Abstract
Objective
The evaluation of mutations in cell free circulating DNA (cfDNA) has recently been used for tracking disease relapse in colorectal cancer (CRC). This approach requires personalized assay design due to the lack of universally mutated genes. In contrast, early methylation alterations are restricted to defined genomic loci allowing comprehensive assay design for population studies. Our objective was to identify cancer specific methylated biomarkers which could be measured longitudinally in cfDNA to monitor therapeutic outcome in metastatic CRC patients (mCRC).
Design
Genome wide methylation microarrays identified a highly cancer specific panel of five methylated loci (EYA4, GRIA4, ITGA4, MAP3K14-AS1, MSC). Digital PCR assays were designed for these genes. Marker prevalence was retrospectively evaluated in tissue DNA (N=85) and cfDNA from mCRC patients (N=182). Longitudinal assessment was performed in a subset of patients treated with chemotherapy or targeted therapy.
Results
One hundred and fifty-nine mCRC patients (87%) showed positivity in at least one marker. Positivity was observed in 67% for EYA4, 71.3% for GRIA4, 69.2% for ITGA4, 69.8% for MAP3K14-AS1 and 62.1% for MSC. Dynamics of methylation markers over time was not affected by treatment type and correlated with objective tumor response and progression-free survival.
Conclusion
Methylation can be used as a universal test to circumvent the absence of patient specific mutations for monitoring tumor burden dynamics via liquid biopsy. The selected biomarkers allowed monitoring of tumor burden under different therapeutic regimens. This method might be proposed for assessing pharmacodynamics in clinical trials or when conventional imaging has limitations.
Introduction
Tumors release fragments of nucleic acids in the circulation, which could provide a minimally invasive surrogate for tissue biopsy as well as offering the opportunity of serial sampling over time [1 2]. Recent liquid biopsy studies have evaluated tumor specific mutations or gene copy number changes in cell free circulating DNA (cfDNA) derived from digestive tract cancer patients for early diagnosis [3] and for monitoring the emergence of disease relapse [1 4–6]. This approach usually relied on alterations in oncogenic drivers such as RAS and BRAF (up to 50% of the population) or mutations within oncosuppressor genes (usually with no variant hotspots), therefore requiring specific assay design for each mutation. Until now, this application has been mainly restricted to follow-up after surgery [5–7] or in individuals treated with targeted therapies (e.g.: EGFR inhibitors) [4 8 9], in which detection of oncogenic alterations such as RAS mutations, are associated with impaired treatment response; however, due to high heterogeneity in the molecular mechanisms of resistance [4 10] a universal tumor marker panel is warranted for the early detection of relapse through liquid biopsy.
An epigenetic profile is determined during embryologic development allowing differentiation of the zygote's cells into the different tissue layers that will compose the individual. Alteration in this epigenetic pattern is often associated with alterations in certain physiological or pathological conditions that cause cell death and release of DNA with specific epigenetic marks [11]. This process has recently prompted exploring liquid biopsy tests to detect specific epigenetic profiles as a surrogate of liver fibrosis severity in individuals affected by non-alcoholic fatty liver disease [12]. Throughout an individual’s lifetime, changes in DNA methylation, one of the epigenetic marks, in response to environmental stimuli may be associated with an increase in the risk of neoplastic transformation and is therefore considered an early event in carcinogenesis [13]. Consequently, DNA methylation markers have already been proposed for early tumor detection in different settings (including CRC), therefore allowing non-invasive screening of the population using stool [14 15] or blood [16–21]. Notably, the American FDA recently approved the first blood test exclusively based on SEPT9 methylation, which might lead to improvements in CRC screening uptake [16 17].
Here we propose the evaluation of DNA methylation markers in cfDNA, not as an early detection method, but rather as a non-invasive treatment-monitoring assay.
Review of methylation markers for early detection of CRC identified genes which were highly tumor specific (not found in normal adjacent mucosa), but rarely validated in large cohorts [22]. While using innovative approaches and being very informative, studies which aimed at identifying cancer specific methylated markers usually relied on platforms with low genomic coverage [21], or small group numbers [23], or exclusively assessed patients tissue [18 24] which might have been partially infiltrated with stroma. We therefore employed genome-wide assessment of DNA methylation in a large cohort of CRC cell lines and compared it to normal mucosa (from non-cancer patients) and blood cells, to minimize false positivity in liquid biopsy tests. A universal five gene signature was defined and validated in-silico using publicly available cohorts of CRC. The performance of two previously reported markers (SEPT9 and C9ORF50) was compared to the signature. Highly quantitative digital PCR-based assays (methyl-BEAMing [19 25]) were designed and evaluated in tissues and then in cfDNA for prevalence assessment of the five new markers in a cohort of unselected metastatic CRC patients and self-declared healthy donors. The assays were then explored longitudinally in cfDNA over diverse treatment courses in the metastatic setting to understand whether our universal DNA methylation signature could be applied to monitor disease burden in CRC patients.
Material and Methods
Additional details are available in Supplementary File 2.
Cell lines and genome wide DNA methylation data processing and retrieval
A collection of 149 cell lines of intestinal origin (Supplementary File 1A) was assembled, as previously described [26].
Infinium HumanMethylation450 BeadChip arrays were performed according to manufacturer’s protocol. Methylation profiles are available on Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/; accession number: GSE86078).
GSE32146 was used as a control set (i.e. cancer unrelated normal mucosa, after removing the ulcerative colitis cases), for comparison with the cell lines. GSE41169 was used as a blood control (i.e. healthy donors) for verification of tissue specificity of the loci of interest.
GSE42752 [23] was used as a test cohort for the establishment of the beta-value threshold used for the in-silico validation. Level 1 data from the TCGA colorectal adenocarcinoma (COAD/READ) (https://gdc-portal.nci.nih.gov) composed the validation cohort used for the in-silico validation. Further validation in normal tissues was performed GSE48684 [24]
Data Preprocessing and Marker Discovery Analysis
All raw data (IDAT files) were preprocessed in R Bioconductor using the minfi package [27]. The individual probe signal was removed when the detection p-value was above 0.05, and probes were removed from the dataset if more than 1% of the dataset contained no data. Our dataset was then merged with the other publicly available cohorts before removal of probes containing SNPs [28], demonstrating sexual dimorphism [29] or located on sex chromosomes.
Differential probe analysis was performed (adjusted p-value ≤ 1×10−35 and a minimum delta beta-value of 0.8). Probes were selected for absence of methylation in leukocytes (GSE41169; maximum beta-value allowed =0.1) minimizing the risk of false positivity in blood tests which could be caused by methylated DNA from blood cells. Differential region analysis was performed using the bumphunter function from minfi (threshold of 0.8 and restricting the regions to those represented by at least two probes (L ε 2)). Differentially methylated probes were limited to those overlapping differentially methylated regions, and “liquid biopsy” assessable loci were defined as regions represented by at least two selected probes distant of a maximum 150bp (average fragment size reported in cfDNA [30]) and not located in centromeres or telomeres.
