Abstract
Legionella pneumophila is a pathogen causing severe pneumonia in humans called Legionnaires’ disease. Lem22 is a previously uncharacterized effector protein conserved in multiple Legionella strains. Here, we report the crystal structure of Lem22 from the Philadelphia strain, also known as lpg2328, at 1.40 Å resolution. The structure shows an up-and-down three-helical bundle with a significant structural similarity to a number of protein-binding domains involved in apoptosis and membrane trafficking. Sequence conservation identifies a putative functional site on the interface of helices 2 and 3. The structure is an important step towards a functional characterization of Lem22.
Keywords: Legionella, lpg2328, X-ray crystallography, virulence factor, pathogen
INTRODUCTION
When engulfed by alveolar macrophages, L. pneumophila uses the Dot/Icm type IV secretion system (T4SS) to translocate bacterial proteins, termed effectors, into the host cell where they manipulate eukaryotic processes to create a replicative niche termed the Legionella-containing vacuole (LCV) and avoid lysosomal fusion1. At least 300 effectors are identified in L. pneumophila, but the function for many of them is still unknown. One of the approaches to learn more about these effectors is through study of their 3D structure. Structural similarity to proteins of known function provides important clues and hypotheses to be tested in subsequent studies. To this end, we have undertaken structural studies of a large number of eukaryotic-like proteins that interfere with the host through molecular mimicry.
Lem22 is a validated substrate of T4SS2,3 and conserved in several strains of L. pneumophila3,4. Lem22 from the Philadelphia strain (NCBI reference YP_096337), also known as lpg2328, is a 127-residue protein with no sequence similarity to proteins of known structure. It shows 49% identity to the N-terminal position of another Legionella protein, lpg2327, suggesting a highly similar structure and possibly overlapping function (Fig. 1A). It also shares a more distant similarity to a region of olfactomedin 4, also known as antiapoptotic protein GW1125. GW112 was previously shown to be involved in the immune response to bacterial infections6, which suggests a possible functional link. The role of lpg2328 in pathogenesis is unknown.
Figure 1.
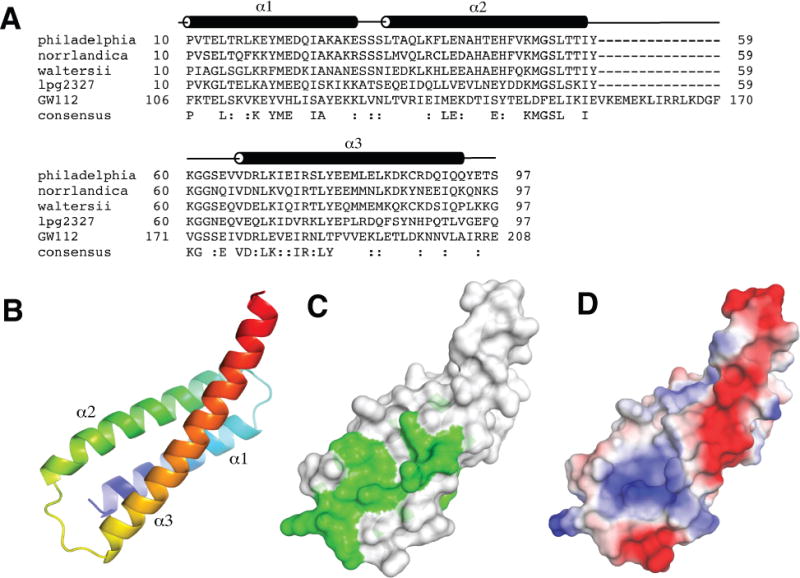
Structure of Lem22. (A) Sequence alignment of Lem22 from Legionella pneumophila (NCBI reference YP_096337), Legionella norrlandica (WP_035889725), Legionella waltersii (WP_058479910), N-terminal domain of lpg2327 from Legionella pneumophila (AEW52564) and a homologous region of human GW112 (XP_014637891). The positions of α-helices (α1-α3) are shown according to the structure from L. pneumophila. (B) Cartoon representation of lpg2328 structure with the molecule colored from blue at the N-terminus to red at the C-terminus. (C) Sequence conservation mapped to the structure. Green color corresponds to highly conserved residues. (D) Surface charge representation of lpg2328. Red color represents negative charge, blue color - positive charge. The surface charge was generated with PyMOL with electrostatic potential values ranging from -67.3 to 67.3.
