin encoding PDC1 orthologs, which coexist with ARO10. In this species, it is possible that the xenologs, and not ARO10, carry out the conversion of pyruvate into acetaldehyde.
The second step in alcoholic fermentation is the conversion of acetaldehyde into ethanol, which is conducted in S. cerevisiae mainly by Adh1. Again, ‘native’ ADH1 genes were absent from all W/S-clade genomes examined, and it seems that the MRCA of the W/S clade acquired a bacterial ADH1 gene, from which extant ADH1 xenologs found in all extant W/S-clade species examined were derived. A similar occurrence was detected involving the loss of the ‘native’ ADH6 ortholog, encoding a NADPH-dependent branched chain alcohol dehydrogenase and the acquisition of bacterial orthologs, although the bacterial donor lineages of the ADH1 and ADH6 xenologs seem to be distinct. We showed that, in St. bombicola, the ADH1 xenolog is absolutely required for growth on ethanol and is also mainly responsible for alcoholic fermentation, thereby performing the functions fulfilled by two different enzymes in S. cerevisiae (where Adh2 catalyzes ethanol assimilation). The two ADH6 xenologs played a minor role, if any, in alcoholic fermentation, similarly to what happens in S. cerevisiae.
All our results are consistent with the hypothesis that alcoholic fermentation was first lost in an ancestral lineage and was subsequently reacquired by W/S-clade yeasts through horizontal acquisition of genes. In one instance, a pre-existing yeast gene (ARO10) also modified its enzymatic specificities to become involved in alcoholic fermentation. On the other hand, no evidence was found for the alternative hypothesis stating that acquisition preceded loss, such as the co-occurrence in the same genome of ‘native’ and bacterial Adh1 or a distinctive role for the bacterial enzyme in yeast metabolism, which might have driven the fixation of the bacterial version of Adh1. Loss and subsequent reacquisition of a metabolic pathway through multiple HGT events was reported previously for the unicellular red algae Galdieria phlegrea where massive gene loss occurred concomitantly with adaptation to a specialized niche in the common ancestor of Cyanidiophytina red algae (Qiu et al., 2013). The lines of evidence supporting a similar event for alcoholic fermentation in the W/S clade are threefold, as follows.
First, one key aspect backing the ‘loss followed by reacquisition’ hypothesis is the identification of a yeast lineage, here represented by C. galacta, which lacks an ADH1 ortholog, either ‘native’ or bacterial. The species phylogeny presented in Figure 8 places this species close to C. infanticola, in a position consistent with our prediction for an extant representative of an Adh1- lineage pre-dating the acquisition of bacterial ADH1 genes. In addition, the two genes forming the alcoholic fermentation pathway seem to have been lost in quick succession because no genome was found that possessed only ‘native ‘ADH1 or only ‘native’ PDC1 genes. This also strongly suggests that loss of the entire pathway pre-dated acquisition of new genes or functions.
While ADH1 and ADH6 xenologs persist in all extant W/S-clade species examined, bacterial PDC1 xenologs, if they were indeed also acquired by the MRCA of the clade, were subsequently lost in most species. These hypothesized losses may have occurred as Aro10 evolved to fulfill the function of Pdc1. C. versatilis, which is the earliest-diverging species in the W/S clade, is a notable exception that possesses two PDC1 xenologs in addition to ARO10. Hence, C. versatilis on the one hand, and the remaining W/S clade species on the other hand, seem to represent two distinct solutions restoring pyruvate decarboxylase activity, which is a second observation in line with gene loss being the ancestral event.
Our assessment of the performance of the Adh1 enzymes in W/S-clade species constitutes a third argument in favor of the ‘loss followed by reacquisition’ hypothesis. We showed that W/S-clade enzymes of bacterial origin possess a potentially advantageous characteristic when compared to their yeast counterparts; they are capable of regenerating both NAD+ and NADP+, while the yeast enzymes accept only NADH as a cofactor. However, at least in St. bombicola, elimination of Adh1 was compensated by an increase in glycerol formation, which is very likely a NAD+ regenerating reaction, and not of mannitol formation, which regenerates NADP+. It is therefore reasonable to infer that the Adh xenologs are playing a similar role to that normally fulfilled by the native Adh enzymes. Morever, complete elimination of alcoholic fermentation, as observed in the aro10Δ mutant, resulted in an even more pronounced compensation at the level of glycerol production.
In conclusion, given the evidence presented here, the presence of xenologs involved in alcoholic fermentation in extant W/S-clade yeasts can be best explained by the need to restore alcoholic fermentation in a lineage that had previously lost it. In this case, the involvement of bacterial genes seems to have been circumstantial and possibly a consequence of the availability of the donor in the same environment. The ecological opportunity for the horizontal exchange of genetic material underlying the acquisition of bacterial ADH1 seems to have existed for a long time, since W/S-clade Adh1 proteins are most closely related to those of a bacterial lineage that is also frequently associated with the floral niche (Acetobacteraceae) (Iino et al., 2012; Suzuki et al., 2010; Tucker and Fukami, 2014).
SUC2 is also among the genes acquired by the MRCA of the W/S clade. Unlike the ‘native’ version of ADH1, which is ubiquitous in the Saccharomycotina except in the W/S clade, evolution of ‘native’ SUC2 seems to be punctuated by multiple loss events resulting in a patchy extant distribution (Carlson et al., 1985), which is probably linked to its role as a ‘social’ gene (Sanchez and Gore, 2013).
Taken together, our results suggest the enticing hypothesis that the basis for fructophily arose in yeasts in a lineage devoid of alcoholic fermentation, but probably well equipped to use fructose as electron acceptor with concomitant production of mannitol (Baek et al., 2010; Lee et al., 2003a), through the horizontal acquisition of the Ffz1 transporter that enabled the efficient utilization of fructose as carbon and energy source. Such a lineage would resemble the aro10∆ mutant in that it would be unable to use glucose in high-sugar conditions. This putative ancestor might subsequently have acquired fermentative capacity to enable efficient glucose consumption and SUC2 to permit the utilization of sucrose, thereby completing the set of tools required to use the sugars abundant in the floral niche.
