Abstract
Metabotropic GABAB receptor is a G protein-coupled receptor (GPCR) that mediates slow and prolonged inhibitory neurotransmission in the brain. It functions as a constitutive heterodimer composed of the GABAB1 and GABAB2 subunits. Each subunit contains three domains; the extracellular Venus flytrap module, seven-helix transmembrane region and cytoplasmic tail. In recent years, the three-dimensional structures of GABAB receptor extracellular and intracellular domains have been elucidated. These structures reveal the molecular basis of ligand recognition, receptor heterodimerization and receptor activation. Here we provide a brief review of the GABAB receptor structures, with an emphasis on describing the different ligand-bound states of the receptor. We will also compare these with the known structures of related GPCRs to shed light on the molecular mechanisms of activation and regulation in the GABAB system, as well as GPCR dimers in general.
Keywords: GABAB receptor, structure, heterodimer, ligand recognition, activation, G protein-coupled receptor
Introduction
First discovered in 1979 by Dr. Norman Bowery (Bowery et al., 1979; Bowery et al., 1981; Bowery et al., 1980; Bowery and Hudson, 1979; Hill and Bowery, 1981), metabotropic gamma-aminobutyric acid (GABA) type B (GABAB) receptor is found on both ends of synapses throughout the central nervous system, playing a vital role in inhibitory neurotransmission (Bowery et al., 1980; Bowery et al., 1987; Chu et al., 1990). It is one of two types of cell surface receptors that are activated by the neurotransmitter GABA. While GABA type A (GABAA) receptor is a ligand-gated ion channel that mediates large and quick neuronal inhibition (Macdonald and Olsen, 1994), GABAB receptor is a G protein-coupled receptor (GPCR) that acts slowly and maintains the inhibitory tone (Bettler et al., 2004; Bowery et al., 2002). The delayed action of GABAB receptor results from relying on a second messenger, G protein, to mediate its response (Bettler et al., 2004; Bowery et al., 2002). The GABAB receptor signaling pathways involve one of three effector proteins: voltage-gated Ca2+ channels, G protein-activated inwardly-rectifying K+ channels (GIRK), and adenylyl cyclase (Bettler et al., 2004; Bowery et al., 2002). The downstream effects of GABAB receptor include blocked neurotransmitter release and hyperpolarization of neurons (Bettler et al., 2004; Bowery et al., 2002).
GABAB receptor is associated with brain and behavioral diseases, including epilepsy, spasticity, anxiety and neuropathic pain (Bettler et al., 2004; Bowery et al., 2002; Froestl, 2010). Baclofen, a clinical drug and selective GABAB receptor agonist, is used to treat muscle spasticity in patients with multiple sclerosis, cerebral palsy, and spinal cord injury (Bettler et al., 2004; Bowery et al., 2002; Froestl, 2010). Knowledge of the GABAB receptor structure has provided a detailed understanding of how current drugs act on the receptor, and will facilitate the design of better candidates for desired regulation.
GABAB receptor is an obligatory heterodimer, with two subunits specialized for different functions (Jones et al., 1998; Kaupmann et al., 1998; Kuner et al., 1999; Martin et al., 1999; Ng et al., 1999; White et al., 1998). The first subunit, known as GABAB1, binds orthosteric ligands (Kaupmann et al., 1997; Malitschek et al., 1999), while the second, GABAB2, couples with G protein (Duthey et al., 2002; Galvez et al., 2001; Havlickova et al., 2002; Margeta-Mitrovic et al., 2001; Monnier et al., 2011; Pin et al., 2004a; Robbins et al., 2001). In addition, a subfamily of the potassium channel tetramerization-domain (KCTD) proteins interact with GABAB2, and act as auxiliary subunits of the receptor to modulate the kinetics of G protein signaling (Bartoi et al., 2010; Schwenk et al., 2010).
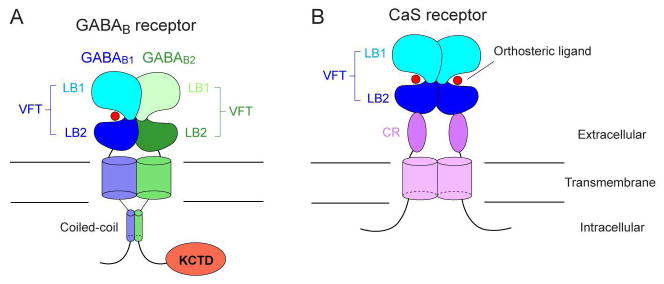
Each GABAB receptor subunit consists of three domains: an N-terminal extracellular domain, a seven-helix transmembrane (TM) domain, and a cytoplasmic tail (Pin and Bettler, 2016) (Fig. 1A). Of these domains, three-dimensional structures have been solved for the extracellular domain and a fragment of the intracellular domain (Blein et al., 2004; Burmakina et al., 2014; Geng et al., 2013; Geng et al., 2012). The ectodomain structure has also been determined in multiple conformations, allowing us to describe the heterodimer interfaces, receptor-ligand interactions, and conformational changes associated with receptor activation (Geng et al., 2013). What remains unknown is the conformational dynamics of GABAB receptor TM domain, and how the individual domains function in an intact receptor. The structures of the full-length receptor in multiple functional states would be required to address these questions.
Fig. 1.
Schematic representation of GABAB and CaS receptors.
Currently, additional insights on GABAB receptor function can be obtained by examining the comparable structures within its related group of GPCRs, called the class C family. GPCRs are divided into four main classes (A, B, C and F) based on the sequence homology of their TM domains (Lagerstrom and Schioth, 2008). Class C GPCRs mediate key biological phenomena, including excitatory and inhibitory neurotransmission, calcium homeostasis, taste and smell (Pin et al., 2003; Pin et al., 2004b). The class C GPCRs are unique in that they require dimerization to function (Pin et al., 2003; Pin et al., 2004b). Within this family, GABAB receptor and taste receptors (TAS1R) are obligatory heterodimers (Jones et al., 1998; Kaupmann et al., 1998; Kuner et al., 1999; Nelson et al., 2002; Nelson et al., 2001; White et al., 1998). While metabotropic glutamate (mGlu) receptors and calcium-sensing (CaS) receptor are traditionally considered to function as homodimers (Bai et al., 1998; El Moustaine et al., 2012; Okamoto et al., 1998; Pidasheva et al., 2006; Ray et al., 1999; Romano et al., 1996; Tsuji et al., 2000; Ward et al., 1998; Zhang et al., 2001), they have recently been discovered to assemble into heterodimers with other class C and class A GPCRs (Doumazane et al., 2011; Gama et al., 2001; Gonzalez-Maeso et al., 2008; Moreno Delgado et al., 2017; Pandya et al., 2016; Yin et al., 2014). Like GABAB receptor, each class C receptor is characterized by a large extracellular domain in addition to the canonical seven-helix TM domain (Pin et al., 2003; Pin et al., 2004b) (Fig. 1B). This extracellular domain is 500–600 amino acids long, and contains the orthosteric ligand-binding site (Pin et al., 2003; Pin et al., 2004b). As of now, no full-length structure of any of these receptors has been solved, but structural information is available for the ectodomains of several family members (Blein et al., 2004; Geng et al., 2013; Geng et al., 2016; Geng et al., 2012; Kunishima et al., 2000; Muto et al., 2007; Nuemket et al., 2017; Tsuchiya et al., 2002; Zhang et al., 2016) as well as the TM domains of two mGlu receptor subtypes (Dore et al., 2014; Wu et al., 2014). In this review, we will summarize our current knowledge of GABAB receptor structure, and compare it with the known structures of class C GPCRs.
Extracellular domain architecture
The extracellular region of GABAB receptor exists in a heterodimeric configuration regardless of interactions within the rest of the protein (Geng et al., 2012; Liu et al., 2004; Nomura et al., 2008). The extracellular domains of the GABAB1 and GABAB2 subunits are each primarily composed of a Venus flytrap (VFT) module (Pin and Bettler, 2016) (Fig. 1A). The structure earns its moniker by resembling the Venus flytrap plant. In addition, molecular cloning identified two main isoforms of the GABAB1 subunit (Kaupmann et al., 1997): GABAB1a, found predominantly in the presynaptic terminal, and GABAB1b, associated with the postsynaptic terminal (Billinton et al., 1999; Gassmann and Bettler, 2012; Kaupmann et al., 1997) (Fig. 2A). These isoforms differ by the presence of two complement control protein (CCP) modules, or sushi domains, in the N-terminus of GABAB1a, but otherwise perform the same ligand-binding function through an identical VFT region (Blein et al., 2004; Hawrot et al., 1998; Kaupmann et al., 1997). The first CCP module of GABAB1a is not compactly folded according to various biophysical measurements such as circular dichroism (CD) spectroscopy (Blein et al., 2004). In contrast, the structure of the second CCP has been solved by nuclear magnetic resonance (NMR) spectroscopy, and shows a well-ordered barrel-like architecture that is stabilized by disulfide bridges (Blein et al., 2004) (Fig. 2B). The CCP domains are attached to proteins containing axonal-sorting signals to localize GABAB1a to the axon terminal (Biermann et al., 2010).
Fig. 2.
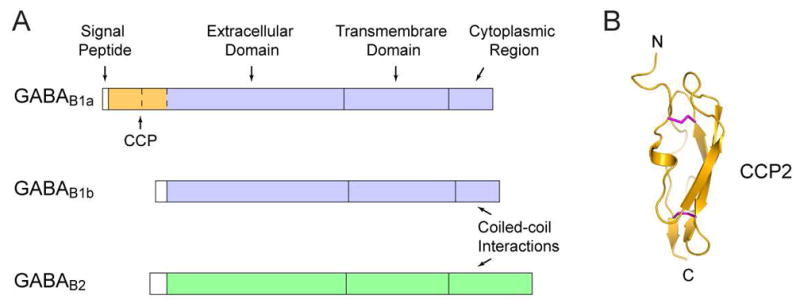
Domain organization of GABAB receptor.
(A) Schematic representation of two isoforms of GABAB1 (GABAB1a and GABAB1b) and GABAB2.
(B) NMR structure of the second CCP module of GABAB1a isoform (PDB code: 1SRZ). Disulfide bridges are in magenta.
