Abstract
The Lactobacillus casei/paracasei group accumulates a high level of manganese, which works to scavenge superoxide anions produced during aerobic growth. The genome of L. casei strain Shirota, however, also codes the gene for superoxide dismutase (SOD), sodA, which catalyzes the dismutation of superoxide anion into hydrogen peroxide and oxygen. We anticipated that the SOD and/or manganese may contribute to the aerobic growth of L. casei Shirota and tried to clarify how L. casei Shirota can eliminate the toxicity of superoxide anion. When the sodA of L. casei Shirota was cloned and expressed in Escherichia coli as well as in L. casei Shirota, there was no increase in SOD activity detected, meaning that the protein is in an inactive form, even if it is produced in L. casei Shirota. We next focused on the role of the manganese transport system of L. casei Shirota. One ABC-type manganese transporter (mtsCBA cluster) and three NRAMP-type manganese transporters (mntH1, mntH2, and mntH3) are coded in the genome. To clarify the role of these genes, we disrupted one or more of these manganese transporter genes in different combinations and analyzed the intracellular manganese levels. As a result, we found that NRAMP-type manganese transporters coded by mntH1 and mntH2 and ABC-type manganese transporter coded by mtsCBA cluster are complementarily involved in the accumulation of intracellular manganese and are necessary for aerobic growth of L. casei Shirota. These results indicate that intracellular manganese accumulated by multiple complementary manganese transporters, but not SOD, plays a pivotal role in tolerance to superoxide in L. casei Shirota.
Keywords: superoxide, superoxide dismutase, manganese transporter, oxidative stress, Lactobacillus casei
INTRODUCTION
Reactive oxygen species (ROS) such as superoxide, hydrogen peroxide, and hydroxyl radicals are produced when microorganisms grow under aerobic conditions. ROS are the main causes of so-called “oxidative stress” and have various adverse effects on the growth and metabolism of the organisms, and therefore they should be eliminated with an appropriate methodology when grown under aerobic growth conditions. Lactic acid bacteria are “aerotolerant anaerobes” that are able to grow in either the presence or absence of oxygen. It is known that individual species and strains of lactic acid bacteria harbor different mechanisms to tolerate oxidative stress under aerobic growth conditions [1,2,3,4,5,6,7,8].
Superoxide dismutase (SOD) catalyzes the dismutation of superoxide into hydrogen peroxide and oxygen [9]. In lactic acid bacteria, lactococci and streptococci are reported to have manganese superoxide dismutase (Mn-SOD) activity [1, 4, 10], while it has been suggested that Lactobacillus casei and L. plantarum do not have SOD activity but instead accumulate millimolar levels of manganese in their cells that play the role of scavenging superoxide [11, 12]. In recent reports, however, there are contradictory results suggesting that L. casei has a sodA gene and SOD activity [13, 14]. When we looked at the genome sequence of Lactobacillus casei strain Shirota, it was clear that it possesses a superoxide dismutase gene, sodA, in its genome (private data).
There have been some studies in L. plantarum strains on the relationship between oxygen tolerance and manganese transporters. L. plantarum has three systems to uptake manganese in the cell [15]. The first is Mn2+- and Cd2+-specific P-type ATPase (MntA), which is a high-affinity uptake system for Mn2+ in Mn2+-starved cells [16]. The second is the ATP-binding cassette-type (ABC-type) transporter, which is composed of solute-binding protein, ATP-binding protein, and integral membrane protein and is widely distributed in bacteria. The growth of solute-binding protein mutants in other bacteria is impaired in Mn-limited medium in the presence of oxygen [17,18,19,20]. The third is the natural resistance-associated macrophage protein (NRAMP) transporter [21]. Homologues of this protein are widely distributed in bacteria and have been characterized [22, 23]. A study using L. plantarum strains where one or two of these uptake genes were disrupted revealed that there was no significant change in growth or manganese uptake, suggesting that Mn2+ homeostasis is regulated by multiple genes [15]. With regard to L. casei, on the other hand, there has been no report of the functions of these manganese transporters.
We previously reported that the thioredoxin–thioredoxin reductase system is essential for aerobic growth of L. casei strain Shirota [24] and discovered a novel hydrogen peroxide resistance gene [25]. In the current study, we further analyzed the contributions of sodA and manganese to superoxide tolerance in L. casei strain Shirota and the uptake of manganese using manganese transporter disruption mutants.