In conclusion, our previous (Gonçalves et al., 2016) and present results uncovered, to our knowledge for the first time, an instance of major remodeling of central carbon metabolism in fungi involving the horizontal acquisition of a multitude of genes mostly from bacteria. Thereby, fructophilic yeasts seem to have been able to overcome the inability to use glucose efficiently, as observed in extant fructophilic bacteria. As progress in the Y1000+ Project provides additional genomic information concerning species within and in the vicinity of the W/S clade, the improved phylogenetic resolution will hopefully help further extricate the complex pattern of acquisitions of genes.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background | Starmerella bombicola PYCC 5882 | Portuguese Yeast Culture Collection | ||
| Strain, strain background | Candida magnoliae PYCC 2903 | Portuguese Yeast Culture Collection | ||
| Strain, strain background | Starmerella bacillaris PYCC 3044 | Portuguese Yeast Culture Collection | ||
| Strain, strain background | Saccharomyces cerevisiae PYCC 7186 (S288C) | Portuguese Yeast Culture Collection | ||
| Genetic reagent (Starmerella bombicola) | adh1∆ (adh1∆::HYG) | This paper | Constructed as described in the materials and methods section; primers used are described in Figure 7—source data 2 | |
| Genetic reagent (Starmerella bombicola) | aro10∆ (aro10∆::HYG) | This paper | Constructed as described in the materials and methods section; primers used are described in Figure 7—source data 2 | |
| Genetic reagent (Starmerella bombicola) | adh6a∆ (adh6a∆::HYG) | This paper | Constructed as described in the materials and methods section; primers used are described in Figure 7—source data 2 | |
| Genetic reagent (Starmerella bombicola) | adh6b∆ (adh6b∆::HYG) | This paper | Constructed as described in the materials and methods section; primers used are described in Figure 7—source data 2 | |
| Genetic reagent (Starmerella bombicola) | ffz1∆ (ffz1∆::HYG) | This paper | Constructed as described in the materials and methods section; primers used are described in Figure 7—source data 2 |
Strains
Yeast strains were obtained from PYCC (Portuguese Yeast Culture Collection, Caparica, Portugal) or NRRL (USDA ARS Culture Collection). All strains were maintained in YPD medium.
Identification of ADH1, PDC1, ADH6, and SUC2 genes in the W/S clade and related species
A custom query consisting of ADH1, ADH6, PDC1, and SUC2 genes from S. cerevisiae was used in a tBLASTx search against W/S-clade genomes (C. versatilis JCM 5958, St. bombicola PYCC 5882, St. bacillaris 3044, C. magnoliae PYCC 2903, C. apicola NRRL Y-50540 and W. domercqiae PYCC 3067). The genomes of closely related species Candida galacta NRRL Y-17645 (Y1000+ Project), Sugiyamaella lignohabitans CBS 10342, Blastobotrys adeninivorans LS3, Trichomonascus petasosporus NRRL YB-2093, and Saprochaete clavata CNRMA 12.647 were also added to the database (Figure 8, see Figure 2—source data 3). The best tBLASTx hit sequences (E-value <e−10) retrieved from the analysis were subsequently blasted against the NCBI non-redundant protein database, and orthology was assumed whenever the best hit in S. cerevisiae was the corresponding protein in the query. Genes related with glycolysis were also inspected (Figure 2—source data 3). For this analysis, distantly related fructophilic species Zygosaccharomyces kombuchaensis (strain CBS 8849) was also added to the database.
For the genes of bacterial origin in the W/S clade, genome location was assessed for each gene in order to discard possible assembly contaminations (as scaffolds containing only bacterial genes). Although assembly artifacts cannot be excluded, all the key genes are located in considerably long scaffolds and flanked by other genes of yeast origin.
Genomes of C. apicola (Vega-Alvarado et al., 2015) and C. versatilis (PRJDB3712) are publicly available, while the draft genomes of the remaining W/S-clade species examined and of Z. kombuchaensis were generated in the course of a previous study (Gonçalves et al., 2016) and are also publicly available (PRJNA416493 and PRJNA416500). The genome assembly of Candida galacta NRRL Y-17645 was obtained in the context of the Y1000+ Project (Hittinger et al., 2015) using standardized sequencing (Hittinger et al., 2010) and assembly (Zhou et al., 2016) protocols and is published here for the first time. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PPSZ00000000. The version described in this paper is version PPSZ01000000.
Protein prediction in newly sequenced genomes
For St. bombicola PYCC 5882, St. bacillaris PYCC 3044, C. magnoliae PYCC 2903, W. domercqiae PYCC 3067, and C. infanticola DS-02 (PRJNA318722) the complete proteome was predicted with AUGUSTUS (Stanke et al., 2008) using the complete model and S. cerevisiae, Scheffersomyces stipitis, and Y. lipolytica as references. To assess the completeness of the predicted proteomes in each case, the number of predicted proteins was compared to the number of proteins reported for the annotated proteomes of W. domercqiae JCM 9478 (PRJDB3620) and S. bombicola JCM 9596 (PRJDB3622). Predictions that used S. cerevisiae as a reference turned out to be the most complete (~4.000 predicted proteins) and were therefore used for all downstream analyses. For C. apicola NRLL Y-50540, C. versatilis JCM 5958, Su. lignohabitans (NCBI) and B. adeninivorans (JGI), publicly available proteomes were used.
High-throughput detection of HGT-derived genes from bacteria in the W/S clade
Query protein sequences were searched against a local copy of the NCBI refseq protein database (downloaded May 5, 2017) using phmmer, a member of the HMMER3 software suite (Eddy, 2009) using acceleration parameters --F1 1e-5 --F2 1e-7 --F3 1e-10. A custom perl script sorted the phmmer results based on the normalized bitscore (nbs), where nbs was calculated as the bitscore of the single best-scoring domain in the hit sequence divided by the best bitscore possible for the query sequence (i.e. the bitscore of the query aligned to itself). The top ≤10,000 hits were retained for further analysis, saving no more than five sequences per unique NCBI Taxonomy ID.
The alien index score (AI) was calculated for each query protein (modified from Gladyshev et al., 2008). Two taxonomic lineages were first specified: the RECIPIENT into which possible HGT events may have occurred (Saccharomycetales, NCBI Taxonomy ID 4892), and a larger ancestral GROUP of related taxa (Fungi, NCBI Taxonomy ID 4751). The AI is given by the formula: , where nbsO is the normalized bitscore of the best hit to a species outside of the GROUP lineage, nbsG is the normalized bitscore of the best hit to a species within the GROUP lineage (skipping all hits to the RECIPIENT lineage). AI is greater than zero if the gene has a better hit to a species outside of the group lineage and can be suggestive of either HGT or contamination. Note that the original Gladyshev et al. (2008) AI calculation was based on relative E-values and ranged from −460 to +460, with AI >45 considered a strong HGT candidate. By converting the AI to a bitscore-based metric, the results are not impacted by BLAST version, database size, or computer hardware. The bitscore-based AI score ranges from −1 to +1, with AI >0.1 considered strong HGT candidates.
Full-length proteins corresponding to the top 200 hits (E-value <1 × 10−10) to each query sequence were extracted from the local database using esl-sfetch (Eddy, 2009). Sequences were aligned with MAFFT v7.310 using the E-INS-i strategy and the BLOSUM30 amino acid scoring matrix (Katoh and Standley, 2013) and trimmed with trimAL v1.4.rev15 using its gappyout strategy (Capella-Gutiérrez et al., 2009). Proteins with trimmed alignments < 150 amino acids in length were excluded. The topologies of the remaining proteins were inferred using maximum likelihood as implemented in IQ-TREE v1.5.4 (Nguyen et al., 2015) using an empirically determined substitution model and rapid bootstrapping (1000 replications). The phylogenies were midpoint rooted and branches with local support less than 95 were collapsed using the ape and phangorn R packages (Paradis et al., 2004; Schliep, 2011). Phylogenies were visualized using ITOL version 3.0 (Letunic and Bork, 2016).
