Abstract
P62 (also named sequestosome-1, SQSTM1) is involved in autophagy regulation through multiple pathways. It interacts with autophagosomes-associated LC3-II and ubiquitinated protein aggregates to engulf the aggregates in autophagosomes, interacts with HDAC6 to inhibit its deacetylase activity to maintain the levels of acetylated α-tubulin and stabilities of microtubules to enhance autophagosome trafficking, and regulates autophagy initiation and cell survival. We performed immunohistochemistry staining of P62 in prostate tissues from prostate cancer patients and found that levels of P62 in patients with prostate adenocarcinomas (PCA) are significantly higher than those in patients with benign prostate hyperplasia (BPH). High levels of P62 predict high tumor grade and high intensity of metastasis. Here we show that P62 increased the levels of HDAC6 and reduced the acetylation of α-tubulin and the stability of microtubules. Consequently, high levels of P62 caused a promotion of epithelial to mesenchymal transition in addition to an impairment of autophagy flux, and further led to an enhancement of proliferation, migration and invasion of prostate cancer cells. Therefore, P62 promotes metastasis of PCA.
Keywords: acetylation, microtubule, prognosis, prostate adenocarcinoma, SQSTM1
Introduction
Autophagy-lysosome pathway is one of the major pathways for mammalian cells to degrade dysfunctional organelles and misfolded/aggregated proteins (1). Autophagy process undergoes multiple stages: its vesicle nucleation is regulated by a class III PI3K complex; autophagic induction by a serine-threonine kinase complex; and the extension and completion of vesicles by a pair of novel ubiquitin-like protein-conjugating systems, the ATG12 and ATG8 systems (1,2). Microtubule-associated protein 1 light chain 3 (LC3) is the mammalian homologue of ATG8, one of the key autophagy markers (4). The LC3 precursor is truncated to form the cytosolic LC3-I and further conjugated with phosphatidylethanolamine to create the membrane-associated LC3-II with the assistance of ATG5 and ATG12 (3,4). The LC3-II-associated isolation membranes target and then completely envelop the autophagy substrates that bind to polyubiquitin-binding protein P62 (SQSTM 1) to form autophagosomes (5). Autophagosomes migrate along acetylated microtubules to fuse with lysosomes to form autolysosomes, in which substrates are degraded (6,7). Microtubule-associated protein MAP1S interacts with LC3 and acts as an activator of autophagy (7–9). Another interactive protein of MAP1S, a mitochondrion-associated protein LRPPRC suppresses both autophagy and mitophagy (10–13).
Autophagy is pro-survival in general. Autophagy defects induce oxidative stresses (1,14) that in turn activate NLRP3 inflammasomes that result in a direct activation of caspase-1 (15). Activation of caspase-1 subsequently induces secretion of potent pro-inflammatory cytokines interleukin-1β (IL-1β) and IL-18, mitochondrial dysfunction and production of reactive oxygen species, and eventually an inflammatory form of cell death referred to as pyroptosis (16–21). The release of immunogenic danger signals or danger-associated molecular patterns (DAMPs) from pyroptotic cells can fuel pro-inflammatory cascades that promote the mortality of host structural, hematopoietic and immune-competent cells (21,22). Chronic inflammation induces tissue regeneration (23) and further leads to tumorigenesis by the generation of oxidative stress to induce genome instability and by reactivating cell division to amplify genome instability (23–27). Thus, enhancing autophagy may prevent cancers and improve patient survivals.
We performed immunohistochemistry analyses on tissue sections collected from patients suffering from prostate adenocarcinomas (PCA) or benign prostatic hyperplasia (BPH) and found that either high levels of autophagy activator MAP1S or low levels of autophagy inhibitor LRPPRC in prostate tumor tissues predict better prognosis (28,29). As an autophagy regulator, P62 is similarly expressed in the luminal epithelial cells of prostate glands in both BPH and PCA patients but levels of P62 are relatively higher in PCA than in BPH (29). However, levels of P62 are poor prognosis marker although they are positively correlated to metastasis (29). Acting as a substrate receptor, P62 accumulates when autophagy initiation is activated or autophagy degradation is impaired. P62 was detected with polyubiquitinated protein aggregates in Mallory bodies in hepatocytes (30), suggesting an inhibited autophagy degradation. To understand how P62 functions in prostate tissues, we were triggered to investigate the roles of P62 in prostate tumorigenesis. Here we show that P62 not only passively acts as an autophagy substrate receptor but also positively functions as an autophagy suppressor similarly as LRPPRC does. P62 destabilizes microtubules by sustaining HDAC6 activity to impair autophagy flux. In addition, P62 promotes epithelial to mesenchymal transition and further enhance tumor growth and metastasis through HDAC6.
