Abstract
Understanding how the first cell fate decision has chosen is a fascinating biological question that was received consider attention over the last decade. Numerous transcription factors are required, and many have been shown to have essential roles in this process. Here we reexamine the function that transcription factors play primarily in the mouse—the model system most thoroughly examined in this process. We address how the first embryonic lineage is established and maintained, with a particular emphasis on subsequent trophectoderm development and the role of the recently established Arid3a transcription factor in this process. In addition, we review relevant aspects of embryonic stem cell reprogramming into trophoblast stem cells —the equivalent of the epiblast (inner cell mass) and the establishment of induced trophoblast stem cells—the in vitro equivalent of the trophectoderm.
Keywords: Embryonic stem cells, Trophectoderm, Transcriptional regulation
3. Introduction
During early embryogenesis, the totipotent single cell zygote undergoes a sequence of cell divisions accompanied by distinct cellular, molecular, and epigenetic changes. This leads to blastocysts that develop according to the cell fates of partitioning cells [1–3]. These cells develop into three morphologically distinct lineages by the late blastocyst stage of preimplantation [4,5]. The outer layer of the late blastocyst, or trophectoderm (TE), will develop into the placenta and invade the maternal endometrium to form the intervillous space that links the growing fetus to the mother [6] (Figure 1). The inner layer of the blastocyst, the inner cell mass (ICM), later differentiates into two distinct lineages: The inner epiblast (EPI) cells and an outerlayer of primitive endodermal (PE) cells facing the blastocoel [7,8]. While PE cells are restricted to the yolk sac, EPI cells can give rise to all adult tissues [6]. From the aforementioned cell lineages, three types of stem cells are established: (1) trophoblast stem (TS) cells from the TE, (2) extra embryonic endoderm stem (XEN) cells from the PE, and (3) embryonic stem (ES) cells from the EPI or earlier stages of the ICM [9,10]. These stem cells serve as invaluable in vitro model systems that help illuminate the mechanisms underlying early embryogenesis, as well as lineage specifications, and ultimately will enable stem cell therapies which can be used to treat numerous disorders and injuries.
Figure 1.
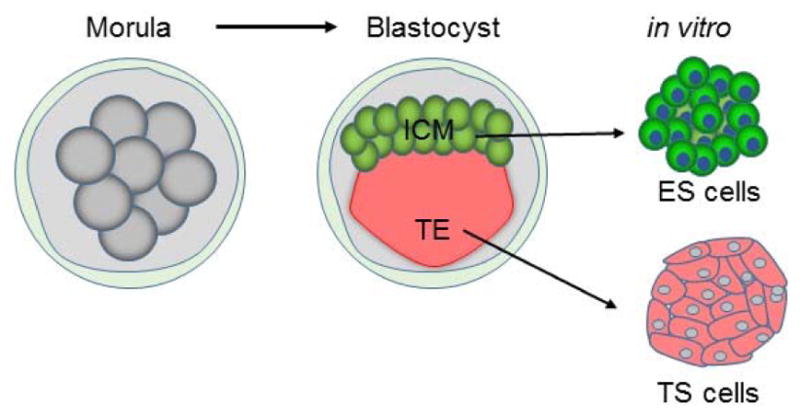
Stem cell cultivation. The zygote undergoes successive divisions to morula and then to blastocyst. In the blastocyst, the inner cell mass (ICM) develops into the fetus, whereas the trophectoderm (TE) at its periphery generates the embryonic membranes and placenta. Embryonic stem (ES) cells and trophoblast stem (TS) cells are derived from the ICM and TE, respectively. These stem cells can self-renew and differentiate into multiple lineages.
4. Overview of Preimplantation Development
During the initial rounds of zygotic cleavage, the cells are morphologically identical and are distributed symmetrically within the embryo until compaction at the eight-cell (morula) stage, during which these cells become adhesive and polarized [11,12]. Junctional complexes are gradually formed at apicolateral and lateral sites, followed by polarization of outer cells [13]. Compaction and polarization during the morula stage generate cellular asymmetry, leading to the regression of totipotency and the formation of the polarized outer and apolar inner compartments—the TE and the ICM, respectively [14]. This segregation process is termed the first cell fate decision, after which these two groups of cells diverge sharply in terms of transcriptional and epigenetic regulation during development [2,15,16]. Mouse and human embryos undergo similar embryonic developmental processes, although the timeline to reach the blastocyst is delayed and in humans this occurs as Embryonic (E) day 6 and in mice, E3.5 [17].
Despite studies that have advanced our understanding of embryogenesis over recent years, it is still not well-understood how this essential cell fate decision is controlled by signaling pathways as well as by global transcriptional and epigenetic regulatory mechanisms.
5. The First Cell Fate Decision: Inner Cell Mass (ICM) and Trophectoderm (TE)
Both cell polarity and position in the embryo influence the first cell fate decision. Upon blastocyst formation, the cleavage plane and polarization axis are perpendicular, resulting in the formation of inner apolar and outer polar cells [18,19]. Inner apolar cells are progenitors of the pluripotent ICM, which can engender all three germ layers; i.e., the mesoderm, endoderm, and ectoderm, whereas the outer polar cells are antecedents of the multipotent TE that can be differentiated into all cell types of the placenta [20]. In addition, cell polarity and cell position cross-regulate one another, as transplantation of inner cells to an outside position results in polarization and adaption to the TE fate. On the other hand, downregulation of key polarity molecules such as aPKC (atypical protein kinase C) and PARD3 (par-3 family cell polarity regulator) promotes allocation of the cells to inner parts of blastocysts [21,22]. Although segregation of the ICM and the TE becomes apparent as polarization of blastomeres occurs, cells are not yet fully committed toward ICM or TE lineages at the 16-cell stage [14]. Manipulation of the cells at this stage can alter their cell fate; thus, they are still plastic and totipotent. Cell fate is further determined by signaling cascades of environmental cues, followed by changes in transcriptional activities coupled with selective epigenetic marks.
The mechanisms underlying the first cell fate decision are remarkably complex and remain poorly understood. Recent studies identified substantial changes in the transcriptome during the first cell fate decision, suggesting important roles for transcription factor (TF) activities [23]. In addition, microRNAs (miRNAs) and epigenetic regulators regulate specification [24,25].
6. Transcriptional Regulation
TFs play crucial roles during the development of the blastocyst. Importantly, some TFs show restricted expression patterns associated with the segregation of the ICM and the TE. For example, Nanog and Oct4 (Pou5f1) are expressed in ICM cells, but not in TE cells, while Cdx2 is exclusively expressed in the TE [26,27]. These exclusively expressed TFs not only serve as markers to distinguish between the ICM and the TE, but also play crucial roles in first cell fate determination.
