Abstract
In aggressive prostate cancers, the oncoprotein STMN1 (also known as stathmin 1 and oncoprotein 18) is often overexpressed. STMN1 is involved in various cellular processes, including cell proliferation, motility, and tumor metastasis. Here, it was found that the expression of STMN1 RNA and protein is elevated in metastatic prostate cancers. Knockdown of STMN1 resulted in reduced proliferation and invasion of cells and tumor growth and metastasis in vivo. Furthermore, miR-34a downregulated STMN1 by directly binding to its 3'UTR. Overexpression of miR-34a in prostate cancer cells reduced proliferation and colony formation, suggesting that it is a tumor suppressor. The transcriptional corepressor C-terminal binding protein 1 (CtBP1) negatively regulated expression of miR-34a. Furthermore, gene expression profiling of STMN1-modulated prostate cancer cells revealed molecular alterations, including elevated expression of growth differentiation factor 15 (GDF15), which is involved in cancer progression and potentially in STMN1-mediated oncogenesis. Thus, in prostate cancer, CtBP1-regulated miR-34a modulates STMN1 expression and is involved in cancer progression through the CtBP1\miR-34a\STMN1\GDF15 axis.
Introduction
Prostate cancer is the third leading cause of death from cancer and the most prevalent cancer in men; over their lifetime, 19% of men in the United States develop prostate cancer (1). Recent advances in genomic technology have allowed molecular characterization of these cancers (2). Understanding the mechanism of action and regulation of differentially expressed genes that regulate growth and progression of metastatic prostate cancer can provide insight into the biology of the disease and offer targets for developing therapeutic interventions for this lethal disease. Despite recent increments in survival of men with metastatic, castration-resistant prostate cancer by use of taxane chemotherapy (docetaxel, cabazitaxel), androgen inhibitors (abiraterone, enzalutamide), a vaccine (sipuleucel-T), and a radiopharmaceutical (radium223), the disease remains incurable (3–8).
Our studies of expression profiling and transcriptomic sequencing using RNA from metastatic prostate cancers showed overexpression of STMN1 (also known as stathmin 1, oncoprotein 18, Op18, and metablastin), which is involved in cancer progression (9). STMN1, a highly conserved, 18-KDa, cytosolic phosphoprotein involved in control of cell proliferation, differentiation, motility, clonogenicity, and survival (10), regulates cell division by destabilizing microtubules in a phosphorylationdependent manner (11). The unphosphorylated form of STMN1 destabilizes microtubules in cell cultures and in animals and binds to soluble β-tubulin dimers; phosphorylation switches off both of these activities. In glioma cells, STMN1 is involved in cellcycle progression and in cell migration and invasion (12). In the STMN1 protein of human esophageal adenocarcinoma, there is a Q18-->E substitution (11). STMN1 is overexpressed in ovarian (13), cervical (14), prostate (15), breast (16), and gallbladder carcinomas (17). During mitosis, serine residues 16, 25, 38, and 63 of STMN1 are phosphorylated (18–25). Protein kinase A, the Ca2+/calmodulin-dependent kinase IV/mitogen-activated protein kinase family (MAP/ERK), and cyclin-dependent kinases phosphorylate STMN1 (26). The efficacy of microtubule-directed chemotherapeutics, such as docetaxel and cabazitaxel, suggests that STMN1 is a therapeutic target. Overexpression of STMN1 is associated with poor outcomes for nasopharyngeal and ovarian cancers; estrogen receptor–positive, tamoxifen-treated breast cancers; and oral squamous cell carcinomas (27–30). In the current study, we investigated the function and regulation of STMN1 during prostate cancer progression.
miRNAs participate in tumorigenesis through modulating the expression of their cognate target genes by binding to the 3'-untranslated regions (UTR) of target mRNAs, causing their translational inhibition or cleavage (31). Thus, it is of interest to identify an miRNA that interferes with STMN1 expression and thereby leads to cancer invasion and metastasis. Our in silico analysis predicted binding of miR-34a to STMN1, which could regulate its expression in prostate cancers. miR-34a is involved in various biological and pathologic processes, including the cell cycle, cell proliferation, and antiapoptosis (32). However, the underlying mechanism for miR-34a– mediated regulation of STMN1 remains poorly understood. However, in cancers, other miRNAs also negatively regulate STMN1 expression. These include miR-223 in malignant pleural mesothelioma (33) and hepatocellular carcinoma (34), miR-101 in hepatocellular carcinoma (35) and breast cancer (36), miR-193b in pancreatic cancer (37), miR-9 in glioma (38), and miR-1247 in non–small cell lung cancer (39). We and others have shown that tumor suppressor miRNAs target oncogenes associated with various cancer pathways. Examples include miR-101 and 26a, which target the histone methyltransferase EZH2 (40, 41); miR-124, which targets collagen prolyl hydroxylase P4HA1 (42); miR-101, which targets SUB1 (43); let-7a (44) and miR-34b (45), which target MYC; miR-29b, which targets MCL1 (46); and let-7, which targets RAS (47). In the current report, we show that miR-34a binds to the 3'UTR of STMN1 and regulates its expression. In addition, miR-34a is a target of the transcriptional corepressor CtBP1. We have earlier shown that, in metastatic prostate cancer, CtBP1 is overexpressed (48). We now show that STMN1 affects proliferation of prostate cancer cells and tumor growth in animals and that reduced expression of miR-34a, a tumor suppressor, negatively regulates STMN1 expression. The oncoprotein GDF15 promotes metastasis of colorectal cancers and radioresistance in head and neck cancers ((49, 50). In summary, our results indicate that, in aggressive prostate cancers, CtBP1 negatively regulates miR-34a, which upregulates STMN1, which, in turn, upregulates GDF15. Through this process, STMN1 is involved in the development and progression of prostate cancers.
Materials and Methods
Cell cultures
The human prostate cancer cell lines, DU145, PC3, LNCaP, and 293T, were from ATCC. Cells were cultured in RPMI1640 (Life Technologies) supplemented with 10% FBS (Life Technologies) and penicillin–streptomycin (100 U/ml) under 5% CO2. Normal human prostate epithelial cells (PrEC, Lonza) were grown in PrEGM™ Prostate Epithelial Cell Growth Medium (Lonza), and human benign prostate cells RWPE-1 (henceforth referred as RWPE; ATCC) were grown in keratinocyte-serum free medium with supplements (Life Technologies) as specified by the supplier. Cells were established as free of mycoplasma and bacteria following instructions of ATCC. Lenti- and adenoviruses were generated by the University of Michigan Vector Core (Ann Arbor, MI). Prostate cancer cells were infected with lentiviruses expressing STMN1 or CtBP1 shRNAs or with nontargeting (NT)-shRNA controls; stable cell lines were generated by selection with 1 ug/mL puromycin (Thermo Fisher Scientific). For transient overexpression, PrEC cells were infected with adenoviruses expressing CtBP1 or lacZ.
Benign and tumor tissues
Tissues were from patients with clinically localized prostate cancer who underwent radical prostatectomy. Samples were also obtained from patients with androgen-independent metastatic prostate cancers from a rapid autopsy program through the University of Michigan Prostate SPORE Tissue Core, as described previously (51, 52). The Institutional Review Board at the University of Michigan Medical School approved this study. All methods were performed in accordance with the university guidelines and regulations.
Gene expression from TCGA
The Cancer Genome Atlas (TCGA) gene expression profile of STMN1 and GDF15 were obtained from a web portal UALCAN (53). In UALCAN (http://ualcan.path.uab.edu/), the clinical data for patients with prostate cancer and Level3 TCGA RNA-seq data (including raw_read_count and scaled_estimate for each sample) for primary tumors and matched normal samples were downloaded using TCGA assembler (54). For each gene, transcript per million values were obtained by multiplying the scaled estimate by 1,000,000. Boxplots were generated by use of R (https://cran.r-project.org/).