For the data included in Figure 2A, the number of genes for which a bacterial origin was confirmed after inspection of the correspondent phylogenetic tree are shown. AI results per species are shown as in Figure 2—source data 2.
We subsequently selected HGT candidates in the W/S clade (Figure 2—source data 4) according to two criteria: the putative bacterial ortholog should be present in at least two W/S-clade species, and it should cluster with a bacterial lineage with strong bootstrap support (>90%). Putative HGT-derived genes shared with non-W/S-clade species were excluded. Application of these criteria yielded 200 strong candidate trees, many of which referred to the same ortholog (genes shared by several W/S species and/or paralogs, as the case of ADH1 which corresponds to 10 trees). A final number of 52 different ortholog groups was established after collapsing the replicate phylogenies and lineage-specific gene duplications. The resulting set of 52 orthologs was subsequently cross-referenced with GO and InterPro annotations provided by InterproScan and the Joint Genome Institute MycoCosm Portal (Grigoriev et al., 2014). KEGG annotation was performed using the KAAS database (Moriya et al., 2007). A BLAST KOALA annotation (Kanehisa et al., 2016) was also conducted in the final dataset (52 proteins).
Phylogenetic analyses and topology tests
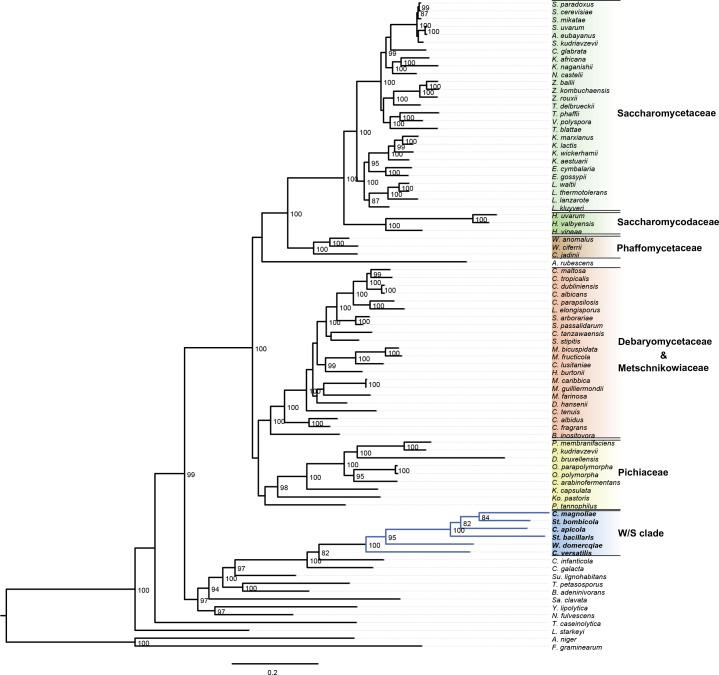
Species phylogenies were constructed according to Gonçalves et al. (2016). The same dataset was used with the addition of W/S-clade species C. apicola NRRL Y-50540; C. versatilis JCM 9598; and close relatives C. infanticola DS-02, Candida galacta NRRL Y-17645 (Y1000+ Project), Sugiyamaella lignohabitans CBS 10342 (Su. lignohabitans), Trichomonascus petasosporus NRRL YB-2093 (T. petasosporus), and Saprochaete clavata CNRMA 12.647 (Sa. clavata). Rpa1, Rpa2, Rpb1, Rpb2, Rpc1 and Rpc2 protein sequences for each species were used to construct the ML tree with RAxML (Stamatakis, 2006) v7.2.8 using the PROTGAMMAILG model of amino acid substitution and 1000 rapid bootstraps. Species names abbreviations and accession numbers of the proteins used to construct the phylogeny are indicated in Figure 2—source data 1. Phylogenetic relationships are in agreement with the recently published phylogeny of 86 yeast species based on genome sequences (Shen et al., 2016).
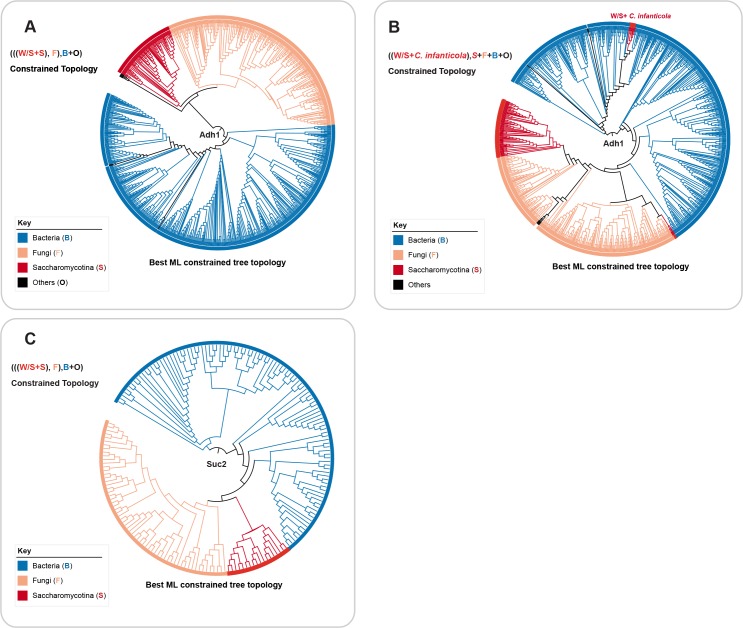
For the construction of the Pdc1 phylogeny, the top 500 NCBI BLASTp hits from searches against the non-redundant database using Pdc1 from S. cerevisiae (CAA97573.1), St. bombicola Pdc1-like, and C. versatilis Pdc1-like from apparent bacterial origin as queries, were selected. Sequences with more than 90% similarity were removed using CD-HIT v 4.6.7 (Li and Godzik, 2006). Pdc1 sequences from the closest relative species T. petasosporus, B. adeninivorans, and Sa. clavata of the W/S clade were also added to the alignment. A total of 479 proteins were aligned using MAFFT v 7.2.15, (Katoh and Standley, 2014) using the fast but progressive method (FFT-NS-2). Poorly aligned regions were removed with trimAl (Capella-Gutiérrez et al., 2009) using the ‘gappyout’ option. The ML phylogeny in Figure 3 was constructed with IQ-TREE v 1.4.3 (Nguyen et al., 2015) using the LG + I + G substitution model. For the Suc2 phylogeny (Figure 4), the top 200 hits from the phmmer search against the local database were selected and the ML phylogeny was constructed as above. Given the absence of other Saccharomycotina sequences in the top 200 hits for Adh1, the top 4000 hit sequences were selected instead. For Adh6, no Saccharomycotina sequences were found even in the top 4000 phmmer hits, so the top 10,000 top hit sequences were used in this case. For both phylogenies, CD-HIT (Li and Godzik, 2006) was used to remove sequences with more than 85% (Adh1) and 80% (Adh6) similarity. For Adh1, a preliminary ML tree was constructed to eliminate sequences outside the Adh1 family. A final set of 976 Adh1 sequences were subsequently aligned and trimmed as aforementioned and used to construct the final Adh1 phylogeny. ML phylogenies were constructed as previously described. Original raw phylogeny files can be accessed using the following links: https://figshare.com/s/c59f135885f31565a864. The likelihood of HGT for ADH1 and SUC2 was investigated assuming monophyly of Saccharomycotina as the constrained topology (Figure 5—figure supplement 1A and Figure 5—figure supplement 1C). The best tree was inferred in RAxML, and ML values for constrained and unconstrained trees were also calculated. The AU test (Shimodaira, 2002) implemented in CONSEL (Shimodaira and Hasegawa, 2001) was used to compare the unconstrained best tree and the best tree given a constrained topology. To test the independence of Adh1 acquisition, monophily of W/S clade and C. infanticola was assumed (Figure 5—figure supplement 1B). Phylogenies were visualized using iTOL version 3.0 (Letunic and Bork, 2016).