Here, we determined a high-resolution crystal structure of lpg2328. The structure shows three long α-helices forming a coiled-coil-like bundle. It displays strong similarity to a number of previously characterized protein-binding domains such as BAG domains that are involved in apoptosis7.
MATERIALS AND METHODS
Protein Expression and Purification
The gene lpg2328 (2-123) from Legionella pneumophila str. Philadelphia was inserted into the pGEX-4T-1 vector (Amersham-Pharmacia) with a TEV cleavage site and expressed in E. coli BL21(DE3) strain as an N-terminal GST-tag fusion. The cells were grown at 37°C in Luria Broth to an optical density of 0.8, and expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 30°C for 4 hours. After centrifugation, the cell pellets were resuspended in phosphate buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mg/ml lysozyme and 0.1% (v/v) deoxyribonuclease (DNAse), and the cells were lysed by sonication. Cell debris was removed by centrifugation, and the GST-fusion protein was purified using Glutathione-Sepharose affinity columns (GE Healthcare). After eluting the protein in PBS containing 20 mM glutathione followed by overnight cleavage with TEV protease, the protein was further purified on a Superdex75 gel filtration column (GE Healthcare) in buffer A (10 mM HEPES, 100 mM NaCl, 2 mM DTT, pH 7.0) before crystallization trials.
The truncated construct lpg2328 (10-97) was cloned into a pET15b vector (Amersham-Pharmacia) with a TEV cleavage site and expressed in E. coli BL21(DE3) as an N-terminal His-tag fusion. The expression conditions were the same as for the full-length protein. The fusion protein was bound to Ni–NTA Agarose (Qiagen) beads, washed with buffer B (50 mM HEPES pH 7.6, 0.5 M NaCl, 5% (v/v) glycerol) containing 30 mM imidazole and eluted with buffer B containing 250 mM imidazole. The N-terminal tag was removed by cleavage with TEV protease, leaving a Ser-Asn-Ala N-terminal extension. The cleaved proteins were additionally purified using Superdex75 gel filtration column (GE Healthcare) in buffer A.
For 15N-labeling, the cells were grown in M9 minimal medium supplemented with 15N-ammonium chloride as the sole source of nitrogen. For selenomethionine labeling, the plasmid was transformed into a methionine-auxotroph DL41 (DE3) E. coli strain, and the cells were grown in LeMaster medium supplemented with selenomethionine. The expression and purification protocols were the same as for the unlabeled protein.
Crystallization
Initial crystallization conditions for lpg2328 (10-97) were identified utilizing hanging drop vapor diffusion at 20°C using the Classics II screen (QIAGEN). The selenomethionine-labeled crystals were obtained at 20°C by mixing a 0.6 mL drop of protein (10 mg/mL) with 0.6 mL of reservoir solution containing 25% (w/v) PEG 3350, 0.2 M ammonium acetate, 0.1 M HEPES, pH 7.5 and suspending over 0.6 mL of reservoir solution. The best native crystals were obtained at 20°C by mixing a 0.6 mL drop of protein (10 mg/mL) with 0.6 mL of reservoir solution containing 25% (w/v) PEG 3350, 0.1 M Bis-Tris, pH 5.5 and suspending over 0.6 mL of reservoir solution. For cryoprotection, the crystals were transferred into reservoir solution containing 20% (v/v) ethylene glycol. For data collection, crystals were picked up in a nylon loop and flash frozen in a N2 cold stream (Oxford Cryosystem).