RNAi and lenti-miRNA overexpression
STMN1 shRNAs (pGreenPuro Vector) were generated by System Biosciences (Supplementary Table S1). Human premiRNA expression constructs lenti-miR-34a (MI0000268), -135b (MI0000810), -193b (MI0003137), and -196a (MI0000279) were purchased from System Biosciences. Lentiviruses for these constructs were generated by the University of Michigan Vector Core. For transient knockdowns, two independent siRNAs specific for STMN1 were purchased from Dharmacon (GE Healthcare). Transfections were performed with Lipofectamine RNAiMAX reagent (Life Technologies). Stable knockdowns of STMN1 and CtBP1, and overexpressions of miRNAs were generated in prostate cancer cells, which were harvested for RNA isolation or Western blot analysis.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from prostate cancer cells and prostate cancer tissue samples using RNeasy Mini Kits (Qiagen). RNA was reverse transcribed into complementary DNA using SuperScript III Reverse Transcriptase (Invitrogen). qRTPCR was performed as described previously (42). SYBR Green was used to determine the mRNA expression level of genes of interest. All primers for SYBR green were synthesized by Integrated DNA Technologies. GAPDH or ACTB was used as a normalized control. Primer sequences used for SYBR Green qRT-PCR are listed in Supplementary Table S2. miRNA was extracted using miRNeasy Mini Kit (Qiagen) and was reverse transcribed using specific RT anchor primers following the manufacturer's instructions. For TaqMan miRNA assays, U6 snRNA (#001973) and hsa-miR-34a (#000426) were purchased from Applied Biosystems, Thermo Fisher Scientific. U6 snRNA was used as a normalized control. All PCR reactions were performed in triplicate.
Western blot analyses
Cells, tissues, and mouse xenograft tumors were harvested following various treatments as described above, lysed in NP-40 lysis buffer (Boston BioProducts), and quantified by a DC Protein Assay (Bio-Rad). For Western blot analyses, protein samples (10 mg) were separated on a NuPAGE 4%–12% Bis-Tris Protein Gel (Invitrogen) and transferred onto Immobilon-P PVDF membranes (EMD Millipore). The membranes were incubated for 1 hour in blocking buffer [5% nonfat dry milk in Tris-buffered saline and 0.1% Tween (TBS-T)], followed by incubation overnight at 4 C with the primary antibody. After three washes with TBS-T, the blots were incubated with horseradish peroxidase– conjugated secondary antibody, and signals were visualized by Luminata Forte Western blot HRP substrate (EMD Millipore) using an Amersham Imager 600RGB (GE Healthcare Life Sciences). Antibodies used are listed in Supplementary Table S3.
Gene expression analysis
Gene expression profiling was accomplished with RNA isolated from STMN1 shRNA knockdown PC3 and LNCaP cells and nontarget control cells. Profiling was performed using the Agilent Whole Human Genome Oligo Microarray (Agilent), and the analyses were performed according to the manufacturer's protocol. A bioconductor package "agilp" (55) was used to extract and normalize raw data from two-channel experiment arrays. Loess normalization was applied on each array. The gene expression profiling data have been deposited at gene expression omnibus (GSE96523). The differential expression of each gene was estimated by subtracting loess-normalized, log-2–transformed signal intensity of control samples from that of knockdown samples. Genes with an absolute fold change of 1.5 were selected as differentially expressed. The heatmap.2 function of R package "gplots" was used to create the heatmap.
Cell proliferation assays
Transient and stable STMN1 knockdown and cells stably overexpressing miR-34a were used. The test cells were seeded at a density of 10,000 cells per well in 24-well plates (n = 3). Cells were harvested and counted at indicated time points by use of a Coulter counter (Beckman Coulter). Untreated, NT-siRNA, shRNA, lenti-miRNA, or scramble miRNA treated cells served as controls. Each experiment was performed with three replicates per sample.
Colony formation assays
For study of STMN1, after 72 hours of transfection, nontargeting and STMN1 siRNA-treated cells were counted and seeded at 800 cells per well of 6-well plates (in triplicates) and incubated at 37 C under 5% CO2 for 10 days. For study of miR-34a, scramble miRNA and miR-34a cells were seeded as above and incubated for 10 days as described earlier (43, 56). The colonies were fixed with 5% (v/v) glutaraldehyde for 15 minutes and stained with crystal violet (Sigma-Aldrich) for 15 minutes. Photographs of the colonies were taken using Amersham Imager 600RGB (GE Healthcare Life Sciences). Colony quantification was accomplished with ImageQuant TL Colony v8.1 software (GE Healthcare Life Sciences).
Cellular migration assays
Scratch wound migration assays of PCA cells were performed as described (42, 43).
Matrigel invasion assays
Cell invasion was assessed by use of BD BioCoat Matrigel matrix (Corning Life Sciences) Transwell chamber plates. Matrigel invasion assays were performed as described earlier (42, 43, 56). Various test cells were seeded onto the Transwell chambers of 24-well culture plates. To the lower chambers, 750 uL of 10% FBS-RPMI1640 medium was added as a chemoattractant. After 48 hours, the noninvading cells and the Matrigel matrix were removed with a cotton swab. The invading cells on bottom surfaces of the membrane were stained with 0.2% crystal violet in methanol for 15 minutes and air-dried. Cell images were obtained with an inverted phase-contrast microscope (X4). Invasion was quantified by a colorimetric assay. The inserts were treated with 150 uL of 10% acetic acid, and the absorbance was measured at 560 nm.
miRNA reporter luciferase assays
3'-UTR luciferase assays were performed as described earlier (42, 43). The wild-type or mutant 3'-UTRs of STMN1 were cloned into the pMIR-REPORT miRNA Expression Reporter Vector (Life Technologies). 293T cells were transfected with either pre–miR-34a (#PM11030) or nontargeting miRNA for 4 hours, followed by transfection with wild-type or mutant 3'UTR-luc, as well as a pRL-TK vector as an internal control for luciferase activity. At 72 hours posttransfection, the cells were lysed, and luciferase assays were conducted using the dual luciferase assay system (Promega) as per the manufacturer's instructions. Each experiment was performed in triplicate.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were accomplished with anti-CtBP1 (#C8741, Sigma-Aldrich) and IgG antibodies using iDeal ChIP-seq Kits for Transcription Factors (#C01010055, Diagenode Inc.) following the manufacturer's instructions. The primer sequences for the MIR34A promoter are provided in Supplementary Table S4.
Chick chorioallantoic membrane assay
In vivo cell invasion, intravasation, metastasis, and tumor (or xenograft) formation were modeled by chick chorioallantoic membrane (CAM) assays (42, 43, 56). Briefly, fertilized chicken eggs (Charles River Laboratories) were incubated for 10 days in a rotary incubator at 38C and humidity of 58% to 60%. CAM, 1 cm away from the branch point of the chorioallantoic vein, was dropped by applying gentle suction created with the aid of an automatic pipette through a small hole in the air sac. The test cells [PC3-scramble, STMN1 shRNA1 and 2 (1 × 106 each)] in a volume of 50 uL of culture medium were applied to the dropped CAM. At 3 days after cell implantation, the lower CAMs were harvested, and extraembryonic tumors were isolated and weighed. For assay of metastasis, the embryonic livers were harvested on day 18 of embryonic growth and analyzed for the presence of tumor cells by quantitative human Alu-specific PCR. Genomic DNA from the lower CAMs and livers was prepared using the Puregene DNA purification system (Qiagen), and quantification of human Alu was performed as described previously (42, 43, 56). An average of eight eggs per group was used in each experiment.
Mouse tumor xenografts
All procedures involving mice were accomplished according to protocols reviewed and approved by the University Committee on Use and Care of Animals at the University of Michigan. Male Hsd: Athymic Nude-Foxn1nu mice (n = 8/group) were purchased from Envigo. To evaluate the role of STMN1 in tumor formation, stable STMN1 knockdown PC3 cells were propagated using two independent shRNAs and non-T shRNA control cells, and inoculated at 1 × 106 cells subcutaneously into the dorsal flanks of 5-weekold male athymic nude mice. The tumor data obtained using scramble cells are the same as that used in an earlier study, as the STMN1 tumor xenograft study was conducted simultaneously with common control animals (42). Tumor sizes were measured biweekly, and tumor volumes were calculated using the formula (π/6) (L× W2), where L is the length and W is the width of the tumor. After 5 weeks, mice from different groups were killed, the tumors were photographed and weighed, and the results were plotted.