GSE42752 data were averaged for each of the genomic regions defined in the previous step according to sample type (normal and normal adjacent versus adenocarcinoma). Thresholds were calculated using Receiver operating characteristic analyses performed with the pROC package in R Bioconductor [31] considering normal and adjacent mucosa as positive outcome and cancer as negative; only loci showing a threshold below 0.35 were kept. Each characterized threshold were used to stratify the TCGA COREAD cohort, defining a positive predictive value (PPV) and negative predictive value (NPV) for discriminating normal adjacent from tumor tissue. Normal healthy and peri-tumoral samples from GSE48684 [24] were also controlled for absence of methylation above the threshold.
Methylation and genetic alteration assays for cfDNA evaluation
Assays were designed to allow methylation independent amplification. Primers, probes and amplicon sizes are detailed in Supplementary File 1C. For further validation, ROC analyses in GSE42752 and stratification of TCGA data were reassessed using solely the probes which were located within the amplicon (Supplementary File 4). Methyl-BEAMing assays was optimized as previously described [25] (Supplementary File 5). In-silico validation results display for the five markers of interest are available in Supplementary File 4.
Assays for SEPT9 and C9ORF50 loci were designed targeting the CpG previously described [18 32].
Evaluation of genetic alterations (KRAS, BRAF mutations and MET gene copy number) in cfDNA was performed as previously reported [4 33]. Assays, commercially available (Biorad), are listed in Supplementary File 1C.
Tissue collection and DNA isolation
Formalin fixed paraffin embedded tissues originate from two different cohorts. One containing macro-dissected tumor and normal adjacent tissues (from diverse hospitals) which were controlled and assembled at Niguarda Cancer Center, Grande Ospedale Metropolitano Niguarda (Milan, Italy) and from which DNA was newly extracted. A second cohort of independent tumor tissue was assembled from remaining DNA extracted during the process of the DETECT (EUDRACT 2011-002080-21) [34] or TEMECT (EUDRACT number 2012-003338-17) [35] trials.
Plasma Collection and DNA isolation
De-identified whole blood samples from healthy donors (N=50) were purchased from the Brigham and Women’s Hospital specimen bank (Boston, USA).
One hundred and eighty two cases of mCRC were retrospectively enrolled in the study. One hundred and thirty-six cases were selected for plasma time-points based on blood sample availability at a time when patients were presenting radiological evidence of disease. The remaining 47 cases were treatment baselines, selected for availability of longitudinal follow-up (additional total of 134 longitudinal samples). Summary of the clinico-pathological features of the two cohorts can be found in Supplementary File 1D (mCRC patients clinical features are presented in Supplementary File 1E). mCRC plasma samples were collected at Niguarda Cancer Center, Grande Ospedale Metropolitano Niguarda (Milan, Italy) or at San Giovanni Battista Hospital (Turin, Italy). The study was conducted according to Good Clinical Practices and was approved by the local ethics committee. Circulating DNA was extracted as previously described [25] from one milliliter of plasma (due to limited amount availability).
Statistics and data analyses
All methylation microarray analyses and figures were carried out in R Bioconductor as previously mentioned. Prevalence and longitudinal representations were assembled in Graph-Pad. Wilcoxon (for matched tissues analysis), Mann-Whitney U and Kruskall-Wallis test (for group prevalence analyses) were performed in Graph-Pad. Scatter matrix for marker correlation was obtained using Origin Pro 2016 (OriginLabs). All expressed p-values were calculated with two-tailed tests and were considered significant when p≤0.05, unless otherwise specified.
Results
Marker Discovery And Assay Optimization
Differential methylation analysis between CRC cell lines and normal mucosa identified 162 CpG dinucleotides representative of a larger genomic region controlled to be unmethylated in blood (Figure 1, Supplementary File 1B and 3). Forty loci defined by 93 probes, were selected as “liquid biopsy” assessable region (Supplementary File 3). An in-silico validation, confirmed that the selected methylated loci were cancer specific and not a consequence of cell line establishment and identified six loci capable of discriminating cancer from normal mucosa with a positive predicted value of 1 with a negative predictive value above 0.5. Short methylation independent amplification was considered feasible in five candidates (EYA4, GRIA4, ITGA4, MAP3K14-AS1, MSC).
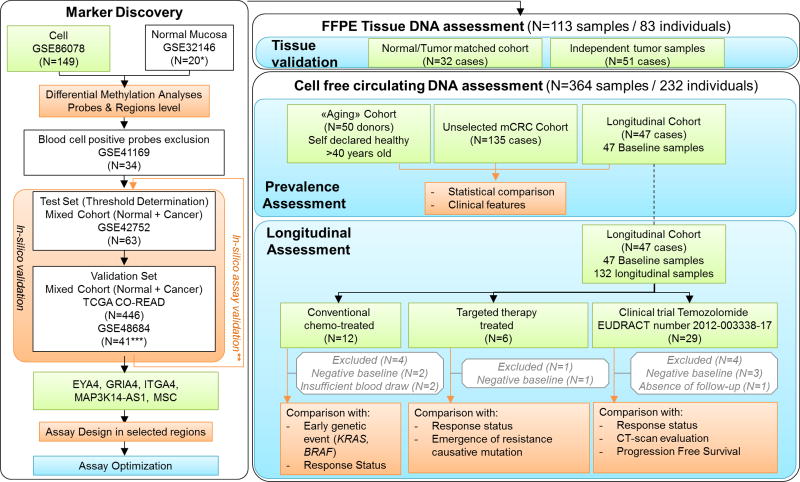
Figure 1. Workflow of the study.
A multistep marker discovery analysis was first performed to identify highly prevalent cancer specific markers. After design, assay probes were further validated in-silico. Assays were optimized to achieve linear quantification over a wide methylation range (0.09% to 100%). Marker prevalence was first evaluated in a total of 83 individuals with mCRC. Then marker prevalence was evaluated in cfDNA in a total of 232 donors enrolled in the study among which 50 were de-identified healthy self-declared donors, and 182 mCRC patients. Among mCRC cases, 47 were followed longitudinally and treated either with conventional chemotherapy, targeted therapy regimen, or with temozolomide (TMZ) as part of a clinical trial. Methylation was analyzed longitudinally for cases with positivity in at least one marker at baseline sample. Methylation dynamics was then compared to additional available clinical or molecular features. In green: Unpublished data; in blue: bench experiments; in orange: bioinformatics or statistical analyses with clinical correlates; in grey: sample exclusion. *GSE32146 was used after removal of ulcerative colitis cases. **in-silico validation was performed again restricting the analysis to the probes included in the assay amplicon. *** Only normal healthy and prei-tumoral tissues were used from GSE48684.
Methyl-BEAMing assays were designed and the probes, which were located within the amplicon, were reassessed via in-silico validation assuring their sensitivity and specificity (Supplementary File 4). The quantitative aspect of the assay was privileged over sensitivity, for better serving the purpose of monitoring tumor burden in advanced disease patients. Digital Miqe checklist can be found in Supplementary File 5.