Oct4 and Cdx2 currently are considered to be the master regulators of the first fate decision based on their respective mutant phenotypes, distinctive gene expression patterns, and differential functions at the molecular level [28]. Initially, Oct4 and Nanog were identified as critical factors required for forming and maintaining the ICM, because they become restricted to the ICM cells of the blastocyst [29,30]. However, subsequent genetic studies in mice revealed that both Oct4 and Nanog knockout (KO) embryos formed blastocysts without obvious developmental defects [26,29]. Yet, ES cells, the in vitro counterpart of the ICM, cannot be easily derived from the ICM of these mutant embryos, nor do they behave as typical ES cells. For example, ES cells from Oct4-KO embryos tend to differentiate into the extraembryonic trophoblast lineages, whereas ES cells from Nanog-KO embryos are prone to death via apoptosis [29,31]. These results suggested that Oct4 and Nanog are, instead, key regulators of pluripotency and self-renewal [32]. Consistent with these observations, recent genome-wide mapping of pluripotency factors, including Oct4, Sox2, and Nanog in ES cells, revealed that these pluripotency ICM factors tightly maintain their expression levels in the interwoven transcriptional gene regulatory network to auto-regulate and control one another through a feed-forward regulatory loop. The pluripotency network requires many additional TFs, epigenetic regulators, and miRNAs [32,33].
On the other hand, Cdx2, a key TF in the generation of the TE, was first detected at the eight-cell stage of the embryo [34]. Cdx2 is exclusively expressed within cells of the outer embryonic segment, while no expression is observed within the ICM. Cdx2-KO embryos can initiate lineage commitment, but cannot maintain a blastocyst due to a lack of epithelial integrity in their outside cells [27]. Thus, Cdx2 is crucial for both epithelial identity and trophoblast multipotency. In addition to Cdx2, other TE-specific TFs, including Eomes, Gata3, and Tfap2c, are also solely expressed in the TE [35–37]. Unlike Cdx2, embryos deficient in any of these factors are able to form a blastocyst, but fail after implantation due to defects in more differentiated placental tissues. These results indicated that these factors are required during later stages of TE development. Deletion of another TE-specific factor, Tead4, resulted in preimplantation lethality, suggesting that its function is upstream of Cdx2 [38]. Tead4-KO embryos failed to activate TE-specific genes, including Cdx2, [38,39]. Thus, Tead4 is crucial for TE development [38]. During the process of TE specification, Tead4 cooperates with its co-activator, Yap1, to activate TE-specific genes [40]. Likewise, Gata3 participates in TE development, downstream of Tead4 but in parallel to Cdx2 [37,41]. In addition to TFs, Fgf signaling, mediated upstream by Ras/Erk signaling, is essential for trophoblast expansion and the sequential induction of TE-specific TFs [42,43]. ICM-specific TFs positively regulate one another, and the same is observed for TE-specific TFs. Indeed, ICM-specific TFs tend to antagonize TE-specific TFs and vice-versa. This is fundamental to the decision of ICM vs. TE cell fate. For instance, Oct4, Nanog, and Cdx2 directly repress each other to allow cells to develop into ICM or TE lineages [2,27].
Although our knowledge of the mechanisms underlying ICM and TE segregation has considerably expanded, many questions remain unanswered. For example, how do the key TFs interact with epigenetic regulators, such as histone-modifying enzymes and/or chromatin remodelers, to activate or suppress gene expression? Which additional TFs are essential for blastocyst development, and what are the critical downstream targets of these TFs? How are these factors themselves regulated within the pluripotency or TE-specific networks? Answers to these central questions are required to fully elucidate the fundamental mechanisms underlying pluripotency of the ICM and multipotency of the TE lineages, as well as to advance stem cell-based future cell therapies.
7. Epigenetic Regulation
From zygote to blastocyst formation, the epigenome, including DNA methylation and histone modifications, changes dramatically, and these changes strongly influence developmental potential [44,45]. Asymmetric epigenetic marks are tightly associated with the segregation between the ICM and TE [46]. Recently, DNA methylation and histone modifications have been shown to be indispensable for growth and differentiation of the extra embryonic lineages [47,48]. In mammals, sperm and egg DNA are highly methylated as compared to somatic cells, yet most of this methylation disappears during preimplantation, after which the embryo undergoes de novo methylation. Methylation-deficient (Dmt1, Dmt3a or Dnmt3b KO) mice are lethal at mid-gestation. An important stem cell mechanism that relies on active DNA demethylation is associated with the global growth arrest and DNA-damage-inducible GADD45A. Indeed, these and other DNA methylation are useful markers in characterizing different types of pluripotent and differentiated cells.
In particular, histone methylation signatures such as H3K4me3 and H3K27me3 are enriched at the promoters of ICM-specific genes exclusively expressed in ICM and TE-specific genes that are highly expressed in TE [49]. Distribution of H3K27me3 and H3K9me3 in ICM and TE are correlated with lineage specification to embryonic versus extra embryonic tissues [50]. For example, a histone methyltransferase enzyme, ESET, maintains pluripotency of the ICM through catalyzing a repressive H3K9me3 mark at the promoter of Cdx2 [51–53]. In contrast, within the TE lineage, methyltransferase Suv39h mediates H3K9me3 at regulatory regions of ICM-specific genes [50,54]. Each of these histone modifications cooperates with developmental-specific TFs to mediate spatial and temporal expression of lineage-specific genes.
There are a number of “outliers” of the above families that have been shown to be essential in establishing the first fate decision. For example, Zhou et al. [55] found that DNA methylation was programmed by the pattern of appearance of DNA 5mC and 5hmC epigenetic modifications. Loss of the catalyst for this reaction, MBD3, retains ES cells in their pluripotent state unable to commit to downstream lineages [56]. Similarly, the NuRD complex this blocks reprogramming of somatic cells to pluripotent stem cells. And indicates a context-dependent manner of MBD3/NuRD collaboration in facilitating pluripotency and stemness [57]. Finally, the CoREST2 complex shown to regulate ESC proliferation and pluripotency, also is critical for during neuronal differentiation [58].
8. Signaling Pathways
The Hippo signaling pathway regulates cell fate during embryonic development in position-, cell-polarity-, and density-dependent manners [59]. After the 16-cell stage, Tead4, a downstream effector of Hippo signaling, is active in outer polar cells to promote TE development [38]. In contrast, cell–cell interactions in the inner apolar cells activate the Hippo pathway, which sequesters Yap1 in the cytoplasm, prevents co-activation with Tead4, and promotes the ICM fate. Although Hippo signaling is spatially regulated at the 16-cell stage, segregation of ICM and TE fate is not clearly patterned until the 32-cell stage. This may be due to another signaling pathway, the Notch pathway, which activates TE-specific genes. A key component of Notch signaling, RBPJ (also known as CBF1), is stochastically activated up to the morula stage and gradually becomes restricted to the outer cells around the 32-cell stage [60].