Materials and Methods
Cell culture and plasmid transfections
The hormone-Independent PCa cell lines (DU145) was purchased from American Type Culture Collection (ATCC, USA) and cultured in 1640 medium (Catalog Number: SH30809.01, HyClone Laboratories, ) supplemented with 10% fetal bovine serum (Catalog Number: 10270-106, Gibco, USA), 100 μ/mL penicillin and 100 μg/mL streptomycin. The cells, placed in 37°C and 5% CO2 humidity incubator, were sub-cultured for passage every 3–4 days. The well-grown logarithmic phase cells were selected in this experiment, and P62 suppression and overexpression plasmids were constructed and transfected into DU145 cell lines with TurboFectTM Transfection Reagent (Catalog Number: #R0531, Thermo Scientific, USA). The plasmids of over-expression P62 and control were made by HYYMed Company based on lentivirus with the ORF sequence of P62 (NM_003900). The gRNA targeted at P62 (gene ID:8878) expression oligos were introduced into the PXC9-puro vector (Addgene). The sgRNA sequences with minimal off-target specificity designed by the optimized online CRISPR design tool (http://crispor.tefor.net/). A mixture of 1 μg of PXC9-puro plasmid (Addgene) DNA containing target gRNA sequence, 3 μg PPACK-HI-GAG, 3 μg PPACK-HI-REV and 3 μg PVSV-G were mixed with 0.6 mL DMEM without FBS. Thirty-six μL PEI was added into the mixture, briefly vortexed and incubated for 15 min before being added to the cells. After 6 hrs, the medium was replaced with fresh DMEM medium. After 24 hrs, the medium was collected and centrifuged at 3,000 rpm for 5 min to pellet cell debris. The supernatant was collected and frozen at −80°C. A total of 1 X 105 cells were transfected at MOI~0.4 by being treated with 2 mL supernatant, 3.0 μg polybrene and then incubated for 8 hrs, then the medium was replaced with fresh DMEM medium. The cells were cultured in medium containing 2.0 μg/ml puromycin for 2 days for selection.
Cell proliferation assay
For cell proliferation assays, 2X103 cells were seeded in 96-well plates and cultured for 24 and 72 hrs. Cells were incubated with 10 μL of CCK-8 (Catalog Number: C0039, Beyotime Biotechnology) for 2 hrs at 37°C. The absorbance was measured at 450 nm wavelength with a spectrophotometer. Cells were then stained by Crystal violet for 1 hr at 37°C for taking pictures. Data were expressed as mean ± SE of three independent experiments.
Cell migration assay
For the scratch wound-healing motility assay, a scratch was made with a pipette tip when the cells reach the confluence. After being cultured under standard conditions with mitomycin for 48 hrs, plates were washed twice with fresh medium to remove non-adherent cells and then photographed. The cell migrated from the wound edge were counted and the data were expressed as mean ± SE of three independent experiments.
Cell invasion assay
The transwell inserts (8-μm pores) were filled with 50 μl of a mixture of serum-free RPMI1640 medium and Matrigel (1:10; catalog number: 354234, BD Biosciences, USA). The inserts were then placed in 24-well tissue culture plates (Transwell, catalog number: #3422, Corning, USA) containing 10% FBS-medium. After solidification by incubation at 37 °C for 4 hrs, 5×104 cells in 200 μl medium were placed in upper chambers. Following 48 hrs of incubation at 37 °C with 5% CO2 and in culture medium with mitomycin to stop the mitosis, the membranes were fixed with 10% formalin and stained with 0.05% Crystal Violet. The number of cells that migrated through the pores was assessed and the data were expressed as mean ± SE of three independent experiments.
Western blot analysis
Cells were collected and lysis in RIPA containing broad protease inhibitor cocktail (EASYpacks, Roche) and then centrifuged to obtain supernatants. The total protein concentration was measured by bicinchonininc acid (BCA). Equal amount of protein (30 μg) was loaded and separated by SDS-PAGE (Bio-Rad), and blotted with primary antibodies and corresponding horseradish peroxidase-conjugated secondary antibodies. GAPDH was used as an internal loading control. The primary antibodies used were as follows: anti-P62 (#8025, dilution 1:1000), anti-α-tubulin (#5335, dilution 1:2000), anti-Ac-α-tubulin (#14652, dilution 1:2000), anti-HDAC6 (#7558, dilution 1:2000) and anti-GAPDH (#5174, dilution 1:2000) polyclonal antibodies were obtained from Cell Signaling Technology (Beverly, MA, USA). The anti-LRPPRC (#ab97505, dilution 1:1000), anti-E-cadherin (#ab1416, dilution 1:1000), anti-fibronectin (#ab2413, dilution 1:1000), anti-vimentin (#ab92547, dilution 1:2000) polyclonal antibodies were obtained from Abcam (Cambridge, MA, USA). The results were visualized with the GelDoc XR+ chemiluminescent detection system (BIO-RAD, USA). Densitometric analysis of the bands was performed using ImageJ free software.