Structure determination and refinement
The selenium SAD dataset was collected on a MarMosaic CCD 300 detector at beamline 08ID-1 at the Canadian Light Source (CLS) (Table 1). Data processing and scaling were performed with HKL20008. The starting phases were obtained by Selenium-SAD using PHENIX9. The native dataset from a single crystal was collected using a single-wavelength (0.9779 Å) regime on an ADSC Quantum-210 CCD detector (Area Detector Systems Corp.) at beamline F1 at the Cornell High-Energy Synchrotron Source (CHESS) (Table 1). The native structure was solved by molecular replacement using coordinates from Se-labeled crystals. The resulting model was extended manually with the help of the program Coot10 and was improved by several cycles of refinement using the program REFMAC511 followed by the translation-libration-screw (TLS) refinement12. Predictions of TLS Motion Determination Server were used to define TLS groups13. During the final refinement, ten cycles of TLS refinement were followed by ten cycles of maximum likelihood restrained refinement. The final model has good stereochemistry with no outliers in the Ramachandran plot. Coordinates have been deposited in the RCSB Protein Data Bank with accession codes 5WD8 and 5WD9.
Table 1.
Statistics of data collection and refinement.
| Data collection | Native | SeMet |
| Space group | C2 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 83.51, 24.85, 41.30 | 23.04, 81.14, 45.68 |
| α, β, γ (°) | 90.0, 114.6, 90.0 | 90.0, 90.8, 90.0 |
| Resolution (Å) | 50–1.40 (1.42–1.40) | 50–1.95 (1.98–1.95) |
| Rsym | 0.102 (0.403) | 0.135 (0.506) |
| I/σI | 19.2 (8.8) | 19.9 (2.5) |
| Completeness (%) | 99.0 (100) | 99.1 (91.6) |
| Redundancy | 5.8 (5.9) | 6.6 (3.8) |
| Refinement | ||
| Resolution (Å) | 22.4 – 1.40 | 40.6 – 1.94 |
| No. reflections | 15317 | 12194 |
| Rwork/Rfree | 0.16/0.20 | 0.22/0.27 |
| No. atoms | ||
| Protein | 726 | 1458 |
| Water | 113 | 174 |
| B-factors | ||
| Protein | 14.3 | 26.9 |
| Water | 26.9 | 34.5 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.006 | 0.001 |
| Bond angles (°) | 0.76 | 0.33 |
| Ramachandran statistics (%) | ||
| Most favored regions | 100.0 | 100.0 |
| Additional allowed regions | 0.0 | 0.0 |
| Disallowed regions | 0.0 | 0.0 |
| PDB code | 5WD9 | 5WD8 |
Highest resolution shell is shown in parentheses.
RESULTS AND DISCUSSION
Crystallization of lpg2328
Our initial attempts to crystallize full-length lpg2328 were unsuccessful. Based on limited proteolysis experiments, we produced several truncated constructs. The construct containing residues 10-97 expressed with high yields and yielded reproducible plate-like crystals which diffracted to < 2 Å. In order to determine the phases, this construct was labeled with selenomethionine. Crystallization trials with labeled protein produced crystals in the P21 space group that diffracted to 1.95 Å using synchrotron radiation. The structure was solved using the single-wavelength anomalous dispersion (SAD) method (Table 1) and contains two lpg2328 molecules in the asymmetric unit with density visible from Pro10 to Thr96 (chain A) or Ser97 (chain B). Along with the P21 crystal form, we also obtained native crystals in the C2 space group that contained one molecule in the asymmetric unit. The native structure was solved by molecular replacement and refined to 1.40 Å. The structure displays residues Pro10-Tyr94, identifying the boundaries of the folded region of the protein. The native and selenomethionine-labeled structures are very similar (RMSD of 0.6 Å over 70 Cα atoms), suggesting that the observed structure is not significantly influenced by crystal contacts (Supporting Information Fig. S1). The largest conformational differences exist in the loop between helices 2 and 3 and in the differing curvature of the C-terminus of helix 3.
Structure of lpg2328
The structure consists of three long helices forming an up-and-down bundle (Fig. 1B). The helices are formed by residues Pro10-Glu30, Leu34-Ile58 and Val66-Gln93 and display some curvature reminiscent of coiled-coil structures. The lpg2328 molecules in the P21 crystals appear to dimerize through the helices 2 and 3 (Supporting Information Fig. S2). Nevertheless, these contacts were not observed in the C2 structure, suggesting that this protein is monomeric in solution. This is in agreement with the protein elution as a monomer on size exclusion chromatography (data not shown).