Validation of the Methylation Markers in tissue from CRC patients
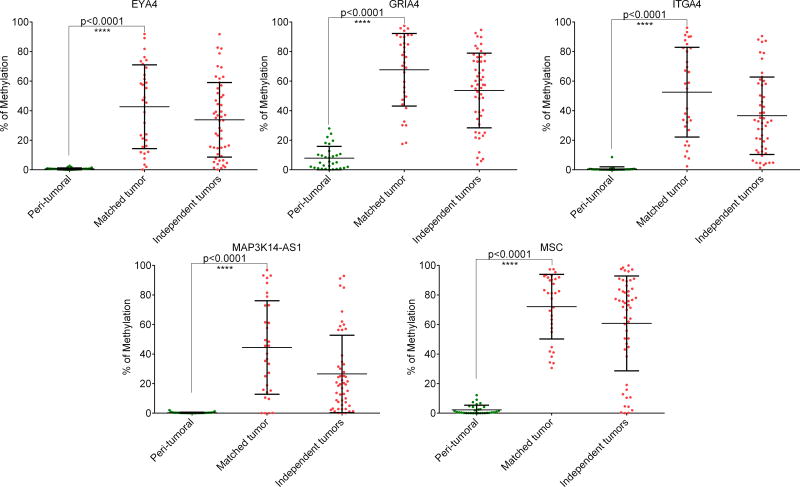
Methylation status was evaluated in tissue DNA for the five markers defined above (EYA4, GRIA4, ITGA4, MAP3K14-AS1, MSC). Amplification was successful in all the cases. Significantly higher methylation level was observed in tumor tissue compared to their normal counterpart (p<0.0001), and remained high in an independent set of non-macro-dissected tumor tissue (Figure 2). Average methylation (and range) for normal tissues was 0.6% [0–3], 7.9% [0–28], 0.5% [0–9], 0.3% [0–2] and 2.3% [0–12] for EYA4, GRIA4, ITGA4, MAP3K14-AS1, MSC respectively. Average methylation (and range) for matched tumor tissues was 42.7% [0–92], 67.8% [18–97], 52.5% [2–96], 44.4% [0–97] and 72.1% [31–97] for EYA4, GRIA4, ITGA4, MAP3K14-AS1, MSC respectively.
Figure 2.
Prevalence of methylated markers in tissue DNA from mCRC patients. A total of 82 cases were analyzed. A first cohort was composed of 32 cases from which tumor and peri-tumoral tissue DNA were available. A second cohort of independent tumor tissue was assembled from remaining DNA extracted during the process of two clinical trials.
Detection Of Methylation Markers In Circulating Tumor DNA Of Healthy Individuals And CRC Patients
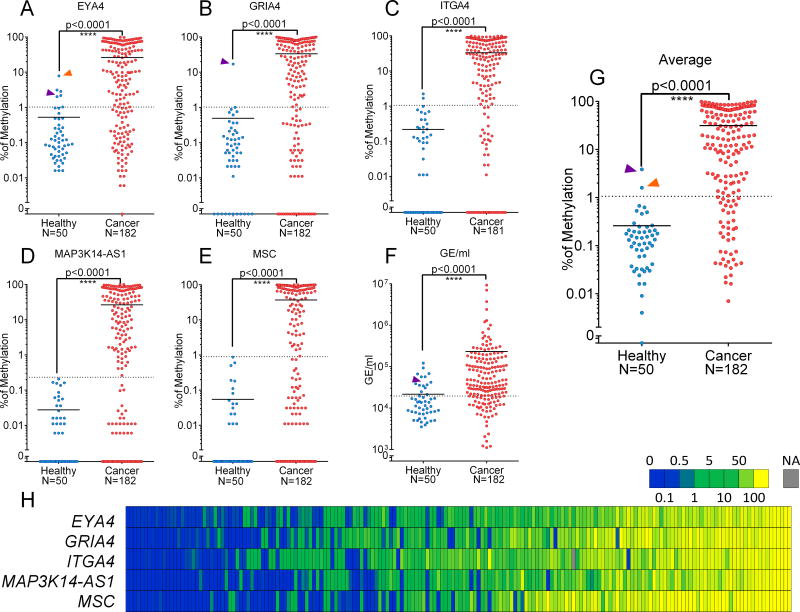
Plasma samples were obtained from a cohort of self-declared healthy individuals above age 40 (N=50), and from a cohort of mCRC patients (N=182). Methylation status was evaluated in a total of 367 cfDNA samples for the five markers defined above (EYA4, GRIA4, ITGA4, MAP3K14-AS1, MSC), as well as two previously reported cancer specific markers (SEPT9 [16] and C9ORF50 [18], Figure 3, and Supplementary File 1G). Amplification was successful in all samples but four for ITGA4 (1.1%), one for GRIA4 (0.3%), and one for C9ORF50 (0.3%) (Supplementary File 1F).
Figure 3. Prevalence of methylated markers in cfDNA and total amount of cfDNA.
Fifty self-declared healthy donors (blue) and 182 mCRC patients (red) were analyzed for the six selected markers. Group mean is represented by a horizontal bar. Mann-Whitney U test was performed to compare distribution in healthy and cancer patients which were all significantly different (with p-value<0.0001). Representation of individual markers: A: EYA4, B: GRIA4, C: ITGA4, D: MAP3K14-AS1E: MSC, F:Genome equivalent/ml (GE/ml). G: Representation of average methylation signal. Two healthy donors presented an average methylation value above positivity threshold (purple, orange and green arrow), which was due to high positivity in GRIA4 and/or EYA4. For each marker, the dashed-line correspond to the threshold established by ROC analyses available in supplementary data XX. H: Heatmap of methylation values in mCRC cases sorted by average methylation.
Marker prevalence
Considering only samples from healthy donors and non-longitudinal mCRC samples for prevalence purposes (Figure 1), all markers showed strong significant differences in methylation distribution between healthy and mCRC patients (u-test: p<0.0001, Figure 3A–F and Supplementary File 1F). ROC analyses were performed for each marker in order to evaluate the performance of the markers and to establish a clinically relevant positivity threshold to discriminate healthy donors from mCRC patients (Supplementary File 6). Cut-off values were 0.7% for EYA4, 1.02% for GRIA4, 2.3% for ITGA4 0.24% for MAP3K14-AS1, 0.9% for MSC and 0.68% considering the average of the five markers. Only two plasma samples from healthy donors showed an average methylation above the threshold (Figure 3H) mostly due to high methylation values in GRIA4 and EYA4 for one individual (43 years old), and EYA4 only for the second (62 years old). One hundred and forty (77%) of the mCRC patients displayed an average methylation above the threshold (Figure 3H). Using the cut-off values established above, positivity was observed in 71.4% for EYA4, 67.6% for GRIA4, 75.1% for ITGA4, 69.8% for MAP3K14-AS1, 61.5% for MSC. This results were similar to those obtained with two previously established markers (SEPT9 and C9ORF50). Considering that positivity in only one marker would be enough to track tumor burden we evaluated that 159 cases (87%) showed positivity in at least one marker. When methylation values were positive, all markers were correlated (Figure 3H and Supplementary File 7).
Association Between cfDNA Methylation And Clinico-Pathological Features
Using univariate analyses, age, treatment status, and BRAF or RAS oncogenic mutations were not associated with different methylation values (Supplementary File 1H and 8). High CEA level (>5ng/ml) showed a non-significant trend for higher methylation values in cfDNA (p=0.13). Male gender was significantly associated with lower methylation values (p=0.029), while the presence of the primary lesion (p=0.001), bulky disease (defined as massive tumor involvement of >50% of liver or lungs; p=0.012) or multiple metastatic lesions (p=0.025) were significantly associated with higher methylation values. The sum of target lesions as per Response Evaluation Criteria for Solid Tumors (RECIST) [36] from available CT-scans was also associated with high methylation in the highest quartiles. Altogether these findings strongly suggest an association between release of methylated cfDNA and tumor burden.