Confocal fluorescent microscopy
Cells were grown on glass coverslips for 6 hrs, washed three times with PBS, fixed in 4% PFA for 20 mins and permeabilized with 0.1% Triton X-100 in PBS for 20 mins, and then washed three times with PBS and blocked with 2% BSA in PBS for 1hr at room temperature. The blocked cells were incubated overnight with anti-Ac-α-tubulin (#14652, dilution 1:800) in 1% BSA in PBS at 4 °C and followed by five-times washing with PBS. Cells were incubated with DYLight649 goat anti-rabbit IgG [H+L] secondary antibody (Multi Sciences, GAR6492) with a 1:500 dilution in PBS at room temperature for 1 hr. After washing with PBS for five times, cells were mounted with Fluoromount-G™ (Invitrogen, 00-4958-02) and visualized under a confocal laser scanning microscope (ZEISS, LSM800) equipped with a 40X objective.
Statistical analysis
Statistical analyzes were performed using SPSS 19.0 (SPSS Inc, IL, USA) statistical software. Two-tailed Student’s t test was used to determine the difference between groups by GraphPad Prism (San Diego, California, USA). A P value ≤ 0.05 was considered statistically significant.
Results
P62 promotes proliferation, migration and invasion of prostate cancer cells
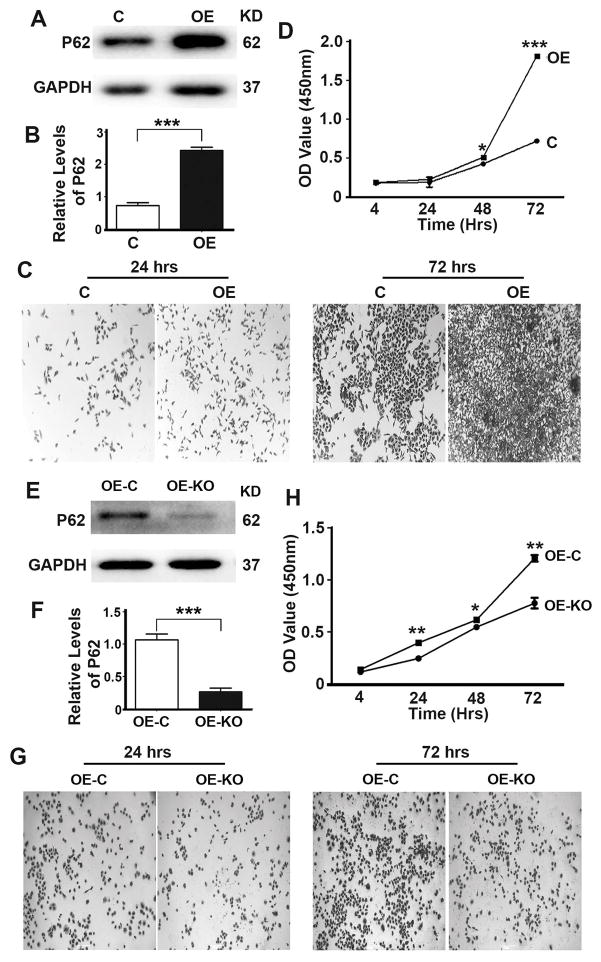
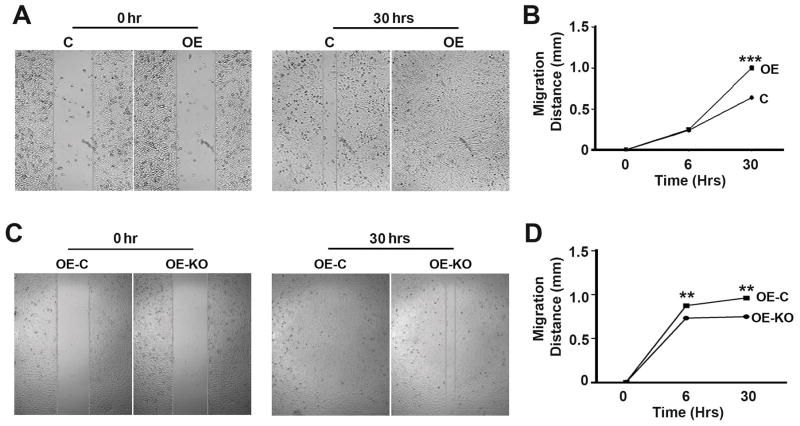
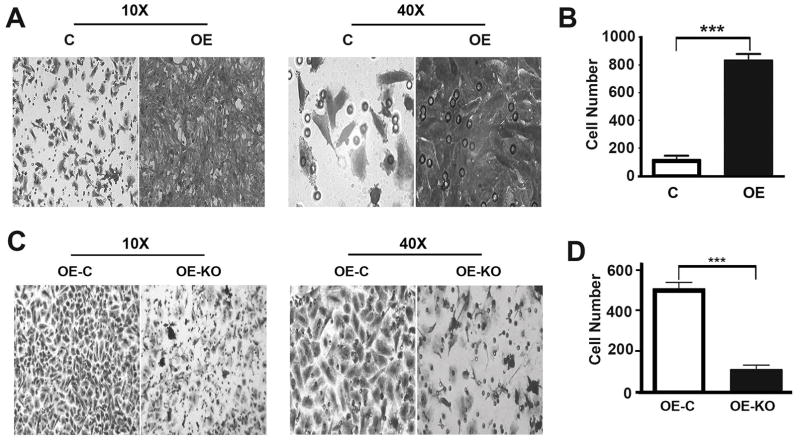
To understand the role of P62 in the development of prostate cancer, we generated OE, a stable DU145 cell line overexpressing P62 (Fig. 1A,B). Overexpression of P62 led to an enhancement of cell proliferation (Fig. 1C,D). To confirm the impact of p62 on cell proliferation, we utilized CRISPR/Cas9 technology to delete the overexpressed P62 (Fig. 1E,F). Depletion of P62 led to a suppression of cell proliferation (Fig. 1G,H). To investigate the role of P62 in cell migration and invasion, we performed migration and invasion assays of cell lines as above (Fig. 1). Overexpression of P62 led to a promotion while depletion of P62 led to a reduction in cell migration (Fig. 2) and invasion (Fig. 3). Therefore, P62 promotes proliferation, migration of invasion of prostate cancer cells.