Statistical analyses
To determine significant differences between two groups, Student two-tailed t test was used for all experiments, except for microarrays. P < 0.05 was considered statistically significant.
Results
Aggressive prostate cancers overexpress STMN1
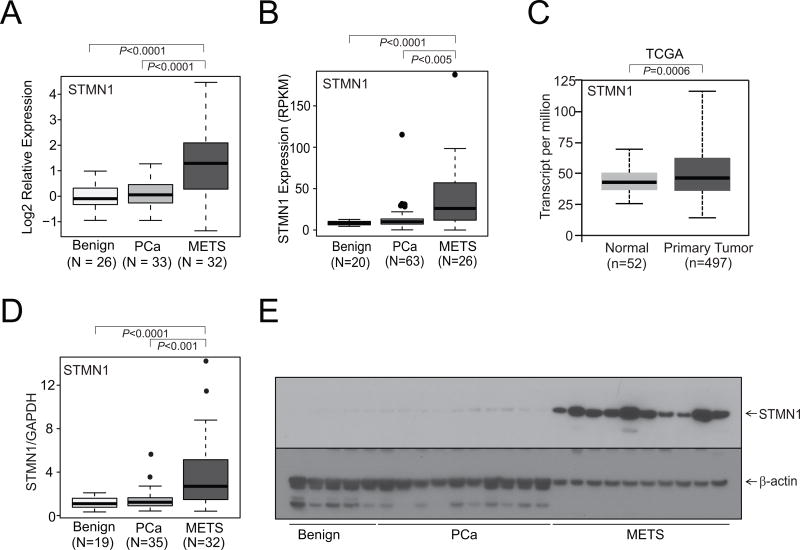
To identify the expression of STMN1 in prostate cancer, we utilized publicly available cancer gene expression profiling and transcriptome sequencing data. In our in silico analysis of metastatic prostate cancer, there was upregulation of STMN1 (Fig. 1A and B). TCGA data for STMN1 were obtained using the "UALCAN" web portal for gene expression analyses, which showed upregulation of STMN1 in prostate cancer (Fig. 1C; ref. 53). Next, to investigate the expression of STMN1 in a cohort of benign and malignant prostate tissue samples, qRTPCR, performed with RNA from these tissues, confirmed increased expression of STMN1 in metastatic prostate cancer tissues relative to benign prostate samples (Fig. 1D). To validate the mRNA results, Western blotting was performed on randomly selected cases (benign, localized prostate cancer, and hormone-refractory, and metastatic prostate cancer) by use of a mAb specific for STMN1. The amounts of STMN1 protein were greater in metastatic prostate cancers relative to localized prostate cancers or benign prostate (Fig. 1E). Thus, in silico, tissue RNA and protein measurements demonstrated elevated expression of STMN1 in prostate cancer tissues.
Figure 1.
STMN1 is overexpressed in prostate cancer. STMN1 gene expression from A, microarray, B and C, next-generation RNA sequencing data from benign tissue, prostate carcinoma tissue, and metastatic prostate cancer (MET) tissue from the University of Michigan and TCGA datasets, respectively (B and C). D, qRT-PCR of STMN1 using RNA from benign, prostate cancer, and MET tissues. E, Western blot analysis of the STMN1 protein from prostate tissue extracts using an STMN1-specific antibody. β-Actin was used as a loading control.
STMN1 knockdown reduces proliferation and invasion of prostate cancer cells
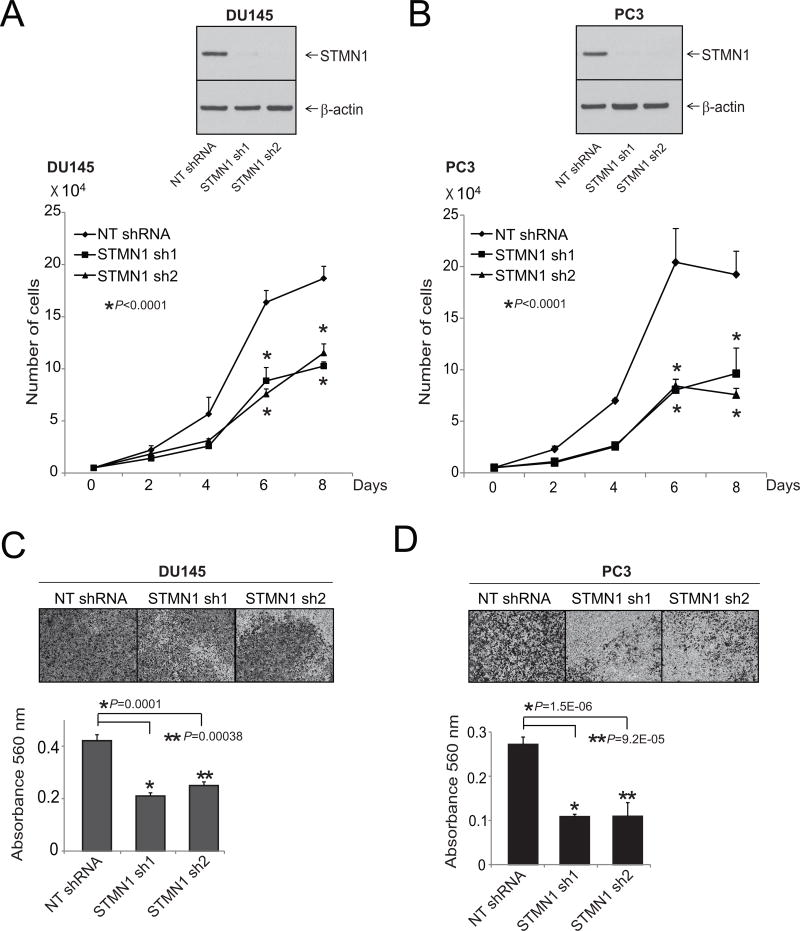
To determine the role of STMN1 in growth of prostate cancer cells, cells with stable STMN1 knockdown were generated by use of two specific and independent shRNAs in aggressive prostate cancer cell lines DU145 and PC3 and were used to conduct assays for cell proliferation and invasion. Western blot analysis confirmed knockdown of STMN1 (Fig. 2A and B). Simultaneously, cell proliferation assays were conducted for DU145 and PC3 cells by counting the cells at various time intervals. STMN1 knockdown reduced proliferation of prostate cancer cells (Fig. 2A and B). To determine whether STMN1 knockdown reduces cell motility and migration, scratch wound migration assays were performed. The capacity of the stable STMN1 knockdown cells to close a wound was lower relative to cells expressing scramble shRNA (Supplementary Fig. S1). In addition, in DU145, PC3, and LNCaP cells, transient knockdowns of STMN1 were achieved by use of two specific and independent siRNA duplexes (Supplementary Fig. S2A). Transient knockdown of STMN1 reduced formation of cell colonies as compared with untreated cells and cells treated with nontargeting siRNA (Supplementary Fig. S2B). As determined by Matrigel assays, the stable knockdown cells also showed lower capacity for cell invasion as measured (Fig. 2C and D). Thus, these experiments demonstrated the function of STMN1 in prostate cancer cell proliferation, invasion, and colony formation.
Figure 2.