Longitudinal assessment
Among the 182 mCRC patients, 47 were followed-up longitudinally (Figure 1); of those, nine were excluded due to absence of any positive markers at baseline or insufficient blood draw or follow-up. Methylation changes, between longitudinal plasma samples (within 20 days from the first radiologic evaluation) and baseline, were annotated with the tumor response status to therapy (Supplementary File 9). Samples collected close to radiologic assessment of a clinical benefit (defined as objective disease stabilization or partial response as per RECIST criteria) were significantly associated with lower methylation values for average and all markers but EYA4. In comparison samples collected close to documented objective tumor progression were associated with non-significant increased methylation values for average and all markers. This suggests that circulating DNA methylation changes could be associated with tumor burden dynamics. Therefore we investigate whether longitudinal follow-up of methylation could track tumor burden overtime. For this purpose, for each longitudinal time-point an average of selected marker (ASM) was calculated (based exclusively on the markers which displayed methylation value over the positivity threshold in the baseline).
Monitoring of mCRC response to conventional chemotherapy regimen
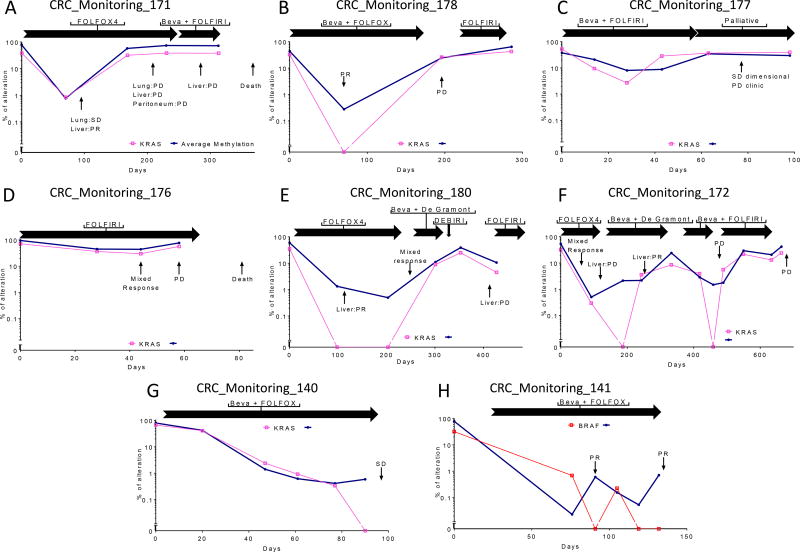
Twelve cases received conventional chemotherapy regimens (FOLFOX, FOLFIRI, plus or minus bevacizumab). Four cases were excluded due to the low amount of plasma available or because they were negative at baseline. All patients carried KRAS or BRAF mutated tumors (considered as early events in tumorigenesis) allowing correlative assessment with methylation (Figure 4). ASM was used for longitudinal monitoring. For most time-points, ASM dynamics recapitulated patient response assessed by imaging, with a decrease in methylation preceding partial response or stable disease, while an increase or stable ASM anticipated progression. For all cases with known mutations in the corresponding tissue, KRAS or BRAF mutant levels in cfDNA paralleled the methylation trend, showing negative results in few time-points, at which methylation was below 1%.
Figure 3. Average of selected markers (ASM) in cfDNA dynamics in eight mCRC cases treated with conventional chemotherapy regimens.
ASM is plotted in blue, while KRAS mutations are plotted in pink and BRAF in red. Methylation and genetic mutations evolve in parallel demonstrating the possibility to use methylation instead of genetic alterations for tracking response. Response status is indicated with arrows and the following abbreviations: PR: Partial Response, SD: Stable Disease, PD: Progressive Disease. Treatment periods are indicated as horizontal black arrows with corresponding chemotherapy regimens.
Monitoring of mCRC response to targeted therapy
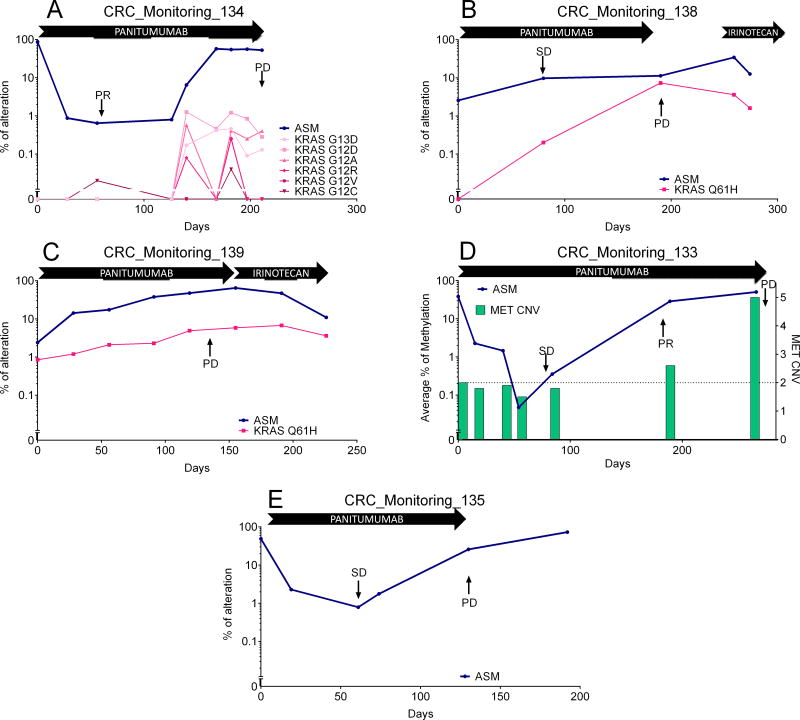
Additional six cases with longitudinal follow-up were treated with the anti EGFR antibody panitumumab based on RAS wild type status (Figure 1). One case displaying a baseline sample negative for methylation was excluded. Four out of five cases (Figure 5) demonstrated emergence of a resistance causative alteration at progression which could be retrospectively analyzed over time. In three cases, progression was associated with the emergence of KRAS alterations (Figure 5A–C) [4], in one by MET amplification [33] (Figure 5D). In all cases, ASM increased in parallel with the emergence of the resistance causative alteration. However, ASM in plasma were much more abundant than the percentage of mutant KRAS alleles in two cases. In two patients, panitumumab was followed at progression by standard chemotherapy (irinotecan) that was associated with decrease of KRAS cfDNA level independently of the methylation dynamics. The remaining case for which the molecular mechanism of resistance to EGFR target therapy remained unexplained could still be monitored in cfDNA with an increase in the ASM value at progression (Figure 5E).
Figure 4.
Average of selected markers (ASM) in cfDNA dynamics in five mCRC cases treated with panitumumab for whom resistance causative mutations were discovered at progression and retrospectively assessed longitudinally. A–C: Resistance was acquired through the emergence of a KRAS alteration, while D: Resistance was acquired through the emergence of amplification of MET. In each case, increase in ASM follows the emergence of resistance alterations. E: A case in which resistance mechanism remained unidentified but for which ASM could detect relapse.