Detection of enzymatic activities in cell-free extracts
Cultures were grown overnight in YPD medium until late exponential phase (OD640nm ~15–25). Cells were then collected by centrifugation (3000 x g for 5 min), washed twice with cold Tris-HCL buffer (pH = 7.6), and disrupted with glass beads in 500 µL of Lysis Buffer (0.1 M triethanolamine hydrochloride, 2 mM MgCl2, 1 mM DTT and 1 µM PMSF) with six cycles of 60 s vortex-ice. Cell debris were removed by centrifugation at 4°C and 16,000 x g for 20 min and the extracts were stored at −20°C. Alcohol dehydrogenase activity (Adh) assays were performed at 25°C in 500 µL reaction mixtures containing 50 mM Potassium Phosphate buffer (pH = 7.5), 1 mM of NADH or NADPH, and 25 µL of cell-free extract. The reaction was started by adding acetaldehyde to a final concentration of 100 mM, and reduction of NADH and NADPH was monitored spectrophotometrically by the decrease in absorbance at 340 nm for two minutes. For St. bombicola PYCC 5882, 5 mM, 12.5 mM, 50 mM, and 100 mM as final concentrations of acetaldehyde were also used. The absence of Adh activity in the adh1∆ mutant was also confirmed with protein extract of adh1∆ cells grown in 20FG medium (conditions where ethanol was detected by HPLC in the mutant, Figure 7A), using up to three times more protein extract.
For the detection of NADP+-dependent glycerol dehydrogenase activity (Klein et al., 2017) in St. bombicola, cultures and cell-free extracts were obtained as above. Glycerol dehydrogenase activity was measured in a reaction mixture containing 50 mM Tris-HCl (pH = 8.5) buffer, 1 mm NADP+ and 25 µL of cell-free extract. The reaction was started by adding glycerol to a final concentration of 100 mM, and NADPH formation was monitored spectrophotometrically for two minutes.
Construction of St. bombicola deletion mutants
Standard molecular biology techniques were performed essentially as described in Sambrook and Russell (2001) using E.coli DΗ5α as host. St. bombicola PYCC 5882 was used in all procedures involving this species. Disruption constructs were designed essentially as outlined by Van Bogaert et al. (2008)(Van Bogaert et al., 2008). The St. bombicola GPD promoter was first amplified by PCR and fused to the hygromycin B phosphotransferase gene (hygB) from E.coli and the CYC1 terminator from S. cerevisiae. Phusion High Fidelity (Thermo Fisher Scientific, Waltham, MA) was used for hygB and GPD promoter amplifications. A 491 bp fragment of the GPD promoter (Van Bogaert et al., 2008) was amplified using the primer pair GPD_SacI_Fw/GPD_Hind III_Rv (Figure 7—source data 2). The TEF1 promoter from p414TEF-CYC (Mumberg et al., 1995) vector was replaced by the GPD promoter using Sac I and Hind III. The primer pair Hyg_Hind III_Fw/Hyg_Xho I_Rv was used to amplify the hygB gene from a commercial plasmid (pBlueScript-hyg, [Niklitschek et al., 2008]). The amplicon was then cloned into the previously obtained p414GPD-CYC plasmid. The resulting plasmid harbors the hygromycin resistance gene controlled by the St. bombicola GPD promoter and followed by the CYC1 terminator of S. cerevisiae.
For disruption of the ADH1, SUC2, and ARO10 genes the coding sequences (CDS) with 1 kb upstream and downstream were amplified from genomic DNA using the primer pairs listed in Figure 7—source data 2. The two fragments, corresponding to each of the genes, were separately cloned into the PJET1.2 plasmid. The GPD-HYG-CYC cassette was then cloned into each of the resulting PJET1.2 plasmids using the restriction enzymes listed in Figure 7—source data 2. For FFZ1, ADH6a, and ADH6b, two sets of primers were used to amplify 1 kb upstream and 1 kb downstream of the CDS. The upstream and downstream fragments of each of the genes were subsequently cloned into the p416GPD-HYG-CYC plasmid using suitable enzymes (Figure 7—source data 2), yielding three plasmids each containing a distinct disruption cassette.
St. bombicola was transformed by electroporation with each gene disruption construct in turn, amplified by PCR from the respective plasmid template using Phusion High Fidelity DNA polymerase and the primers listed in (Figure 7—source data 2), using the protocol described by Saerens et al. (2011) (Saerens et al., 2011). Two different transformants from each gene disruption transformation were subsequently used for all phenotypic assays.
Sugar assimilation, consumption, and metabolite production assessments
For metabolite and sugar consumption profiling (Figure 7A and B), 10 mL cultures of St. bombicola (wild type and mutants) were grown overnight at 30°C with orbital shacking (180 rpm) in YP medium supplemented with 10% (w/v) of fructose and 10% (w/v) of glucose. The overnight culture was used to inoculate a fresh culture in 30 mL of the same medium to an OD640nm of 0.2, which was incubated under the same conditions. Growth was monitored until late stationary phase was reached (typically after 150 hr), and 2 mL samples were taken at several time points, centrifuged at 12,000 x g for 5 min, and analyzed by HPLC, as previously described (Gonçalves et al., 2016). Statistical significance was tested using a one way ANOVA using the Bonferroni’s correction for multiple testing, implemented in GraphPad Prism v5. Growth on sucrose and on ethanol was assessed in wild type and mutants (suc2∆, adh1∆ and aro10∆) cultivated overnight in YP medium supplemented with 2% (w/v) sucrose (suc2∆) or 2% (v/v) ethanol (adh1∆ and aro10∆) with orbital shacking (180 rpm) at 30°C. Cultures were transferred into the same medium (OD640nm = 0.2), and growth was monitored over time. Consumption of sucrose at different time points was monitored by HPLC.