The crystal structures of a heterodimeric complex of GABAB1b VFT and GABAB2 VFT have been solved by x-ray crystallography (Geng et al., 2013). Within each complex, the two subunits bind in a side-by-side manner while facing opposite directions, as if they are “dancing cheek-to-cheek” (Fig. 3). The VFT module of each GABAB receptor subunit contains two domains or lobes, LB1 and LB2, with LB1 resting atop LB2 and reaching further into the extracellular space (Fig 1A, 3). These domains are joined by three loops on a single end to form a VFT-like configuration that can oscillate between open and closed conformations. The VFT module is a shared structural feature among all class C GPCRs (Pin et al., 2003). It is also found in bacterial periplasmic binding proteins (Sack et al., 1989) and ionotropic glutamate receptors (Jin et al., 2009; Karakas et al., 2009; Kumar et al., 2009).
Fig. 3.
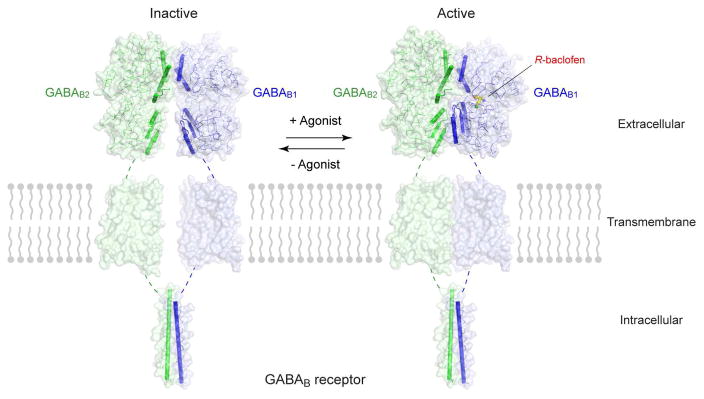
Conformational equilibrium between the inactive and active states of GABAB receptor.
Full-length GABAB receptor models were constructed using the extracellular VFT structures of human GABAB receptor (inactive: apo form, PDB code: 4MQE; active: agonist R-baclofen-bound, PDB code: 4MS4), the TM domain structure of mGlu1 (PDB code: 4OR2), and the intracellular coiled-coil structure of human GABAB receptor (PDB code: 4PAS).
In addition, x-ray imaging has resolved the dimeric extracellular region in various ligand-bound states: ligand-free, in complex with six different antagonists and in complex with two different agonists (Geng et al., 2013). The apo- and antagonist-bound structures have nearly identical conformations and represent the receptor in the resting state. The agonist-bound structures correspond to the active receptor state.
GABAB1 and GABAB2 display different conformational dynamics consistent with their distinct functional roles. It is within the interdomain crevice of GABAB1 VFT that orthosteric ligands bind (Kaupmann et al., 1997; Malitschek et al., 1999) (Fig. 3, 4). Binding of an agonist causes the GABAB1b VFT to close 29° compared with the inactive state (Geng et al., 2013). On the other hand, GABAB2 VFT is not involved in ligand recognition, despite a sequence homology of 33% with GABAB1b VFT (Kniazeff et al., 2002). As a result, the GABAB2 VFT is perpetually vacant and its interdomain cleft remains open (Geng et al., 2013; Geng et al., 2012).
Fig. 4.
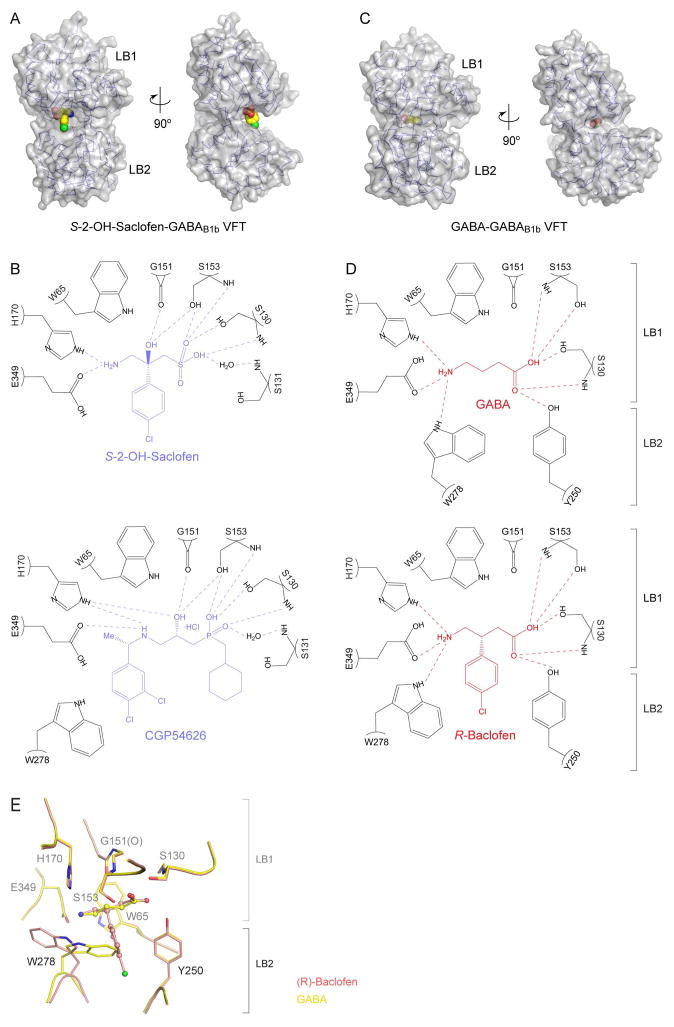
Ligand recognition by GABAB1b VFT.
(A) Molecular surface of antagonist S-2-OH-saclofen-bound GABAB1b VFT (PDB code: 4MQF). The ligand is shown in ball-and-stick model. (B) Schematic diagram showing the interactions between GABAB1b VFT (black) and two antagonists: S-2-OH-saclofen and CGP54626. Hydrogen bonds are represented as dotted lines.
(C) Molecular surface of agonist GABA-bound GABAB1b VFT (PDB code: 4MS3). The ligand is shown in ball-and-stick model. (D) Schematic diagram showing the interactions between GABAB1bVFT (black) and two agonists: GABA and R-baclofen. Hydrogen bonds are represented as dotted lines.
(E) Overlay of the binding sites of GABA and (R)-baclofen in GABAB1b VFT.
Despite these differences, both subunits cooperate with each other to perform signal activation. The inactive structures indicate that GABAB1b and GABAB2 subunits attach at the N-terminal LB1 domain through noncovalent interactions (Geng et al., 2013). This interface is largely conserved in the active state, suggesting that the LB1-LB1 interaction mostly serves to facilitate heterodimer formation.
Based on the extracellular domain structures, we can infer that the dimer arrangement of GABAB receptor will undergo substantial changes upon receptor activation (Geng et al., 2013). First, agonist-stimulated closure of GABAB1 VFT would cause an upward rotation of its LB2 domain, coupled with an inward movement toward its GABAB2 counterpart. Second, the LB2 domain of GABAB2 subunit would also twist toward its GABAB1 counterpart, resulting in the formation of a novel interface that is sustained by polar contacts between the LB2 domains of both VFTs. The LB2-LB2 interaction allows GABAB2 VFT to stabilize the closed conformation of GABAB1 VFT, thus enhancing receptor activity. The importance of LB2-LB2 association is demonstrated by experiments where insertion of a large glycan at this interface prevents agonist-induced receptor activation (Rondard et al., 2008). Furthermore, locking this LB2-LB2 interface through a disulfide bond is sufficient to confer constitutive activity to the receptor (Geng et al., 2013). These observations indicate that the LB2-LB2 interface found in the active-state GABAB1b VFT-GABAB2 VFT structures is physiologically relevant. However, the interface is not unbreakable, thereby allowing the receptor to return to its resting state. Structural observations suggest that GABAB receptor exists in a dynamic equilibrium between inactive and active conformations (Geng et al., 2013), like other GPCRs including β2-adrenergic receptor (Rosenbaum et al., 2011). Finally, the conformational changes reduce the distance between the C-termini of the two VFT modules. The decrease in the separation between VFTs is expected to be sensed by the TM domains.
Orthosteric ligand recognition
The ligand-binding subunit GABAB1 has its active and closed conformation stabilized by an agonist bound to VFT. In contrast, an antagonist would limit the receptor subunit to an inactive and open conformation. The open or closed configuration of GABAB1 VFT also determines whether the ligand in its interdomain crevice is exposed to the surrounding solvent, and could potentially influence the on- and off-rate of ligand binding.
The orthosteric ligands of GABAB receptor are usually derivatives of GABA (Bowery et al., 2002; Froestl, 2010). Antagonists also feature a bulky substituent at either end that hinders closure of GABAB1 VFT (Geng et al., 2013) (Fig. 4A, B). Antagonists bind to LB1 domain via an interface composed primarily of hydrogen bonds. A majority of the antagonists studied, such as S-2-OH-saclofen, do not interact with the lower LB2 lobe at all. Two exceptions are CGP54626 and SCH50911, which manage to form direct contact with Trp278 on LB2 (Geng et al., 2013). Antagonists that maintain this interaction have stronger binding affinity than other candidates (Froestl, 2010; Geng et al., 2013). The role of LB2 in variable antagonist affinity makes it an important domain to consider when determining antagonist selectivity for pharmacological purposes.
GABA and the clinical drug R-baclofen are among the best-understood GABAB receptor agonists (Bowery et al., 2002; Froestl, 2010). An agonist closes the groove of GABAB1 by interacting with both LB1 and LB2 domains of the VFT (Geng et al., 2013) (Fig. 4C, D). The two ends of an agonist are each held by a network of hydrogen bonds. An identical set of LB1 residues mediate the binding of agonists and antagonists. However, only agonists interact with both LB2 residues Tyr250 and Trp278. The difference between GABA and R-baclofen-bound GABAB1 is a complete flip of the indole ring found in Trp278, indicating that the orthosteric ligand-binding site of GABAB receptor exhibits plasticity and can accommodate various agonist structures without altering ligand affinity (Fig. 4E).
The interdomain cleft of VFT is also the orthosteric ligand-binding site of all class C GPCRs (Kniazeff et al., 2011). These receptors have evolved an affinity for amino acids and their analogs (Bowery et al., 2002; Chandrashekar et al., 2006; Conigrave and Hampson, 2010; Conigrave and Ward, 2013; Froestl, 2010; Kniazeff et al., 2011; O’Hara et al., 1993). This is true even in the case of CaS receptor. Although it is traditionally held that Ca2+ is the principal agonist of CaS receptor, recent research shows that the VFT cleft of activated CaS receptor is solely occupied by an L-amino acid, L-Trp, instead of Ca2+ (Geng et al., 2016). Indeed, Ca2+ and L-amino acids serve as co-agonists to jointly trigger CaS receptor response (Geng et al., 2016).