STMN1 expression is essential for prostate cancer cell proliferation and invasion. A and B, Stable knockdown of STMN1 in prostate cancer cell lines reduced cell proliferation. Western blot analysis of protein lysates from prostate cancer cell lines DU145 and PC3 stably transfected with two specific and independent STMN1 shRNAs. β-Actin was used as a loading control. Cell proliferation assays using cells stably transfected with either STMN1 or NT-shRNA. C and D, Matrigel invasion assays were performed with STMN1 knockdown cells. NT-shRNA–treated cells served as control. Invading cells were stained with crystal violet, and the absorbance was measured at 560 nm.
miR-34a downregulates STMN1
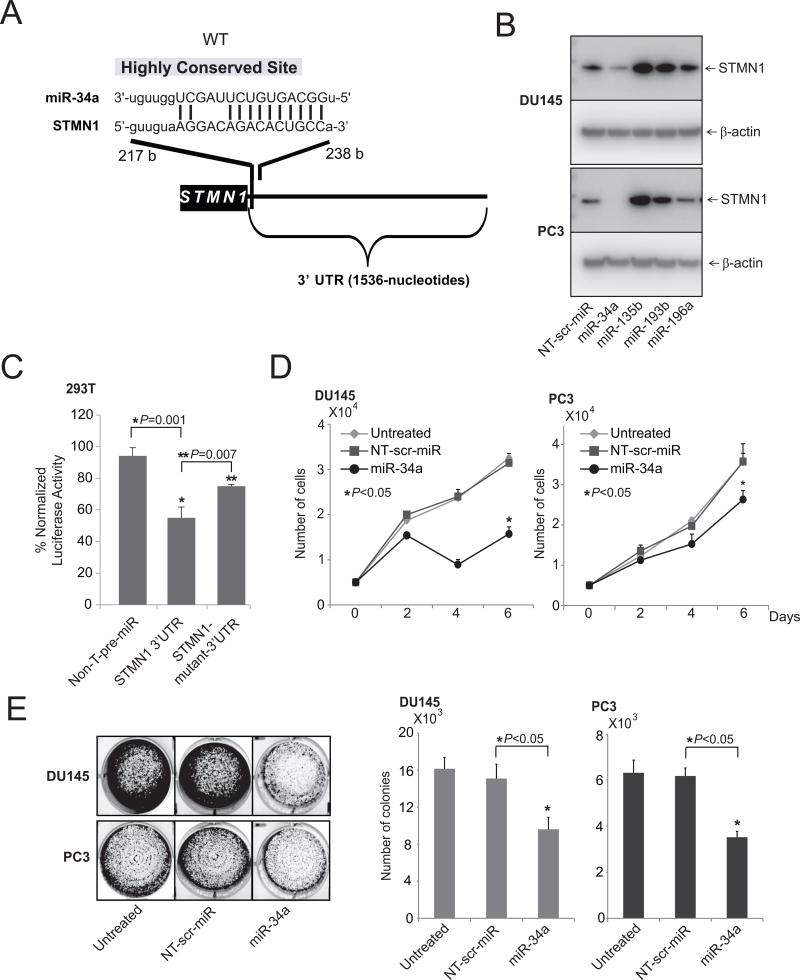
Earlier studies demonstrated that, in prostate cancer cells, miRNAs negatively regulate various oncogenes (40–44, 46, 47). To investigate the upstream regulators of STMN1, freely available web-based miR target prediction resources, miRNA.org (57) and miRSearch V3.0 (58, 59), were used. With these, we established that miR-34a could target STMN1. By targeting various oncogenes, including ERBB2 in breast cancer (60), EGFR in glioblastoma multiforme (61), and SIRT1 in colon cancer, miR-34a exerts tumor suppressor activity (62). A binding site for miR-34a in the 3'-UTR ofSTMN1was present (Fig. 3A). Next, the role of miR-34a in regulation of STMN1 was investigated by stably overexpressing nontargeting miRNA, miR-34a, -135b, -193b, or -196a in DU145 and PC3 cells. STMN1 protein expression, evaluated by Western blot analyses, showed downregulation of STMN1 protein levels (Fig. 3B). Cells treated with miR-34a showed reductions in STMN1 protein levels, whereas the control and other miRNAs did not alter expression (Fig. 3B). Downregulation of STMN1 by miR-34a was due to an miRNA:mRNA interaction, as miR-34a inhibited the expression of a luciferase reporter gene fused to a fragment of the STMN1 3'UTR containing the target site (Fig. 3C). Mutation of the miR-34a target site reversed this effect (Fig. 3C and Supplementary Fig. S3). Furthermore, the inhibitory effect of miR-34a on cell growth was assessed by use of cell proliferation (Fig. 3D) and colony formation assays (Fig. 3E). Thus, STMN1 is a target of miR-34a.
Figure 3.
miR-34a regulates STMN1 expression. A, The predicted miR-34a–binding site at the 3’-UTR of STMN1. B, Western blot analysis showing STMN1 in lysates of DU145 and PC3 cells with lentiviral-mediated stable miR-34a, 135b, 193b, and 196a overexpression and control stable lenti-miR-expression. β-Actin was used as a loading control. C, STMN1-3’-UTR luciferase reporter assay. For this, 293T cells were transfected either with pre–miR-34a or nontargeting pre-miR (NT-pre-miR) along with either STMN1-3’UTR wild-type or mutant luciferase constructs. A pRL-TK vector was used as an internal control. D and E, Cell proliferation (D) and colony formation assays of untreated, lenti-scramble, and miR-34a overexpressing DU145 and PC3 cells (E).
The corepressor CtBP1 modulates STMN1 expression by downregulating miR-34a
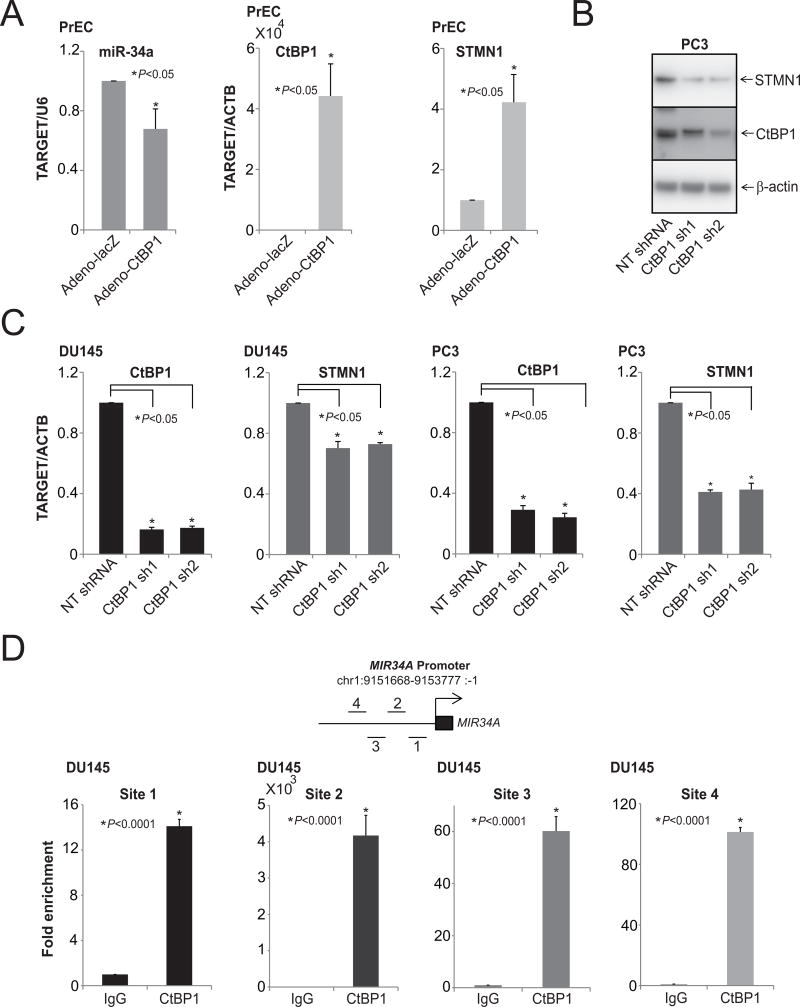
The transcriptional corepressor CtBP1 acts as an oncogene and is involved in PCA progression; it can also act as tumor suppressor in context-dependent manner (48, 63, 64). Moreover, CtBP1 mediates suppression of various tumor suppressors, such as the epithelial cell markers, E-cadherin and LCN2, suggesting its role in the epithelial-to-mesenchymal transition (48). In breast cancers, CtBP1 modulates various miRNAs involved in metabolic processes, the cell cycle, and cell communication (63), and miR-124 is involved in progression of PCA (42). Earlier studies, including ours, have demonstrated the role of miRNAs in cellular processes, such as cell proliferation, invasion, and metastasis by repressing oncogenes (40, 41), including P4HA1 (42), SUB1 (43), MYC (44, 45), MCL1 (46), and RAS (47). To investigate the potential role of CtBP1 on miR-34a expression, Taqman qRTPCR analyses were performed with normal prostate epithelial cells over-expressing CtBP1 (Fig. 4A). CtBP1 negatively regulated miR-34a, leading to overexpression of STMN1 (Fig. 4A). Next, protein and RNA levels of STMN1 were measured in cells with stable knockdowns of CtBP1 In CtBP1 knockdown cells, STMN1 expression was lower at both protein (Fig. 4B) and RNA (Fig. 4C) levels. Thus, by repressing miR-34a expression, CtBP1 enhanced expression of STMN1. To validate the binding of CtBP1 to the MIR34A promoter, chromatin immunoprecipitation (ChIP) assays were conducted with DU145 cells by use of a commercially available anti-CtBP1 antibody. CtBP1 was enriched at the MIR34A promoter region (Fig. 4D). These data demonstrate that, in PCA cells, CtBP1 transcriptionally represses the MIR34A expression.