Growth assays
St. bombicola wild type and adh1∆ mutants were tested for growth aerobically (favoring respiration) and in microaerophily (favoring fermentation) conditions. For the mutants, two biological replicates from independent transformations were used. For microaerophily experiments, a 24 hr pre-culture in SC medium supplemented with 0.2% glucose was performed. These cultures were used to inoculate 200 µL of SC medium supplemented with the desired carbon source (2% (w/v) glucose or 2% (w/v) fructose), at a 1:40 ratio in a 96 well plate. The absorbance of each well was read by an unshaken BMG FLUOstar Omega plate reader (Kuang et al., 2016) every 120 min at 600 nm for five days. For aerobic growth, a 10 mL pre-culture in SC medium supplemented with 0.2% glucose was performed overnight with shaking (200 rpm). Cells were transferred to 30 mL (in a 250 mL flask) of SC medium supplemented with 2% (w/v) glucose or 2% (w/v) fructose until a final OD640nm=0.2. Cultures were grown for 5 days, with shaking.
Acknowledgements
The authors thank PYCC for providing strains, members of the Yeast Genomics Lab for helpful discussions, Jeremy DeVirgilio and Amanda Beth Hulfachor for technical assistance, the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing Illumina facilities and services, and Lucigen Corporation (Middleton, WI) for use of their Covaris sonicator. Draft genome data from C. versatilis JCM 5958 and W. domercqiae JCM 9478 were obtained by RIKEN BioResource Center and RIKEN Center for Life Science Technologies through the Genome Information Upgrading Program of the National Bio-Resource Project of the MEXT, Japan. The authors also thank the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov) for providing access to genomic data produced in collaboration with the user community. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer. This work was mainly supported by Unidade de Ciências Biomoleculares Aplicadas-UCIBIO which is financed by national funds from FCT/MEC (UID/Multi/04378/2013) and co-financed by the ERDF under the PT2020 Partnership Agreement (POCI-01–0145-FEDER-007728). This work was also supported by grant PTDC/AGR-ALI/112802/2009 from Fundação para a Ciência e a Tecnologia, Portugal (http://www.fct.pt). CG and MJL are recipients of grants (SFRH/BD/89489/2012 and SFRH/BPD/102803/2014, respectively) from Fundacão para a Ciência e Tecnologia, Portugal. This work was also supported by the National Science Foundation under Grant Nos. IOS-1401682 (JHW), DEB-1253634 (CTH), DEB-1442148 (CTH), and DEB-1442113 (AR); by USDA National Institute of Food and Agriculture Hatch Project 1003258 (CTH); the Robert Draper Technology Innovation Fund from the Wisconsin Alumni Research Foundation (CTH); and funded in part by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494 to Timothy J. Donohue). The work was also in part supported through computational resources provided by Information Technology at Purdue, West Lafayette, Indiana; the Center for High Throughput Computing at UW-Madison; and the Advanced Computing Center for Research and Education at Vanderbilt University. CTH is a Pew Scholar in the Biomedical Sciences and a Vilas Faculty Early Career Investigator, supported by the Pew Charitable Trusts and the Vilas Trust Estate. DP is a Marie Sklodowska-Curie fellow of the European Union’s Horizon 2020 research and innovation programme, grant agreement No. 747775.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Paula Gonçalves, Email: pmz@fct.unl.pt.
Paul B Rainey, Massey University, New Zealand.
Funding Information
This paper was supported by the following grants:
Fundação para a Ciência e a Tecnologia PTDC/AGR-ALI/112802/2009 to Paula Gonçalves.
National Science Foundation IOS-1401682 to Jennifer H Wisecaver.
National Institute of Food and Agriculture Hatch Project 1003258 to Chris Todd Hittinger.
Wisconsin Alumni Research Foundation to Chris Todd Hittinger.
DOE Great Lakes Bioenergy Research Center BER DE-FC02-07ER64494 to Chris Todd Hittinger.
Pew Charitable Trusts to Chris Todd Hittinger.
National Science Foundation DEB-1253634 to Chris Todd Hittinger.
National Science Foundation DEB-1442148 to Chris Todd Hittinger.
National Science Foundation DEB-1442113 to Antonis Rokas.
Fundação para a Ciência e a Tecnologia UID/Multi/04378/2013 and to Paula Gonçalves.
European Regional Development Fund POCI-01-0145-FEDER-007728 to Paula Gonçalves.
Fundação para a Ciência e a Tecnologia SFRH/BD/89489/2012 to Carla Gonçalves.
Fundação para a Ciência e a Tecnologia SFRH/BPD/102803/2014 to Maria José Leandro.
H2020 Marie Skłodowska-Curie Actions 747775 to David Peris.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing—original draft, Writing—review and editing.
Resources, Formal analysis, Investigation, Writing—review and editing.
Formal analysis, Investigation, Data acquisition and analysis concerning the Candida galacta genome.
Supervision, Writing—review and editing.
Resources, Supervision, Writing—review and editing.
Formal analysis, Contributed to topology tests and ontology analysis.
Investigation, Data acquisition concerning the Candida galacta genome.
Formal analysis, Data acquisition concerning the Candida galacta genome.
Investigation, Writing—review and editing, Data acquisition for Figure 5- figure supplement 3B.
Resources, Funding acquisition, Data acquisition concerning the Candida galacta genome.
Resources, Supervision, Funding acquisition, Writing—review and editing.
Resources, Supervision, Funding acquisition, Writing—review and editing.
Conceptualization, Resources, Supervision, Funding acquisition, Writing—original draft, Project administration, Writing—review and editing.
Additional files
Major datasets
The following datasets were generated:
Gonçalves C, author; Gonçalves P, author. Comparative genomics of fructophilic ascomycetous yeasts. 2017 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA416493 PRJNA416493.
Gonçalves C, author; Gonçalves P, author. Comparative genomics of fructophilic ascomycetous yeasts. 2017 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA416500 PRJNA416500.