The class C receptors also share a common agonist-binding mode. In all cases, an agonist induces VFT closure by forming intermolecular bonds with surface residues in both the LB1 and LB2 domains (Geng et al., 2013; Geng et al., 2016; Kunishima et al., 2000; Muto et al., 2007; Nuemket et al., 2017; Tsuchiya et al., 2002; Zhang et al., 2016). Key agonist-binding residues are conserved among the class C receptors. For example, a conserved Ser residue is responsible for anchoring the carboxylate of endogenous agonist in GABAB, mGlu, CaS and TAS1R2/TAS1R3 receptors (Geng et al., 2013; Geng et al., 2016; Kunishima et al., 2000; Muto et al., 2007; Nuemket et al., 2017; Tsuchiya et al., 2002; Zhang et al., 2016). An antagonist, on the other hand, depends on its bulky size to obstruct VFT closure, thereby keeping the receptor in its open conformation (Geng et al., 2013; Tsuchiya et al., 2002).
Allosteric ligand recognition
Like other class C GPCRs, the activity of GABAB receptor can be regulated by allosteric modifiers (Brauner-Osborne et al., 2007; Conn et al., 2009; Kniazeff et al., 2011; Pin and Prezeau, 2007; Urwyler, 2011). These ligands usually fall under three categories: positive, negative and neutral. Positive allosteric modulators (PAMs) increase the efficacy of orthosteric ligands, while negative allosteric modulators (NAMs) decrease it (Brauner-Osborne et al., 2007; Conn et al., 2009; Kniazeff et al., 2011; Pin and Prezeau, 2007; Urwyler, 2011). At the same time, neutral or silent allosteric modifiers (SAMs) compete with PAMs and NAMs for specific allosteric sites, but do not alter receptor activity (Conn et al., 2009; Engers and Lindsley, 2013; Gregory et al., 2011; Urwyler, 2011).
The first PAM of GABAB receptor, CGP7930, was discovered through drug screening (Urwyler et al., 2001). Since then, PAMs with different structural folds and improved potency have been developed, including GS39783, rac-BHFF, and BHF177 (Guery et al., 2007; Malherbe et al., 2008; Urwyler et al., 2003). Recently, a NAM called CLH304a, was reported to non-competitively inhibit GABAB receptor signaling (Chen et al., 2014). A neutral allosteric modulator of GABAB receptor has yet to be discovered.
Studies employing mutations and chimeric receptors indicate that the known allosteric modulators of GABAB receptor bind within the TM domain of the G protein-coupling subunit, GABAB2 (Binet et al., 2004; Dupuis et al., 2006; Sun et al., 2016). Most PAMs do not activate full-length GABAB receptor on their own, but enhance the receptor response initiated by an agonist through the VFT (Brauner-Osborne et al., 2007; Kniazeff et al., 2011; Pin and Prezeau, 2007; Urwyler, 2011). Nevertheless, PAMs have been shown to induce G protein coupling when the extracellular domain of GABAB receptor is excised, implying that the unbound ectodomain constitutively maintains the inactive conformation of the receptor TM region (Binet et al., 2004). Fluorescence resonance energy transfer (FRET) experiments further revealed that the main GABAB receptor PAMs CGP7930, GS39783 and rac-BHFF differ in their modes of action, with CGP7930 and rac-BHFF displaying intrinsic agonist activity (Lecat-Guillet et al., 2017). However, the molecular basis of this variety of mechanisms is unknown since the PAM-binding site within the GABAB2 TM domain remains uncharacterized. On the other hand, the NAM CLH304a performs the opposite function, suppressing receptor stimulation by a GABAB receptor agonist (Chen et al., 2014). It also displays inverse agonist property by decreasing the basal activity of the receptor (Sun et al., 2016).
In addition to allosteric interactions within the GABAB2 TM domain, GABAB receptor has been shown to be susceptible to allosteric modification by extracellular Ca2+ bound within the GABAB1 VFT module (Galvez et al., 2000; Wise et al., 1999). Ca2+ increases the affinity of GABA to the receptor, and enhances G protein activation as a result (Galvez et al., 2000; Wise et al., 1999). However, the allosteric effect of Ca2+ is not observed for baclofen (Galvez et al., 2000; Wise et al., 1999). Although mutational analysis points to a Ca2+-binding site within the GABAB1 VFT (Galvez et al., 2000; Wise et al., 1999), Ca2+ ion has not been identified in the known crystal structures of GABAB receptor extracellular domain (Geng et al., 2013).
The allosteric site of class C receptors is generally located on the extracellular side of the TM domain (Brauner-Osborne et al., 2007; Christopher et al., 2015; Dore et al., 2014; Gregory et al., 2011; Harpsoe et al., 2017; Kniazeff et al., 2011; Pin and Prezeau, 2007; Urwyler, 2011; Wu et al., 2014). Recently, crystal structures of mGlu1 and mGlu5 TM domains have been solved, each in the presence of a NAM (Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014). These structures reveal that the NAMs for mGlu1 (FITM) and mGlu5 (mavoglurant; HTL14242 and analogue) have overlapping but distinct binding pockets, with the mGlu5 modulators bound deeper into the TM core (Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014). In addition, the FITM-binding site is analogous to the orthosteric ligand-binding site of class A GPCRs (Rosenbaum et al., 2009; Wu et al., 2014).
Previous mutagenesis studies indicate that different allosteric modifiers of a particular class C GPCR often occupy similar spaces and bind to an overlapping set of residues, as mutations that decrease the binding of one modifier can also hinder others (Conn et al., 2009; Gregory et al., 2011; Jensen and Brauner-Osborne, 2007; Urwyler, 2011). Furthermore, many of the known PAMs and NAMs of class C receptors share a common site within the overlapping FITM/mavoglurant binding pocket that is enclosed by TM helices 2, 3, 5, 6 and 7 (TM2, TM3, TM5, TM6 and TM7) (Christopher et al., 2015; Conn et al., 2009; Dore et al., 2014; Gregory et al., 2011; Harpsoe et al., 2017; Jensen and Brauner-Osborne, 2007; Urwyler, 2011; Wu et al., 2014). At the same time, class C TM domains are known to possess regions that serve as distinct binding sites for specific allosteric modulators (Conn et al., 2009; Gregory et al., 2011; Jensen and Brauner-Osborne, 2007; Urwyler, 2011). It has been shown that CPPHA, a PAM of mGlu5, binds within the TM domain at an unrelated site from most known mGlu5 allosteric modulators (Chen et al., 2008). In another example, CaS receptor contains separate allosteric sites within its TM domain for phenylalkylamines like the PAM Calindol, and structurally different molecules like the NAM ‘BMS compound 1’ (Arey et al., 2005; Hu et al., 2006; Jensen and Brauner-Osborne, 2007).
Allosteric modulators are attractive drug candidates because of their potential for adjusting receptor action without risking side effects from full activation or complete inhibition (Brauner-Osborne et al., 2007; Christopoulos, 2002; Pin and Prezeau, 2007; Urwyler, 2011). The various PAMs of GABAB receptor have shown therapeutic effects in pre-clinical studies on anxiety (Cryan et al., 2004), depression (Cryan et al., 2004), and drug addiction (Filip and Frankowska, 2007; Lhuillier et al., 2007; Mombereau et al., 2004; Slattery et al., 2005; Smith et al., 2004; Sturchler et al., 2017). Furthermore, these allosteric modulators lack the muscle-relaxing and sedative effects associated with the orthosteric drug baclofen (Kniazeff et al., 2011; Pin and Prezeau, 2007; Urwyler, 2011). These side effects also limit the use of baclofen in behavioral pharmacological studies and help to further research interests in PAMs and NAMs (Dalvi and Rodgers, 1996). However, detailed receptor-allosteric modulator interactions still await structural elucidation of GABAB receptor TM domain.
Trafficking control
The intracellular domain of GABAB receptor controls the surface expression of the intact receptor (Margeta-Mitrovic et al., 2000). The GABAB1 subunit is retained inside the cell when expressed alone (Couve et al., 1998), and is only transported to the cell surface when it is chaperoned by GABAB2 in a heterodimeric pair (Margeta-Mitrovic et al., 2000; Pagano et al., 2001). Recently it has been discovered that the endoplasmic reticulum (ER) membrane protein, PRAF2, directly binds to GABAB1, and is responsible for trapping the subunit within the ER (Doly et al., 2016). GABAB1 exits the ER only after the bound PRAF2 is displaced by GABAB2. The assembled heterodimer then progresses to the Golgi apparatus, and in turn the cell surface where the receptor can function (Doly et al., 2016).
Two critical determinants of receptor trafficking are found within the GABAB1 cytoplasmic tail: the di-leucine internalization signal (EKSRLL) (Margeta-Mitrovic et al., 2000; Restituito et al., 2005) and the ER retention signal (RSRR) (Calver et al., 2001; Margeta-Mitrovic et al., 2000; Pagano et al., 2001). The former reduces the level of GABAB1 expression on the cell surface, while the latter completely eliminates it (Margeta-Mitrovic et al., 2000). The internalization sequence lies within a coiled-coil region in the intracellular domain (Fig. 1A). It is directly blocked by the formation of a coiled-coil heterodimer between the GABAB1 and GABAB2 subunits as shown by the heterodimer crystal structure (Burmakina et al., 2014) (Fig. 3). The retention signal does not exist inside the coiled-coil region itself, but rather a few residues away from the C-terminal end of the motif. Nevertheless, mutations of the coiled-coil interfacial residues in either subunit prevent GABAB1 from expressing on the surface (Burmakina et al., 2014; Margeta-Mitrovic et al., 2000). Although the heterodimeric coiled-coil interface does not seal the retention signal, it prevents access to the motif through steric hindrance (Burmakina et al., 2014).