Figure 1. P62 promotes proliferation of prostate cancer cells.
(A,B) Representative immunoblot (A) and quantification (B) showing levels of P62 in DU145 cells (Control, C) and DU145 cells stably expressing P62 (overexpression, OE). C cells were stable DU145 cells carrying the control plasmid vector. (C,D) Representative images (C) and quantification (D) showing cell densities of DU145 cells (C and OE) at 24 and 72 hrs after seeding. (E,F) Representative immunoblot (E) and quantification (F) showing levels of P62 in DU145 cells stably expressing P62 (Control, OE-C) and DU145 cells stably expressing P62 with P62 depleted by through CRISPR/Cas9 (OE-KO). OE-C cells were stable DU145 cells expressing P62 carrying the control plasmid vector. (G,H) Representative images (G) and quantification (H) showing cell densities of DU145 cells (OE-C and OE-KO) at 24 and 72 hrs after seeding. Tissue culture plates were seeded with equal number of cells. *, p ≤ 0.05; **, p ≤ 0.01; and ***, p ≤ 0.001.
Figure 2. P62 promotes the migration of prostate cancer cells.
(A,B) Representative images (A) and quantification (B) showing migration distances of DU145 cells (C and OE) at 30 hrs after scratching. (C,D) Representative images (C) and quantification (D) showing migration distances of DU145 cells (OE-C and OE-KO) at 30 hrs after scratching. **, p ≤ 0.01; and ***, p ≤ 0.001.
Figure 3. P62 promotes the invasion of prostate cancer cells.
(A,B) Representative images (A) and quantification (B) showing invaded DU145 cells (C and OE) which passed at 45 hrs after seeding. (C,D) Representative images (C) and quantification (D) showing invaded DU145 cells (OE-C and OE-KO) at 45 hrs after seeding. ***, p ≤ 0.001.
P62 promotes epithelial to mesenchymal transition
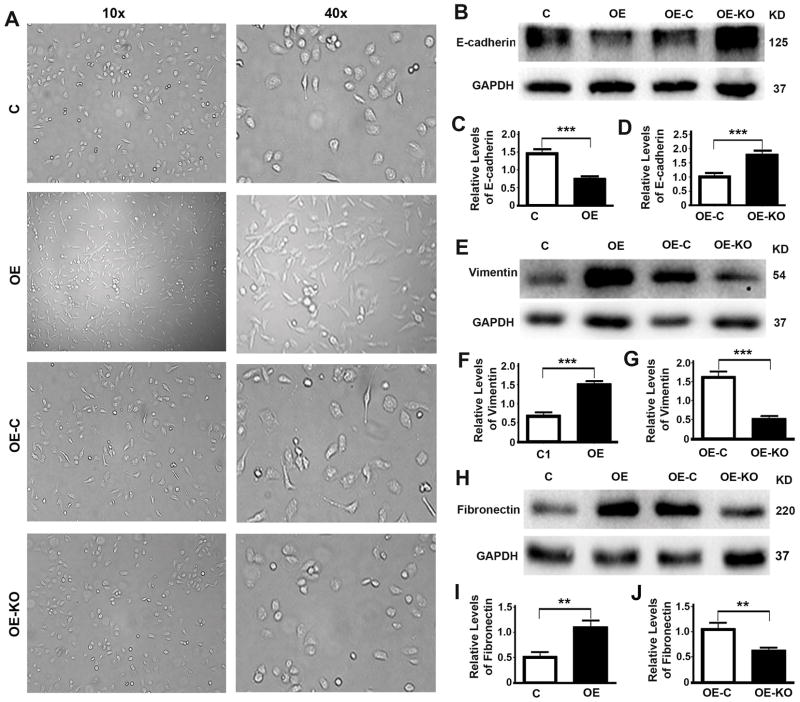
We further examine the impact of P62 on cell morphology. Overexpression of P62 led to a conversion of DU145 cells from a round to spindle-like shape while further depletion of P62 led to a reversal of cell shapes (Fig. 4A), suggesting a epithelia to mesenchymal transition. Further examining the changes of related markers revealed that levels of epithelial marker E-cadherin were reduced when P62 was overexpressed (Fig. 4B,C) but increased when P62 was depleted (Fig. 4B,D). Levels of mesenchymal marker vimentin were increased when P62 was overexpressed (Fig. 4E,F) but decreased when P62 was depleted (Fig. 4E,G). Levels of another mesenchymal marker fibronectin were also increased when P62 was overexpressed (Fig. 4H,I) but decreased when P62 was depleted (Fig. 4H,J). Therefore, P62 promotes epithelial to mesenchymal transition.