MATERIALS AND METHODS
Strains, plasmids, media, and growth conditions
The strains and plasmids used are shown in Table 1. L. casei strain Shirota (YIT 9029) was used as the wild-type strain. Escherichia coli JM109 was purchased from Toyobo (Osaka, Japan) as competent cells for DNA transformation. E. coli BLR(DE3) was purchased from Merck (Tokyo, Japan). L. casei ATCC334, L. lactis ATCC19435, and E. coli K-12 HfrH were used for cloning the sodA genes. Plasmid pYSSE3 [26] was used as a cloning vector for deletion mutagenesis. Plasmid pYAP300 [26], which enables gene integration into the L. casei chromosome at the attB site for phage phiFSW and has a constitutive expression promoter, was used for expression of various sodA genes. Plasmid pET-21(+) (Merck) was used as an expression vector for recombinant proteins in E. coli. Lactic acid bacteria were grown at 37°C in MRS medium or Mn-free MRS medium supplemented with various concentrations of manganese. E. coli strains were grown at 37°C in LB medium [27]. Erythromycin (Abbott Japan, Tokyo, Japan) was added to a final concentration of 20 µg/ml for lactic acid bacteria and 500 µg/ml for E. coli if needed. The turbidity of the cultures was measured with a Klett-Summerson photoelectric colorimeter (Klett MFG, New York, USA) or a DU 800 spectrophotometer (Beckman Coulter, Tokyo, Japan).
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype or phenotype | Source or reference | ||
|---|---|---|---|---|
| Strains | ||||
| Lactobacillus casei | ||||
| Shirota (YIT 9029) | Wild-type | Our collection | ||
| TM1002 | Deletion in mntH1 gene | This study | ||
| TM1003 | Deletion in mntH2 gene | This study | ||
| TM1004 | Deletion in mtsA gene | This study | ||
| TM1005 | Deletion in mntH1 and mntH2 gene | This study | ||
| TM1006 | Deletion in mntH1 and mtsA gene | This study | ||
| TM1010 | Deletion in mntH2 and mtsA gene | This study | ||
| TM1011 | Deletion in mntH1,mntH2, and mtsA gene | This study | ||
| TM1012 | Deletion in mntH1,mntH2, and mntH3 gene | This study | ||
| MS1001 | attB::pYAP300–LlsodA, Emr | This study | ||
| MS1002 | attB::pYAP300–sodA, Emr | This study | ||
| ATCC334 | Strain isolated from cheese | ATCC | ||
| Lactococcus lactis | ||||
| ATCC19435 | Type strain | ATCC | ||
| Escherichia coli | ||||
| JM109 | Commercial strain purchased from Toyobo Co., Ltd. | |||
| K-12 HfrH | High-frequency recombination strain of K-12 | Our collection | ||
| BLR(DE3) | Commercial strain purchased from Merck Ltd. | |||
| BSM4 | BLR(DE3) harboring pET-21(+)–sodAM | This study | ||
| BES3 | BLR(DE3) harboring pET-21(+)–EcsodA | This study | ||
| Plasmids | ||||
| pET-21(+) | Commercial plasmid purchased from Merck Ltd. | |||
| pET-21(+)–sodAM | pET-21(+) carrying wild-type sodA with the E. coli Shine-Dalgarno sequence | This study | ||
| pET-21(+)–EcsodA | pET-21(+) carrying E. coli sodA with the E. coli Shine-Dalgarno sequence | This study | ||
| pYSSE3 | E. coli cloning vector carrying the pUC19 ori region, pAMβ1 erythromycin resistance gene, and multicloning sites | [26] | ||
| pYSSE3–ΔmntH1-del | pYSSE3 carrying an upstream region with the N terminus of mntH1and downstream region with the C terminus of mntH1 | This study | ||
| pYSSE3–ΔmntH2-del | pYSSE3 carrying an upstream region with the N terminus of mntH2and downstream region with the C terminus of mntH2 | This study | ||
| pYSSE3–ΔmntH3-del | pYSSE3 carrying an upstream region with the N terminus of mntH3and downstream region with the C terminus of mntH3 | This study | ||
| pYSSE3–ΔmtsA-del | pYSSE3 carrying an upstream region with the N terminus of mtsA and downstream region with the C terminus of mtsA | This study | ||
| pYAP300 | E. coli cloning vector carrying the p15A ori region, pAMβ1 erythromycin resistance gene, phiFSW attP site and int, multicloning site, and synthetic promoter sequence active in lactobacilli upstream of the multicloning site | [26] | ||
| pYAP300–LlsodA | pYAP300 carrying L. lactis ATCC19435 sodA | This study | ||
| pYAP300–sodA | pYAP300 carrying wild-type sodA | This study | ||
Construction of gene disruption mutants
Gene disruption mutants were constructed by deletion of targeted genes as reported previously [24]. The primer pairs used to amplify fragments containing the 5′- and 3′- terminal ends of target genes are listed in Table 2. Recombinant plasmids were constructed with these two fragments and pYSSE3. These plasmids were introduced into L. casei strain Shirota, and gene deletion mutants were constructed using a stepwise double-crossover method.