Signaling complex
GABAB receptor is unique in possessing auxiliary KCTD subunits to generate functional diversity in neurons (Gassmann and Bettler, 2012). Four different KCTD molecules (numbered 8, 12, 12b and 16) affect GABAB receptor activity (Bartoi et al., 2010; Schwenk et al., 2010). All of these KCTD proteins are constitutively bound to the intracellular domain of GABAB2 subunit, and expedite agonist-dependent receptor activation by pre-anchoring the G protein to the GABAB receptor TM domain (Schwenk et al., 2010; Turecek et al., 2014). Aside from this function, each KCTD has its own role within the GABAB receptor signaling complex (Gassmann and Bettler, 2012). Within seconds after receptor activation, KCTD12 and KCTD12b attach to the βγ subunits of G protein, and disengage the structures from their target GIRK channel (Turecek et al., 2014). This action desensitizes the receptor and ceases hyperpolarization of the neuron (Schwenk et al., 2010; Seddik et al., 2012; Turecek et al., 2014). In addition, KCTD8 decreases basal G protein activation and agonist-independent GABAB receptor signaling (Rajalu et al., 2015). Finally, KCTD16 provides a template for binding effector channels such as the hyperpolarization-activated cyclic nucleotide gated (HCN) channels (Schwenk et al., 2016). Three-dimensional structures of GABAB receptor-associated KCTD proteins are not yet known. Once available, the structural information will provide important insights on how KCTD subunits allosterically modulate agonist affinity, G protein activation, and receptor desensitization.
Activation Mechanism
Like their class A counterparts (Manglik and Kobilka, 2014), class C GPCRs exist in a conformational equilibrium between resting and active states in the absence of ligands (Kniazeff et al., 2011; Pin and Bettler, 2016). Agonists drive the equilibrium toward active states, while antagonists enhance the inactive states. The ensemble of conformations can be further stabilized by various allosteric modulators. Activation is associated with conformational transitions within the receptor structure (Fig. 3). In this section, we discuss the similarities and differences in the conformational states of various class C GPCRs.
For all class C GPCRs, the first step of receptor activation involves agonist-triggered closure of the extracellular VFT module. For GABAB receptor, this event only occurs in the ligand-binding subunit, GABAB1 (Geng et al., 2013). In the case of mGlu receptors, closure of one protomer is sufficient to induce the active conformation, although full activation requires both VFT modules of the homodimer to be occupied and closed (Kniazeff et al., 2004; Kunishima et al., 2000; Tsuchiya et al., 2002). The significance of dual-VFT closure on mGlu receptor activation is further demonstrated in mGlu2-mGlu4 heterodimers, where mutations that prevent agonist binding to either subunit only result in partial activation (Moreno Delgado et al., 2017). Structural evidence also suggests that the active state of CaS receptor homodimer (Geng et al., 2016; Zhang et al., 2016) and TAS1R2-TAS1R3 heterodimer (Nuemket et al., 2017) involve both their protomers in the closed conformation.
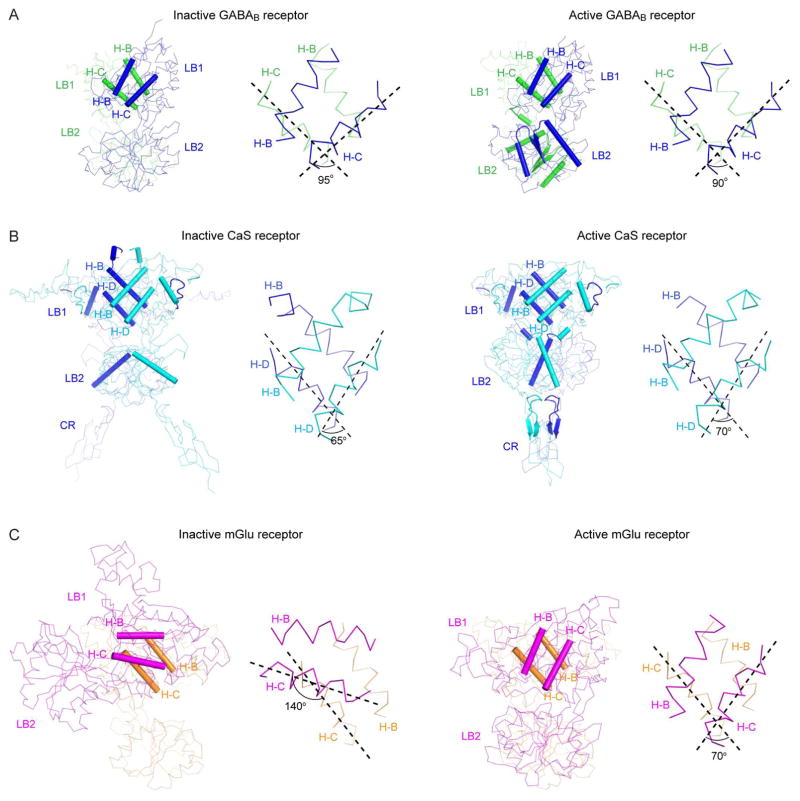
The known crystal structures of class C GPCR VFTs do not include attached TM domains, therefore current conformational data may not accurately reflect the dynamics of extracellular domains in full-length receptors. Nevertheless, compelling mechanisms of signal transduction can still be postulated. Structural comparison indicates that GABAB and CaS receptors share a common agonist-dependent activation mechanism, although the design of CaS receptor is typically homodimeric (Geng et al., 2013; Geng et al., 2016). First, the extracellular portions of both receptors dimerize through their N-terminal LB1 domains (Fig. 5A, B). The LB1-LB1 dimer interface remains relatively static throughout the activation process since agonist binding only induces a small 5°-rotation in the orientation of this interface (Fig. 5A, B). The near-constant angle between LB1 domains in GABAB receptor heterodimer is further evidenced in recent FRET studies of full-length receptor, where a negligible change in distance between the N-termini of GABAB1 and GABAB2 was detected (Lecat-Guillet et al., 2017; Scholler et al., 2017). These observations suggest that the functional role of LB1-LB1 interaction in GABAB and CaS receptors is to facilitate dimerization between receptor subunits.
Fig. 5.
Dimer Interface.
(A, B) Cα traces of inactive (A; PDB code: 4MQE) and active (B; PDB code: 4MS4) GABAB receptor VFT structures.
(C, D) Cα traces of inactive (C; PDB code: 5K5T) and active (D; PDB code: 5K5S) CaS receptor extracellular domain structures.
(E, F) Cα traces of inactive (E; PDB code: 1EWT) and active (F; PDB code: 1EWK) mGlu1 VFT structures.
For each structure, the structural elements at the dimer interface is highlighted by cartoons. The right panel shows a detailed view of the LB1-LB1 dimer interface; the angle between a pair of helices at this interface is marked.
Second, receptor activation is associated with a wider range of movements between the membrane proximal domains. In the GABAB receptor heterodimer, the LB2 domains of both subunits approach each other until a critical heterodimer interface forms (Fig. 5A). Functional analysis indicates that development of the LB2-LB2 interface is both necessary and sufficient to initiate G protein coupling (Geng et al., 2013; Rondard et al., 2008). CaS receptor enters a similar arrangement, but its interface is more extensive (Fig. 5B). CaS receptor possesses a cysteine-rich (CR) domain immediately below the LB2 domain, as do all class C GPCRs except GABAB receptor (Kniazeff et al., 2011; Pin and Bettler, 2016). In the active state, the LB2 domain of each CaS receptor protomer forms contacts with residues on the LB1, LB2, and CR of the other protomer. Contraction of LB2 domains toward each other causes the CR domains to dimerize as well. The CR domain mediates signal transduction from the VFT to the TM domain, and the CR-CR interface is mandatory for CaS receptor activation (Hauache et al., 2000; Hu et al., 2000).
The mGlu group of receptors consists of eight subtypes, and has been the best-studied of all class C GPCRs (Niswender and Conn, 2010; Pin and Acher, 2002). Extensive knowledge of mGlu receptor conformational dynamics have been obtained through x-ray crystallography and FRET spectroscopy (Christopher et al., 2015; Dore et al., 2014; Doumazane et al., 2013; Hlavackova et al., 2012; Huang et al., 2011; Kunishima et al., 2000; Levitz et al., 2016; Marcaggi et al., 2009; Muto et al., 2007; Olofsson et al., 2014; Scholler et al., 2017; Tateyama et al., 2004; Tsuchiya et al., 2002; Vafabakhsh et al., 2015; Wu et al., 2014; Xue et al., 2015). The mGlu receptors differ from GABAB and CaS receptors in the role of LB1-LB1 interaction during receptor activation. Crystallographic analysis finds that transition between the resting and active configurations causes a 70°-rotation in the LB1-LB1 homodimer interface of mGlu receptors (Kunishima et al., 2000; Muto et al., 2007; Tsuchiya et al., 2002) (Fig. 5C). This large relative movement of mGlu LB1 domains is independently detected in FRET studies where a donor or acceptor fluorophore is tagged onto the N-terminal end of each protomer (Doumazane et al., 2013; Levitz et al., 2016; Olofsson et al., 2014; Scholler et al., 2017; Vafabakhsh et al., 2015). The inactive state produces a high FRET level, while the active state shows low FRET efficiency. These findings indicate an increase in the distance between the N-termini of protomers upon receptor activation.
The contrast in LB1-LB1 movement of mGlu and GABAB receptors may be explained by the different properties of their dimer interfaces. The mGlu LB1 domains are largely joined together by hydrophobic contacts (Kunishima et al., 2000). GABAB receptor, however, employs hydrogen bonds in addition to nonpolar contacts (Geng et al., 2013). The combination of ionic and hydrophobic interactions may have limited reorientation of the LB1-LB1 interface in GABAB receptor. On the other hand, the LB1-LB1 interface of CaS receptor may be locked through additional interaction that is unique to this receptor. In addition to the interface formed by two central LB1 helices, as in mGlu and GABAB receptors, the LB1-LB1 homodimer of CaS receptor is held in place by an “embrace” extended from an arm-like long loop of each subunit to its partner (Geng et al., 2016).
Although the LB2 domains of mGlu receptors have not been shown to form an extensive interface during activation, they do draw closer to each other, as demonstrated in crystal structures (Kunishima et al., 2000; Muto et al., 2007; Tsuchiya et al., 2002) and by FRET analysis (Doumazane et al., 2013; Vafabakhsh et al., 2015) (Fig. 5C). Electrostatic interactions between the LB2 domains help stabilize the active conformation (Levitz et al., 2016; Vafabakhsh et al., 2015). Furthermore, the potentially repulsive interaction between an acidic patch on the surface of this domain is alleviated by metal cation in the active state (Tsuchiya et al., 2002). Finally, similar to CaS receptor, a precise association between the CR domains of mGlu receptors has been shown by disulfide crosslinking experiments to lead to full receptor activation (Huang et al., 2011).