Figure 4. P62 promotes epithelial to mesenchymal transition.
(A) Representative images showing the morphology of DU145 cells (C, OE, OE-C and OE-KO). (B–J) Representative immunoblots (B,E,H) and quantification (C,D,F,G,I,J) showing levels of epithelial marker E-cadherin (B–D), Vimentin (E–G) and Fibronectin (H–J) in DU145 cells (C, OE) (C,F,I) and DU145 cells stably expressing P62 (OE-C, OE-KO) (D,G,J). **, p ≤ 0.01; and ***, p ≤ 0.001.
P62 inhibits autophagy autophagy flux
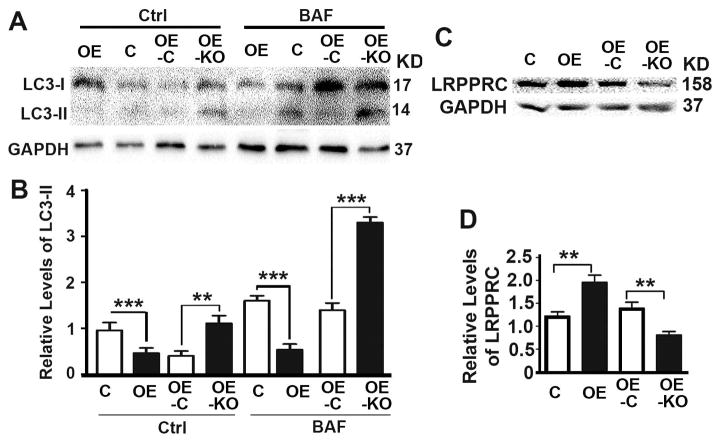
To investigate the impact of P62 on autophagy, we applied lysosomal inhibitor bafilomycin A1 to treat cell lines described above to measure autophagy flux (Fig. 5). Overexpression of P62 caused a reduction while depletion of P62 caused an increase in autophagy flux as reflected by the levels of LC3-II in the presence of lysosomal inhibitor (Fig. 5A,B). LRPPRC was characterized as an autophagy inhibitor in cancer cells.(10,11) Overexpression of P62 led to increases while depletion of P62 led to decreases of levels of LRPPRC (Fig. 5C,D), suggesting that P62 inhibits autophagy.
Figure 5. P62 inhibits autophagy flux.
(A,B) Representative immunoblot (A) and quantification (B) showing levels of autophagy marker LC3-II in DU145 cells (C, OE, OE-C, OE-KO) in the absence (Ctrl) or presence of bafilomycin A1 (BAF). (C,D) Representative immunoblot (C) and quantification (D) showing levels of autophagy marker LRPPRC in DU145 cells (C, OE, OE-C, OE-KO). **, p ≤ 0.01; and ***, p ≤ 0.001.