Table 2. Primers used in this study.
| Target gene | Primer sequence (5´→3´) |
Restriction enzyme site | |
|---|---|---|---|
| Forward | Reverse | ||
| Construction of deletion mutants | |||
| mntH1-5´ | TATTCTAGAAGTGAGCGGTAAGCAG | TATCTGCAGGCGTATCTTCGAAGTGAAT | Xba I/Pst I |
| mntH1-3´ | AATCTGCAGTGCTCATCTTCGCGAT | AATAAGCTTTTGATGCATGGACA | Pst I/Hind III |
| mntH2-5´ | AACAAGCTTCTCCAGTGAGGACAG | TAAGAGCTCACAATAACTCTCCCCT | Hind III/SacI |
| mntH2-3´ | ATAGAGCTCACATCTTGCAGCTGTAGC | TAATCTAGAACGGTGCCAGCCGCC | SacI/Xba I |
| mntH3-5´ | CGCGGATCCGTCATCACAATGCCTAAAG | CCGGAATTCATAACCAACAGCAACCAAGG | BamH I/EcoR I |
| mntH3-3´ | CCGGAATTCATGTGACGGCTTTTCAAGG | GTTGCTGCAGCATTGGCAGG | EcoR I/Pst I |
| mtsA-5´ | AATCTGCAGGCTTGATCGGAAGCT | TATGTCGACCCCTGCAACCATGGTT | Pst I/Sal I |
| mtsA-3´ | ATAGTCGACGGTTTGGCGGAATA | TAATCTAGAGCAACGTTCCAGTCTC | Sal I/Xba I |
| Expression in L. casei | |||
| sodA | TCCCCCGGGGAAAGGTTGATTCCTATGAC | GCTCTAGATCAGGCGTTTGTATCGGG | Sma I/Xba I |
| LlsodA | TCCCCCGGGTAAAAGGAGAAATTACTATGG | GCTCTAGATTATTTTGCTTTAGCATAAAG | Sma I/Xba I |
| Expression in E. coli | |||
| sodA | CGGGATCCaagaaggagatatacatATGACATTTGTTTTGCCAGAT | CCGCTCGAGGGCGTTTGTATCGGGATGC | BamH I/Xho I |
| EcsodA | CGGGATCCaagaaggagatatacatATACATATGAGCTATACCCTGCCAT | CCGCTCGAGTTTTTTCGCCGCAAAACGTG | BamH I/Xho I |
The restriction enzyme sites are indicated (underlined).
The sequences derived from pET21a(+) containing the Shine-Dalgarno sequence are indicated by lowercase letters.
Preparation of cell-free extracts
Cells from 2 ml of overnight cultures of lactic acid bacteria and E. coli were harvested by centrifugation. They were washed twice with 0.1 M phosphate buffer (pH 7.0). They were then suspended in 500 µl of the same buffer, to which had been added 0.3 g of glass beads (0.1 mm in diameter), and were disrupted by a FastPrep FP120 cell disrupter (Thermo, Tokyo, Japan) at speed of 6.5 m/sec for 30 sec. The solution was centrifuged at 18,000 g for 10 min, and the supernatant was used as a cell-free extract. The protein concentration was determined with a Bio-Rad Protein assay (Bio-Rad Laboratories, Tokyo, Japan).
Measurement of SOD activity
SOD activity was measured by the method of McCord and Fridovich [9] with some modifications. A mixture of 1.1 ml of 50 mM phosphate buffer (pH 7.8), 0.6 ml of 0.5 mM EDTA in phosphate buffer, 0.5 ml of 0.06 mM cytochrome c in phosphate buffer, 0.5 ml of 0.3 mM xanthine in phosphate buffer, and 0.1 ml of sample solution with or without 0.1 mM EDTA was prepared. The reaction was initiated by adding to the mixture 0.2 ml of xanthine oxidase solution at a concentration to produce reduction of cytochrome c at a rate of 0.025 absorbance units per minute at 550 nm. The mixture was then incubated at 25°C, and the increase in absorbance at 550 nm during 1 min incubation was measured. The amount of superoxide dismutase required to inhibit the rate of reduction of cytochrome c by 50% was defined as 1 U of activity. The activity was measured without EDTA when we evaluated the effect of manganese.
Cloning and expression of sodA genes from different bacteria
Expression in L. casei
The sodA gene from L. lactis ATCC19435 or L. casei strain Shirota was amplified from the respective genomic DNA by PCR using KOD Plus DNA polymerase (Toyobo) with the primers indicated in Table 2 and was cloned into pYAP300 containing a constitutive expression promoter. pYAP300–LlsodA or pYAP300–sodA were introduced into L. casei strain Shirota in a manner similar to that for gene disruption.
Expression in E. coli
The sodA gene from L. casei strain Shirota or E. coli K-12 HfrH was amplified from the respective genomic DNA by PCR using KOD Plus DNA polymerase (Toyobo). Forward primers were designed containing the pET-21a(+) Shine-Dalgarno sequence (Table 2). The fragments of PCR products were digested at BamH I/Xho I and cloned into pET-21(+), which had previously been digested with the same restriction enzymes. These plasmids (pET-21(+)–sodAM and pET-21(+)–EcsodA) were amplified in E. coli JM109 and transferred into E. coli BLR(DE3).