The conformational phases of class C TM domains are still at the frontier of research. Our current knowledge is mostly drawn from crystal structures of mGlu1 and mGlu5 TM domains in the inactive state (Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014), as well as FRET analysis on GABAB and mGlu receptors (Hlavackova et al., 2012; Lecat-Guillet et al., 2017; Marcaggi et al., 2009; Matsushita et al., 2010; Tateyama et al., 2004; Xue et al., 2015). Like their class A counterparts, the TM domains of class C GPCRs consist of a seven-helix bundle. Despite many similarities, one major distinction is observed between the solved TM structures of class A and C. The extracellular opening of class C TM domains possesses a narrower ligand-binding cavity than is found in the class A receptors (Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014). This discrepancy stems from an inward shift of TM5 and TM7 helices compared to those in class A structures (Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014). In general, class C TM domains have extracellular regions that lack congruency with class A, as opposed to their intracellular ends which more closely match class A when superimposed (Dore et al., 2014). This may be related to the structural constraints imposed by conserved G protein coupling on the intracellular portion (Dore et al., 2014).
The class C GPCRs also possess variants of the structural motifs found in class A receptors that regulate receptor function. One of the conserved motifs is the ionic lock (Hofmann et al., 2009; Rosenbaum et al., 2009). In both receptor families, it involves a salt bridge between a pair of basic and acidic residues that tethers TM3 and TM6 at the intracellular end (Dore et al., 2014; Hofmann et al., 2009; Rosenbaum et al., 2009; Wu et al., 2014). The ionic lock maintains the inactive conformation of the TM domain by preventing outward movement of TM6 to expose the G protein-binding site at the intracellular end. The interaction in class A receptors is formed by Arg of the D/ERY motif in TM3 with an acidic residue in TM6 (Hofmann et al., 2009; Rosenbaum et al., 2009). In class C receptors, the salt bridge is observed between a Lys residue in TM3 and a Glu or Asp residue in TM6 (Binet et al., 2007; Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014). The ionic lock is further secured by a hydrogen bond between the Lys residue and a Ser residue from intracellular loop 1 (Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014).
A second conserved motif in class A GPCRs is the NPxxY sequence found at the juncture of TM7 and TM8 (Hofmann et al., 2009; Rosenbaum et al., 2009). The conserved Tyr residue in this motif undergoes conformational change to fill the gap generated by the lateral movement of TM6 during activation (Hofmann et al., 2009; Rosenbaum et al., 2009). The class C receptors feature an analogous motif, F/Y/HxPKxY, at the intracellular end of TM7 (Dore et al., 2014). The conserved Lys and Phe residues in the class C sequence are located at positions equivalent to Tyr in class A motif, and may play a similar role in stabilizing the active conformation (Dore et al., 2014).
A third motif involves the toggle switch and consists of the conserved FxxCWxP sequence in TM6 of class A GPCRs (Hofmann et al., 2009; Rosenbaum et al., 2009). Rotamerization of Trp in combination with a kink caused by Pro facilitates the outward movement of TM6 upon activation (Hofmann et al., 2009; Rosenbaum et al., 2009). A conserved Trp residue is found at the corresponding position in class C receptors, and lines the allosteric site in mGlu1 and mGlu5 TM structures (Christopher et al., 2015; Dore et al., 2014; Wu et al., 2014). The conformation of this Trp residue is a determining factor of the size and shape of the allosteric pocket (Christopher et al., 2015; Dore et al., 2014).
During activation of class C GPCRs, the contraction of extracellular structure would force the TM domains to rearrange within the dimer. FRET measurements reveal a shift in the orientation and distance between TM domains in both GABAB and mGlu receptors by tracking the intersubunit movement between intracellular loops (Hlavackova et al., 2012; Marcaggi et al., 2009; Matsushita et al., 2010; Tateyama et al., 2004) and between extracellular loops (Lecat-Guillet et al., 2017). In addition, a recent study uses a combination of disulfide crosslinking and FRET techniques to determine the location of dimer interfaces within the mGlu receptor TM domain (Xue et al., 2015). It shows that covalently crosslinking TM4 and TM5 together results in a non-functioning receptor, suggesting that TM4 and TM5 form a dimer interface in the inactive state. On the other hand, a receptor with a permanently attached TM6 pair displays constitutive activity, indicating that TM6 mediates dimer interaction in the active state.
Taken together, a common theme of class C GPCR activation involves agonist-induced closure of extracellular VFT followed by union of membrane-proximal domains. These changes are accompanied by a decrease in separation between the C-terminal ends of the extracellular domains, which subsequently modify the relationship between the TM regions for receptor activation. In addition, an intermediate state has been detected in mGlu receptor by FRET, and likely involves one agonist-bound VFT (Vafabakhsh et al., 2015). It remains to be seen whether this is a universal feature of class C GPCRs.
Conclusion
The recent structures of GABAB receptor components combined with extensive functional studies have improved our understanding of the relationship between ligand binding and conformational changes in the heterodimeric complex. To visualize how these events are eventually translated into transmembrane signaling, we will ultimately need three-dimensional structures of the full-length receptor in multiple conformational states, and in complex with auxiliary subunits and downstream signaling molecules. The insights obtained from these studies will aid the design of valuable therapeutic agents for GABAB receptor-related neurological diseases.
Highlights.
Review of current knowledge of GABAB receptor structure
Analysis of ligand-binding states and conformational dynamics of GABAB receptor
Comparison of GABAB receptor with related G protein-coupled receptors
Acknowledgments
We thank Jonathan Liu for critical reading of the manuscript.
Funding
This work was supported by the National Institute of Health [R01GM088454, R01GM112973] and American Heart Association [SDG0835183N, 15GRNT25420002]. QRF was an Irma Hirschl Career Scientist, Pew Scholar, McKnight Scholar and Schaefer Scholar.
Abbreviations
- GPCR
G protein-coupled receptor
- VFT
Venus flytrap
- TM
transmembrane
- PAM
positive allosteric modulator
- NAM
negative allosteric modulator
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arey BJ, Seethala R, Ma Z, Fura A, Morin J, Swartz J, Vyas V, Yang W, Dickson JK, Jr, Feyen JH. A novel calcium-sensing receptor antagonist transiently stimulates parathyroid hormone secretion in vivo. Endocrinology. 2005;146:2015–2022. doi: 10.1210/en.2004-1318. [DOI] [PubMed] [Google Scholar]
- Bai M, Trivedi S, Brown EM. Dimerization of the extracellular calcium-sensing receptor (CaR) on the cell surface of CaR-transfected HEK293 cells. J Biol Chem. 1998;273:23605–23610. doi: 10.1074/jbc.273.36.23605. [DOI] [PubMed] [Google Scholar]
- Bartoi T, Rigbolt KT, Du D, Kohr G, Blagoev B, Kornau HC. GABAB receptor constituents revealed by tandem affinity purification from transgenic mice. J Biol Chem. 2010;285:20625–20633. doi: 10.1074/jbc.M109.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettler B, Kaupmann K, Mosbacher J, Gassmann M. Molecular structure and physiological functions of GABA(B) receptors. Physiol Rev. 2004;84:835–867. doi: 10.1152/physrev.00036.2003. [DOI] [PubMed] [Google Scholar]
- Biermann B, Ivankova-Susankova K, Bradaia A, Abdel Aziz S, Besseyrias V, Kapfhammer JP, Missler M, Gassmann M, Bettler B. The Sushi domains of GABAB receptors function as axonal targeting signals. J Neurosci. 2010;30:1385–1394. doi: 10.1523/JNEUROSCI.3172-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billinton A, Upton N, Bowery NG. GABA(B) receptor isoforms GBR1a and GBR1b, appear to be associated with pre- and post-synaptic elements respectively in rat and human cerebellum. Br J Pharmacol. 1999;126:1387–1392. doi: 10.1038/sj.bjp.0702460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet V, Brajon C, Le Corre L, Acher F, Pin JP, Prezeau L. The heptahelical domain of GABA(B2) is activated directly by CGP7930, a positive allosteric modulator of the GABA(B) receptor. J Biol Chem. 2004;279:29085–29091. doi: 10.1074/jbc.M400930200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet V, Duthey B, Lecaillon J, Vol C, Quoyer J, Labesse G, Pin JP, Prezeau L. Common structural requirements for heptahelical domain function in class A and class C G protein-coupled receptors. J Biol Chem. 2007;282:12154–12163. doi: 10.1074/jbc.M611071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein S, Ginham R, Uhrin D, Smith BO, Soares DC, Veltel S, McIlhinney RA, White JH, Barlow PN. Structural analysis of the complement control protein (CCP) modules of GABA(B) receptor 1a: only one of the two CCP modules is compactly folded. J Biol Chem. 2004;279:48292–48306. doi: 10.1074/jbc.M406540200. [DOI] [PubMed] [Google Scholar]
- Bowery NG, Bettler B, Froestl W, Gallagher JP, Marshall F, Raiteri M, Bonner TI, Enna SJ. International Union of Pharmacology. XXXIII Mammalian gamma-aminobutyric acid(B) receptors: structure and function. Pharmacol Rev. 2002;54:247–264. doi: 10.1124/pr.54.2.247. [DOI] [PubMed] [Google Scholar]
- Bowery NG, Doble A, Hill DR, Hudson AL, Shaw JS, Turnbull MJ. Baclofen: a selective agonist for a novel type of GABA receptor proceedings. Br J Pharmacol. 1979;67:444P–445P. [PMC free article] [PubMed] [Google Scholar]