Thus, the Notch and Hippo pathways cooperate to establish segregation of the ICM and TE—events observed at the 32-cell stage [61]. Ultimately, crosstalk between multiple signaling pathways such as Hippo and Notch, as well as the MAPK pathway, regulates cell fate determination in preimplantation embryos. However, further studies are needed to fully appreciate the constellation of regulatory inputs required for lineage specification at this critical developmental time point.
9. Second Lineage Specification in Preimplantation
Shortly after the first cell fate decision, ICM cells differentiate into the EPI and PE lineages—a transition known as the second lineage specification. This differentiation is mediated through FGF/MAPK signaling and differential expression of key TFs, including Nanog and Gata6 [62–64]. FGF signaling orchestrates reciprocal repression of TFs specific for the EPI and PE lineages [65]. Nanog and Gata6, which are expressed in all ICM cells until the 32-cell stage, become restricted to either the EPI or PE, respectively. Additional TFs, including Gata4 and Sox17, also contribute to specification of EPI versus PE lineages within the mouse ICM [66,67]. While the expression profiles of these TFs are conserved between human and mouse [68], human embryos are not dependent upon FGF signaling. This suggests that alternative, as yet undiscovered, signaling pathways are critical for second fate specification in human embryos.
10. Terminal Trophectoderm Differentiation
The TE ultimately differentiates into all the specialized cells of the placenta. The placenta nourishes and safeguards the fetus during pregnancy by mediating exchange of nutrients, gases, and waste between mother and fetus, as well as providing immune protection and secretion of growth factors and pregnancy-dependent hormones [69,70]. Placental health is essential for a successful pregnancy. In mice, placental tissues are composed of multiple specialized cell types, including trophoblast giant cells, spongiotrophoblasts, and syncytial trophoblasts [69,71]. The failure of normal TE development can cause mortality and developmental defects not only during pregnancy, but also after birth [72].
Despite its crucial importance to pregnancy health, the molecular basis of preimplantation is poorly understood. Thus, the discovery of biomarkers of early placental development will not only serve as diagnostic markers in gynecology, but also will catalyze the development of treatments to save maternal and fetal lives from pregnancy-related disorders.
11. Embryonic Stem (ES) Cells: In Vitro Model Systems of Early Embryogenesis
Stem cells are self-renewing (i.e., capable of continual duplication) and either multipotentor pluripotent; i.e., capable of differentiation into other, more specialized cell types. Each of the blastocyst lineages discussed above can be used to derive stem cells that can be grown in culture, which under appropriate culture conditions, can indefinitely self-renew and be induced to differentiate in vitro or be incorporated into a developing embryo ex-vivo.
Embryonic stem (ES) cells were first derived from the ICM of mouse blastocysts [73,74]. To maintain their self-renewal and pluripotency, they are cultured either on feeder layers of mouse embryonic fibroblasts (MEFs), without feeders in the presence of leukemia inhibitory factor (LIF) or under serum-free conditions supplemented with Mek and GSK3 kinase inhibitors (this latter condition is termed “2i”) [74,75]. The combination of 2i and LIF sustains the self-renewing “ground state”; i.e, a basal proliferative state that is free of epigenetic restriction and has minimal requirements for extrinsic stimuli. Intensive studies have revealed that mouse ES cells express many critical factors, including stage-specific embryonic antigens (SSEA-1 and SSEA-3) and TFs that are required for early ICM development, such as Oct4, Sox2, and Nanog [76,77]. ES cells are karyotypically stable, capable of generating chimeras in transgenic mice, and able to form ectoderm, endoderm, and mesoderm in culture; but they cannot contribute to the TE lineages of the placenta [78].
Several years after mouse ES cells were established, human ES cell lines were derived that share some of the conserved features [73]. Similar to mouse ES cells, human ES cells require an OCT4/SOX2/NANOG regulatory module [32]. Both cells also express surface markers such as SSEA-3, SSEA-4, and TRA-1, as well as other critical, conserved components, including epigenetic regulators, miRNAs, and signaling molecules. However, key differences exist. For example, human ES cells do not require the LIF growth factor for self-renewal [79,80]. More importantly, human ES cells display transcriptional and epigenetic profiles more similar to those of mouse epiblast stem cells (EpiSC) which are derived from the epiblast of the inner cell mass [81]. Although controversial, it has been suggested that human ES cells, are able to differentiate into TE/trophoblast stem (TS)-like cells under appropriate culture conditions [82–84]. Further studies are required to establish interspecies differences between mouse and human.
12. The Pluripotency Transcriptional Network
Genetic studies in combination with high-throughput knockdown (KD) screening revealed that many critical regulators (including sequence-specific TFs, chromatin modifying enzymes, chromatin remodelers, miRNAs, and non-coding RNAs) are indispensable for maintaining self-renewal and pluripotency [85,86]. Genome-wide target mapping of sequence-specific TFs has unveiled a “core transcriptional network” of essential pluripotency TFs (Oct4, Sox2, and Nanog) as well as additional TFs with partially redundant activities (e.g., Klf4, Esrrb, and Tbx3) [77,87–89].
Oct4 is the key master regulator that sits atop of the hierarchy of the pluripotency regulatory network. Both self-renewal and developmental potential of ES cells are highly sensitive to either up-or down-modulation of Oct4 expression. For example, Oct4-“high” cells are more prone to differentiate into the primitive endoderm, whereas Oct4-“low” cells tend to differentiate into TE lineages [31]. Sox2 is a well-known interaction partner of Oct4, and alteration of Sox2 levels also disrupts the pluripotency of ES cells. For instance, Oct4- and Sox2-KO ES cells neither self-renew nor trans-differentiate/reprogram to TE lineages [90]. Within the pluripotency network, Oct4 and Sox2 positively regulate one other and co-occupy the vast majority of active transcriptional enhancers of ES cell-specific genes.
In addition to the activation of pluripotency genes, development-related genes must be suppressed to maintain self-renewal in ES cells. Some pluripotency factors exert such repression by occupying key gene regulatory regions. In addition, histone modifying enzymes and histone remodeling complexes such as Hdac1, Mta1, and Mbd3 (the NuRD complex) as well as the Polycomb (PRC1 and PRC2) and SWI/SNF (Brg1 and Baf155) complexes are required for this critical developmental gene repression [91,92]. In particular, most developmental genes are marked by a “bivalent” histone signature consisting of both H3K4me3 and H3K27me3; this is considered to be the “poised status” for rapid activation upon differentiation [93].
In summary, key ES cell-specific TFs orchestrate transcriptional gene activation and repression in conjunction with chromatin regulators within the pluripotency network to sustain self-renewal and pluripotency of ES cells.
13. Trophoblast Stem (TS) Cells
TS cells can be isolated from the polar TE of mouse embryos and maintained in a proliferative and undifferentiated state in vitro in the presence of Fgf4 (fibroblast growth factor 4) along with its cofactor, heparin and a feeder layer of MEFs [94]. TS cells are both self-renewing and multipotent. Upon withdrawal of Fgf4, the vast majority of cells undergo terminal differentiation to trophoblast giant cells, while a small portion of cells differentiate into other trophoblast lineages, including spongiotrophoblasts and labyrinthine cells. Following their injection into an early embryo, TS cells can participate in the development of chimeras in which they contribute exclusively to trophoblastic components of the placenta and parietal yolk sac [95]. This is in stark contrast to ES cells, whose contributions are restricted to the three germ layers.