Figure 4.
CtBP1 modulates STMN1 levels by downregulating miR-34a expression. A, qRT-PCR analysis of CtBP1, STMN1, and miR-34a in PrEC-adeno CtBP1 overexpressing cells. B, Western blot analysis of CtBP1 and STMN1 in lysates of PC3-CtBP1 stable knockdown cells. β-Actin was used as a loading control. C, qRT-PCR analysis of CtBP1 and STMN1 in CtBP1-stable knockdown cells. D, miR-34a is a target of CtBP1. Schematic of amplicon locations on chromosome 1, analyzed via ChIP-qPCR assays as described. ChIP assays were performed with antibodies specific to CtBP1 and IgG. The DNA purified after ChIP assays was evaluated by semiquantitative PCR with primers specific for regions of the MIR34A promoter. IgG was used as a negative control.
In prostate cancer cells, STMN1 modulates gene expression
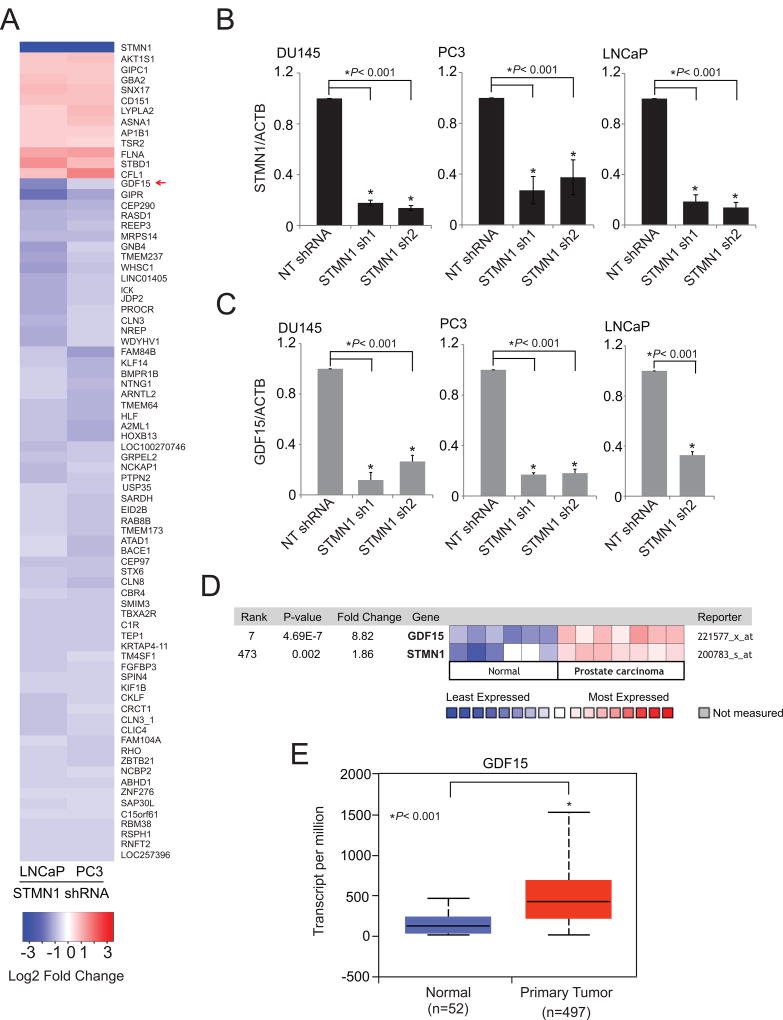
To assess STMN1-mediated effects in prostate cancer progression, gene expression analyses were conducted by use of a microarray with RNA from STMN1 knockdown PC3 and LNCaP cells. Genes modulated by STMN1 knockdown included growth differentiation factor GDF15 (Fig. 5A). GDF15, a secreted ligand of the TGFβ superfamily of proteins, shows elevated expression or secretion in colorectal (49), pancreatic (65), esophageal (66), ovarian (67), and endometrial (68) cancers. This elevated expression is often associated with diseases that are more aggressive. GDF15 also contributes to the radioresistance of head and neck cancer (50). However, GDF15 has different functions, depending on the specific tissues and disease conditions. STMN1 knockdown downregulated GDF15 at the mRNA level (Fig. 5B and C). Furthermore, coexpression analyses between STMN1 and GDF15 using the Oncomine gene expression analyses platform (Life Technologies) showed that these genes are overexpressed in prostate cancers (Fig. 5D) compared with normal tissues (69). TCGA data showed that aggressive primary tumors overexpress GDF15 (Fig. 5E). Thus, GDF15 acts downstream to the STMN1-mediated proliferation and invasion of prostate cancer cells.
Figure 5.
Gene expression profiling of STMN1-modulated cells reveals regulation of GDF15 expression. A, Heatmap of selected genes from expression profiling of stable STMN1 knockdown PC3 and LNCaP cells. B and C, qRT-PCR analysis of STMN1 and GDF15 in RNA of DU145, PC3, and LNCaP-STMN1 cells with stable knockdown. D, Expression of GDF15 and STMN1 are positively correlated. Heat map of GDF15 and STMN1 expression levels across normal and prostate carcinoma tissues. The data was retrieved from Oncomine (69). Blue and red color bars signify the lowest and highest levels of expression respectively (log2 median-centered ratio). E, The TCGA gene expression profile of GDF15 in benign and prostate primary tumor tissues (53).