- Bowery NG, Doble A, Hill DR, Hudson AL, Shaw JS, Turnbull MJ, Warrington R. Bicuculline-insensitive GABA receptors on peripheral autonomic nerve terminals. Eur J Pharmacol. 1981;71:53–70. doi: 10.1016/0014-2999(81)90386-1. [DOI] [PubMed] [Google Scholar]
- Bowery NG, Hill DR, Hudson AL, Doble A, Middlemiss DN, Shaw J, Turnbull M. (−)Baclofen decreases neurotransmitter release in the mammalian CNS by an action at a novel GABA receptor. Nature. 1980;283:92–94. doi: 10.1038/283092a0. [DOI] [PubMed] [Google Scholar]
- Bowery NG, Hudson AL. gamma-Aminobutyric acid reduces the evoked release of [3H]-noradrenaline from sympathetic nerve terminals [proceedings] Br J Pharmacol. 1979;66:108P. [PMC free article] [PubMed] [Google Scholar]
- Bowery NG, Hudson AL, Price GW. GABAA and GABAB receptor site distribution in the rat central nervous system. Neuroscience. 1987;20:365–383. doi: 10.1016/0306-4522(87)90098-4. [DOI] [PubMed] [Google Scholar]
- Brauner-Osborne H, Wellendorph P, Jensen AA. Structure, pharmacology and therapeutic prospects of family C G-protein coupled receptors. Curr Drug Targets. 2007;8:169–184. doi: 10.2174/138945007779315614. [DOI] [PubMed] [Google Scholar]
- Burmakina S, Geng Y, Chen Y, Fan QR. Heterodimeric coiled-coil interactions of human GABAB receptor. Proc Natl Acad Sci U S A. 2014;111:6958–6963. doi: 10.1073/pnas.1400081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calver AR, Robbins MJ, Cosio C, Rice SQ, Babbs AJ, Hirst WD, Boyfield I, Wood MD, Russell RB, Price GW, et al. The C-terminal domains of the GABA(b) receptor subunits mediate intracellular trafficking but are not required for receptor signaling. J Neurosci. 2001;21:1203–1210. doi: 10.1523/JNEUROSCI.21-04-01203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Chen LH, Sun B, Zhang Y, Xu TJ, Xia ZX, Liu JF, Nan FJ. Discovery of a Negative Allosteric Modulator of GABAB Receptors. ACS Med Chem Lett. 2014;5:742–747. doi: 10.1021/ml500162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Goudet C, Pin JP, Conn PJ. N-{4-Chloro-2-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-hydroxybe nzamide (CPPHA) acts through a novel site as a positive allosteric modulator of group 1 metabotropic glutamate receptors. Mol Pharmacol. 2008;73:909–918. doi: 10.1124/mol.107.040097. [DOI] [PubMed] [Google Scholar]
- Christopher JA, Aves SJ, Bennett KA, Dore AS, Errey JC, Jazayeri A, Marshall FH, Okrasa K, Serrano-Vega MJ, Tehan BG, et al. Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile) J Med Chem. 2015;58:6653–6664. doi: 10.1021/acs.jmedchem.5b00892. [DOI] [PubMed] [Google Scholar]
- Christopoulos A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nature reviews Drug discovery. 2002;1:198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- Chu DC, Albin RL, Young AB, Penney JB. Distribution and kinetics of GABAB binding sites in rat central nervous system: a quantitative autoradiographic study. Neuroscience. 1990;34:341–357. doi: 10.1016/0306-4522(90)90144-s. [DOI] [PubMed] [Google Scholar]
- Conigrave AD, Hampson DR. Broad-spectrum amino acid-sensing class C G-protein coupled receptors: molecular mechanisms, physiological significance and options for drug development. Pharmacol Ther. 2010;127:252–260. doi: 10.1016/j.pharmthera.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Conigrave AD, Ward DT. Calcium-sensing receptor (CaSR): pharmacological properties and signaling pathways. Best Pract Res Clin Endocrinol Metab. 2013;27:315–331. doi: 10.1016/j.beem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nature reviews Drug discovery. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couve A, Filippov AK, Connolly CN, Bettler B, Brown DA, Moss SJ. Intracellular retention of recombinant GABAB receptors. J Biol Chem. 1998;273:26361–26367. doi: 10.1074/jbc.273.41.26361. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Kelly PH, Chaperon F, Gentsch C, Mombereau C, Lingenhoehl K, Froestl W, Bettler B, Kaupmann K, Spooren WP. Behavioral characterization of the novel GABAB receptor-positive modulator GS39783 (N,N′-dicyclopentyl-2-methylsulfanyl-5-nitro-pyrimidine-4,6-diamine): anxiolytic-like activity without side effects associated with baclofen or benzodiazepines. J Pharmacol Exp Ther. 2004;310:952–963. doi: 10.1124/jpet.104.066753. [DOI] [PubMed] [Google Scholar]
- Dalvi A, Rodgers RJ. GABAergic influences on plus-maze behaviour in mice. Psychopharmacology (Berl) 1996;128:380–397. doi: 10.1007/s002130050148. [DOI] [PubMed] [Google Scholar]
- Doly S, Shirvani H, Gata G, Meye FJ, Emerit MB, Enslen H, Achour L, Pardo-Lopez L, Yang SK, Armand V, et al. GABAB receptor cell-surface export is controlled by an endoplasmic reticulum gatekeeper. Mol Psychiatry. 2016;21:480–490. doi: 10.1038/mp.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. doi: 10.1038/nature13396. [DOI] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Fabre L, Zwier JM, Trinquet E, Pin JP, Rondard P. Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2013;110:E1416–1425. doi: 10.1073/pnas.1215615110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Zwier JM, Trinquet E, Rondard P, Pin JP. A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 2011;25:66–77. doi: 10.1096/fj.10-163147. [DOI] [PubMed] [Google Scholar]
- Dupuis DS, Relkovic D, Lhuillier L, Mosbacher J, Kaupmann K. Point mutations in the transmembrane region of GABAB2 facilitate activation by the positive modulator N,N′-dicyclopentyl-2-methylsulfanyl-5-nitro-pyrimidine-4,6-diamine (GS39783) in the absence of the GABAB1 subunit. Mol Pharmacol. 2006;70:2027–2036. doi: 10.1124/mol.106.028183. [DOI] [PubMed] [Google Scholar]
- Duthey B, Caudron S, Perroy J, Bettler B, Fagni L, Pin JP, Prezeau L. A single subunit (GB2) is required for G-protein activation by the heterodimeric GABA(B) receptor. J Biol Chem. 2002;277:3236–3241. doi: 10.1074/jbc.M108900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Moustaine D, Granier S, Doumazane E, Scholler P, Rahmeh R, Bron P, Mouillac B, Baneres JL, Rondard P, Pin JP. Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc Natl Acad Sci U S A. 2012;109:16342–16347. doi: 10.1073/pnas.1205838109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engers DW, Lindsley CW. Allosteric modulation of Class C GPCRs: a novel approach for the treatment of CNS disorders. Drug discovery today Technologies. 2013;10:e269–276. doi: 10.1016/j.ddtec.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filip M, Frankowska M. Effects of GABA(B) receptor agents on cocaine priming, discrete contextual cue and food induced relapses. Eur J Pharmacol. 2007;571:166–173. doi: 10.1016/j.ejphar.2007.05.069. [DOI] [PubMed] [Google Scholar]
- Froestl W. Chemistry and pharmacology of GABAB receptor ligands. Adv Pharmacol. 2010;58:19–62. doi: 10.1016/S1054-3589(10)58002-5. [DOI] [PubMed] [Google Scholar]
- Galvez T, Duthey B, Kniazeff J, Blahos J, Rovelli G, Bettler B, Prezeau L, Pin JP. Allosteric interactions between GB1 and GB2 subunits are required for optimal GABA(B) receptor function. EMBO J. 2001;20:2152–2159. doi: 10.1093/emboj/20.9.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez T, Urwyler S, Prezeau L, Mosbacher J, Joly C, Malitschek B, Heid J, Brabet I, Froestl W, Bettler B, et al. Ca(2+) requirement for high-affinity gamma-aminobutyric acid (GABA) binding at GABA(B) receptors: involvement of serine 269 of the GABA(B)R1 subunit. Mol Pharmacol. 2000;57:419–426. doi: 10.1124/mol.57.3.419. [DOI] [PubMed] [Google Scholar]
- Gama L, Wilt SG, Breitwieser GE. Heterodimerization of calcium sensing receptors with metabotropic glutamate receptors in neurons. J Biol Chem. 2001;276:39053–39059. doi: 10.1074/jbc.M105662200. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Bettler B. Regulation of neuronal GABA(B) receptor functions by subunit composition. Nat Rev Neurosci. 2012;13:380–394. doi: 10.1038/nrn3249. [DOI] [PubMed] [Google Scholar]
- Geng Y, Bush M, Mosyak L, Wang F, Fan QR. Structural mechanism of ligand activation in human GABA(B) receptor. Nature. 2013;504:254–259. doi: 10.1038/nature12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Mosyak L, Kurinov I, Zuo H, Sturchler E, Cheng TC, Subramanyam P, Brown AP, Brennan SC, Mun HC, et al. Structural mechanism of ligand activation in human calcium-sensing receptor. Elife. 2016:5. doi: 10.7554/eLife.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Xiong D, Mosyak L, Malito DL, Kniazeff J, Chen Y, Burmakina S, Quick M, Bush M, Javitch JA, et al. Structure and functional interaction of the extracellular domain of human GABA(B) receptor GBR2. Nat Neurosci. 2012;15:970–978. doi: 10.1038/nn.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Maeso J, Ang RL, Yuen T, Chan P, Weisstaub NV, Lopez-Gimenez JF, Zhou M, Okawa Y, Callado LF, Milligan G, et al. Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature. 2008;452:93–97. doi: 10.1038/nature06612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Dong EN, Meiler J, Conn PJ. Allosteric modulation of metabotropic glutamate receptors: structural insights and therapeutic potential. Neuropharmacology. 2011;60:66–81. doi: 10.1016/j.neuropharm.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guery S, Floersheim P, Kaupmann K, Froestl W. Syntheses and optimization of new GS39783 analogues as positive allosteric modulators of GABAB receptors. Bioorg Med Chem Lett. 2007;17:6206–6211. doi: 10.1016/j.bmcl.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpsoe K, Boesgaard MW, Munk C, Brauner-Osborne H, Gloriam DE. Structural insight to mutation effects uncover a common allosteric site in class C GPCRs. Bioinformatics. 2017;33:1116–1120. doi: 10.1093/bioinformatics/btw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauache OM, Hu J, Ray K, Spiegel AM. Functional interactions between the extracellular domain and the seven-transmembrane domain in Ca2+ receptor activation. Endocrine. 2000;13:63–70. doi: 10.1385/ENDO:13:1:63. [DOI] [PubMed] [Google Scholar]