Multiple TFs, serum factors, and signaling pathways, such as Ras/Mapk signaling, maintain the self-renewal and multipotency of TS cells by upregulating TS-specific TFs such as Eomes (which only requires one of the two signaling pathways) and Cdx2 (which requires both) [96]. In contrast to the molecular details underlying ICM and ES cell biology, placentation, including TE differentiation, is poorly understood, despite its critical importance for successful mammalian reproduction. Importantly, human TS cells are not yet available, mainly due to ethical issues, such as the necessity of destroying human embryos.
Spontaneous differentiation of human ES cells to the TE lineage (termed “trans-differentiation”) can occur at only low frequency. Thus, a common method to induce trans-differentiation is via BMP4 treatment of ES cells [82]. BMP4 down-regulates the expression of OCT4, SOX2, and NANOG in ES cells, thus promoting their conversion to TS-like cells through the induction of TS cell-specific TFs, including TEAD4 and GATA3. Despite dramatic changes in cell morphology and global gene expression that are similar to the human TE, these TS-like cells are mixed cultures that express TE, mesodermal and vascular endothelial cell markers [84]. These data suggest that BMP4 treatment alone is insufficient to induce complete trans-differentiation to the TE lineage.
Although as yet not fully successful, a number of attempts have been made to induce human TS-like cells from ES cells by inhibiting the Activin/Nodal pathway or by the addition of Fgf2 [97,98]. Success in this area is essential for developing a well-characterized model of human TS cell differentiation to understand early human placentation and inform therapeutic interventions for pathological pregnancies.
14. Core TS Cell-specific Transcription Factors
A group of key TS-specific TFs, including Tead4, Cdx2, Gata3, Eomes, Elf5, and Tfap2c, positively regulate one another to orchestrate TS cell-specific gene expression [41,99]. In contrast to ES cells, TS cells do not require the LIF-Stat3 signaling pathway; instead, they need Ras/Mapk and Nodal signaling to maintain self-renewal [100]. Notably, depletion of any one of the above subset of TFs leads to lethality of the mouse embryo due to defects in proper TE differentiation [37,38,101–103]. In addition, enforced expression of Gata3 and Elf5 in TS cells abrogated TS self-renewal. Thus, the precise levels are crucial for the balance between self-renewal and differentiation.
Genetic studies in mice further revealed regulatory hierarchy among several TS cell-specific TFs. Tead4, the most “upstream” factor of this series, controls the level of Cdx2, Eomes, and Elf5 [28,41,104]. Functional and mechanistic studies further disclosed that Cdx2 not only auto-regulates itself but also is controlled by multiple TS cell-specific TFs, such as Gata3, Eomes, and Tfap2c. Unfortunately, only a few TS cell-specific regulators have been identified. Furthermore, exhaustive mapping of genome-wide DNA binding sites of TS cell-specific TFs has not been achieved, and epigenetic regulation of TE gene expression remains relatively uncharacterized. Many questions remain unanswered, including: How do these TS cell-specific TFs control self-renewal and modulate differentiation of TS cells toward more specialized placental cell types? How do these TFs interact with their DNA targets or with histone modifying enzymes and remodelers? These central questions constitute a major challenge for future investigation.
15. Cross Talk between Pluripotency and Epigenetic Factors in ES and TS Cells
Balancing pluripotency TF expression with their epigenetic modification is critical in maintaining “stemness” of ES cells. Epigenetic regulators, such as the Polycomb (eg., Eed, Ezh2 and Suz12) and MLL (Wdr5 and Ash2l) complexes are positively regulated by Oct4/Sox2/Nanog [105–107]. Both complexes are required to keep lineage-specific genes silenced. Genome-wide DNA methylation patterns mediated by the Ten-Eleven Translocation (TET) DNA methyltransferases (DNMTs) also maintain ES cell identity [108].
Unlike ES cells, only a few histone modifications have been globally investigated in TS cells. Among those, the levels of H3K27me3 appear to be globally lower in TS cells than in ES cells [109]. This is probably linked to the low expression levels of Eed, a component of the Polycomb complex responsible for H3K27me3 signatures. In addition, ChIP-seq analyses revealed that the few TS-cell genomic regions enriched for H3K27me3 are only rarely localized near transcriptional start sites. Moreover, H3K27me3/H3K4me3 bivalent domains, which restrain the expression of developmental genes in ES cells, are relatively rare in TS cells [109]. However, a recent study did show the existence of bivalency in TS cells [110]. Thus, further investigation is needed to understand TS cell biology at an epigenetic level.
16. Extraembryonic Endoderm Stem (XEN) Cells
For many years, mouse ES and TS cells have been used as model systems for ICM/EPI or TE biology to study mechanisms of embryonic and placental development, respectively [74,94]. Recently, XEN cells have been successfully derived from mouse blastocysts at E3.5 [111]. As with other stem cells, XEN cells can propagate indefinitely in vitro, while maintaining their ability to contribute to extraembryonic endodermal lineages after injection into blastocysts ex vivo. Although Gata6 and Gata4 are known to be the key XEN-specific TFs, these TFs are not homogeneously expressed within populations of XEN cells [112,113]. It remains unclear whether this heterogeneity represents distinct XEN cell types. XEN cells are emerging as a useful stem cell model and provide another layer of control in developmental biology.
17. Reprogramming
Cellular reprogramming is the process that allows the conversion of one specific cell type to another. Nuclear reprogramming, first established using frog embryos, ultimately enabled animal cloning via somatic cell nuclear transfer [114,115]. This technique relies on de-differentiation of (assumedly) terminally differentiated cells into a stem-like state; thus, refuting the principle of irreversible cellular differentiation. Though powerful, nuclear reprogramming has extremely low efficiency [116]. Whereas direct reprogramming, in which target cells are delivered as TFs, miRNAs, or chemical compounds, can cause dramatic changes in morphology, global gene expression, and epigenetic marks [117,118]. This technique is far more efficient than nuclear reprogramming, although its mechanism(s) remains incompletely understood.