STMN1 is involved in growth and metastasis of prostate cancers
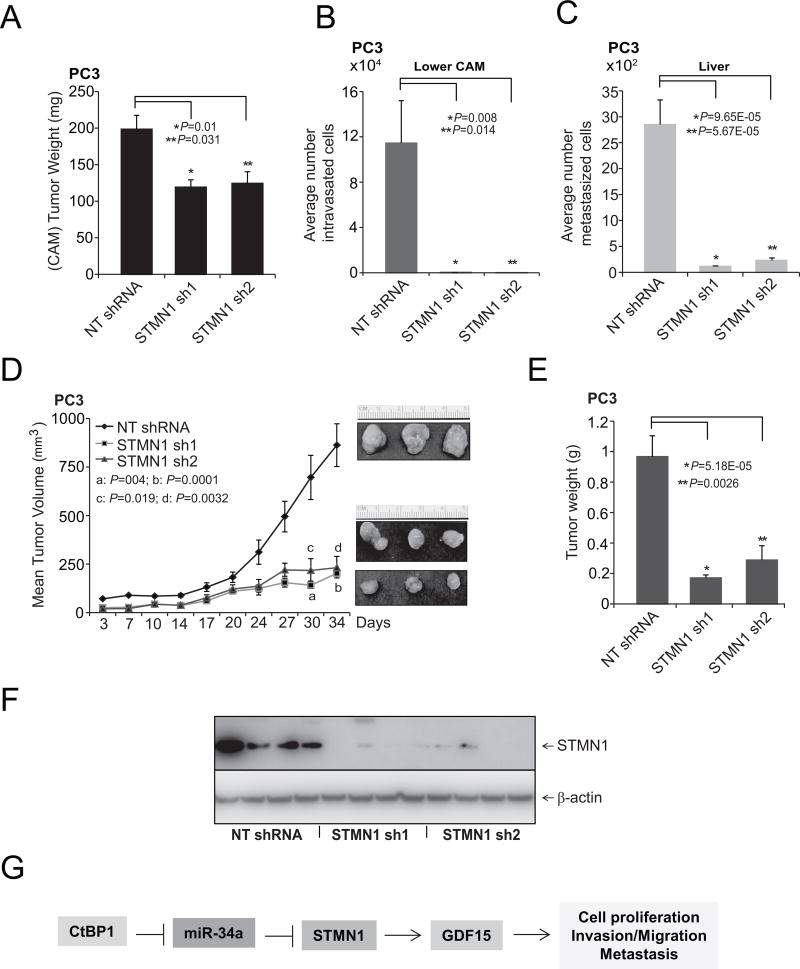
To determine the effects of STMN1 on tumor growth in vivo, a CAM assay was used to measure local intravasation and metastasis to distant organs. The CAM assay was performed as described previously with prostate cancer PC3-STMN1 knockdown cells (42, 43, 56). Knockdown of STMN1 resulted in reduced tumor weights compared with control cells expressing nontargeting shRNA (Fig. 6A). Compared with control cells, STMN1 knockdown in PC3 cells impaired their capacity to invade the CAM basement membrane and resulted in fewer numbers of intravasated cells in the lower CAM (Fig. 6B). In addition, STMN1 knockdown reduced the numbers of metastatic cells in the chick livers compared with control cells expressing nontargeting shRNA (Fig. 6C). STMN1-mediated tumorigenesis in a mouse xenograft model was examined using NT-shRNA and two independent STMN1 stable knockdown PC3 cells. Relative to controls, cells treated with STMN1-shRNA 1 or 2 showed less tumor growth and lower tumor weights (Fig. 6D and E). Furthermore, STMN1 expression in the tumor tissues was determined by Western blot analysis. STMN1 expression was lower in the STMN1 sh1 and 2 groups compared with the level in the NT-shRNA group (Fig. 6F), demonstrating that STMN1 inhibition attenuates tumor growth (CAM and mouse models) and metastasis (CAM model). These observations suggest a role for STMN1 in prostate tumor growth.
Figure 6.
STMN1 is required for growth of prostate tumor cells. A, Tumor growth of stable STMN1 knockdown or control non-T shRNA PC3 cells in the chick CAM tumor assay. Tumors from the upper CAM were harvested and weighed at 72 hours after implantation of cells. B and C, STMN1 knockdown reduced intravasation and metastasis of PC3 cells in the lower CAM and liver, respectively. Cells that moved to the lower CAMs and livers of chicken embryos were quantified with human Alu-specific PCR. D, STMN1 knockdown in PC3 cells inhibited tumor growth in mouse xenografts. A plot of mean tumor volume at indicated time points for mice inoculated with cells treated with STMN1 stable knockdown shRNA 1 (solid line with filled squares) or 2 (solid line with filled triangles) or with NT-shRNA (solid line with filled diamonds). Inset, photomicrographs of xenograft tumors. E, Tumor weights for the NT-shRNA, STMN1 shRNA 1, and STMN1 shRNA 2 groups (n = 8 mice/group). F, Western blot analyses showing STMN1 expression in mouse xenograft tumors. G, A proposed model of CtBP1, miR-34a, STMN1, and GDF15 regulation in prostate cancer. STMN1, regulated by miR-34a, is involved in cell proliferation, invasion, metastasis, and tumor growth.
Discussion
The STMN1 protein, which has functions in various cellular processes, including cell proliferation, differentiation, motility, clonogenicity, and survival (10), is linked to breast (16, 30), ovarian (13), cervical (14), prostate (15), and gallbladder (17) cancers. However, the function of STMN1 in prostate cancer is not clear. In the current study, we confirmed that prostate cancer tissues frequently overexpress STMN1 mRNA and protein relative to noncancerous prostate tissues. Knockdown of STMN1 inhibited cell proliferation, invasion, and colony formation and tumor growth of prostate cancer cells in the CAM assay and in mice. The elevated expression of STMN1 in aggressive prostate cancer tissues was confirmed. Earlier, STMN1 expression was measured in prostate cancer (70). Such expression of STMN1 is associated with the proliferation and metastasis of a variety of cancer cells (71). The silencing of STMN1 causes a decrease in cell proliferation, migration, and invasion and growth of human gallbladder carcinomas (17). Likewise, our results showed that knockdown of STMN1 caused less tumor growth, invasion, and metastasis.
In human cancers, there is often downregulation of miRNAs, which are related to tumor progression and negatively regulate expression of their target mRNAs (40–47). miR-34a, a master tumor suppressor downstream of p53 (72), is downregulated in various cancers (73). miR-34a antagonizes oncogenic processes by regulating genes that function in various cellular pathways, including those that relate to the cell cycle and to proliferation, antiapoptosis, cancer stemness, metastasis, oncogenic transcription, and chemoresistance (32). MRX34, a synthetic miR-34a mimic loaded in liposomal nanoparticles, is the first miRNA-based therapy specifically for cancer (74). For lung cancer, local and systematic administration of the lipid-based miR-34a formulation to animals leads to accumulation of miR-34a in the tumor tissues and to downregulation of miR-34a targets (75). In esophageal carcinoma cells, miR-34a is regulated at the transcriptional level by NF-kB (76) and, in acute myeloid leukemia, by the transcription factor, CCAAT enhancer binding protein alpha (C/EBPα), which in turn leads to E2F3 repression and inhibition of cell-cycle progression (77). Recently, it was shown that miR-34a regulates stathmin 1 via 3'UTR in osteosarcoma (78).
In the current study, we showed that, in prostate cancer cells, miR-34a is repressed by the transcriptional corepressor CtBP1, thus elevating STMN1 expression. The oncoprotein CtBP1, overexpressed in prostate cancer (48), represses miR-124 expression, thereby sustaining activity of collagen prolyl hydroxylase P4HA1 (42). In various cancers, reduced expression of miR-34a leads to overexpression of oncogenes (79–82). Our investigations demonstrated the role of an miR-34a target, STMN1, in proliferation and invasion of prostate cancer cells by regulating GDF15, a member of the TGFβ superfamily. Although earlier studies showed the involvement of STMN1 in cancer, its mechanistic role in oncogenesis was not fully understood. The current study shows increased expression of STMN1 in prostate cancer. Moreover, it is involved in tumorigenesis, potentially through the activation of GDF15 during cancer progression. GDF15 is involved in various cellular processes, including radioresistance of head and neck cancer (50). GDF15 is upregulated in cancers (50, 83), and elevated expression is associated with cancers that are more aggressive. In the current work, we demonstrated that STMN1 upregulates expression of GDF15.
In summary, elevated expression of CtBP1 reduced miR-34a expression, and downregulation of miR-34a resulted in overexpression of STMN1. STMN1 was found to be involved in regulation of prostate cancer cell proliferation, invasion, and metastasis and modulated expression of various genes, including GDF15 (Fig. 6G). The current results show STMN1 overexpression in aggressive prostate cancers and reveal therapeutic options to block STMN1-mediated oncogenesis.
Supplementary Material
Implications.
The CtBP1\miR-34a\STMN1\GDF15 axis is a potential therapeutic target for treatment of aggressive prostate cancer.
Acknowledgments
Grant Support
This work was supported, in part, by National Institutes of Health grant R01CA154980. S. Varambally is also supported by grant R01CA157845 and a University of Michigan Prostate Cancer SPORE Career Development award. The authors thank the University of Michigan Vector Core for generating lentivirus. Gene expression data has been deposited in the GEO database under accession number GSE96523. We thank Dr. Donald Hill for his help in editing.