The recombinant SOD proteins were overproduced in E. coli in accordance with the pET system manual (Merck), with some modifications. E. coli BSM4 (harboring pET-21(+)–sodAM) and BES3 (harboring pET-21(+)–EcsodA) were grown at 30°C in LB medium containing 100 µg/ml ampicillin until OD600 = 0.6. Then, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added for SOD induction, and the culture was incubated overnight at 20°C or 30°C. Cell-free extracts and whole cells were analyzed by SDS-PAGE with Perfect NT Gels (5–20%) (DRC, Tokyo, Japan). Concentrated protein precipitated from 50 µl of cell-free extracts by trichloroacetic acid or whole cells from 100 µl of culture were loaded onto the gels.
Treatment with paraquat
Overnight cultures grown in Mn-free MRS medium were inoculated at 1% (v/v) into Mn-free MRS medium with or without 1 µg/ml manganese. Paraquat at 0.5, 1, or 2 mM was added to the respective cultures. The cultures were grown at 37°C and the turbidity of the cultures was measured by using a Klett-Summerson photoelectric colorimeter (Klett MFG).
Measurement of manganese concentration
Overnight cultures grown in Mn-free MRS medium supplemented with 1 µg/ml manganese were inoculated at 1% (v/v) into 30 ml of Mn-free MRS medium supplemented with various concentrations of manganese. Cells were grown at 37°C for 24 hr and harvested by centrifugation at 5,000 g for 10 min. Cells were washed twice with H2O and suspended in the same volume of H2O. The amount of intracellular manganese was measured by inductively coupled plasma optical emission spectrometry (ICP-OES) (Vista-PRO, Agilent Technologies, Tokyo, Japan).
Multiple alignment
Multiple alignment was performed by using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
RESULTS
SOD of L. casei strain Shirota
L. casei strain Shirota has a sodA gene in its genome. The predicted amino acid sequence of SOD of L. casei strain Shirota was of high similarity to Mn-, Fe-, and Fe/Mn-SOD of other organisms (Fig. 1). The amino acid residues at the active sites and metal ligands [4, 28] were fully conserved (Fig. 1). The identity with Mn-SOD was slightly higher than that with Fe-SOD (e.g., Bacillus subtilis Mn-SOD, 56%; L. lactis Mn-SOD, 53%; E. coli Mn-SOD, 50%; E. coli Fe-SOD, 41%; and P. aeruginosa Fe-SOD, 41%). In addition, since the SOD of L. casei strain Shirota has the typical amino acid residues of Mn-SOD [28], L. casei SOD was predicted to be Mn-SOD. Further, the amino acid sequence of L. casei SOD also has high similarity with B. anthracis Fe/Mn-SOD (55% identity), which was recently reported as cambialistic SOD [29].
Fig. 1.
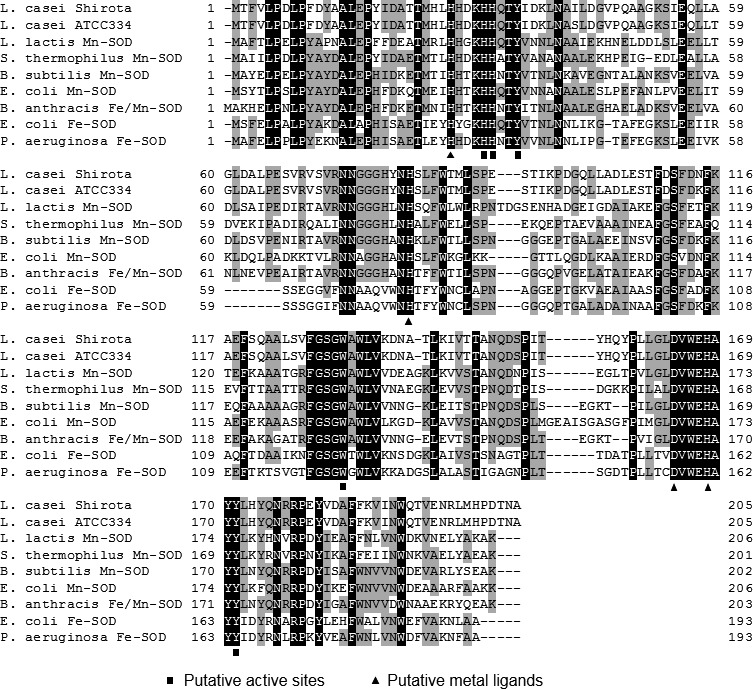
Multiple alignment of the amino acid sequences of superoxide dismutase in various species. The residues with a black background represent residues conserved in all sequences; those with a grey background represent residues conserved in at least 50% of all sequences. Putative active sites and putative metal ligands are indicated by square and triangle symbols, respectively.
SOD activity of wild-type strain and recombinants
Table 3 shows the SOD activity of wild-type strain and recombinants. While the SOD activity of L. lactis was 45.1 U/mg protein, L. casei wild-type strains did not show any detectable activity. When we expressed L. lactis sodA and L. casei sodA in L. casei strain Shirota, L. lactis sodA showed SOD activity in L. casei strain Shirota (MS1001), while L. casei sodA did not show any activity even under the constitutively active promoter (MS1002).