18. Transcription Factor-Mediated Direct Reprogramming
The first instance of successful direct reprogramming was the conversion of fibroblasts into muscle cells by ectopic expression of the MyoD TF [119]. Subsequently, Yamanaka’s group in Japan established a protocol for the generation of “induced” pluripotent stem (iPS) cells by the overexpression in fibroblasts of a handful of TFs (Oct4, Sox2, Klf4, and c-Myc ) [120]. As with ICM-derived ES cells, iPS cells are self-renewing and pluripotent. Their successful exploitation overcomes the ethical issues in deriving ES cells from developing human embryos, thereby paving the way to develop future patient-specific stem cell therapies. For instance, patient-specific iPS cells may be generated from fibroblasts of a patient donor (e.g., a sufferer of Alzheimer’s disease) followed by the correction of genomic mutations by genome editing approaches. These genome-edited “normal” cells then can be re-introduced into patients. In addition to iPS cells, numerous studies have recently reported defined TF-mediated cell type conversions in a broad range of cell types, including fully functional induced neural (iN) cells and induced cardiomyocytes (iCMs) [118,121].
Mouse ES cells can be directly reprogrammed into extraembryonic lineages by the over expression of single TFs, such as Cdx2 or Gata6, to derive TS- and XEN-like cells, respectively [28,122]. The morphology and global gene expression patterns of TS-and XEN-like cells are highly similar to their counterpart genuine cells, suggesting that TF-mediated reprogrammed cells may acquire proper epigenetic characteristics in vitro.
Although TF-mediated reprogramming from numerous cell types is possible, substantial obstacles remain, including inconsistencies in differentiation potential. This suggests that there are still yet-to-be discovered TFs and/or epigenetic regulators necessary to augment the efficiency and speed of reprogramming. In this regard, it has become possible to reprogram mouse fibroblasts into induced trophoblast stem (iTS) cells by introducing a handful of TFs, including Eomes, Gata3, Tfap2c, and Ets2 or Myc [123,124]. It is critical that further development of mouse model protocols are efficiently translated to the human system.
19. Mechanisms of Direct Reprogramming
Reprogramming requires that cell-type specific genes of the original cell type must be repressed, while those of the target cell type must be activated. The mechanism of how this occurs is poorly understood; particularly the repression aspect. Reprogramming to iPS cells can be broadly divided into two gene activation phases: a long, stochastic phase followed by a shorter deterministic phase [125]. At first, cells undergo increased proliferation and alteration in histone modification to initiate mesenchymal-to-epithelial transition. Later, cells enter an intermediate phase followed by long latency of the process. In this late phase, cells stochastically undergo an epigenetic “reset”, as well as activation of their target-specific transcriptional network to stabilize the reprogrammed cells.
Another well-established gene activation mechanism accompanying reprogramming is the ability by which ectopically expressed TFs act as “Pioneer” Factors [126]. Pioneer factors initially bind to closed chromatin of genes specific to the target cell type. Once bound, pioneer factors interact with chromatin modifying enzymes or remodeling complexes to convert closed chromatin into open chromatin, thereby de-repressing target cell-specific genes. For example, in reprogramming, Oct4, Sox2, and Klf4 function as pioneer factors by accessing closed chromatin of distal regulatory elements of pluripotency genes, such as Esrrb and Sall4 [127]. Ascl1, a TF capable of converting fibroblasts to iN cells, also functions as a pioneer factor [118]. Thus, the pioneer concept extends beyond the reprogramming of a single target.
Recently insights have been gained into the process of reprogramming, including the finding that cells undergo defined sequential molecular events in an apparently stochastic manner. These events are influenced by the choice and number of TFs, as well as the starting cell type. Many questions remain unanswered; in particular, how do ectopically expressed TFs mediate gene repression?
20. ES to TS-Like Cell Fate Conversion Under Arid3a Transcriptional Control
ES cells can be trans-differentiated to TS-like cells that are highly similar to genuine multipotent TS cells with respect to morphology and global gene expression [128]. This provides an informative in vitro model for the investigation of factors and the mechanisms underlying the first cell fate decision. Several methods promoting ES to TS-like cell conversion have been reported, including KD of Oct4 and induction of trophoblast-specific TFs in ES cells [31]. Ectopic expression of components implicated in the Ras/Erk signaling pathway also induces conversion of ES to TS-like cells [96]. However, the mechanisms by which ES cell-specific genes are repressed, while cell-specific genes are activated by these diverse manipulations, have remained elusive. Unfortunately, TS-like cells obtained through KD of Oct4 or induction of TE-specific TFs in ES cells do not recapitulate all properties of genuine TS cells. However, such TS-like cells do show upregulation of core TS cell-specific regulators and incorporation into the TE of developing embryos, where they contribute to placental lineages ex vivo [28,129].
Work from our own group demonstrated that over expression of a single TF, Arid3a results in trans-differentiation of ES cells to TS-like cells, even under ES cell culture conditions [129]. Arid3a was initially identified as a transcriptional activator of B-lymphocyte development [130]. However, several somatic cell types from rare survivors of conventional Arid3a KO mice were developmentally plastic with the capacity to differentiate into multiple lineages [131]. We later observed that Arid3a enhanced standard four factor reprogramming of MEFs and was alone capable of spontaneously forming stable, fully reprogrammed iPS-like cells [132]. Subsequently Rhee, et al. [129] observed that Arid3a is not only required for the maintenance of self-renewal of TS cells, but also promoted further trophoblastic lineage differentiation. Consistent with these in vitro results, Arid3a−/− mice suffer severely impaired post-implantation development, which results in early embryonic lethality [129]. Mechanisms underlying this essential Arid3a function include regulated nuclear entry-export, HDAC-mediated deacetylation of target genes, and regulation of embryonic hematopoiesis (Figure 2).
Figure 2.
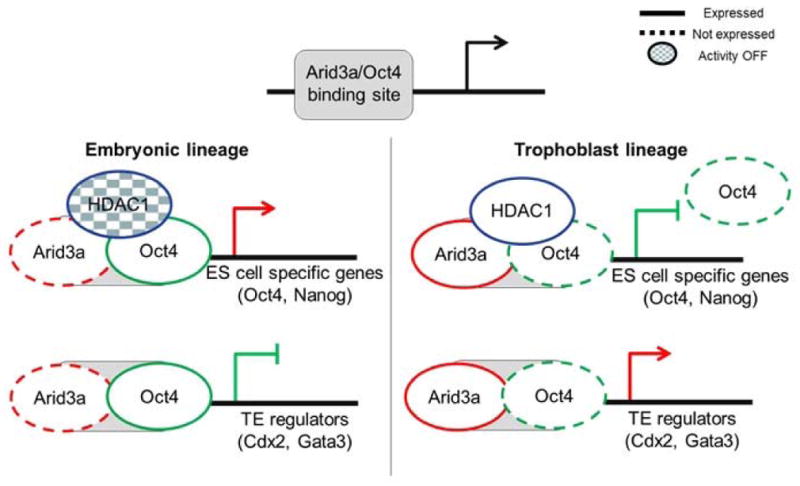
Embryonic versus trophoblastic lineage choice depends on differential nuclear import and histone deacetylases (HDAC) association of Arid3a. High level of Arid3a–HDAC1 co-occupancy is observed at pluripotency-associated loci in Arid3a-overexpressing TS-like cells. Arid3a–HDAC1 interaction alters the activity/specificity of HDAC1, leading to deacetylation of pluripotency-associated nucleosomal histone tails.