Kominek J, author; Opulente DA, author; Shen XX, author; Zhou X, author; Kurtzman CP, author; Rokas A, author; Hittinger CT, author. Candida galacta Whole Genome Shotgun project. 2017 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA431085 Publicly available at NCBI BioProject (Accession no. PRJNA431085)
References
- Alexander WG, Wisecaver JH, Rokas A, Hittinger CT. Horizontally acquired genes in early-diverging pathogenic fungi enable the use of host nucleosides and nucleotides. PNAS. 2016;113:4116–4121. doi: 10.1073/pnas.1517242113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek H, Lee YB, Hyun HH. Molecular cloning and sequence analysis of a mannitol dehydrogenase gene and isolation of mdh promoter from Candida magnoliae. Biotechnology Letters. 2010;32:1089–1094. doi: 10.1007/s10529-010-0257-1. [DOI] [PubMed] [Google Scholar]
- Cabral S, Prista C, Loureiro-Dias MC, Leandro MJ. Occurrence of FFZ genes in yeasts and correlation with fructophilic behaviour. Microbiology. 2015;161:2008–2018. doi: 10.1099/mic.0.000154. [DOI] [PubMed] [Google Scholar]
- Campbell MA, Rokas A, Slot JC. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biology and Evolution. 2012;4:289–293. doi: 10.1093/gbe/evs011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto A, Herrera CM, Rodriguez R. Nectar-living yeasts of a tropical host plant community: diversity and effects on community-wide floral nectar traits. PeerJ. 2017;5:e3517. doi: 10.7717/peerj.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Celenza JL, Eng FJ. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Molecular and Cellular Biology. 1985;5:2894–2902. doi: 10.1128/MCB.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Osmond BC, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman K, Ropars J, Renault P, Dupont J, Gouzy J, Branca A, Abraham AL, Ceppi M, Conseiller E, Debuchy R, Malagnac F, Goarin A, Silar P, Lacoste S, Sallet E, Bensimon A, Giraud T, Brygoo Y. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nature Communications. 2014;5:2876. doi: 10.1038/ncomms3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JY, Jeffries TW. Pichia stipitis genes for alcohol dehydrogenase with fermentative and respiratory functions. Applied and Environmental Microbiology. 1998;64:1350–1358. doi: 10.1128/aem.64.4.1350-1358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MA, Gonçalves C, Sampaio JP, Gonçalves P. Extensive intra-kingdom horizontal gene transfer converging on a fungal fructose transporter gene. PLoS Genetics. 2013;9:e1003587. doi: 10.1371/journal.pgen.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashko S, Zhou N, Compagno C, Piškur J. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Research. 2014;14:826–832. doi: 10.1111/1567-1364.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smidt O, du Preez JC, Albertyn J. The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Research. 2008;8:967–978. doi: 10.1111/j.1567-1364.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- de Smidt O, du Preez JC, Albertyn J. Molecular and physiological aspects of alcohol dehydrogenases in the ethanol metabolism of Saccharomyces cerevisiae. FEMS Yeast Research. 2012;12:33–47. doi: 10.1111/j.1567-1364.2011.00760.x. [DOI] [PubMed] [Google Scholar]
- de Vega C, Albaladejo RG, Guzmán B, Steenhuisen SL, Johnson SD, Herrera CM, Lachance MA. Flowers as a reservoir of yeast diversity: description of Wickerhamiella nectarea f.a. sp. nov., and Wickerhamiella natalensis f.a. sp. nov. from South African flowers and pollinators, and transfer of related candida species to the genus wickerhamiella as new combinations. FEMS Yeast Research. 2017;17 doi: 10.1093/femsyr/fox054. [DOI] [PubMed] [Google Scholar]
- Deng X, Petitjean M, Teste MA, Kooli W, Tranier S, François JM, Parrou JL. Similarities and differences in the biochemical and enzymological properties of the four isomaltases from Saccharomyces cerevisiae. FEBS Open Bio. 2014;4:200–212. doi: 10.1016/j.fob.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewke C, Thielen J, Ciriacy M. Ethanol formation in adh0 mutants reveals the existence of a novel acetaldehyde-reducing activity in Saccharomyces cerevisiae. Journal of Bacteriology. 1990;172:3909–3917. doi: 10.1128/jb.172.7.3909-3917.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome informatics. International Conference on Genome Informatics. 2009;23:205–211. doi: 10.1142/9781848165632_0019. [DOI] [PubMed] [Google Scholar]
- Endo A, Futagawa-Endo Y, Dicks LM. Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Systematic and Applied Microbiology. 2009;32:593–600. doi: 10.1016/j.syapm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Endo A, Salminen S. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Systematic and Applied Microbiology. 2013;36:444–448. doi: 10.1016/j.syapm.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Endo A, Tanaka N, Oikawa Y, Okada S, Dicks L. Fructophilic characteristics of Fructobacillus spp. may be due to the absence of an alcohol/acetaldehyde dehydrogenase gene (adhE) Current Microbiology. 2014;68:531–535. doi: 10.1007/s00284-013-0506-3. [DOI] [PubMed] [Google Scholar]
- Englezos V, Rantsiou K, Torchio F, Rolle L, Gerbi V, Cocolin L. Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (synonym Candida zemplinina) in wine fermentation: physiological and molecular characterizations. International Journal of Food Microbiology. 2015;199:33–40. doi: 10.1016/j.ijfoodmicro.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Flikweert MT, Van Der Zanden L, Janssen WM, Steensma HY, Van Dijken JP, Pronk JT. Pyruvate Decarboxylase: An Indispensable Enzyme for Growth of Saccharomyces cerevisiae on Glucose. Yeast. 1996;12:247–257. doi: 10.1002/(SICI)1097-0061(19960315)12:3<247::AID-YEA911>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Ganzhorn AJ, Green DW, Hershey AD, Gould RM, Plapp BV. Kinetic characterization of yeast alcohol dehydrogenases. Amino acid residue 294 and substrate specificity. The Journal of Biological Chemistry. 1987;262:3754–3761. [PubMed] [Google Scholar]
- Gibbons JG, Salichos L, Slot JC, Rinker DC, McGary KL, King JG, Klich MA, Tabb DL, McDonald WH, Rokas A. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Current Biology. 2012;22:1403–1409. doi: 10.1016/j.cub.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- Gojković Z, Knecht W, Zameitat E, Warneboldt J, Coutelis JB, Pynyaha Y, Neuveglise C, Møller K, Löffler M, Piskur J. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Molecular Genetics and Genomics. 2004;271:387–393. doi: 10.1007/s00438-004-0995-7. [DOI] [PubMed] [Google Scholar]