P62 increases HDAC6 activity and reduces microtubule acetylation
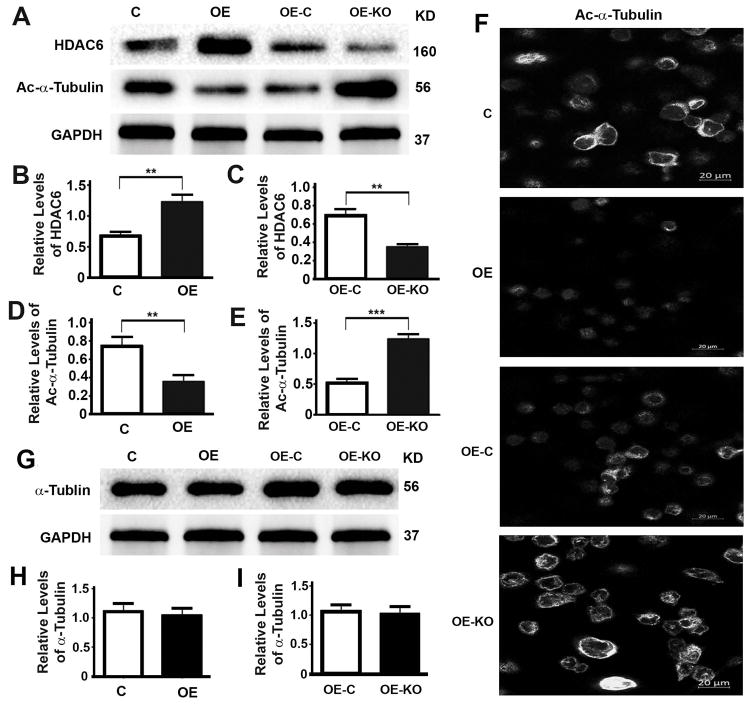
To understand the mechanism by which P62 promotes cell proliferation, migration, invasion and the epithelial to mesenchymal transition of prostate cancer cells, we examined the impact of P62 on the levels of HDAC6. Overexpression of P62 led to increases (Fig. 6A,B) while depletion of P62 led to decreases of HDAC6 levels (Fig. 6A,C). Levels of acetylated α-tubulin were reduced because of the increases of HDAC6 levels (Fig. 6A,D) or increased because of the decreases of HDAC6 levels (Fig. 6A,E). Such impacts of P62 on levels of acetylated α-tubulin were further confirmed by fluorescent immunostaining (Fig. 6F). However, levels of total α-Tubulin remained constant no matter how levels of P62 were changed (Fig. 6G–I). Therefore, P62 sustains levels of HDAC6 and inhibits the acetylation of α-Tubulin and microtubules.
Figure 6. P62 increases HDAC6 activity and reduces microtubular acetylation.
(A–E) Representative immunoblot (A) and quantification (B–E) showing levels of HDAC6 (A,B,C) and acetylated α-tubulin (Ac-α-Tubulin) (A,D,E) in DU145 cells (C, OE) (B,D) and DU145 cells stably expressing P62 (OE-C, OE-KO) (C,E). (F) Representative images showing the immunostaining intensities of Ac-α-tubulin in DU145 cells (C, OE) and DU145 cells stably expressing P62 (OE-C, OE-KO). All images were captured under an identical setting. (G–I) Representative immunoblot (G) and quantification (H,I) showing levels of total α-tubulin in DU145 cells (C, OE) (H) and DU145 cells stably expressing P62 (OE-C, OE-KO) (I). **, p ≤ 0.01, and ***, p ≤ 0.001.
Discussion
Autophagy defects induce oxidative stresses, inflammation and pyroptosis (1,14,16–21). Oxidative stress induces DNA double strand breakage that initiates the cascade of chromosomal breakage-fusion-bridge cycles and results in genome instability, the origin of most types of solid tumors (14). Chronic inflammation induces tissue regeneration that re-activates cell division to amplify genome instability (23–27). Therefore, autophagy defects lead to tumorigenesis and activating autophagy results in tumor suppression (9).
We previously showed that active autophagy reflected by high levels of autophagy activator MAP1S and low levels of autophagy inhibitor LRPPRC is associated with high malignancy of PCA and poor prognosis of PCA patients (28,29). P62 functions as an autophagy substrate receptor through its association with polyubiquitinated proteins and LC3-II (5,31), or as an autophagy regulator through its association with HDAC6 to control the deacetylation of α-tubulin (32,33). High levels of P62 indicating an accumulation of protein aggregates may suggest either that the formation of autophagosomes is stimulated by sequestering more autophagy substrates, or that the degradation of autophagosomes is blocked due to a defective autophagy. Our results that overexpression of P62 caused a suppression of autophagy flux in DU145 cells suggests that it is more likely that the degradation is inhibited. Acetylated microtubules are required for the trafficking of autophagosomes to fuse with lysosomes (7). P62 interacts with HDAC6 and reduces the acetylation and stability of microtubules, which causes an impairment of the trafficking of autophagosomes to fuse with lysosome and a blockade of autophagosomal degradation. HDAC4 specifically interacts with MAP1S to regulate the acetylation of MAP1S to control the trafficking of autophagosomes. Although both of them belong to the same subgroup of histone deacetylases, HDAC6 and HDAC4 were confirmed not to compensate with each other for the deacetylation of MAP1S and are predicted not to compensate with each other for the deacetylation of α-Tubulin (8,9). Similar to LRPPRC, P62 inhibits autophagy flux and consequently enhance malignancies and metastasis of PCA.