Application of Methylated Circulating DNA Monitoring in a Clinical Trial with temozolomide in chemorefractory mCRC with MGMT hypermethylation
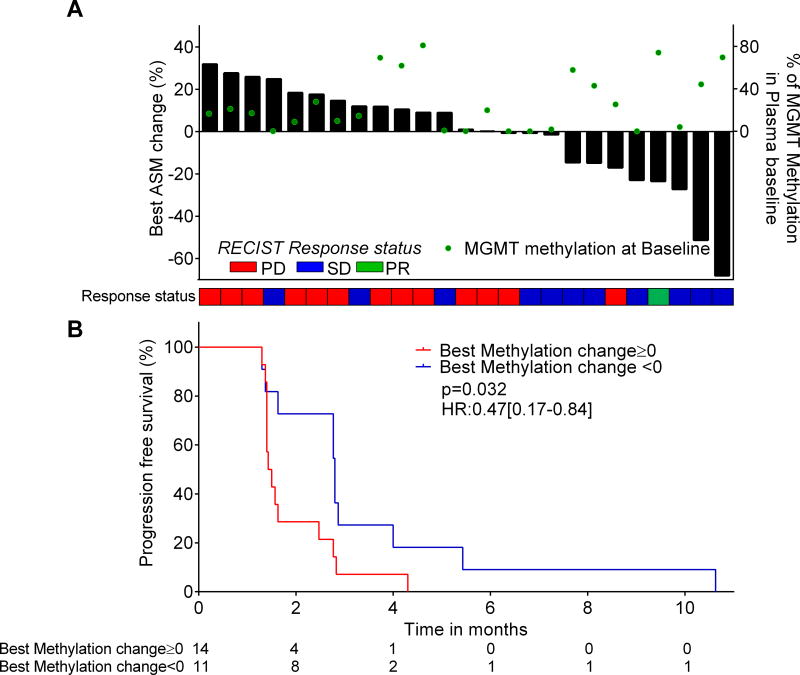
We investigated the use of methylated cfDNA in 29 cases from the TEMECT trial (EUDRACT number 2012-003338-17), which assessed efficacy of temozolomide treatment in chemorefractory mCRC patients selected based on their MGMT methylated status [35]. Three cases without any positive methylated marker at baseline and one case without longitudinal collection were excluded (Figure 1). To explore whether liquid biopsy using methylation markers could be used as a surrogate to imaging, we considered the best methylation change over time, similarly to what is usually performed with imaging-based RECIST criteria. To this aim, the ASM at a longitudinal time-point was subtracted from the ASM value at baseline, and the best methylation change (lowest) over the course of treatment was selected for correlative assessment with radiological response (Supplementary File 1I). A decrease in methylation was associated with clinical benefit as evaluated by RECIST (stable disease or partial response; Figure 6A, PPV=0.8; NPV=0.73, p= 0.015). Considering the lowest methylation change over time, a decrease in ASM was associated with improved progression-free survival (p=0.032, HRrelapse=0.47 [0.17–0.84]; Figure 6B).
Figure 5. Average of selected markers (ASM) incfDNA dynamics assessment in mCRC patients treated with temozolomide within a clinical trial.
A: Comparison of ASM changes to response status and RECIST. The best ASM changes were plotted as a waterfall plot. Response status of patients evaluated by RECIST is plotted as a heatmap. B: Progression free survival according to best ASM change. Negative ASM change shows a trend for improved PFS.
Discussion
The current use of imaging for follow-up in mCRC suffers from limitations potentially leading to overtreatment, delays in treatment reorientation and potential side effects of exposure to imaging contrast agents. The evaluation of serum protein levels such as CEA offers a rapid and cost effective way to measure disease evolution, but is impaired by limited specificity, especially during treatment courses [37] due to inflammation and release of protein in the bloodstream. Furthermore, a fraction of mCRC patients does not show detectable plasmatic CEA levels during the natural history of the disease [38]. Longitudinal evaluation of cancer specific mutations in cfDNA has been exploited to identify relapse after surgery [5] or during treatment with both standard chemotherapies and targeted agents (e.g.: EGFR inhibitors) [1 4], and it demonstrated great specificity and sensitivity. However, these studies must rely on either mutational hotspots with partial prevalence (only up to 50% considering all alterations of KRAS and BRAF) or on personalized assay design after identification of a variant through massive parallel sequencing. Alterations in methylation patterns present an advantage, as they are limited to specific regions of the genome, allowing for a universal assay design compatible for population studies. Moreover, their prevalence is usually high, which triggered their implementation as early diagnostic assays [39].
We confirmed the proof of concept from Garrigou and colleagues that DNA methylation in cfDNA may be employed to track response during therapy in mCRC, enabling non-invasive monitoring of tumor burden. To go beyond previous efforts in this area, we decided to perform a marker discovery analysis using CRC cell lines. This strategy allowed removal of background signal coming from stroma, which has recently been described to impair cancer signal specificity in genome wide analyses [40]. Further validation in independent cohorts (in-silico) or in tissue samples confirmed that these markers were cancer specific and not a consequence of cell line establishment.
To our knowledge, only one study evaluated methylation markers dynamics in mCRC cases upon treatment [41]. In their manuscript, using the methylation of WIF1 and NPY, Garrigou and colleagues found a 80% prevalence in mCRC, and evaluated the changes in cfDNA in three mCRC cases under chemotherapy treatment. While these markers were identified by Roperch et al. using the Illumina goldengate methylation arrays (lower coverage than the Illumina Infinium), these markers were sorted out from our pipeline due to positivity in blood (for NPY) and/or high methylation in normal healthy and normal adjacent tissue.
While EYA4 and ITGA4 are known or putative tumor suppressor gene [42–45], the functional role of the other markers −GRIA4, MSC and MAP3K14-AS1- in carcinogenesis remains to be elucidated; however, their in-silico validation in independent cohorts as well as in tissue demonstrated their reliability in identifying tumor cells in tissue.
When evaluating cancer patient plasma samples, we established positivity as methylation value above a threshold defined by ROC analysis (which allowed the best discrimination between healthy and cancer patients). While these positivity threshold were efficient to define which marker to follow over-time, additional validation such as calculation of the limit of blank are warranted to use these assay for other purposes such as early detection or minimal residual disease.
The high prevalence of at least one methylated marker validated the use of this panel as a universal blood test for detection of plasma DNA of tumor origin. In the current setting, all cases with intact primary CRC in situ displayed at least one positive methylation marker, confirming that cases in which primary lesions were not resected could be more efficiently tracked through liquid biopsy.
As seen for SEPT9 [46], there was a correlation between cfDNA concentrations (here measured by genome equivalent per milliliter of plasma) and average methylation. However, in some samples with low DNA content (below 20000 genome equivalent) high methylation values were recorded, while a subset a sample with high genome equivalent did not display detectable methylation in the selected loci. We hypothesize that these discrepancies could be explained by hemolysis during sample preparation (and consequent contamination of plasma by leukocyte DNA), or by release of DNA from non-cancerous tissues during treatment (possibly due to inflammation or hepatotoxicity).