Table 3. SOD activity of lactic acid bacteria and E. coli.
| Strain | SOD activity (U/mg protein) |
||
|---|---|---|---|
| IPTG+ | IPTG– | ||
| L. casei Shirota | ND | - | - |
| L. casei ATCC334 | ND | - | - |
| L. lactis ATCC19435 | 45.1 | - | - |
| L. casei MS1001 | 11.0 | - | - |
| L. casei MS1002 | ND | - | - |
| E. coli BES3 (20°C) | - | 100.4 | 55.6 |
| E. coli BSM4 (20°C) | - | 32.8 | 38.4 |
ND: not detected.
Temperatures in parentheses are those when the gene expression was induced.
The production of sodA protein was further confirmed in E. coli using the pET system. The SOD protein was not found in cell-free extract when the recombinant cells were cultured at or higher than at 30°C, probably due to the formation of inclusion bodies. The SOD of L. casei strain Shirota remained in soluble form when expression of recombinant protein was induced overnight in the presence of 1 mM IPTG at 20°C (Fig. 2). However, the SOD activity in the cell-free extract of E. coli BSM4 (L. casei SOD-expressing recombinant) did not vary regardless of induction by IPTG (Table 3). In contrast, the SOD activity in the cell-free extract of E. coli BES3 (E. coli SOD-overexpressing recombinant) was increased by IPTG induction (Table 3). From these results, it was suggested that the SOD protein of L. casei strain Shirota, even if it is produced, is in an inactive form.
Fig. 2.
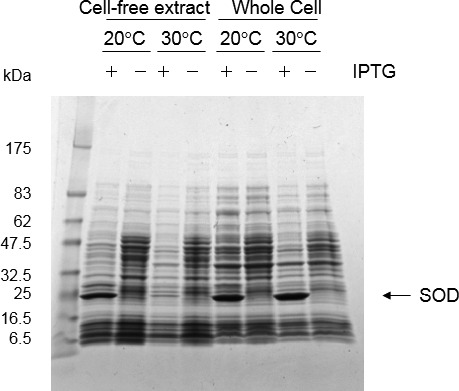
SDS-PAGE analysis of L. casei strain Shirota SOD expression in E. coli. E. coli BSM4 (harboring pET-21(+)–sodAM) was grown at 30°C in LB medium containing 100 µg/ml ampicillin until OD600=0.6. Then, IPTG (1 mM) was added for SOD induction, and the culture was grown overnight at 20°C or 30°C. Concentrated protein was precipitated from 50 µL of cell-free extracts by trichloroacetic acid, and whole cells from 100 µL cultures were loaded onto the gel. The arrow indicates SOD proteins.
Effect of SOD and manganese on SOD activity and oxidative stress resistance
We evaluated the effect of SOD and manganese with respect to oxidative stress. We examined the effect of paraquat, which is a superoxide generator, on growth by using the wild-type L. casei strain Shirota and the recombinant strain MS1001 (L. casei expressing L. lactis sodA), with or without 1 µg/ml manganese (Fig. 3). Growth of both strains was partly inhibited in Mn-free medium. Further, the growth of the wild-type strain in the absence of manganese was inhibited by the addition of paraquat, whereas the growth inhibition caused by paraquat was partially recovered by expression of the L. lactis sodA gene (Fig. 3c, 3d). However, in the presence of 1 µg/ml manganese, the addition of paraquat did not affect growth in either strain (Fig. 3a, 3b).
Fig. 3.
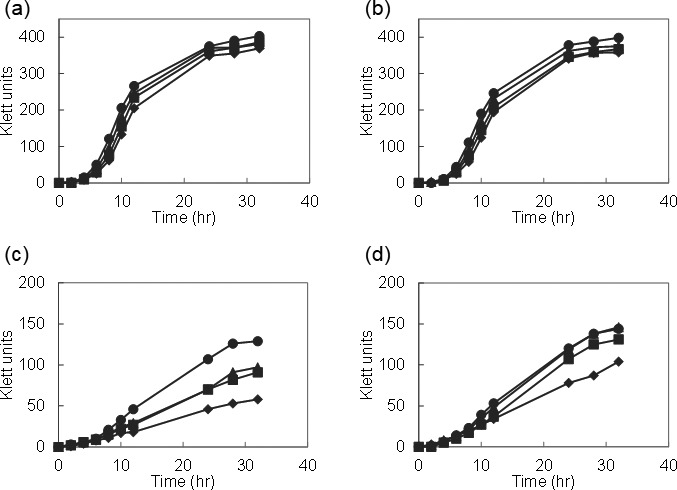
Effect of paraquat on the growth of L. casei strain Shirota and L. casei MS1001 (expressing L. lactis sodA). Overnight cultures grown in Mn-free MRS medium were inoculated at 1% (v/v) into Mn-free MRS medium with or without 1 µg/ml manganese. The growth of L. casei strain Shirota with manganese (a) or without manganese (c). The growth of L. casei MS1001 with manganese (b) or without manganese (d). ●, no paraquat; ▲, 0.5 mM paraquat; ■, 1.0 mM paraquat; and ♦, 2 mM paraquat.