P62 binds with HDAC6 to maintain its deacetylase activity and destabilize microtubules. In addition to the impact on autophagosome trafficking, microtubule deacetylation promotes cytoskeletal re-organization and epithelial-mesenchymal transition (EMT) (34). EMT is a biologic process that allows a polarized epithelial cell to assume a mesenchymal cell phenotype that exhibits an enhanced migratory capacity and invasiveness (35). Since epithelial cells normally interact with basement membranes via its basal surface while mesenchymal cells lose such attachment, those cancer cells that undergo EMT enter the circulation and exit the blood stream to become metastatic (35). P62 increased levels of epithelial marker E-cadherin and decreased the levels of mesenchymal markers fibronectin and vimentin, suggesting an EMT was promoted. Consequently, P62 promotes the migration and invasion of DU145 cells, indicating an enhancement of metastatic capacity. Although it is still not conclusive that EMT is directly related to the metastasis of prostate cancer (36), we show here a close association of P62-promoted EMT with the metastasis of PCA. Therefore, high levels of P62 promote EMT to induce metastasis of PCA patients.
Although it is well recognized that P62 acts as a receptor of autophagy substrates, the accumulated P62 in PCA tissues due to autophagy defects binds with HDAC6 and sustains its deacetylase activity, leading to the deacetylation of α-tubulin and microtubules. Deacetylated microtubules cause an impairment of autophagy flux and a promotion of EMT. Autophagy defects and EMT eventually promote the initiation and development of PCA and the migration and invasiveness of cancer cells. Therefore, high levels of P62 promotes the metastasis of PCA (Fig. 7). To further confirm the impact of P62 on the metastasis of PCA, we are going to use the P62 overexpression and knockdown stable cell lines to inject non-specific pathogen free male immune-deficient NOD/SCID mice to establish a mouse model of metastasis of prostate cancer to lung tissues as we previously reported (37).
Figure 7.
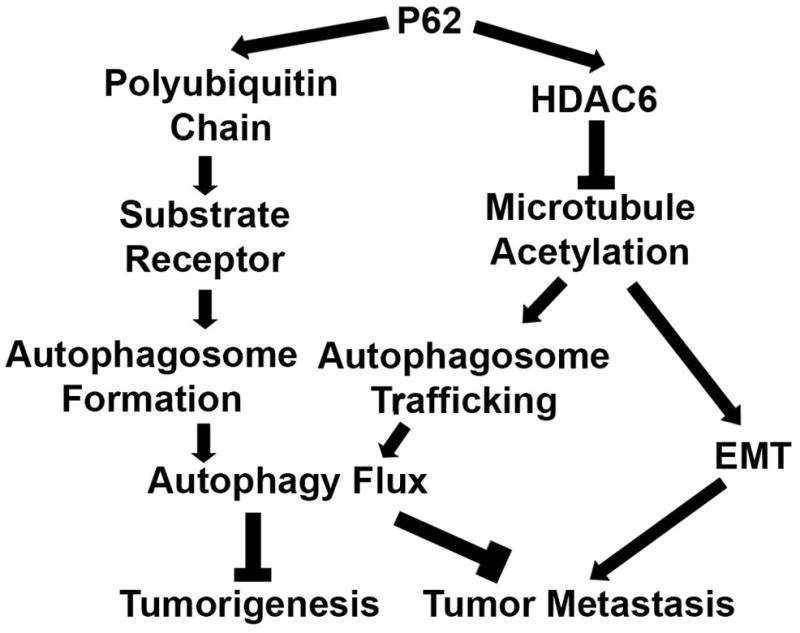
A diagram showing how P62 impacts tumorigenesis and metastasis of PCA.
Acknowledgments
This work was supported by Guangzhou Municipal Science and Technology Project (1563000448) to Xianhan Jiang, and National Natural Science Foundation of China (81772931) and NIH NCI CA142862 to Leyuan Liu.
Footnotes
CONFLICT OF INTEREST
The Authors have no conflict to report that they believe could be construed as resulting in an actual, potential, or perceived conflict of interest with regard to the manuscript submitted for review.