Combining the positivity of five highly cancer-specific methylated markers allowed a slightly higher but similar prevalence (87%) of methylated cfDNA than previously shown [41]. The fact that there was still 13% of mCRC with no observed circulating DNA methylation warrants further studies to evaluate whether this is a biological or technical phenomenon. None of the recorded clinicopathological features correlated with lack of detectable methylation levels in cfDNA. However, since tissue and microarray data displayed a lower rate of non-methylated template, we expect that technical improvements at the detection level as well as at the DNA isolation level (e.g.: using higher plasma volume), will be required to achieve full penetrance of the assay.
One self-declared healthy donor displayed positivity in two markers and high GE content, possibly suggesting either a false positive result or that this individual had an asymptomatic neoplastic lesion. However, the sample was collected through a de-identified process and it was not possible to verify this hypothesis. It should be acknowledged that our digital PCR based approach was not designed as a cancer diagnostic test, but rather optimized for its linearity and quantification ability of methylated DNA in advanced disease. Nevertheless, future studies are warranted to establish the methylation status and prevalence of the five novel markers identified by our study in the earlier stages of colorectal neoplastic disease. This knowledge together with the development of assays that would privilege sensitivity over quantification are key to establish whether our findings could be relevant also in the setting of early detection.
There was good correlation between levels of circulating DNA methylation and early genetic events in colorectal tumorigenesis such as KRAS or BRAF mutations, which validates the possibility to use methylation without prior knowledge of the tumor genetic pattern as previously shown [41]. Of note, four samples involved in the study were also assessed by massive parallel sequencing (data not shown) and presented genetic mutations at an allelic frequency comparable to the percentage of methylated markers, which highlights a possible role of our panel for plasma quality assessment prior to sequencing. In fact, massive parallel sequencing sometimes fails to detect tumor specific somatic mutations due to low tumor content in cfDNA and methylation assay could therefore be used for checking tumor DNA enrichment in plasma as a quality control step.
When evaluating methylation markers in cfDNA of patients treated with EGFR inhibitors, we observed a good correlation between their dynamics and the emergence of resistance causative genetic alterations. This suggests the possibility to use these methylation markers as whole tumor DNA content normalizer. This will be of particular importance when qualitative assessment (presence or absence) of individual mutation variants in the blood is not enough to predict response. Interestingly, in two cases, the expected resistance mechanism (KRAS alteration) showed very low mutant allelic frequency in comparison to the expected amount of tumor DNA (as judged by the average methylation levels). We hypothesized that either the tumor harbored additional unknown mechanisms of resistance or a small fraction of KRAS altered cells was enough to protect the main bulk of the neoplastic lesion via paracrine effectors as previously demonstrated [47]. In two cases that progressed through emergence of a KRAS mutation, levels of mutant alleles decreased upon treatment with irinotecan, while methylation increased, indicating that the fitness of KRAS mutant clones is dependent on the presence of anti EGFR antibodies, as previously suggested [4].
To our knowledge, very few studies evaluated longitudinally methylated cfDNA [41 48], most of them being essentially focused on analysis of pre-treatment samples to find predictive markers of response. Here, we showed that evolution of the methylation abundance over time demonstrated good prediction of response status during treatment with some of the most used conventional therapies for mCRC. This suggests that longitudinal assessment of methylation could be used in between radiologic assessments for more accurate follow-up of the disease.
We were also able to retrospectively assess a batch of samples that were collected in a clinical trial with temozolomide for the treatment chemorefractory mCRC. Samples were not collected with the aim to directly compare cfDNA to CT-scans. Imaging was not usually performed at the very same time-points when blood was drawn. Despite this limitation and the low response rate of mCRC upon temozolomide treatment, dynamics of methylation could predict clinical benefit. This implicates that the monitoring of methylated circulating DNA might be used as a surrogate to imaging in order to evaluate treatment efficacy and might help reducing delays in therapeutic reorientation. In fact, with the emergence of concepts such as early tumor shrinkage associated with long-term outcome [49], short-term evaluation of pharmacodynamic response using liquid biopsy might become common practice.
In summary, we presented here some novel epigenetic universal markers of circulating DNA of tumor origin that can be efficiently used to monitor mCRC upon most currently available treatments. We hypothesize that combining radiologic and non-invasive and repetitive cfDNA assessments will improve monitoring of mCRC patients and would help clinicians to adjust treatment more efficiently by adopting more timely surgical intervention or early therapeutic reorientation.
Supplementary Material
Supplementary File 1: A: List of samples used in the marker discovery analysis, B: Differentially methylated probes identified with exclusion criteria; C: digital MiqE: Assay description and conditions; D: Clinical data summary for the two cohorts of donors involved in the study; E: Clinical information on individual mCRC cases; F: raw data for methylation markers obtained in tissue. G: raw data for methylation and genetic alterations obtained in cfDNA, H: Association between methylation prevalence and clinico-pathological features; I: longitudinal radiologic assessment and methylation value in the TEMECT trial.
Supplementary File 2: More detailed material and method section.
Supplementary File 3: Marker discovery pipeline. A: using GSE32146 as control: Differentially methylated probes (probes with abs(logFC)>0.8 & adj pvalue<10−35. Differentially Methylated regions (region with Beta-difference >0.8 & L ε 2). Blood discrimination set (Removal of probes with maximum beta-value >0.1 in blood). «Liquid biopsy» assessable regions (Removal of regions located in centromere or telomere and segregation of probes within regions of max 150bp). ROC analysis (establishment of threshold to stratify cancer tissue from normal mucosa (removal of loci with threshold above 0.35). TCGA COREAD – Data, Stratification based on threshold (selection of only loci displaying a PPV of 1 and NPV above 0.5). Methylation independent assay design (selection of four loci for which methylation independent amplification would be possible). B: Validation of the results using normal healthy tissue from GSE48684 as controls.
Supplementary File 4: In-silico validation results for the five selected markers, using the probes overlapping our assays. Each dot corresponds to on sample. Color code is as follow: Crimson: blood; Cyan: normal tissue (cancer unrelated); Green: normal peri-tumoral tissue; Red: Tumor tissue; Purple: cell lines. Dashed red line correspond to threshold obtained by ROC analysis in GSE42752.
Supplementary File 5: MethylBEAMing assay descriptions for A: GRIA4; B: EYA4; C: MSC; and D: MAP3K14-AS1. Average methylation beta-values in cell lines (red) and normal mucosae (blue) from the microarray data. Areas in green define CpG Islands while orange identify the assay amplicon. ROC curve using GSE42752 for threshold establishment using methylation value average of the probes located within the amplicon. Stratification of TCGA samples validated the sensitivity and specificity of the markers.
Supplementary File 7: Scatter plot of average methylation (calculated on the five markers) in cfDNA of mCRC patients according to clinico-pathological features.
Supplementary File 8: Scatter box-plot of methylation, CEA and circulating DNA amount (genome equivalent [GE] per milliliter) in baseline and longitudinal time-point closest to first radiologic evaluation. In red: cases with progressive disease (PD); in blue: clinical benefit (partial response: PR, stable disease: SD).
Supplementary File 5: Receiver operating characteristic curves for positivity threshold estimation in cfDNA.
Supplementary File 6: Scatter matrix of the methylation and genetic alterations in the mCRC prevalence cohort.
What is already known about this subject
Cancer mutations (e.g.: BRAF, KRAS, TP53…), in cell-free circulating DNA could be used as markers of relapse or response in colorectal cancer patients but require prior knowledge of individual gene variants.