Footnotes
Disclosure of Potential Conflicts of Interest: G. Sonpavde reports receiving commercial research grants from Bayer, Boehringer-Ingelheim, Celgene, Merck, Onyx-Amgen, and Pfizer, has received speakers bureau honoraria from Georgia Society of Clinical Oncology, Physicians Education Resource (PER), Renal Cell Carcinoma Preceptor-ship at Singapore, Singapore Society of Oncology, and Uptodate, and is a consultant/advisory board member for Amgen, AstraZeneca Exelixis, Bristol-Myers Squibb, Genentech, Janssen, Merck, NCCN, Novartis, Pfizer, and Sanofi. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Conception and design: B.V.S.K. Chakravarthi, R. Mehra
Development of methodology: B.V.S.K. Chakravarthi, R. Mehra, I.A. Asangani, U. Manne
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): B.V.S.K. Chakravarthi, S. Agarwal, S.A. Hodigere Balasubramanya, S.S. Pathi, X. Jing, I.A. Asangani, A.M. Chinnaiyan
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): D.S. Chandrashekar, S.S. Pathi, I.A. Asangani, A.M. Chinnaiyan
Writing, review, and/or revision of the manuscript: B.V.S.K. Chakravarthi, D.S. Chandrashekar, S.S. Pathi, U. Manne, G. Sonpavde, G.J. Netto, J. Gordetsky, S. Varambally
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S.A. Hodigere Balasubramanya, X. Jing, R. Wang
Study supervision: B.V.S.K. Chakravarthi, S. Varambally
Other (animal studies): S.S. Pathi
Other (helped in conducting in vivo experiments): M.T. Goswami
Other (helped to generate gene expression profile microarray data): X. Jing
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarthi BV, Nepal S, Varambally S. Genomic and Epigenomic Alterations in Cancer. Am J Pathol. 2016;186:1724–35. doi: 10.1016/j.ajpath.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–33. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bono JS, Oudard S, Ozguroglu M, Hansen S, Machiels JP, Kocak I, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010;376:1147–54. doi: 10.1016/S0140-6736(10)61389-X. [DOI] [PubMed] [Google Scholar]
- 5.Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–22. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 6.Parker C, Nilsson S, Heinrich D, Helle SI, O'Sullivan JM, Fossa SD, et al. Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013;369:213–23. doi: 10.1056/NEJMoa1213755. [DOI] [PubMed] [Google Scholar]
- 7.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–48. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–12. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh R, Gu G, Tillman E, Yuan J, Wang Y, Fazli L, et al. Increased expression and differential phosphorylation of stathmin may promote prostate cancer progression. Prostate. 2007;67:1038–52. doi: 10.1002/pros.20601. [DOI] [PubMed] [Google Scholar]
- 10.Biaoxue R, Xiguang C, Hua L, Shuanying Y. Stathmin-dependent molecular targeting therapy for malignant tumor: the latest 5 years' discoveries and developments. J Transl Med. 2016;14:279. doi: 10.1186/s12967-016-1000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misek DE, Chang CL, Kuick R, Hinderer R, Giordano TJ, Beer DG, et al. Transforming properties of a Q18-->E mutation of the microtubule regulator Op18. Cancer cell. 2002;2:217–28. doi: 10.1016/s1535-6108(02)00124-1. [DOI] [PubMed] [Google Scholar]
- 12.Song Y, Mu L, Han X, Liu X, Fu S. siRNA targeting stathmin inhibits invasion and enhances chemotherapy sensitivity of stem cells derived from glioma cell lines. Acta Biochim Biophys Sin (Shanghai) 2014;46:1034–40. doi: 10.1093/abbs/gmu099. [DOI] [PubMed] [Google Scholar]
- 13.Balachandran R, Welsh MJ, Day BW. Altered levels and regulation of stathmin in paclitaxel-resistant ovarian cancer cells. Oncogene. 2003;22:8924–30. doi: 10.1038/sj.onc.1207060. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Ren JH, Lin F, Wei JX, Long M, Yan L, et al. Stathmin is involved in arsenic trioxide-induced apoptosis in human cervical cancer cell lines via PI3K linked signal pathway. Cancer Biol Ther. 2010;10:632–43. doi: 10.4161/cbt.10.6.12654. [DOI] [PubMed] [Google Scholar]
- 15.Mistry SJ, Atweh GF. Therapeutic interactions between stathmin inhibition and chemotherapeutic agents in prostate cancer. Mol Cancer Ther. 2006;5:3248–57. doi: 10.1158/1535-7163.MCT-06-0227. [DOI] [PubMed] [Google Scholar]
- 16.Kuang XY, Chen L, Zhang ZJ, Liu YR, Zheng YZ, Ling H, et al. Stathmin and phospho-stathmin protein signature is associated with survival outcomes of breast cancer patients. Oncotarget. 2015;6:22227–38. doi: 10.18632/oncotarget.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Yao Y, Ming Y, Shen S, Wu N, Liu J, et al. Downregulation of stathmin 1 in human gallbladder carcinoma inhibits tumor growth in vitro and in vivo. Sci Rep. 2016;6:28833. doi: 10.1038/srep28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beretta L, Dobransky T, Sobel A. Multiple phosphorylation of stathmin. Identification of four sites phosphorylated in intact cells and in vitro by cyclic AMP-dependent protein kinase and p34cdc2. J Biol Chem. 1993;268:20076–84. [PubMed] [Google Scholar]
- 19.Brattsand G, Marklund U, Nylander K, Roos G, Gullberg M. Cell-cycle-regulated phosphorylation of oncoprotein 18 on Ser16, Ser25 and Ser38. Eur J Biochem. 1994;220:359–68. doi: 10.1111/j.1432-1033.1994.tb18632.x. [DOI] [PubMed] [Google Scholar]
- 20.Labdon JE, Nieves E, Schubart UK. Analysis of phosphoprotein p19 by liquid chromatography/mass spectrometry. Identification of two proline-directed serine phosphorylation sites and a blocked amino terminus. J Biol Chem. 1992;267:3506–13. [PubMed] [Google Scholar]
- 21.Leighton IA, Curmi P, Campbell DG, Cohen P, Sobel A. The phosphorylation of stathmin by MAP kinase. Mol Cell Biochem. 1993;127–128:151–6. doi: 10.1007/BF01076766. [DOI] [PubMed] [Google Scholar]
- 22.Luo XN, Mookerjee B, Ferrari A, Mistry S, Atweh GF. Regulation of phosphoprotein p18 in leukemic cells. Cell cycle regulated phosphorylation by p34cdc2 kinase. J Biol Chem. 1994;269:10312–8. [PubMed] [Google Scholar]
- 23.Marklund U, Brattsand G, Osterman O, Ohlsson PI, Gullberg M. Multiple signal transduction pathways induce phosphorylation of serines 16, 25, and 38 of oncoprotein 18 in T lymphocytes. J Biol Chem. 1993;268:25671–80. [PubMed] [Google Scholar]
- 24.Marklund U, Brattsand G, Shingler V, Gullberg M. Serine 25 of oncoprotein 18 is a major cytosolic target for the mitogen-activated protein kinase. J Biol Chem. 1993;268:15039–47. [PubMed] [Google Scholar]
- 25.Marklund U, Larsson N, Brattsand G, Osterman O, Chatila TA, Gullberg M. Serine 16 of oncoprotein 18 is a major cytosolic target for the Ca2+/calmodulin-dependent kinase-Gr. Eur J Biochem. 1994;225:53–60. doi: 10.1111/j.1432-1033.1994.00053.x. [DOI] [PubMed] [Google Scholar]
- 26.Lawler S. Microtubule dynamics: if you need a shrink try stathmin/Op18. Curr Biol. 1998;8:R212–4. doi: 10.1016/s0960-9822(98)70128-9. [DOI] [PubMed] [Google Scholar]
- 27.Hassan MK, Watari H, Mitamura T, Mohamed Z, El-Khamisy SF, Ohba Y, et al. P18/Stathmin1 is regulated by miR-31 in ovarian cancer in response to taxane. Oncoscience. 2015;2:294–308. doi: 10.18632/oncoscience.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu HP, Li CF, Lee SW, Wu WR, Chen TJ, Chang KY, et al. Overexpression of stathmin 1 confers an independent prognostic indicator in nasopharyngeal carcinoma. Tumour Biol. 2014;35:2619–29. doi: 10.1007/s13277-013-1345-3. [DOI] [PubMed] [Google Scholar]
- 29.Kouzu Y, Uzawa K, Koike H, Saito K, Nakashima D, Higo M, et al. Overexpression of stathmin in oral squamous-cell carcinoma: correlation with tumour progression and poor prognosis. Br J Cancer. 2006;94:717–23. doi: 10.1038/sj.bjc.6602991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miceli C, Tejada A, Castaneda A, Mistry SJ. Cell cycle inhibition therapy that targets stathmin in in vitro and in vivo models of breast cancer. Cancer Gene Ther. 2013;20:298–307. doi: 10.1038/cgt.2013.21. [DOI] [PubMed] [Google Scholar]