The SOD activity of wild-type L. casei and MS1001 was measured with or without EDTA in the reaction mixture (Fig. 4). The SOD activity with EDTA indicates the activity caused by an enzymatic reaction, while that without EDTA indicates the activity caused by both enzymatic and nonenzymatic (manganese) reactions. We did not detect SOD activity in the wild-type L. casei in the presence of EDTA in the reaction mixture, while the SOD activity of MS1001 was detected after adding EDTA to the reaction mixture. This SOD activity remained the same regardless of the manganese concentration of the medium. However, when the reaction mixture contained no EDTA, the SOD activities of both strains increased in proportion to the concentration of manganese in the medium. In Mn-rich medium in particular, the SOD activity derived from manganese was markedly higher than the activity derived from the enzyme.
Fig. 4.
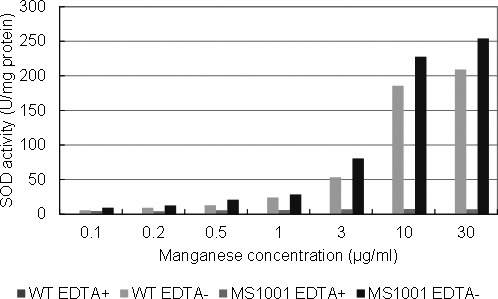
SOD activity of L. casei strain Shirota (WT) and L. casei MS1001 (expressing L. lactis sodA). Cell-free extracts of cells grown overnight were measured with or without EDTA in the mixture.
Effect of manganese transporters on manganese accumulation
We found one ABC-type manganese transporter (mtsCBA) and three NRAMP-type manganese transporters (mntH1, mntH2, and mntH3) in the genome of L. casei strain Shirota. However, we did not find the P-type ATPase mntA. In order to clarify the role of these transporters, we disrupted mntH1, mntH2, mntH3, and mtsA, which codes a solute-binding protein of the ABC-type transporter, in different combinations. Then, we measured the intracellular manganese concentrations in the wild-type strain and in manganese transporter–deficient mutants grown overnight in Mn-free MRS medium with various concentrations of manganese (Fig. 5). The wild-type cells incorporated manganese in a concentration-dependent manner, and almost all manganese in the medium up to 1 µg/ml was incorporated into the cells. However, at higher concentration of manganese, the uptake increased gently. The uptake of manganese was not affected in the mutant cells in which only one of the four predicted manganese transporter genes was disrupted (TM1002, TM1003, TM1004), even though a variation in uptake was seen at high concentrations of manganese. On the other hand, TM1005, which had deletion mutations in two genes (∆mntH1/∆mntH2), showed marked reduction of manganese uptake. Furthermore, TM1011, a triple mutant (∆mntH1/∆mntH2/∆mtsA), could not grow and could not accumulate manganese in the medium supplemented with lower manganese concentrations (≤3 µg/ml). However, TM1011 grew marginally and showed residual manganese incorporation activity in the medium supplemented with higher manganese concentrations. On the other hand, the manganese uptake pattern of another triple mutant, TM1012 (∆mntH1/∆mntH2/∆mntH3), was quite similar to that of TM1005 (∆mntH1/∆mntH2).
Fig. 5.
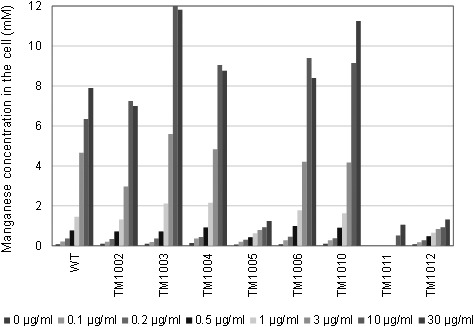
Intracellular manganese accumulation by L. casei strain Shirota and manganese transporter–disrupted mutants. Cells were grown in Mn-free MRS medium supplemented with various concentrations of manganese. Manganese concentrations in the cells are represented as mmol per whole washed pellet volume.
DISCUSSION
SOD is an enzyme that catalyzes dismutation of superoxide anion and is important for defense against oxygen toxicity. Many lactobacilli do not have SOD activity; however, oxygen-tolerant L. casei and L. plantarum are able to scavenge superoxide with high intracellular levels of accumulated manganese [11, 12]. Lin et al. [13] and Liu et al. [14] recently reported that some strains of L. casei encode a sodA gene in the genome and have SOD activity. As we describe in this report, it is true that L. casei strain Shirota possesses the sodA gene sequence in its genome. The predicted amino acid sequence of the sodA gene was highly similar to those of Mn-, Fe-, and Fe/Mn-SOD of other bacterial species, and the amino acid residues of putative metal ligands and putative active sites were conserved, although we could not identify its cofactor through homology analysis. When the sodA of L. casei strain Shirota was expressed in L. casei strain Shirota or in E. coli, we failed to detect its activity. Because the activity of SOD derived from L. lactis used as a positive control could be detected in L. casei strain Shirota and because the SOD protein of L. casei strain Shirota expressed in E. coli could be detected in soluble form, it is most probable that the product is not functional even though the protein is synthesized.