Similar to what we observed in the mouse, transient induction of ARID3A into human ES cells up-regulated TE-associated factors such as GATA3 and TFAP2C while down-regulating levels of OCT4 and NANOG [133]. Thus, the regulatory function of Arid3a during placental development appears to be conserved in humans, and Arid3a has emerged as a pivotal regulator of TE and placental development by regulating commitment to the first cell fate as well as by the execution of TE lineage differentiation.
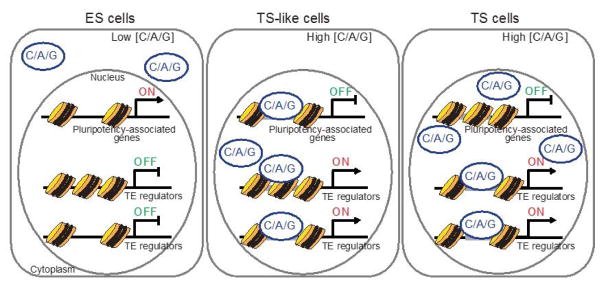
Our current studies have been focused on the mechanisms underlying Arid3A-mediated trans-differentiation/direct reprogramming of mouse ES to TS-like cells. We addressed this mechanism specifically via the TS cell-specific TFs Cdx2, Arid3a, and Gata3 (CAG factors) [134]. We carried out time–course profiling of chromatin accessibility, transcriptomes, and occupancy of reprogramming factors during direct ES to TS-like cell conversion. We discovered that CAG factors orchestrate conversion via a two-step mechanism (Figure 3): 1) initial repression of ES cell-specific genes by decommissioning of active enhancers and 2) direct activation of TS cell-specific genes via accession of closed chromatin by employing different DNA binding motifs during reprogramming. Our data [134] support a sequential gene regulation mechanism in which the exit of ES cells from pluripotency states is followed by the activation of TS cell-specific genes, leading to the conversion of ES cells to multipotent TS-like cells.
Figure 3.
Two-step mechanism of the CAG factor-mediated reprogramming of ES cells to TS-like cells. OE of CAG in ES cells initiates ES to TS-like cell reprogramming through changes in histone signatures on the enhancers. This process is reversible at least at the early stage of reprogramming.
21. Conclusions and Future Perspectives
Although it has been established that at least 9 TFs play critical roles in the first cell fate decision, it is unknown when and how epigenetic marks arise and fade during this crucial point in development. It is also unknown what role such marks play during ES to TS-like cell reprogramming. This is vital to consider, as histone modifications have strong implications for gene expression by promoter silencing, RNA polymerase recruitment, and other mechanisms. This question can be addressed by identifying essential epigenetic modifiers during the ES to TS-like cell transition, including their target sites within the genome, and determination of the mechanism(s) by which they are recruited or suppressed by cell type-specific TFs. Understanding such developmentally regulated epigenetic signatures will be crucial in unraveling key features of the previously observed epigenetic plasticity during this transition. Progress in this area is essential for the ultimate goal of developing safe cell therapies and regenerative medicines.
A more thorough understanding of the signaling pathways governing self-renewal and multipotency in mouse TS cells also remains a crucial issue. Although the Ras/Mapk and Nodal signaling pathways are strongly implicated, each participates in “cross-talk” with other pathways. Thus, it is vital to establish a signaling network to better understand how distinct trophoblast lineages are specified—for example, what TF-associated signaling pathways regulate differentiation to giant cells vs. labyrinthine cells. The benefits of constructing such a network are numerous. After all, the use of chemical inhibitors that target specific signaling pathways has allowed the derivation of pluripotent ES cells across species and facilitated creative approaches to cell reprogramming. Additionally, understanding signaling in the ES to TS-like cell reprogramming context may enable development of a chemically defined media to culture and maintain TS cells. This would eliminate the variability and ethical issues inherent in serum use, as well as link individual signaling components to vital transcriptional targets.
Derivation of human TS cells, which to date has been unsuccessful, would provide a highly valuable tool to study genetic and epigenetic control mechanisms of trophoblast proliferation, differentiation, and function in normal and pathological placentation during human embryogenesis. Unfortunately, the human implantation process is still poorly understood. The loss of potential embryos occurs relatively frequently due to the loss of blastocysts before or around implantation. Thus, it will be essential to decipher how TS cell-specific TFs are conserved in the human placentation process and to examine how their deregulation affects serious pregnancy diseases, such as pre-eclampsia. Once viable human TS cells are generated, they may be used in the future to treat various pregnancy-related disorders that appear to involve dysregulated trophoblast differentiation.
Paradoxically, despite its fundamental importance, the placenta remains one of the least understood organs in mammals. There are only a few TFs identified to date that are implicated in trophoblast lineage specification [134]. This short list includes ARID3A. Although the ES cell pluripotency network is well-characterized, TS cell regulatory networks remain insufficiently understood. It remains unknown as to how TS cell-specific TFs regulate one another to maintain self-renewal and multipotency. We hypothesize that, analogous to ES cells, TS cells are maintained via positively regulated feedback loops governed by multiple TS cell-specific master TFs. Perturbations in this network then cause an exit from multipotency and result in terminal differentiation into specialized placental cells. This is crucial to consider, as improper or unbalanced differentiation of placental lineages is a hallmark in placental disorders. Indeed, such differentiation defects also pose risks for congenital malformations, pre-eclampsia, and miscarriage.
In the age of genome sequencing and massive biological data sets, systems-level approaches have become increasingly critical for understanding biological processes. Construction of a TS cell-specific regulatory network would be a great asset to the fields of development, differentiation, and medicine. It is our great hope that the present studies delineated in this review may catalyze further advances in placental biology and trophoblast development.
Acknowledgments
We thank members of our laboratories for helpful suggestions. The work was supported by National Institutes of Health (NIH) [R01GM112722]; Burroughs Welcome Fund (to J.K.); NIH [R01CA31534]; Cancer Prevention and Research Institute of Texas [RP16704, RP100612, RP120348]; Marie Betzner Morrow Centennial Endowment (to H.O.T.). Funding for open access charge: NIH [R01GM112722].