- González-Siso MI, García-Leiro A, Tarrío N, Cerdán ME. Sugar metabolism, redox balance and oxidative stress response in the respiratory yeast Kluyveromyces lactis. Microbial Cell Factories. 2009;8:46. doi: 10.1186/1475-2859-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves C, Coelho MA, Salema-Oom M, Gonçalves P. Stepwise functional evolution in a fungal sugar transporter family. Molecular Biology and Evolution. 2016;33:352–366. doi: 10.1093/molbev/msv220. [DOI] [PubMed] [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research. 2014;42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Gonçalves P, Sampaio JP, Dover J, Johnston M, Rokas A. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–58. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Rokas A, Bai FY, Boekhout T, Gonçalves P, Jeffries TW, Kominek J, Lachance MA, Libkind D, Rosa CA, Sampaio JP, Kurtzman CP. Genomics and the making of yeast biodiversity. Current Opinion in Genetics & Development. 2015;35:100–109. doi: 10.1016/j.gde.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Cederberg H. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. European Journal of Biochemistry. 1990;188:615–621. doi: 10.1111/j.1432-1033.1990.tb15442.x. [DOI] [PubMed] [Google Scholar]
- Ida Y, Furusawa C, Hirasawa T, Shimizu H. Stable disruption of ethanol production by deletion of the genes encoding alcohol dehydrogenase isozymes in Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering. 2012;113:192–195. doi: 10.1016/j.jbiosc.2011.09.019. [DOI] [PubMed] [Google Scholar]
- Iino T, Suzuki R, Kosako Y, Ohkuma M, Komagata K, Uchimura T. Acetobacter okinawensis sp. nov., Acetobacter papayae sp. nov., and Acetobacter persicus sp. nov.; novel acetic acid bacteria isolated from stems of sugarcane, fruits, and a flower in Japan. The Journal of General and Applied Microbiology. 2012;58:235–243. doi: 10.2323/jgam.58.235. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Journal of Molecular Biology. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods in molecular biology. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Wolfe KH. Evolutionary origins of the fumonisin secondary metabolite gene cluster in fusarium verticillioides and Aspergillus niger. International Journal of Evolutionary Biology. 2011;2011:1–7. doi: 10.4061/2011/423821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Swinnen S, Thevelein JM, Nevoigt E. Glycerol metabolism and transport in yeast and fungi: established knowledge and ambiguities. Environmental Microbiology. 2017;19:878–893. doi: 10.1111/1462-2920.13617. [DOI] [PubMed] [Google Scholar]
- Kneen MM, Stan R, Yep A, Tyler RP, Saehuan C, McLeish MJ. Characterization of a thiamin diphosphate-dependent phenylpyruvate decarboxylase from Saccharomyces cerevisiae. FEBS Journal. 2011;278:1842–1853. doi: 10.1111/j.1742-4658.2011.08103.x. [DOI] [PubMed] [Google Scholar]
- Kuang MC, Hutchins PD, Russell JD, Coon JJ, Hittinger CT. Ongoing resolution of duplicate gene functions shapes the diversification of a metabolic network. eLife. 2016;5:e19027. doi: 10.7554/eLife.19027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP, Price NP, Ray KJ, Kuo TM. Production of sophorolipid biosurfactants by multiple species of the Starmerella (Candida) bombicola yeast clade. FEMS Microbiology Letters. 2010;311:140–146. doi: 10.1111/j.1574-6968.2010.02082.x. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. New anamorphic yeast species: Candida infanticola sp. nov., Candida polysorbophila sp. nov., Candida transvaalensis sp. nov. and Trigonopsis californica sp. nov. Antonie van Leeuwenhoek. 2007;92:221–231. doi: 10.1007/s10482-007-9150-x. [DOI] [PubMed] [Google Scholar]
- Lachance MA, Starmer WT, Rosa CA, Bowles JM, Barker JS, Janzen DH. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Research. 2001;1:1–8. doi: 10.1111/j.1567-1364.2001.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Leandro MJ, Cabral S, Prista C, Loureiro-Dias MC, Sychrová H. The high-capacity specific fructose facilitator ZrFfz1 is essential for the fructophilic behavior of Zygosaccharomyces rouxii CBS 732T. Eukaryotic Cell. 2014;13:1371–1379. doi: 10.1128/EC.00137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Koo BS, Kim SY, Hyun HH. Purification and characterization of a novel mannitol dehydrogenase from a newly isolated strain of Candida magnoliae. Applied and Environmental Microbiology. 2003a;69:4438–4447. doi: 10.1128/AEM.69.8.4438-4447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Song JY, Kim SY. Controlling substrate concentration in fed-batch Candida magnoliae culture increases mannitol production. Biotechnology Progress. 2003b;19:768–775. doi: 10.1021/bp034025o. [DOI] [PubMed] [Google Scholar]
- Leskovac V, Trivić S, Pericin D. The three zinc-containing alcohol dehydrogenases from baker's yeast, Saccharomyces cerevisiae. FEMS Yeast Research. 2002;2:481–494. doi: 10.1111/j.1567-1364.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Maeno S, Tanizawa Y, Kanesaki Y, Kubota E, Kumar H, Dicks L, Salminen S, Nakagawa J, Arita M, Endo A. Genomic characterization of a fructophilic bee symbiont Lactobacillus kunkeei reveals its niche-specific adaptation. Systematic and Applied Microbiology. 2016;39:516–526. doi: 10.1016/j.syapm.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Magyar I, Tóth T. Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiology. 2011;28:94–100. doi: 10.1016/j.fm.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends in Genetics. 2010;26:5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Marsit S, Mena A, Bigey F, Sauvage FX, Couloux A, Guy J, Legras JL, Barrio E, Dequin S, Galeote V. Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Molecular Biology and Evolution. 2015;32:1695–1707. doi: 10.1093/molbev/msv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Débarbouillé M, Ferrari E, Klier A, Rapoport G. Characterization of the levanase gene of Bacillus subtilis which shows homology to yeast invertase. MGG Molecular & General Genetics. 1987;208:177–184. doi: 10.1007/BF00330439. [DOI] [PubMed] [Google Scholar]
- Masud U, Matsushita K, Theeragool G. Molecular cloning and characterization of two inducible NAD⁺-adh genes encoding NAD⁺-dependent alcohol dehydrogenases from Acetobacter pasteurianus SKU1108. Journal of Bioscience and Bioengineering. 2011;112:422–431. doi: 10.1016/j.jbiosc.2011.07.020. [DOI] [PubMed] [Google Scholar]
- Mittelbach M, Yurkov AM, Nocentini D, Nepi M, Weigend M, Begerow D. Nectar sugars and bird visitation define a floral niche for basidiomycetous yeast on the Canary Islands. BMC Ecology. 2015;15:2. doi: 10.1186/s12898-015-0036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklitschek M, Alcaíno J, Barahona S, Sepúlveda D, Lozano C, Carmona M, Marcoleta A, Martínez C, Lodato P, Baeza M, Cifuentes V. Genomic organization of the structural genes controlling the astaxanthin biosynthesis pathway of Xanthophyllomyces dendrorhous. Biological Research. 2008;41:93–108. doi: 10.4067/S0716-97602008000100011. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pina C, Gonçalves P, Prista C, Loureiro-Dias MC. Ffz1, a new transporter specific for fructose from Zygosaccharomyces bailii. Microbiology. 2004;150:2429–2433. doi: 10.1099/mic.0.26979-0. [DOI] [PubMed] [Google Scholar]