The surface formed by helix 2 and the N-terminal half of helix 3 has the most sequence conservation across Legionella species (Fig. 1C). This is the same surface involved in dimer contacts in the P21 crystals and suggests a propensity of this region for protein interactions. Intriguingly, this surface also overlaps with the regions of highest sequence identity with the eukaryotic protein GW112. We should note that the comparison with GW112 should be taken with caution, as the sequence alignment of helix 2 is rather poor and is followed by a 15-residue insert in GW112. On the other hand, the sequence similarity in helices 1 and 3 is unmistakable, and suggests that at least this part of the structure is conserved between Lem22 and GW112. Unlike the N-terminus of helix 3, the other half of this helix is poorly conserved and is unlikely to contain functional sites. The α2-α3 surface is mostly positively charged (Fig. 1D), which suggests it could be binding to a negatively charged binding partner.
Structural comparison to other proteins
A structural similarity search using DALI14 identified a large number of hits. This perhaps is not surprising due to a simplicity of the fold and its similarity to a coiled-coil fold. Some of the hits were to coiled-coil-like regions in larger folds (such as in protein kinase YopO, glycine receptor subunit alpha 1, polyphosphate kinase, vacuolar protein sorting-associated protein 35, among others), which makes it difficult to infer any functional implications in those cases. For Lem22-like stand-alone domains, there are three types of proteins that have a very similar fold. These are N, N′-diacetylchitobiose-specific phosphotransferase enzyme IIA component, and two families of protein-binding domains in Bcl-associated athanogene (BAG) proteins and syntaxin proteins (Fig. 2A). The most structurally similar protein is N, N′-diacetylchitobiose-specific phosphotransferase enzyme IIA component15 (PDB 2LRL, Z-score 10.6) with an RMSD of 2.6 Å for 86 Cα atoms. Among the other top hits are the BAG domain from silencer of death domains (SODD)16 (PDB 1M7K, Z-score 9.2), the BAG domain from BAG417 (PDB 4HWH, Z-score 9.0), and the N-terminal domain of syntaxin 10 (PDB 4DND, Z-score 8.7) (Fig. 2B). Among those, lpg2328 has 16% sequence identity to SODD and only 8-10% identity to other mentioned proteins. BAG domains are protein interaction domains implicated in apoptosis regulation18. Many known BAG domains interact with the ATPase domain of HSP70 as nucleotide exchange factors and with Bcl2, a regulator of apoptosis7,19. In particular, SODD interacts with Bcl2 using its BAG domain and demonstrates antiapoptotic properties7,20. It is worth noting that the α2-α3 surface of lpg2328 has similar surface charge distribution to SODD among other structural homologs (data not shown).
Figure 2.
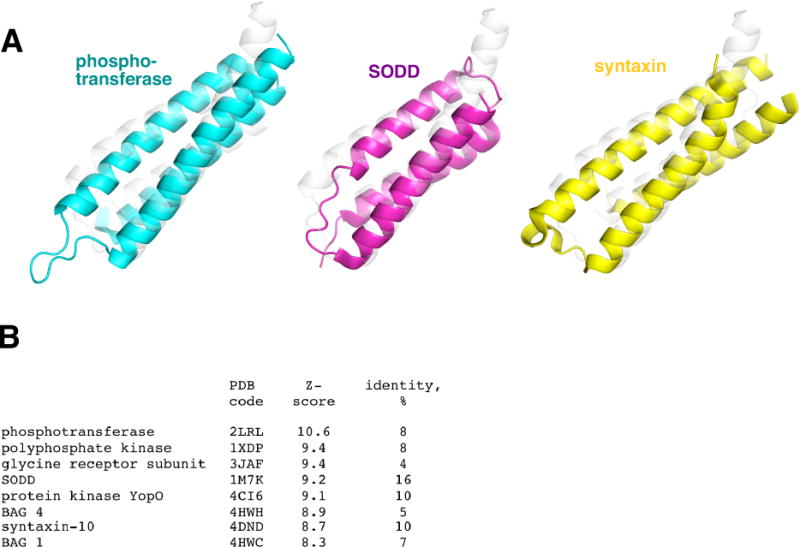
(A) Structural similarity of Lem22 to N, N′-diacetylchitobiose-specific phosphotransferase enzyme IIA component (PDB 2LRL), SODD BAG domain (PDB 1M7K), and syntaxin 10 (PDB 4DND). Overlaid structure of lpg2328 is shown in gray. (B) Table of the top structural similarity hits from the DALI database.