References
- 1.TARKOWSKI AK. Experiments on the development of isolated blastomers of mouse eggs. Nature. 1959;184:1286–7. doi: 10.1038/1841286a0. [DOI] [PubMed] [Google Scholar]
- 2.Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nature reviews Nat Rev Genet. 2009;10(7):467–77. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]
- 3.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nature reviews Nat Rev Mol Cell Biol. 2009;10(8):526–37. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 4.Kelly SJ. Studies of the developmental potential of 4- and 8-cell stage mouse blastomeres. J Exp Zool. 1977;200(3):365–76. doi: 10.1002/jez.1402000307. [DOI] [PubMed] [Google Scholar]
- 5.Ralston A, Rossant J. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet. 2005;68(2):106–12. doi: 10.1111/j.1399-0004.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- 6.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nature reviews Nat Rev Mol Cell Biol. 2009;10(2):91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 7.Dyce J, George M, Goodall H, Fleming TP. Do trophectoderm and inner cell mass cells in the mouse blastocyst maintain discrete lineages? Development. 1987;100(4):685–98. doi: 10.1242/dev.100.4.685. [DOI] [PubMed] [Google Scholar]
- 8.Grabarek JB, Zyzyńska K, Saiz N, Piliszek A, Frankenberg S, Nichols J, et al. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012 Jan;139(1):129–39. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat Rev Mol Cell Biol. 2013;14(6):357–68. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossant J. Stem cells and early lineage development. Cell. 2008;132(4):527–31. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- 11.Rossant J. Lineage development and polar asymmetries in the peri-implantation mouse blastocyst. Semin Cell Dev Biol. 2004;15(5):573–81. doi: 10.1016/j.semcdb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Johnson MH. From mouse egg to mouse embryo: polarities, axes, and tissues. Annu Rev Cell Dev Biol. 2009;25:483–512. doi: 10.1146/annurev.cellbio.042308.113348. [DOI] [PubMed] [Google Scholar]
- 13.Bischoff M, Parfitt DE, Zernicka-Goetz M. Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions. Development. 2008;135(5):953–62. doi: 10.1242/dev.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134(23):4219–31. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 15.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136(5):701–13. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 16.Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24(1):71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- 17.Niakan KK, Han J, Pedersen RA, Simon C, Pera RA. Human pre-implantation embryo development. Development. 2012;139(5):829–41. doi: 10.1242/dev.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, et al. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22(19):2692–706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suwińska A, Czołowska R, Ozdzeński W, Tarkowski AK. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol. 2008;322(1):133–44. doi: 10.1016/j.ydbio.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139(1):3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- 21.Plusa B, Frankenberg S, Chalmers A, Hadjantonakis AK, Moore CA, Papalopulu N, et al. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci. 2005;118(Pt 3):505–15. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- 22.Pauken CM, Capco DG. Regulation of cell adhesion during embryonic compaction of mammalian embryos: roles for PKC and beta-catenin. Mol Reprod Dev. 1999;54(2):135–44. doi: 10.1002/(SICI)1098-2795(199910)54:2<135::AID-MRD5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 23.Moignard V, Gottgens B. Transcriptional mechanisms of cell fate decisions revealed by single cell expression profiling. Bioessays. 2014;36(4):419–426. doi: 10.1002/bies.201300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Bai W, Zhang L, Yin G, Wang X, Wang J, et al. Determination of microRNAs in mouse preimplantation embryos by microarray. Dev Dyn. 2008;237(9):2315–27. doi: 10.1002/dvdy.21666. [DOI] [PubMed] [Google Scholar]
- 26.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 27.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132(9):2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123(5):917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 29.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoproteinNanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 30.Palmieri SL, Peter W, Hess H, Schöler HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166(1):259–67. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 31.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 32.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck F, Erler T, Russell A, James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204(3):219–27. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- 35.Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404(6773):95–9. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 36.Weber S, Eckert D, Nettersheim D, Gillis AJ, Schäfer S, Kuckenberg P, et al. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod. 2010;82(1):214–23. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- 37.Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284(42):28729–37. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125(3–4):270–83. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134(21):3827–36. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 40.Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16(3):398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137(3):395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- 42.Yamauchi N, Kiessling AA, Cooper GM. The Ras/Raf signaling pathway is required for progression of mouse embryos through the two-cell stage. Mol Cell Biol. 1994;14(10):6655–62. doi: 10.1128/mcb.14.10.6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maekawa M, Yamamoto T, Tanoue T, Yuasa Y, Chisaka O, Nishida E. Requirement of the MAP kinase signaling pathways for mouse preimplantation development. Development. 2005;132(8):1773–83. doi: 10.1242/dev.01729. [DOI] [PubMed] [Google Scholar]
- 44.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 45.Rougier N, Bourc’his D, Gomes DM, Niveleau A, Plachot M, Pàldi A, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12(14):2108–13. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reik W, Santos F, Mitsuya K, Morgan H, Dean W. Epigenetic asymmetry in the mammalian zygote and early embryo: relationship to lineage commitment? Philos Trans R Soc Lond B Biol Sci. 2003;358(1436):1403–9. doi: 10.1098/rstb.2003.1326. discussion 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127(6):643–51. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- 48.Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, et al. Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol Cell. 2016;63(6):1066–79. doi: 10.1016/j.molcel.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 49.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 50.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci U S A. 2010;107(24):10783–90. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6(12B):2536–41. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- 52.Yeap LS, Hayashi K, Surani MA. ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin. 2009;2(1):12. doi: 10.1186/1756-8935-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, et al. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23(21):2507–20. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, et al. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137(15):2483–92. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou FC. DNA methylation program during development. Front Biol (Beijing) 2012;7(6):485–494. doi: 10.1007/s11515-012-9246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8(3):285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 57.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, et al. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31(7):1278–86. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 58.Yang P, Wang Y, Chen J, Li H, Kang L, Zhang Y, et al. RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem Cells. 2011;29(5):791–801. doi: 10.1002/stem.634. [DOI] [PubMed] [Google Scholar]
- 59.Harvey KF, Hariharan IK. The hippo pathway. Cold Spring Harb Perspect Biol. 2012;4:a011288. doi: 10.1101/cshperspect.a011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cormier S, Vandormael-Pournin S, Babinet C, Cohen-Tannoudji M. Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr Patterns. 2004;4(6):713–7. doi: 10.1016/j.modgep.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, et al. Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell. 2014;30(4):410–22. doi: 10.1016/j.devcel.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12(22):3579–90. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126(9):723–32. doi: 10.1242/dev.126.9.723. [DOI] [PubMed] [Google Scholar]
- 64.Baussano I, Tardivo I, Bellezza-Fontana R, Forneris MP, Lezo A, Anfossi L, et al. Neonatal screening for cystic fibrosis does not affect time to first infection with Pseudomonas aeruginosa. Pediatrics. 2006;118(3):888–95. doi: 10.1542/peds.2004-2599. [DOI] [PubMed] [Google Scholar]