- Qiu H, Price DC, Weber AP, Reeb V, Yang EC, Lee JM, Kim SY, Yoon HS, Bhattacharya D. Adaptation through horizontal gene transfer in the cryptoendolithic red alga Galdieria phlegrea. Current Biology. 2013;23:R865–R866. doi: 10.1016/j.cub.2013.08.046. [DOI] [PubMed] [Google Scholar]
- Richards TA, Soanes DM, Jones MD, Vasieva O, Leonard G, Paszkiewicz K, Foster PG, Hall N, Talbot NJ. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. PNAS. 2011;108:15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Talbot NJ. Horizontal gene transfer in osmotrophs: playing with public goods. Nature Reviews Microbiology. 2013;11:720–727. doi: 10.1038/nrmicro3108. [DOI] [PubMed] [Google Scholar]
- Richards TA. Genome evolution: horizontal movements in the fungi. Current Biology. 2011;21:R166–R168. doi: 10.1016/j.cub.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Romagnoli G, Luttik MA, Kötter P, Pronk JT, Daran JM. Substrate specificity of thiamine pyrophosphate-dependent 2-oxo-acid decarboxylases in Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2012;78:7538–7548. doi: 10.1128/AEM.01675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J, Rodríguez de la Vega RC, López-Villavicencio M, Gouzy J, Sallet E, Dumas É, Lacoste S, Debuchy R, Dupont J, Branca A, Giraud T. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Current Biology. 2015;25:2562–2569. doi: 10.1016/j.cub.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y-W, Park CY, Park JB, Kim S-Y, Seo J-H. Optimization of erythritol production by Candida magnoliae in fed-batch culture. Journal of Industrial Microbiology and Biotechnology. 2000;25:100–103. doi: 10.1038/sj.jim.7000039. [DOI] [Google Scholar]
- Saerens KM, Saey L, Soetaert W. One-step production of unacetylated sophorolipids by an acetyltransferase negative Candida bombicola. Biotechnology and Bioengineering. 2011;108:2923–2931. doi: 10.1002/bit.23248. [DOI] [PubMed] [Google Scholar]
- Samad A, Zhang J, Chen D, Liang Y. Sophorolipid production from biomass hydrolysates. Applied Biochemistry and Biotechnology. 2015;175:2246–2257. doi: 10.1007/s12010-014-1425-x. [DOI] [PubMed] [Google Scholar]
- Sambrook J. Russell D. Molecular Cloning: A Laboratory Manual. 3 ed. Cold Spring Harbor,NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sanchez A, Gore J. feedback between population and evolutionary dynamics determines the fate of social microbial populations. PLoS Biology. 2013;11:e1001547. doi: 10.1371/journal.pbio.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Zhou X, Kominek J, Kurtzman CP, Hittinger CT, Rokas A. Reconstructing the backbone of the saccharomycotina yeast phylogeny using genome-scale data. G3: Genes|Genomes|Genetics. 2016;6:3927–3939. doi: 10.1534/g3.116.034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Systematic Biology. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Slot JC, Rokas A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. PNAS. 2010;107:10136–10141. doi: 10.1073/pnas.0914418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JC, Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Current Biology. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Sousa-Dias S, Goncalves T, Leyva JS, Peinado JM, Loureiro-Dias MC. Kinetics and regulation of fructose and glucose transport systems are responsible for fructophily in Zygosaccharomyces bailii. Microbiology. 1996;142:1733–1738. doi: 10.1099/13500872-142-7-1733. [DOI] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Zhang Y, Iino T, Kosako Y, Komagata K, Uchimura T. Asaia astilbes sp. nov., Asaia platycodi sp. nov., and Asaia prunellae sp. nov., novel acetic acid bacteria isolated from flowers in Japan. The Journal of General and Applied Microbiology. 2010;56:339–346. doi: 10.2323/jgam.56.339. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Morita T, Wada K, Hirose N, Fukuoka T, Imura T, Kitamoto D. Production of sophorolipid glycolipid biosurfactants from sugarcane molasses using Starmerella bombicola NBRC 10243. Journal of Oleo Science. 2011;60:267–273. doi: 10.5650/jos.60.267. [DOI] [PubMed] [Google Scholar]
- Tofalo R, Schirone M, Torriani S, Rantsiou K, Cocolin L, Perpetuini G, Suzzi G. Diversity of candida zemplinina strains from grapes and italian wines. Food Microbiology. 2012;29:18–26. doi: 10.1016/j.fm.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Tucker CM, Fukami T. Environmental variability counteracts priority effects to facilitate species coexistence: evidence from nectar microbes. Proceedings of the Royal Society B: Biological Sciences. 2014;281:20132637. doi: 10.1098/rspb.2013.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bogaert IN, De Maeseneire SL, Develter D, Soetaert W, Vandamme EJ. Cloning and characterisation of the glyceraldehyde 3-phosphate dehydrogenase gene of Candida bombicola and use of its promoter. Journal of Industrial Microbiology & Biotechnology. 2008;35:1085–1092. doi: 10.1007/s10295-008-0386-x. [DOI] [PubMed] [Google Scholar]
- Vega-Alvarado L, Gómez-Angulo J, Escalante-García Z, Grande R, Gschaedler-Mathis A, Amaya-Delgado L, Sanchez-Flores A, Arrizon J. High-quality draft genome sequence of Candida apicola NRRL Y-50540. Genome Announcements. 2015;3:e00437-15. doi: 10.1128/genomeA.00437-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, Maere S, Verstrepen KJ. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biology. 2012;10:e1001446. doi: 10.1371/journal.pbio.1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuralhan Z, Luttik MA, Tai SL, Boer VM, Morais MA, Schipper D, Almering MJ, Kötter P, Dickinson JR, Daran JM, Pronk JT. Physiological characterization of the ARO10-dependent, broad-substrate-specificity 2-oxo acid decarboxylase activity of Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2005;71:3276–3284. doi: 10.1128/AEM.71.6.3276-3284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Rokas A. Fungal metabolic gene clusters-caravans traveling across genomes and environments. Frontiers in Microbiology. 2015;6:161. doi: 10.3389/fmicb.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaunmüller T, Eichert M, Richter H, Unden G. Variations in the energy metabolism of biotechnologically relevant heterofermentative lactic acid bacteria during growth on sugars and organic acids. Applied Microbiology and Biotechnology. 2006;72:421–429. doi: 10.1007/s00253-006-0514-3. [DOI] [PubMed] [Google Scholar]
- Zhou X, Peris D, Kominek J, Kurtzman CP, Hittinger CT, Rokas A. in silico Whole Genome Sequencer & Analyzer (iWGS): A Computational Pipeline to Guide the Design and Analysis of de novo Genome Sequencing Studies. G3: Genes|Genomes|Genetics. 2016;6:3655–3662. doi: 10.1534/g3.116.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]