DNA methylation alteration is a common early event in colorectal carcinogenesis, is detectable in cell-free circulating DNA and can also be used for early detection or tumor monitoring (e.g.: SEPT9, VIM, NPY, WIF1,…).
Previous identification of methylated marker most often relied on assessment of few samples with low genome coverage methods and might have omitted important putative cancer specific markers.
What are the new findings
Genome wide DNA methylation analysis from stroma free colorectal cancer cells identified highly prevalent and specific methylated loci (EYA4, GRIA4, ITGA4, MAP3K14-AS1, and MSC).
Assessment of methylated markers in cell-free circulating DNA allows non-invasive monitoring of disease
DNA shedding of methylated markers is not impaired by treatment type or duration.
Methylation changes over time correlate with tumor response evaluated by CT-scan in metastatic colorectal cancer patients treated with chemotherapy or targeted agents.
How might it impact on clinical practice in the foreseeable future?
This specific panel of methylated markers was able to monitor tumor burden in colorectal cancer patients treated with different conventional treatment regimens, including chemotherapy, anti-angiogenic agents and targeted agents. This non-invasive method could be coupled with imaging to improve timely therapeutic changes. It might also be particularly useful for early pharmacodynamic assessments in clinical trials.
Acknowledgments
We are grateful to the members of the Core Facilities (Candiolo Cancer Institute-FPO, IRCCS, Candiolo, Italy), for their wonderful support and dedication. We thank the different members of the lab for their invaluable comments discussion, in particular Beth Van Emburgh for editing the manuscript and Pamela Arcella for her technical support. We thank Simonetta Guarrera and Professor Matullo from Hugef for providing technical support and allowing us to use their facility for the microarray assessment of part of samples. We thank the study subjects and staff of the University of Washington Medical School’s GICaRes Biorepository and the Fred Hutchinson Cancer Research Center’s ColoCare study for providing the some of the samples used in this study. We also thank the Fred Hutchinson Cancer Research Center’s Genomics Shared Resource for generating some of the methylation array data in this study. We thank Professor Béla Molnár for critical feedback. The results presented here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Funding: Research in the authors’ laboratory was supported by grants AIRC IG n. 17707 (F.D.N.); AIRC IG n. 16788 (A.B.); Fondo per la Ricerca Locale (ex 60%), Università di Torino, 2014 (F.D.N.); grant Fondazione Piemontese per la Ricerca sul Cancro—ONLUS Farmacogenomica 5 per mille 2009 MIUR (F.D.N.) and grant Fondazione Piemontese per la Ricerca sul Cancro-ONLUS 5 per mille 2011 Ministero della Salute (A.B. and F.D.N.). Investigators at Niguarda Cancer Centers are supported by the following grants: Terapia Molecolare dei Tumori (A.S-B, S. S.) and Dynamic of Tumor Evolution & Therapy (A. S-B) from Fondazione Oncologia Niguarda Onlus; Associazione Italiana per la Ricerca sul Cancro (AIRC) 2010 Special Program Molecular Clinical Oncology 5×1000, project 9970 (S.S. ; A.B.). Partly supported also by NIH grants 1R01CA194663, 1R01CA189184, U01CA152756, and P30CA15704 to W.M.G. Ludovic Barault was the recipient of a post-doctoral fellowship from Fondazione Umberto Veronesi in 2013 and 2015, and of ‘Assegno di Ricerca’ from the University of Torino in 2016.
Role of the Funding Sources: the sponsors had no role in study design, in the collection, analysis and interpretation of data, in the writing of the report or in the decision to submit this manuscript.
Footnotes
Contributions:
LB, AB, ASB and FDN participated in the design of the study. AA, AP, AC, ABi, CCr, SB, KB, PR, KBM, SS and ASB contributed to the collection and retrieval of clinical data of blood samples. AA, AC, AV, MT, SS and ASB contributed to the collection and retrieval of tissue samples.LB, AP, CF, CCr, AC, BM, GS and DO contributed to blood sample preparation and processing. LB, SMo, SMa, WG and ME contributed to the genome wide methylation experiments. PZ contributed to establishing the pipeline for in silico and wet validation of GRIA4. LB performed the bioinformatics analyses of the genome wide methylation experiments. LB, CF and DO performed methylation analyses in cfDNA samples. BM and GS performed genetic alteration analyses in cfDNA samples. LB, BM and GS interpreted the results in cfDNA. LB and FDN wrote the manuscript. All authors critically reviewed and commented the manuscript.
References
- 1.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Science translational medicine. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature reviews. Clinical oncology. 2013;10(8):472–84. doi: 10.1038/nrclinonc.2013.110. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 3.Berger AW, Schwerdel D, Costa IG, et al. Detection of Hot-Spot Mutations in Circulating Cell-Free DNA From Patients With Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology. 2016;151(2):267–70. doi: 10.1053/j.gastro.2016.04.034. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 4.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nature medicine. 2015;21(7):827. doi: 10.1038/nm0715-827b. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 5.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Science translational medicine. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625–34. doi: 10.1136/gutjnl-2014-308859. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 7.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nature medicine. 2008;14(9):985–90. doi: 10.1038/nm.1789. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spindler KL, Pallisgaard N, Appelt AL, et al. Clinical utility of KRAS status in circulating plasma DNA compared to archival tumour tissue from patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor therapy. European journal of cancer. 2015;51(17):2678–85. doi: 10.1016/j.ejca.2015.06.118. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 9.Taly V, Pekin D, Benhaim L, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clinical chemistry. 2013;59(12):1722–31. doi: 10.1373/clinchem.2013.206359. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 10.Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526(7572):263–7. doi: 10.1038/nature14969. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(13):E1826–34. doi: 10.1073/pnas.1519286113. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy T, Zeybel M, Day CP, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2016 doi: 10.1136/gutjnl-2016-311526. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews. Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. Journal of the National Cancer Institute. 2005;97(15):1124–32. doi: 10.1093/jnci/dji204. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 15.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. The New England journal of medicine. 2014;370(14):1287–97. doi: 10.1056/NEJMoa1311194. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 16.Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC medicine. 2011;9:133. doi: 10.1186/1741-7015-9-133. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317–25. doi: 10.1136/gutjnl-2012-304149. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange CP, Campan M, Hinoue T, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PloS one. 2012;7(11):e50266. doi: 10.1371/journal.pone.0050266. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nature biotechnology. 2009;27(9):858–63. doi: 10.1038/nbt.1559. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young GP, Pedersen SK, Mansfield S, et al. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer medicine. 2016;5(10):2763–72. doi: 10.1002/cam4.868. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roperch JP, Incitti R, Forbin S, et al. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC cancer. 2013;13:566. doi: 10.1186/1471-2407-13-566. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue M, Lai SC, Xu ZP, Wang LJ. Noninvasive DNA methylation biomarkers in colorectal cancer: A systematic review. Journal of digestive diseases. 2015;16(12):699–712. doi: 10.1111/1751-2980.12299. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 23.Naumov VA, Generozov EV, Zaharjevskaya NB, et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics. 2013;8(9):921–34. doi: 10.4161/epi.25577. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, Wong CJ, Kaz AM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014;147(2):418–29. e8. doi: 10.1053/j.gastro.2014.04.039. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015;26(9):1994–9. doi: 10.1093/annonc/mdv272. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 26.Medico E, Russo M, Picco G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nature communications. 2015;6:7002. doi: 10.1038/ncomms8002. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 27.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. doi: 10.1093/bioinformatics/btu049. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price ME, Cotton AM, Lam LL, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics & chromatin. 2013;6(1):4. doi: 10.1186/1756-8935-6-4. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–9. doi: 10.4161/epi.23470. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature reviews. Cancer. 2011;11(6):426–37. doi: 10.1038/nrc3066. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 31.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wasserkort R, Kalmar A, Valcz G, et al. Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC cancer. 2013;13:398. doi: 10.1186/1471-2407-13-398. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer discovery. 2013;3(6):658–73. doi: 10.1158/2159-8290.CD-12-0558. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amatu A, Sartore-Bianchi A, Moutinho C, et al. Promoter CpG island hypermethylation of the DNA repair enzyme MGMT predicts clinical response to dacarbazine in a phase II study for metastatic colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(8):2265–72. doi: 10.1158/1078-0432.CCR-12-3518. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 35.Amatu A, Barault L, Moutinho C, et al. Tumor MGMT promoter hypermethylation changes over time limit temozolomide efficacy in a phase II trial for metastatic colorectal cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2016 doi: 10.1093/annonc/mdw071. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 36.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European journal of cancer. 2009;45(2):228–47. doi: 10.1016/j.ejca.2008.10.026. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 37.Lech G, Slotwinski R, Slodkowski M, Krasnodebski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World journal of gastroenterology. 2016;22(5):1745–55. doi: 10.3748/wjg.v22.i5.1745. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson BD, Shinkins B, Pathiraja I, et al. Blood CEA levels for detecting recurrent colorectal cancer. The Cochrane database of systematic reviews. 2015;(12):CD011134. doi: 10.1002/14651858.CD011134.pub2. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laird PW. The power and the promise of DNA methylation markers. Nature reviews. Cancer. 2003;3(4):253–66. doi: 10.1038/nrc1045. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 40.Isella C, Terrasi A, Bellomo SE, et al. Stromal contribution to the colorectal cancer transcriptome. Nature genetics. 2015;47(4):312–9. doi: 10.1038/ng.3224. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 41.Garrigou S, Perkins G, Garlan F, et al. A Study of Hypermethylated Circulating Tumor DNA as a Universal Colorectal Cancer Biomarker. Clinical chemistry. 2016;62(8):1129–39. doi: 10.1373/clinchem.2015.253609. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 42.De Carvalho DD, Sharma S, You JS, et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer cell. 2012;21(5):655–67. doi: 10.1016/j.ccr.2012.03.045. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SJ, Tae CH, Hong SN, et al. EYA4 Acts as a New Tumor Suppressor Gene in Colorectal Cancer. Molecular carcinogenesis. 2015;54(12):1748–57. doi: 10.1002/mc.22247. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 44.Ausch C, Kim YH, Tsuchiya KD, et al. Comparative analysis of PCR-based biomarker assay methods for colorectal polyp detection from fecal DNA. Clinical chemistry. 2009;55(8):1559–63. doi: 10.1373/clinchem.2008.122937. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 45.Chang E, Park DI, Kim YJ, et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepato-gastroenterology. 2010;57(101):720–7. [PubMed] [Google Scholar]
- 46.Toth K, Wasserkort R, Sipos F, et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PloS one. 2014;9(12):e115415. doi: 10.1371/journal.pone.0115415. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hobor S, Van Emburgh BO, Crowley E, Misale S, Di Nicolantonio F, Bardelli A. TGFalpha and amphiregulin paracrine network promotes resistance to EGFR blockade in colorectal cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(24):6429–38. doi: 10.1158/1078-0432.CCR-14-0774. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 48.Fackler MJ, Lopez Bujanda Z, Umbricht C, et al. Novel methylated biomarkers and a robust assay to detect circulating tumor DNA in metastatic breast cancer. Cancer research. 2014;74(8):2160–70. doi: 10.1158/0008-5472.CAN-13-3392. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cremolini C, Loupakis F, Antoniotti C, et al. Early tumor shrinkage and depth of response predict long-term outcome in metastatic colorectal cancer patients treated with first-line chemotherapy plus bevacizumab: results from phase III TRIBE trial by the Gruppo Oncologico del Nord Ovest. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015;26(6):1188–94. doi: 10.1093/annonc/mdv112. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: A: List of samples used in the marker discovery analysis, B: Differentially methylated probes identified with exclusion criteria; C: digital MiqE: Assay description and conditions; D: Clinical data summary for the two cohorts of donors involved in the study; E: Clinical information on individual mCRC cases; F: raw data for methylation markers obtained in tissue. G: raw data for methylation and genetic alterations obtained in cfDNA, H: Association between methylation prevalence and clinico-pathological features; I: longitudinal radiologic assessment and methylation value in the TEMECT trial.
Supplementary File 2: More detailed material and method section.
Supplementary File 3: Marker discovery pipeline. A: using GSE32146 as control: Differentially methylated probes (probes with abs(logFC)>0.8 & adj pvalue<10−35. Differentially Methylated regions (region with Beta-difference >0.8 & L ε 2). Blood discrimination set (Removal of probes with maximum beta-value >0.1 in blood). «Liquid biopsy» assessable regions (Removal of regions located in centromere or telomere and segregation of probes within regions of max 150bp). ROC analysis (establishment of threshold to stratify cancer tissue from normal mucosa (removal of loci with threshold above 0.35). TCGA COREAD – Data, Stratification based on threshold (selection of only loci displaying a PPV of 1 and NPV above 0.5). Methylation independent assay design (selection of four loci for which methylation independent amplification would be possible). B: Validation of the results using normal healthy tissue from GSE48684 as controls.
Supplementary File 4: In-silico validation results for the five selected markers, using the probes overlapping our assays. Each dot corresponds to on sample. Color code is as follow: Crimson: blood; Cyan: normal tissue (cancer unrelated); Green: normal peri-tumoral tissue; Red: Tumor tissue; Purple: cell lines. Dashed red line correspond to threshold obtained by ROC analysis in GSE42752.
Supplementary File 5: MethylBEAMing assay descriptions for A: GRIA4; B: EYA4; C: MSC; and D: MAP3K14-AS1. Average methylation beta-values in cell lines (red) and normal mucosae (blue) from the microarray data. Areas in green define CpG Islands while orange identify the assay amplicon. ROC curve using GSE42752 for threshold establishment using methylation value average of the probes located within the amplicon. Stratification of TCGA samples validated the sensitivity and specificity of the markers.
Supplementary File 7: Scatter plot of average methylation (calculated on the five markers) in cfDNA of mCRC patients according to clinico-pathological features.
Supplementary File 8: Scatter box-plot of methylation, CEA and circulating DNA amount (genome equivalent [GE] per milliliter) in baseline and longitudinal time-point closest to first radiologic evaluation. In red: cases with progressive disease (PD); in blue: clinical benefit (partial response: PR, stable disease: SD).
Supplementary File 5: Receiver operating characteristic curves for positivity threshold estimation in cfDNA.
Supplementary File 6: Scatter matrix of the methylation and genetic alterations in the mCRC prevalence cohort.