References
- 1.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 2.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Internat J Biochem Cell Biol. 2004;36(12):2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2(2):138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010;11(1):89. doi: 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue F, Li W, Zou J, Chen Q, Xu G, Huang H, Xu Z, Zhang S, Gallinari P, Wang F, McKeehan WL, Liu L. Blocking the association of HDAC4 with MAP1S accelerates autophagy clearance of mutant Huntingtin. Aging. 2015;7(10):839–853. doi: 10.18632/aging.100818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue F, Li W, Zou J, Jiang X, Xu G, Huang H, Liu L. Spermidine Prolongs Lifespan and Prevents Liver Fibrosis and Hepatocellular Carcinoma by Activating MAP1S-Mediated Autophagy. Can Res. 2017;77(11):2938–2951. doi: 10.1158/0008-5472.CAN-16-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou J, Yue F, Jiang X, Li W, Yi J, Liu L. Mitochondrion-associated protein LRPPRC suppresses the initiation of basal levels of autophagy via enhancing Bcl-2 stability. Biochem J. 2013;454(3):447–457. doi: 10.1042/BJ20130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou J, Yue F, Li W, Song K, Jiang X, Yi J, Liu L. Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PloS One. 2014;9(4):e94903. doi: 10.1371/journal.pone.0094903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Amy V, Liu G, McKeehan WL. Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. In vitro Cell Develop Biol Ani. 2002;38(10):582–594. doi: 10.1290/1543-706x(2002)38<582:ncimat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Vo A, Liu G, McKeehan WL. Putative tumor suppressor RASSF1 interactive protein and cell death inducer C19ORF5 is a DNA binding protein. Biochem Biophys Res Commun. 2005;332(3):670–676. doi: 10.1016/j.bbrc.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, McKeehan WL, Wang F, Xie R. MAP1S enhances autophagy to suppress tumorigenesis. Autophagy. 2012;8(2):278–280. doi: 10.4161/auto.8.2.18939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 17.Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Ann Rev Cell Develop Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 18.Ryter SW, Mizumura K, Choi AM. The Impact of Autophagy on Cell Death Modalities. Internat J Cell Biol. 2014;2014:502676. doi: 10.1155/2014/502676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 20.Gurung P, Lukens JR, Kanneganti TD. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trend Mol Med. 2015;21(3):193–201. doi: 10.1016/j.molmed.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, Horng T. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Nat Acad Sci USA. 2014;111(43):15514–15519. doi: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terlizzi M, Casolaro V, Pinto A, Sorrentino R. Inflammasome: cancer’s friend or foe? Pharmacol Therap. 2014;143(1):24–33. doi: 10.1016/j.pharmthera.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Zhang DY, Friedman SL. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology. 2012;56(2):769–775. doi: 10.1002/hep.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie R, Wang F, McKeehan WL, Liu L. Autophagy enhanced by microtubule- and mitochondrion-associated MAP1S suppresses genome instability and hepatocarcinogenesis. Can Res. 2011;71(24):7537–7546. doi: 10.1158/0008-5472.CAN-11-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundu JK, Surh YJ. Emerging avenues linking inflammation and cancer. Free Rad Biol Med. 2012;52(9):2013–2037. doi: 10.1016/j.freeradbiomed.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 26.Moss SF, Blaser MJ. Mechanisms of disease: Inflammation and the origins of cancer. Nat Clin Pract Oncol. 2005;2(2):90–97. doi: 10.1038/ncponc0081. quiz 91 p following 113. [DOI] [PubMed] [Google Scholar]
- 27.Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Cur Biol. 2012;22(13):1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X, Li X, Huang H, Jiang F, Lin Z, He H, Chen Y, Yue F, Zou J, He Y, You P, Wang W, Yang W, Zhao H, Lai Y, Wang F, Zhong W, Liu L. Elevated levels of mitochondrion-associated autophagy inhibitor LRPPRC are associated with poor prognosis in patients with prostate cancer. Cancer. 2014;120(8):1228–1236. doi: 10.1002/cncr.28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X, Zhong W, Huang H, He H, Jiang F, Chen Y, Yue F, Zou J, Li X, He Y, You P, Yang W, Lai Y, Wang F, Liu L. Autophagy defects suggested by low levels of autophagy activator MAP1S and high levels of autophagy inhibitor LRPPRC predict poor prognosis of prostate cancer patients. Mol Carcinog. 2015;54(10):1194–1204. doi: 10.1002/mc.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Amer J Pathol. 2002;160(1):255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 32.Chen Q, Yue F, Li W, Zou J, Xu T, Huang C, Zhang Y, Song K, Huang G, Xu G, Huang H, Li J, Liu L. Potassium Bisperoxo (1,10-phenanthroline) Oxovanadate (bpV(phen)) Induces Apoptosis and Pyroptosis and Disrupts the P62-HDAC6 Interaction to Suppress the Acetylated Microtubule-dependent Degradation of Autophagosomes. J Biol Chem. 2015;290(43):26051–26058. doi: 10.1074/jbc.M115.653568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J, Seibenhener ML, Calderilla-Barbosa L, Diaz-Meco MT, Moscat J, Jiang J, Wooten MW, Wooten MC. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PloS One. 2013;8(9):e76016. doi: 10.1371/journal.pone.0076016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu S, Liu Y, Zhu B, Ding K, Yao TP, Chen F, Zhan L, Xu P, Ehrlich M, Liang T, Lin X, Feng XH. Loss of alpha-Tubulin Acetylation Is Associated with TGF-beta-induced Epithelial-Mesenchymal Transition. J Biol Chem. 2016;291(10):5396–5405. doi: 10.1074/jbc.M115.713123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat Rev Urol. 2011;8(8):428–439. doi: 10.1038/nrurol.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Du T, Zhang Y, Lai Y, Li K, Fan X, Zhu D, Lin T, Xu K, Huang J, Liu L, Guo Z. Elevation of SHARPIN Protein Levels in Prostate Adenocarcinomas Promotes Metastasis and Impairs Patient Survivals. Prostate. 2017;77(7):718–728. doi: 10.1002/pros.23302. [DOI] [PubMed] [Google Scholar]