- Havlickova M, Prezeau L, Duthey B, Bettler B, Pin JP, Blahos J. The intracellular loops of the GB2 subunit are crucial for G-protein coupling of the heteromeric gamma-aminobutyrate B receptor. Mol Pharmacol. 2002;62:343–350. doi: 10.1124/mol.62.2.343. [DOI] [PubMed] [Google Scholar]
- Hawrot E, Xiao Y, Shi QL, Norman D, Kirkitadze M, Barlow PN. Demonstration of a tandem pair of complement protein modules in GABA(B) receptor 1a. FEBS Lett. 1998;432:103–108. doi: 10.1016/s0014-5793(98)00794-7. [DOI] [PubMed] [Google Scholar]
- Hill DR, Bowery NG. 3H-baclofen and 3H-GABA bind to bicuculline-insensitive GABAB sites in rat brain. Nature. 1981;290:149–152. doi: 10.1038/290149a0. [DOI] [PubMed] [Google Scholar]
- Hlavackova V, Zabel U, Frankova D, Batz J, Hoffmann C, Prezeau L, Pin JP, Blahos J, Lohse MJ. Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Science signaling. 2012;5:ra59. doi: 10.1126/scisignal.2002720. [DOI] [PubMed] [Google Scholar]
- Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, Ernst OP. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Hu J, Hauache O, Spiegel AM. Human Ca2+ receptor cysteine-rich domain. Analysis of function of mutant and chimeric receptors. J Biol Chem. 2000;275:16382–16389. doi: 10.1074/jbc.M000277200. [DOI] [PubMed] [Google Scholar]
- Hu J, Jiang J, Costanzi S, Thomas C, Yang W, Feyen JH, Jacobson KA, Spiegel AM. A missense mutation in the seven-transmembrane domain of the human Ca2+ receptor converts a negative allosteric modulator into a positive allosteric modulator. J Biol Chem. 2006;281:21558–21565. doi: 10.1074/jbc.M603682200. [DOI] [PubMed] [Google Scholar]
- Huang S, Cao J, Jiang M, Labesse G, Liu J, Pin JP, Rondard P. Interdomain movements in metabotropic glutamate receptor activation. Proc Natl Acad Sci U S A. 2011;108:15480–15485. doi: 10.1073/pnas.1107775108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AA, Brauner-Osborne H. Allosteric modulation of the calcium-sensing receptor. Curr Neuropharmacol. 2007;5:180–186. doi: 10.2174/157015907781695982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, Yao WJ, Johnson M, Gunwaldsen C, Huang LY, et al. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature. 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupmann K, Huggel K, Heid J, Flor PJ, Bischoff S, Mickel SJ, McMaster G, Angst C, Bittiger H, Froestl W, et al. Expression cloning of GABA(B) receptors uncovers similarity to metabotropic glutamate receptors. Nature. 1997;386:239–246. doi: 10.1038/386239a0. [DOI] [PubMed] [Google Scholar]
- Kaupmann K, Malitschek B, Schuler V, Heid J, Froestl W, Beck P, Mosbacher J, Bischoff S, Kulik A, Shigemoto R, et al. GABA(B)-receptor subtypes assemble into functional heteromeric complexes. Nature. 1998;396:683–687. doi: 10.1038/25360. [DOI] [PubMed] [Google Scholar]
- Kniazeff J, Bessis AS, Maurel D, Ansanay H, Prezeau L, Pin JP. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nature Structural and Molecular Biology. 2004;11:706–713. doi: 10.1038/nsmb794. [DOI] [PubMed] [Google Scholar]
- Kniazeff J, Galvez T, Labesse G, Pin JP. No ligand binding in the GB2 subunit of the GABA(B) receptor is required for activation and allosteric interaction between the subunits. J Neurosci. 2002;22:7352–7361. doi: 10.1523/JNEUROSCI.22-17-07352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeff J, Prezeau L, Rondard P, Pin JP, Goudet C. Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol Ther. 2011;130:9–25. doi: 10.1016/j.pharmthera.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner R, Kohr G, Grunewald S, Eisenhardt G, Bach A, Kornau HC. Role of heteromer formation in GABAB receptor function. Science. 1999;283:74–77. doi: 10.1126/science.283.5398.74. [DOI] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nature reviews Drug discovery. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- Lecat-Guillet N, Monnier C, Rovira X, Kniazeff J, Lamarque L, Zwier JM, Trinquet E, Pin JP, Rondard P. FRET-Based Sensors Unravel Activation and Allosteric Modulation of the GABAB Receptor. Cell Chem Biol. 2017;24:360–370. doi: 10.1016/j.chembiol.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Levitz J, Habrian C, Bharill S, Fu Z, Vafabakhsh R, Isacoff EY. Mechanism of Assembly and Cooperativity of Homomeric and Heteromeric Metabotropic Glutamate Receptors. Neuron. 2016;92:143–159. doi: 10.1016/j.neuron.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhuillier L, Mombereau C, Cryan JF, Kaupmann K. GABA(B) receptor-positive modulation decreases selective molecular and behavioral effects of cocaine. Neuropsychopharmacology. 2007;32:388–398. doi: 10.1038/sj.npp.1301102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Maurel D, Etzol S, Brabet I, Ansanay H, Pin JP, Rondard P. Molecular determinants involved in the allosteric control of agonist affinity in the GABAB receptor by the GABAB2 subunit. J Biol Chem. 2004;279:15824–15830. doi: 10.1074/jbc.M313639200. [DOI] [PubMed] [Google Scholar]
- Macdonald RL, Olsen RW. GABAA receptor channels. Annu Rev Neurosci. 1994;17:569–602. doi: 10.1146/annurev.ne.17.030194.003033. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Masciadri R, Norcross RD, Knoflach F, Kratzeisen C, Zenner MT, Kolb Y, Marcuz A, Huwyler J, Nakagawa T, et al. Characterization of (R,S)-5,7-di-tert-butyl-3-hydroxy-3-trifluoromethyl-3H-benzofuran-2-one as a positive allosteric modulator of GABAB receptors. Br J Pharmacol. 2008;154:797–811. doi: 10.1038/bjp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malitschek B, Schweizer C, Keir M, Heid J, Froestl W, Mosbacher J, Kuhn R, Henley J, Joly C, Pin JP, et al. The N-terminal domain of gamma-aminobutyric Acid(B) receptors is sufficient to specify agonist and antagonist binding. Mol Pharmacol. 1999;56:448–454. doi: 10.1124/mol.56.2.448. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kobilka B. The role of protein dynamics in GPCR function: insights from the beta2AR and rhodopsin. Curr Opin Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcaggi P, Mutoh H, Dimitrov D, Beato M, Knopfel T. Optical measurement of mGluR1 conformational changes reveals fast activation, slow deactivation, and sensitization. Proc Natl Acad Sci U S A. 2009;106:11388–11393. doi: 10.1073/pnas.0901290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY. Function of GB1 and GB2 subunits in G protein coupling of GABA(B) receptors. Proc Natl Acad Sci U S A. 2001;98:14649–14654. doi: 10.1073/pnas.251554498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SC, Russek SJ, Farb DH. Molecular identification of the human GABABR2: cell surface expression and coupling to adenylyl cyclase in the absence of GABABR1. Mol Cell Neurosci. 1999;13:180–191. doi: 10.1006/mcne.1999.0741. [DOI] [PubMed] [Google Scholar]
- Matsushita S, Nakata H, Kubo Y, Tateyama M. Ligand-induced rearrangements of the GABA(B) receptor revealed by fluorescence resonance energy transfer. J Biol Chem. 2010;285:10291–10299. doi: 10.1074/jbc.M109.077990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombereau C, Kaupmann K, Froestl W, Sansig G, van der Putten H, Cryan JF. Genetic and pharmacological evidence of a role for GABA(B) receptors in the modulation of anxiety- and antidepressant-like behavior. Neuropsychopharmacology. 2004;29:1050–1062. doi: 10.1038/sj.npp.1300413. [DOI] [PubMed] [Google Scholar]
- Monnier C, Tu H, Bourrier E, Vol C, Lamarque L, Trinquet E, Pin JP, Rondard P. Trans-activation between 7TM domains: implication in heterodimeric GABA(B) receptor activation. EMBO J. 2011;30:32–42. doi: 10.1038/emboj.2010.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Delgado D, Moller TC, Ster J, Giraldo J, Maurel D, Rovira X, Scholler P, Zwier JM, Perroy J, Durroux T, et al. Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells. Elife. 2017:6. doi: 10.7554/eLife.25233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Tsuchiya D, Morikawa K, Jingami H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2007;104:3759–3764. doi: 10.1073/pnas.0611577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Ng GY, Clark J, Coulombe N, Ethier N, Hebert TE, Sullivan R, Kargman S, Chateauneuf A, Tsukamoto N, McDonald T, et al. Identification of a GABAB receptor subunit, gb2, required for functional GABAB receptor activity. J Biol Chem. 1999;274:7607–7610. doi: 10.1074/jbc.274.12.7607. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura R, Suzuki Y, Kakizuka A, Jingami H. Direct detection of the interaction between recombinant soluble extracellular regions in the heterodimeric metabotropic gamma-aminobutyric acid receptor. J Biol Chem. 2008;283:4665–4673. doi: 10.1074/jbc.M705202200. [DOI] [PubMed] [Google Scholar]
- Nuemket N, Yasui N, Kusakabe Y, Nomura Y, Atsumi N, Akiyama S, Nango E, Kato Y, Kaneko MK, Takagi J, et al. Structural basis for perception of diverse chemical substances by T1r taste receptors. Nat Commun. 2017;8:15530. doi: 10.1038/ncomms15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara PJ, Sheppard PO, Thogersen H, Venezia D, Haldeman BA, McGrane V, Houamed KM, Thomsen C, Gilbert TL, Mulvihill ER. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Sekiyama N, Otsu M, Shimada Y, Sato A, Nakanishi S, Jingami H. Expression and purification of the extracellular ligand binding region of metabotropic glutamate receptor subtype 1. J Biol Chem. 1998;273:13089–13096. doi: 10.1074/jbc.273.21.13089. [DOI] [PubMed] [Google Scholar]
- Olofsson L, Felekyan S, Doumazane E, Scholler P, Fabre L, Zwier JM, Rondard P, Seidel CA, Pin JP, Margeat E. Fine tuning of sub-millisecond conformational dynamics controls metabotropic glutamate receptors agonist efficacy. Nat Commun. 2014;5:5206. doi: 10.1038/ncomms6206. [DOI] [PubMed] [Google Scholar]