In L. casei and L. plantarum, the ability of manganese to scavenge superoxide was determined from the results of experiments in which the SOD activity was inhibited by dialysis and treatment with EDTA [11, 12]. In this study, although the SOD activity of L. casei strain Shirota was lost when EDTA was added, the activity was increased depending on the manganese concentration added to the medium in the absence of EDTA, and a correlation between the SOD activity and the concentration of manganese in the cells was found (Fig. 4). Furthermore, when we examined tolerance to superoxide of the wild-type strain and a recombinant strain that express L. lactis sodA, suppression of growth by paraquat was observed only when they were cultured in Mn-free medium. The presence of L. lactis sodA partially recovered their growth (Fig. 3), meaning that both manganese and SOD are involved in quenching superoxide toxicity. Because the SOD activity derived from L. lactis sodA was less than the superoxide elimination activity caused by manganese in the recombinant (Fig. 4), it is considered that the effect of L. lactis SOD was not detected when enough manganese was added to the medium. Although there are reports that heterologous SOD expression improves oxidative stress resistance in some lactic acid bacteria [30, 31], most of the species examined in those studies were sensitive to oxidative stress or were species that did not have SOD activity. Meanwhile, although L. casei strain Shirota has a sodA gene, its SOD activity is suspected to have been lost in the course of evolution because the lost SOD activity is sufficiently compensated for by the much stronger superoxide elimination activity of intracellular manganese.
Recently, some L. casei strains have been reported to have SOD activity [13, 14]. Our results supported the results of Archibald et al. indicating that L. casei had no SOD activity [11, 12]. On the other hand, the sodA sequences of L. casei strain Shirota and L. casei Lc18 are perfectly identical, and it is not clear what caused the difference in the results of Liu et al. [14]. One possibility is that the SOD assay methods used by the two groups were different from each other. In particular, since free manganese has superoxide elimination activity and has a strong influence on the apparent SOD activity, it is important to eliminate the possible influence of manganese by adding EDTA in the assay system. Therefore, we think that there is a necessity to reevaluate whether those activities were derived from true SOD or from superoxide scavenging activity of intracellular manganese.
We further investigated the mechanism of manganese uptake of L. casei strain Shirota. Because single-gene disruption mutants did not affect the phenotype, multiple genes were suggested to have complementary functions (Fig. 5). The quantity of manganese uptake of the ∆mntH1/∆mntH2 double disruption mutant was markedly reduced, and the triple disruption mutant with additional mtsA disruption was not able to take up manganese at concentrations of up to 3 µg/ml in the medium (Fig. 5). Therefore, these three genes are suggested to have a pivotal role in enabling sufficient accumulation of intracellular manganese. However, the ∆mntH1/∆mntH2/∆mtsA triple disruption mutant can grow and take up manganese slightly under high manganese concentration conditions (Fig. 5). Thus, other relevant genes might still function. However, as the triple gene disruption mutant could hardly grow at all, we conclude that NRAMP-type manganese transporters coded by mntH1and mntH2 and an ABC-type manganese transporter coded by the gene cluster of mtsCBA are the transporters that are responsible for accumulating all or majority of the intracellular manganese in L casei strain Shirota.
Taken together, SOD is defective in L. casei strain Shirota and intracellular manganese accumulated by multiple manganese transporters mainly functions to scavenge superoxide.
Acknowledgments
We thank Kosuke Kondo and Eriko Nishi of Yakult Central Institute for their help with the ICP-OES measurements.