- 65.Frum T, Ralston A. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 2015;31(7):402–10. doi: 10.1016/j.tig.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24(3):312–26. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10(5):615–24. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 68.Artus J, Chazaud C. A close look at the mammalian blastocyst: epiblast and primitive endoderm formation. Cell Mol Life Sci. 2014;71(17):3327–38. doi: 10.1007/s00018-014-1630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2(7):538–48. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 70.Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266(5190):1508–18. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 71.Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice--a review. Placenta. 2005;26(Suppl A):S3–9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 72.Farquharson RG, Jauniaux E, Exalto N ESHRE Special Interest Group for Early Pregnancy (SIGEP) Updated and revised nomenclature for description of early pregnancy events. Hum Reprod. 2005;20(11):3008–11. doi: 10.1093/humrep/dei167. [DOI] [PubMed] [Google Scholar]
- 73.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 74.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 75.Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6(10):e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 77.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 78.Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22(15):1987–97. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hirai H, Karian P, Kikyo N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem J. 2011;438(1):11–23. doi: 10.1042/BJ20102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12(13):2048–60. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107(20):9222–7. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20(12):1261–4. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 83.Li Y, Moretto-Zita M, Soncin F, Wakeland A, Wolfe L, Leon-Garcia S, et al. BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a DeltaNp63+ cytotrophoblast stem cell state. Development. 2013;140(19):3965–76. doi: 10.1242/dev.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9(2):144–55. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang JZ, Gao W, Yang HB, Zhang B, Zhu ZY, Xue YF. Screening for genes essential for mouse embryonic stem cell self-renewal using a subtractive RNA interference library. Stem Cells. 2006;24(12):2661–8. doi: 10.1634/stemcells.2006-0017. [DOI] [PubMed] [Google Scholar]
- 86.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4(5):403–15. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 87.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–61. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, et al. The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Symp Quant Biol. 2008;73:195–202. doi: 10.1101/sqb.2008.72.001. [DOI] [PubMed] [Google Scholar]
- 89.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 90.Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22(2):225–35. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- 91.Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15(2):93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–32. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006 Apr;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 94.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282(5396):2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 95.Tam PP, Rossant J. Mouse embryonic chimeras: tools for studying mammalian development. Development. 2003;130(25):6155–63. doi: 10.1242/dev.00893. [DOI] [PubMed] [Google Scholar]
- 96.Lu CW, Yabuuchi A, Chen L, Viswanathan S, Kim K, Daley GQ. Ras-MAPK signaling promotes trophectoderm formation from embryonic stem cells and mouse embryos. Nat Genet. 2008;40(7):921–6. doi: 10.1038/ng.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sarkar P, Randall SM, Collier TS, Nero A, Russell TA, Muddiman DC, et al. Activin/nodal signaling switches the terminal fate of human embryonic stem cell-derived trophoblasts. J Biol Chem. 2015;290(14):8834–48. doi: 10.1074/jbc.M114.620641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8(3):326–34. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuckenberg P, Buhl S, Woynecki T, van Fürden B, Tolkunova E, Seiffe F, et al. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol. 2010;30(13):3310–20. doi: 10.1128/MCB.01215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roberts RM, Fisher SJ. Trophoblast stem cells. Biol Reprod. 2011;84(3):412–21. doi: 10.1095/biolreprod.110.088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Donnison M, Beaton A, Davey HW, Broadhurst R, L’Huillier P, Pfeffer PL. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132(10):2299–308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- 102.Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development. 2002;129(11):2733–47. doi: 10.1242/dev.129.11.2733. [DOI] [PubMed] [Google Scholar]
- 103.Okada Y, Ueshin Y, Isotani A, Saito-Fujita T, Nakashima H, Kimura K, et al. Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat Biotechnol. 2007;25(2):233–7. doi: 10.1038/nbt1280. [DOI] [PubMed] [Google Scholar]
- 104.Latos PA, Sienerth AR, Murray A, Senner CE, Muto M, Ikawa M, et al. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 2015;29(23):2435–48. doi: 10.1101/gad.268821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27(10):3769–79. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26(6):1496–505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9(3):193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45(3):325–9. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu X, Wang C, Liu W, Li J, Li C, Kou X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 111.Niakan KK, Schrode N, Cho LT, Hadjantonakis AK. Derivation of extraembryonic endoderm stem (XEN) cells from mouse embryos and embryonic stem cells. Nat Protoc. 2013;8(6):1028–41. doi: 10.1038/nprot.2013.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, et al. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002;16(7):784–9. doi: 10.1101/gad.968802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Capo-Chichi CD, Rula ME, Smedberg JL, Vanderveer L, Parmacek MS, Morrisey EE, et al. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev Biol. 2005;286(2):574–86. doi: 10.1016/j.ydbio.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 114.Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322(5909):1811–5. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- 115.Wilmut I, Beaujean N, de Sousa PA, Dinnyes A, King TJ, Paterson LA, et al. Somatic cell nuclear transfer. Nature. 2002;419(6907):583–6. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 116.Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24(9):2007–13. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133(2):250–64. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155(3):621–35. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 120.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 121.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shimosato D, Shiki M, Niwa H. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev Biol. 2007;7:80. doi: 10.1186/1471-213X-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kubaczka C, Senner CE, Cierlitza M, Araúzo-Bravo MJ, Kuckenberg P, Peitz M, et al. Direct Induction of Trophoblast Stem Cells from Murine Fibroblasts. Cell Stem Cell. 2015;17(5):557–68. doi: 10.1016/j.stem.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 124.Benchetrit H, Herman S, van Wietmarschen N, Wu T, Makedonski K, Maoz N, et al. Extensive Nuclear Reprogramming Underlies Lineage Conversion intoFunctional Trophoblast Stem-like Cells. Cell Stem Cell. 2015;17(5):543–56. doi: 10.1016/j.stem.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 125.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2(3):230–40. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161(3):555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuckenberg P, Peitz M, Kubaczka C, Becker A, Egert A, Wardelmann E, et al. Lineage conversion of murine extraembryonic trophoblast stem cells to pluripotent stem cells. Mol Cell Biol. 2011;31(8):1748–56. doi: 10.1128/MCB.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rhee C, Lee BK, Beck S, Anjum A, Cook KR, Popowski M, et al. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genes Dev. 2014;28(20):2219–32. doi: 10.1101/gad.247163.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R, Tucker PW. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9(24):3067–82. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 131.An G, Miner CA, Nixon JC, Kincade PW, Bryant J, Tucker PW, et al. Loss of Bright/ARID3a Function Results in Developmental Plasticity. Stem Cells. 2010;28(9):1560–7. doi: 10.1002/stem.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Popowski M, Templeton TD, Lee BK, Rhee C, Li H, Miner C, et al. Bright/Arid3A acts as a barrier to somatic cell reprogramming through direct regulation of Oct4, Sox2, and Nanog. Stem Cell Reports. 2014;2(1):26–35. doi: 10.1016/j.stemcr.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rhee C, Edwards M, Dang C, Harris J, Brown M, Kim J, et al. Arid3a is required for mammalian placenta development. Dev Biol. 2017;422(2):83–91. doi: 10.1016/j.ydbio.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rhee C, Lee BK, Beck S, LeBlanc L, Tucker HO, Kim J. Mechanisms of transcription factor-mediated direct reprogramming of mouse embryonic stem cells to trophoblast stem-like cells. Nucleic Acids Res. 2017;45(17):10103–10114. doi: 10.1093/nar/gkx692. [DOI] [PMC free article] [PubMed] [Google Scholar]