- 31.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–9. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birnie KA, Yip YY, Ng DC, Kirschner MB, Reid G, Prele CM, et al. Loss of miR-223 and JNK Signaling Contribute to Elevated Stathmin in Malignant Pleural Mesothelioma. Mol Cancer Res. 2015;13:1106–18. doi: 10.1158/1541-7786.MCR-14-0442. [DOI] [PubMed] [Google Scholar]
- 34.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–69. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Zheng F, Liao YJ, Cai MY, Liu TH, Chen SP, Wu PH, et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet. 2015;11:e1004873. doi: 10.1371/journal.pgen.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R, Wang HB, Hao CJ, Cui Y, Han XC, Hu Y, et al. MiR-101 is involved in human breast carcinogenesis by targeting Stathmin1. PLoS One. 2012;7:e46173. doi: 10.1371/journal.pone.0046173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Kong F, Wu K, Song K, He J, Sun W. miR-193b directly targets STMN1 and uPA genes and suppresses tumor growth and metastasis in pancreatic cancer. Mol Med Rep. 2014;10:2613–20. doi: 10.3892/mmr.2014.2558. [DOI] [PubMed] [Google Scholar]
- 38.Song Y, Mu L, Han X, Li Q, Dong B, Li H, et al. MicroRNA-9 inhibits vasculogenic mimicry of glioma cell lines by suppressing Stathmin expression. J Neurooncol. 2013;115:381–90. doi: 10.1007/s11060-013-1245-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Fu J, Pan Y, Zhang X, Shen L. Silencing of miR-1247 by DNA methylation promoted non-small-cell lung cancer cell invasion and migration by effects of STMN1. Onco Targets Ther. 2016;9:7297–307. doi: 10.2147/OTT.S111291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–33. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 42.Chakravarthi BV, Pathi SS, Goswami MT, Cieslik M, Zheng H, Nallasivam S, et al. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5:6654–69. doi: 10.18632/oncotarget.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakravarthi BV, Goswami MT, Pathi SS, Robinson AD, Cieslik M, Chandrashekar DS, et al. MicroRNA-101 regulated transcriptional modulator SUB1 plays a role in prostate cancer. Oncogene. 2016;35:6330–40. doi: 10.1038/onc.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 45.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Wang R, Asangani IA, Chakravarthi BV, Ateeq B, Lonigro RJ, Cao Q, et al. Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia. 2012;14:905–14. doi: 10.1593/neo.121192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Wang X, Casal I, Wang J, Li P, Zhang W, et al. Growth differentiation factor 15 is a promising diagnostic and prognostic biomarker in colorectal cancer. J Cell Mol Med. 2016;20:1420–6. doi: 10.1111/jcmm.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li YL, Chang JT, Lee LY, Fan KH, Lu YC, Li YC, et al. GDF15 contributes to radioresistance and cancer stemness of head and neck cancer by regulating cellular reactive oxygen species via a SMAD-associated signaling pathway. Oncotarget. 2017;8:1508–28. doi: 10.18632/oncotarget.13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–9. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 52.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 53.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11:599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chain B, Bowen H, Hammond J, Posch W, Rasaiyaah J, Tsang J, et al. Error, reproducibility and sensitivity: a pipeline for data processing of Agilent oligonucleotide expression arrays. BMC Bioinformatics. 2010;11:344. doi: 10.1186/1471-2105-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chakravarthi BV, Goswami MT, Pathi SS, Dodson M, Chandrashekar DS, Agarwal S, et al. Expression and Role of PAICS, a De Novo Purine Biosynthetic Gene in Prostate Cancer. Prostate. 2017;77:10–21. doi: 10.1002/pros.23243. [DOI] [PubMed] [Google Scholar]
- 57.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–46. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Zhang X, Chao Z, Kung HF, Lin MC, Dress A, et al. MiR-34a modulates ErbB2 in breast cancer. Cell Biol Int. 2017;41:93–101. doi: 10.1002/cbin.10700. [DOI] [PubMed] [Google Scholar]
- 61.Yin D, Ogawa S, Kawamata N, Leiter A, Ham M, Li D, et al. miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene. 2013;32:1155–63. doi: 10.1038/onc.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Luca P, Dalton GN, Scalise GD, Moiola CP, Porretti J, Massillo C, et al. CtBP1 associates metabolic syndrome and breast carcinogenesis targeting multiple miRNAs. Oncotarget. 2016;7:18798–811. doi: 10.18632/oncotarget.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–80. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Li Y, Tian H, Qi J, Li M, Fu C, et al. Macrophage inhibitory cytokine 1 (MIC-1/GDF15) as a novel diagnostic serum biomarker in pancreatic ductal adenocarcinoma. BMC Cancer. 2014;14:578. doi: 10.1186/1471-2407-14-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fisher OM, Levert-Mignon AJ, Lord SJ, Lee-Ng KK, Botelho NK, Falkenback D, et al. MIC-1/GDF15 in Barrett's oesophagus and oesophageal adenocarcinoma. Br J Cancer. 2015;112:1384–91. doi: 10.1038/bjc.2015.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Hua W, Niu LC, Li SM, Wang YM, Shang L, et al. Elevated growth differentiation factor 15 expression predicts poor prognosis in epithelial ovarian cancer patients. Tumour Biol. 2016;37:9423–31. doi: 10.1007/s13277-015-4699-x. [DOI] [PubMed] [Google Scholar]
- 68.Staff AC, Trovik J, Eriksson AG, Wik E, Wollert KC, Kempf T, et al. Elevated plasma growth differentiation factor-15 correlates with lymph node metastases and poor survival in endometrial cancer. Clin Cancer Res. 2011;17:4825–33. doi: 10.1158/1078-0432.CCR-11-0715. [DOI] [PubMed] [Google Scholar]
- 69.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Martin NE, Gerke T, Sinnott JA, Stack EC, Andren O, Andersson SO, et al. Measuring PI3K Activation: Clinicopathologic, Immunohistochemical, and RNA Expression Analysis in Prostate Cancer. Mol Cancer Res. 2015;13:1431–40. doi: 10.1158/1541-7786.MCR-14-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Belletti B, Baldassarre G. Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets. 2011;15:1249–66. doi: 10.1517/14728222.2011.620951. [DOI] [PubMed] [Google Scholar]
- 72.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 73.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 75.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Wang K, Chen X, Meng H, Song M, Wang Y, et al. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol Biol. 2012;13:4. doi: 10.1186/1471-2199-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C, Meshinchi S, et al. C/EBPalpha regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638–49. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vetter NS, Kolb EA, Mills CC, Sampson VB. The Microtubule Network and Cell Death Are Regulated by an miR-34a/Stathmin 1/betaIII-Tubulin Axis. Mol Cancer Res. 2017;15:953–64. doi: 10.1158/1541-7786.MCR-16-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adams BD, Wali VB, Cheng CJ, Inukai S, Booth CJ, Agarwal S, et al. miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer. Cancer Res. 2016;76:927–39. doi: 10.1158/0008-5472.CAN-15-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beard JA, Tenga A, Hills J, Hoyer JD, Cherian MT, Wang YD, et al. The orphan nuclear receptor NR4A2 is part of a p53-microRNA-34 network. Sci Rep. 2016;6:25108. doi: 10.1038/srep25108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dong P, Xiong Y, Watari H, Hanley SJ, Konno Y, Ihira K, et al. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer Res. 2016;35:132. doi: 10.1186/s13046-016-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao X, Huang X, Ye F, Chen B, Song C, Wen J, et al. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci Rep. 2016;6:21735. doi: 10.1038/srep21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li C, Wang J, Kong J, Tang J, Wu Y, Xu E, et al. GDF15 promotes EMT and metastasis in colorectal cancer. Oncotarget. 2016;7:860–72. doi: 10.18632/oncotarget.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.