REFERENCES
- 1.Chang SK, Hassan HM. 1997. Characterization of superoxide dismutase in Streptococcus thermophilus. Appl Environ Microbiol 63: 3732–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kono Y, Fridovich I. 1983. Isolation and characterization of the pseudocatalase of Lactobacillus plantarum. J Biol Chem 258: 6015–6019. [PubMed] [Google Scholar]
- 3.Miyoshi A, Rochat T, Gratadoux JJ, Le Loir Y, Oliveira SC, Langella P, Azevedo V. 2003. Oxidative stress in Lactococcus lactis. Genet Mol Res 2: 348–359. [PubMed] [Google Scholar]
- 4.Sanders JW, Leenhouts KJ, Haandrikman AJ, Venema G, Kok J. 1995. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol 177: 5254–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higuchi M, Shimada M, Yamamoto Y, Hayashi T, Koga T, Kamio Y. 1993. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J Gen Microbiol 139: 2343–2351. [DOI] [PubMed] [Google Scholar]
- 6.Higuchi M, Yamamoto Y, Poole LB, Shimada M, Sato Y, Takahashi N, Kamio Y. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol 181: 5940–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knauf HJ, Vogel RF, Hammes WP. 1992. Cloning, sequence, and phenotypic expression of katA, which encodes the catalase of Lactobacillus sake LTH677. Appl Environ Microbiol 58: 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abriouel H, Herrmann A, Stärke J, Yousif NM, Wijaya A, Tauscher B, Holzapfel W, Franz CM. 2004. Cloning and heterologous expression of hematin-dependent catalase produced by Lactobacillus plantarum CNRZ 1228. Appl Environ Microbiol 70: 603–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCord JM, Fridovich I. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244: 6049–6055. [PubMed] [Google Scholar]
- 10.Vance PG, Keele BB, Jr, Rajagopalan KV. 1972. Superoxide dismutase from Streptococcus mutans. Isolation and characterization of two forms of the enzyme. J Biol Chem 247: 4782–4786. [PubMed] [Google Scholar]
- 11.Archibald FS, Fridovich I. 1981. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J Bacteriol 146: 928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archibald FS, Fridovich I. 1981. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J Bacteriol 145: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Zou Y, Cao K, Ma C, Chen Z. 2016. The impact of heterologous catalase expression and superoxide dismutase overexpression on enhancing the oxidative resistance in Lactobacillus casei. J Ind Microbiol Biotechnol 43: 703–711. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Hang X, Liu X, Tan J, Li D, Yang H. 2012. Cloning and heterologous expression of the manganese superoxide dismutase gene from Lactobacillus casei Lc18. Ann Microbiol 62: 129–137. [Google Scholar]
- 15.Groot MN, Klaassens E, de Vos WM, Delcour J, Hols P, Kleerebezem M. 2005. Genome-based in silico detection of putative manganese transport systems in Lactobacillus plantarum and their genetic analysis. Microbiology 151: 1229–1238. [DOI] [PubMed] [Google Scholar]
- 16.Hao Z, Chen S, Wilson DB. 1999. Cloning, expression, and characterization of cadmium and manganese uptake genes from Lactobacillus plantarum. Appl Environ Microbiol 65: 4746–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crump KE, Bainbridge B, Brusko S, Turner LS, Ge X, Stone V, Xu P, Kitten T. 2014. The relationship of the lipoprotein SsaB, manganese and superoxide dismutase in Streptococcus sanguinis virulence for endocarditis. Mol Microbiol 92: 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janulczyk R, Ricci S, Björck L. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun 71: 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston JW, Myers LE, Ochs MM, Benjamin WH, Jr, Briles DE, Hollingshead SK. 2004. Lipoprotein PsaA in virulence of Streptococcus pneumoniae: surface accessibility and role in protection from superoxide. Infect Immun 72: 5858–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik S, Brown A, Munro CL, Cornelissen CN, Kitten T. 2003. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J Bacteriol 185: 5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forbes JR, Gros P. 2001. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol 9: 397–403. [DOI] [PubMed] [Google Scholar]
- 22.Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35: 1454–1468. [DOI] [PubMed] [Google Scholar]
- 23.Kehres DG, Zaharik ML, Finlay BB, Maguire ME. 2000. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol 36: 1085–1100. [DOI] [PubMed] [Google Scholar]
- 24.Serata M, Iino T, Yasuda E, Sako T. 2012. Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. Microbiology 158: 953–962. [DOI] [PubMed] [Google Scholar]
- 25.Serata M, Kiwaki M, Iino T. 2016. Functional analysis of a novel hydrogen peroxide resistance gene in Lactobacillus casei strain Shirota. Microbiology 162: 1885–1894. [DOI] [PubMed] [Google Scholar]
- 26.Yasuda E, Serata M, Sako T. 2008. Suppressive effect on activation of macrophages by Lactobacillus casei strain Shirota genes determining the synthesis of cell wall-associated polysaccharides. Appl Environ Microbiol 74: 4746–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook JW, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 28.Parker MW, Blake CC. 1988. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett 229: 377–382. [DOI] [PubMed] [Google Scholar]
- 29.Tu WY, Pohl S, Gray J, Robinson NJ, Harwood CR, Waldron KJ. 2012. Cellular iron distribution in Bacillus anthracis. J Bacteriol 194: 932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruno-Bárcena JM, Andrus JM, Libby SL, Klaenhammer TR, Hassan HM. 2004. Expression of a heterologous manganese superoxide dismutase gene in intestinal lactobacilli provides protection against hydrogen peroxide toxicity. Appl Environ Microbiol 70: 4702–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo F, Yu R, Feng X, Khaskheli GB, Chen L, Ma H, Chen S. 2014. Combination of heterogeneous catalase and superoxide dismutase protects Bifidobacterium longum strain NCC2705 from oxidative stress. Appl Microbiol Biotechnol 98: 7523–7534. [DOI] [PubMed] [Google Scholar]