Multiple clues hint at the possible involvement of Lem22 in antiapoptotic pathways. It has a sequence similarity to antiapoptotic protein GW112. At least some of its structural homologs are antiapoptotic. This would be beneficial for Legionella, as it will allow more time for pathogenic bacteria to multiply in the host cell. It would be interesting to test for interactions between Lem22 and Bcl2 and whether Lem22 could be antiapoptotic via cellular pathways involving Bcl2. Lys70, Arg74 and Tyr77 would be attractive mutagenesis targets in these experiments as highly conserved and accessible residues on the α2-α3 surface of lpg2328.
Supplementary Material
Acknowledgments
We thank Dr. Miroslaw Cygler (University of Saskatchewan) for the lpg2328 plasmids. This work was funded by a CIHR Genomics grant GSP-48370 to Miroslaw Cygler and Kalle Gehring. Data were acquired at the Canadian Light Source (CLS) and at the Macromolecular Diffraction (MacCHESS) facility at the Cornell High Energy Synchrotron Source (CHESS). CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-0225180, and the MacCHESS resource is supported by NIH/NCRR award RR-01646.
Footnotes
The authors declare no conflict of interests.
References
- 1.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 2.Aurass P, Gerlach T, Becher D, Voigt B, Karste S, Bernhardt J, Riedel K, Hecker M, Flieger A. Life Stage-specific Proteomes of Legionella pneumophila Reveal a Highly Differential Abundance of Virulence-associated Dot/Icm effectors. Molecular & cellular proteomics : MCP. 2016;15(1):177–200. doi: 10.1074/mcp.M115.053579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5(7):e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. Comparative and functional genomics of legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Frontiers in microbiology. 2011;2:208. doi: 10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Huang Q, Yang Z, Li Y, Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer research. 2004;64(7):2474–2481. doi: 10.1158/0008-5472.can-03-3443. [DOI] [PubMed] [Google Scholar]
- 6.Liu W, Yan M, Liu Y, Wang R, Li C, Deng C, Singh A, Coleman WG, Jr, Rodgers GP. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc Natl Acad Sci U S A. 2010;107(24):11056–11061. doi: 10.1073/pnas.1001269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antoku K, Maser RS, Scully WJ, Jr, Delach SM, Johnson DE. Isolation of Bcl-2 binding proteins that exhibit homology with BAG-1 and suppressor of death domains protein. Biochemical and biophysical research communications. 2001;286(5):1003–1010. doi: 10.1006/bbrc.2001.5512. [DOI] [PubMed] [Google Scholar]
- 8.Otwinowski ZMW. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 9.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The Phenix software for automated determination of macromolecular structures. Methods (San Diego, Calif) 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 13.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 14.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung YS, Cai M, Clore GM. Solution structure of the IIAChitobiose-HPr complex of the N,N′-diacetylchitobiose branch of the Escherichia coli phosphotransferase system. J Biol Chem. 2012;287(28):23819–23829. doi: 10.1074/jbc.M112.371492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockmann C, Leitner D, Labudde D, Diehl A, Sievert V, Bussow K, Kuhne R, Oschkinat H. The solution structure of the SODD BAG domain reveals additional electrostatic interactions in the HSP70 complexes of SODD subfamily BAG domains. FEBS letters. 2004;558(1–3):101–106. doi: 10.1016/S0014-5793(03)01490-X. [DOI] [PubMed] [Google Scholar]
- 17.Fang S, Li L, Cui B, Men S, Shen Y, Yang X. Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 6):934–945. doi: 10.1107/S0907444913003624. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic acids research. 2002;30(1):276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274(2):781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 20.Ozawa F, Friess H, Zimmermann A, Kleeff J, Buchler MW. Enhanced expression of Silencer of death domains (SODD/BAG-4) in pancreatic cancer. Biochemical and biophysical research communications. 2000;271(2):409–413. doi: 10.1006/bbrc.2000.2610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