- Pagano A, Rovelli G, Mosbacher J, Lohmann T, Duthey B, Stauffer D, Ristig D, Schuler V, Meigel I, Lampert C, et al. C-terminal interaction is essential for surface trafficking but not for heteromeric assembly of GABA(b) receptors. J Neurosci. 2001;21:1189–1202. doi: 10.1523/JNEUROSCI.21-04-01189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya NJ, Klaassen RV, van der Schors RC, Slotman JA, Houtsmuller A, Smit AB, Li KW. Group 1 metabotropic glutamate receptors 1 and 5 form a protein complex in mouse hippocampus and cortex. Proteomics. 2016;16:2698–2705. doi: 10.1002/pmic.201500400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidasheva S, Grant M, Canaff L, Ercan O, Kumar U, Hendy GN. Calcium-sensing receptor dimerizes in the endoplasmic reticulum: biochemical and biophysical characterization of CASR mutants retained intracellularly. Hum Mol Genet. 2006;15:2200–2209. doi: 10.1093/hmg/ddl145. [DOI] [PubMed] [Google Scholar]
- Pin JP, Acher F. The metabotropic glutamate receptors: structure, activation mechanism and pharmacology. Curr Drug Targets CNS Neurol Disord. 2002;1:297–317. doi: 10.2174/1568007023339328. [DOI] [PubMed] [Google Scholar]
- Pin JP, Bettler B. Organization and functions of mGlu and GABAB receptor complexes. Nature. 2016;540:60–68. doi: 10.1038/nature20566. [DOI] [PubMed] [Google Scholar]
- Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003;98:325–354. doi: 10.1016/s0163-7258(03)00038-x. [DOI] [PubMed] [Google Scholar]
- Pin JP, Kniazeff J, Binet V, Liu J, Maurel D, Galvez T, Duthey B, Havlickova M, Blahos J, Prezeau L, et al. Activation mechanism of the heterodimeric GABA(B) receptor. Biochem Pharmacol. 2004a;68:1565–1572. doi: 10.1016/j.bcp.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Pin JP, Kniazeff J, Goudet C, Bessis AS, Liu J, Galvez T, Acher F, Rondard P, Prezeau L. The activation mechanism of class-C G-protein coupled receptors. Biol Cell. 2004b;96:335–342. doi: 10.1016/j.biolcel.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Pin JP, Prezeau L. Allosteric modulators of GABA(B) receptors: mechanism of action and therapeutic perspective. Curr Neuropharmacol. 2007;5:195–201. doi: 10.2174/157015907781695919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalu M, Fritzius T, Adelfinger L, Jacquier V, Besseyrias V, Gassmann M, Bettler B. Pharmacological characterization of GABAB receptor subtypes assembled with auxiliary KCTD subunits. Neuropharmacology. 2015;88:145–154. doi: 10.1016/j.neuropharm.2014.08.020. [DOI] [PubMed] [Google Scholar]
- Ray K, Hauschild BC, Steinbach PJ, Goldsmith PK, Hauache O, Spiegel AM. Identification of the cysteine residues in the amino-terminal extracellular domain of the human Ca(2+) receptor critical for dimerization. Implications for function of monomeric Ca(2+) receptor. J Biol Chem. 1999;274:27642–27650. doi: 10.1074/jbc.274.39.27642. [DOI] [PubMed] [Google Scholar]
- Restituito S, Couve A, Bawagan H, Jourdain S, Pangalos MN, Calver AR, Freeman KB, Moss SJ. Multiple motifs regulate the trafficking of GABA(B) receptors at distinct checkpoints within the secretory pathway. Mol Cell Neurosci. 2005;28:747–756. doi: 10.1016/j.mcn.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Robbins MJ, Calver AR, Filippov AK, Hirst WD, Russell RB, Wood MD, Nasir S, Couve A, Brown DA, Moss SJ, et al. GABA(B2) is essential for g-protein coupling of the GABA(B) receptor heterodimer. J Neurosci. 2001;21:8043–8052. doi: 10.1523/JNEUROSCI.21-20-08043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C, Yang WL, O’Malley KL. Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J Biol Chem. 1996;271:28612–28616. doi: 10.1074/jbc.271.45.28612. [DOI] [PubMed] [Google Scholar]
- Rondard P, Huang S, Monnier C, Tu H, Blanchard B, Oueslati N, Malhaire F, Li Y, Trinquet E, Labesse G, et al. Functioning of the dimeric GABA(B) receptor extracellular domain revealed by glycan wedge scanning. EMBO J. 2008;27:1321–1332. doi: 10.1038/emboj.2008.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack JS, Saper MA, Quiocho FA. Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. J Mol Biol. 1989;206:171–191. doi: 10.1016/0022-2836(89)90531-7. [DOI] [PubMed] [Google Scholar]
- Scholler P, Moreno-Delgado D, Lecat-Guillet N, Doumazane E, Monnier C, Charrier-Savournin F, Fabre L, Chouvet C, Soldevila S, Lamarque L, et al. HTS-compatible FRET-based conformational sensors clarify membrane receptor activation. Nat Chem Biol. 2017;13:372–380. doi: 10.1038/nchembio.2286. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Metz M, Zolles G, Turecek R, Fritzius T, Bildl W, Tarusawa E, Kulik A, Unger A, Ivankova K, et al. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465:231–235. doi: 10.1038/nature08964. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Perez-Garci E, Schneider A, Kollewe A, Gauthier-Kemper A, Fritzius T, Raveh A, Dinamarca MC, Hanuschkin A, Bildl W, et al. Modular composition and dynamics of native GABAB receptors identified by high-resolution proteomics. Nat Neurosci. 2016;19:233–242. doi: 10.1038/nn.4198. [DOI] [PubMed] [Google Scholar]
- Seddik R, Jungblut SP, Silander OK, Rajalu M, Fritzius T, Besseyrias V, Jacquier V, Fakler B, Gassmann M, Bettler B. Opposite effects of KCTD subunit domains on GABA(B) receptor-mediated desensitization. J Biol Chem. 2012;287:39869–39877. doi: 10.1074/jbc.M112.412767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery DA, Markou A, Froestl W, Cryan JF. The GABAB receptor-positive modulator GS39783 and the GABAB receptor agonist baclofen attenuate the reward-facilitating effects of cocaine: intracranial self-stimulation studies in the rat. Neuropsychopharmacology. 2005;30:2065–2072. doi: 10.1038/sj.npp.1300734. [DOI] [PubMed] [Google Scholar]
- Smith MA, Yancey DL, Morgan D, Liu Y, Froestl W, Roberts DC. Effects of positive allosteric modulators of the GABAB receptor on cocaine self-administration in rats. Psychopharmacology (Berl) 2004;173:105–111. doi: 10.1007/s00213-003-1706-5. [DOI] [PubMed] [Google Scholar]
- Sturchler E, Li X, de Lourdes Ladino M, Kaczanowska K, Cameron M, Griffin PR, Finn MG, Markou A, McDonald P. GABAB receptor allosteric modulators exhibit pathway-dependent and species-selective activity. Pharmacology research & perspectives. 2017;5:e00288. doi: 10.1002/prp2.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Chen L, Liu L, Xia Z, Pin JP, Nan F, Liu J. A negative allosteric modulator modulates GABAB-receptor signalling through GB2 subunits. Biochem J. 2016;473:779–787. doi: 10.1042/BJ20150979. [DOI] [PubMed] [Google Scholar]
- Tateyama M, Abe H, Nakata H, Saito O, Kubo Y. Ligand-induced rearrangement of the dimeric metabotropic glutamate receptor 1alpha. Nat Struct Mol Biol. 2004;11:637–642. doi: 10.1038/nsmb770. [DOI] [PubMed] [Google Scholar]
- Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K. Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+ Proc Natl Acad Sci U S A. 2002;99:2660–2665. doi: 10.1073/pnas.052708599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, Shimada Y, Takeshita T, Kajimura N, Nomura S, Sekiyama N, Otomo J, Usukura J, Nakanishi S, Jingami H. Cryptic dimer interface and domain organization of the extracellular region of metabotropic glutamate receptor subtype 1. J Biol Chem. 2000;275:28144–28151. doi: 10.1074/jbc.M003226200. [DOI] [PubMed] [Google Scholar]
- Turecek R, Schwenk J, Fritzius T, Ivankova K, Zolles G, Adelfinger L, Jacquier V, Besseyrias V, Gassmann M, Schulte U, et al. Auxiliary GABA Receptor Subunits Uncouple G Protein betagamma Subunits from Effector Channels to Induce Desensitization. Neuron. 2014 doi: 10.1016/j.neuron.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Urwyler S. Allosteric modulation of family C G-protein-coupled receptors: from molecular insights to therapeutic perspectives. Pharmacol Rev. 2011;63:59–126. doi: 10.1124/pr.109.002501. [DOI] [PubMed] [Google Scholar]
- Urwyler S, Mosbacher J, Lingenhoehl K, Heid J, Hofstetter K, Froestl W, Bettler B, Kaupmann K. Positive allosteric modulation of native and recombinant gamma-aminobutyric acid(B) receptors by 2,6-Di-tert-butyl-4-(3-hydroxy-2,2-dimethyl-propyl)-phenol (CGP7930) and its aldehyde analog CGP13501. Mol Pharmacol. 2001;60:963–971. [PubMed] [Google Scholar]
- Urwyler S, Pozza MF, Lingenhoehl K, Mosbacher J, Lampert C, Froestl W, Koller M, Kaupmann K. N,N′-Dicyclopentyl-2-methylsulfanyl-5-nitro-pyrimidine-4,6-diamine (GS39783) and structurally related compounds: novel allosteric enhancers of gamma-aminobutyric acidB receptor function. J Pharmacol Exp Ther. 2003;307:322–330. doi: 10.1124/jpet.103.053074. [DOI] [PubMed] [Google Scholar]
- Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524:497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DT, Brown EM, Harris HW. Disulfide bonds in the extracellular calcium-polyvalent cation-sensing receptor correlate with dimer formation and its response to divalent cations in vitro. J Biol Chem. 1998;273:14476–14483. doi: 10.1074/jbc.273.23.14476. [DOI] [PubMed] [Google Scholar]
- White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, Barnes AA, Emson P, Foord SM, Marshall FH. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- Wise A, Green A, Main MJ, Wilson R, Fraser N, Marshall FH. Calcium sensing properties of the GABA(B) receptor. Neuropharmacology. 1999;38:1647–1656. doi: 10.1016/s0028-3908(99)00119-7. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Rovira X, Scholler P, Zhao H, Liu J, Pin JP, Rondard P. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat Chem Biol. 2015;11:134–140. doi: 10.1038/nchembio.1711. [DOI] [PubMed] [Google Scholar]
- Yin S, Noetzel MJ, Johnson KA, Zamorano R, Jalan-Sakrikar N, Gregory KJ, Conn PJ, Niswender CM. Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J Neurosci. 2014;34:79–94. doi: 10.1523/JNEUROSCI.1129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang T, Zou J, Miller CL, Gorkhali R, Yang JY, Schilmiller A, Wang S, Huang K, Brown EM, et al. Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist. Sci Adv. 2016;2:e1600241. doi: 10.1126/sciadv.1600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sun S, Quinn SJ, Brown EM, Bai M. The extracellular calcium-sensing receptor dimerizes through multiple types of intermolecular interactions. J Biol Chem. 2001;276:5316–5322. doi: 10.1074/jbc.M005958200. [DOI] [PubMed] [Google Scholar]