Abstract
Purpose
Oxidized low-density lipoprotein (oxLDL) plays a key role in endothelial dysfunction, vascular inflammation, and atherogenesis. The aim of this study was to assess blood clearance and in vivo kinetics of radiolabeled oxLDL in mice.
Methods
We synthesized 123I–oxLDL by the iodine monochloride method, and performed an uptake study in CHO cells transfected with lectin-like oxLDL receptor-1 (LOX-1). In addition, we evaluated the consistency between the 123I–oxLDL autoradiogram and the fluorescence image of DiI-oxLDL after intravenous injection for both spleen and liver. Whole-body dynamic planar images were acquired 10 min post injection of 123I–oxLDL to generate regional time-activity curves (TACs) of the liver, heart, lungs, kidney, head, and abdomen. Regional radioactivity for those excised tissues as well as the bladder, stomach, gut, and thyroid were assessed using a gamma counter, yielding percent injected dose (%ID) and dose uptake ratio (DUR). The presence of 123I–oxLDL in serum was assessed by radio-HPLC.
Results
The cellular uptakes of 123I–oxLDL were identical to those of DiI-oxLDL, and autoradiograms and fluorescence images also exhibited consistent distributions. TACs after injection of 123I–oxLDL demonstrated extremely fast kinetics. The radioactivity uptake at 10 min post-injection was highest in the liver (40.8 ± 2.4% ID). Notably, radioactivity uptake was equivalent throughout the rest of the body (39.4 ± 2.7% ID). HPLC analysis revealed no remaining 123I–oxLDL or its metabolites in the blood.
Conclusion
123I–OxLDL was widely distributed not only in the liver, but also throughout the whole body, providing insight into the pathophysiological effects of oxLDL.
Keywords: Oxidized low-density lipoprotein (oxLDL), Radiolabeled ligand, Dynamic planar imaging, Biodistribution
Introduction
Oxidized low-density lipoprotein (oxLDL) is involved in vascular endothelial dysfunction, injury, and inflammation, and plays an important role in atherogenesis [1, 2]. When deposited in the aortic subendothelial layer of atherosclerotic lesions, oxLDL is endocytosed by macrophages, which become foam cells, leading to atherogenesis [3]. oxLDL is also closely associated with diabetes [4], preeclampsia [5], and osteoarthritis [6]. Epidemiological studies of oxLDL revealed that an elevated concentration of oxLDL in the blood is associated with an increased risk of cardiovascular diseases, including coronary disease [7], ischemic stroke [8], and metabolic syndrome [9].
Once oxLDL enters the circulation, the liver is the major organ responsible for its removal from the blood. Specifically, Kupffer cells in the liver play an important role in removing oxLDL [10], as demonstrated by studies using Kupffer cells isolated from human liver [11] and rabbit liver [12]. Therefore, the lifetime of oxLDL in the circulation is very short, and consequently, under normal physiological conditions, the oxLDL concentration in the blood is low. Clearance of oxLDL from the blood is also associated with protection against the pathological alterations caused by this protein [10]. The system that prevents oxLDL accumulation can deteriorate as a consequence of aging or disease, causing higher levels of oxLDL to remain in the blood [7–9]. Thus, the kinetics of oxLDL, including blood clearance and accumulation in various organs (not limited to the liver), might contribute to assessments of risk status for vascular diseases.
Several radiolabeled tracers have been applied to the in vivo kinetic studies of oxLDL. Ling et al. utilized iodine-125-labeled oxLDL (125I–oxLDL) and demonstrated using extracted tissue samples that over 90% ID was cleared from the blood at 5 min post-injection in normal mice, and that 50% ID of 125I–oxLDL accumulated in the liver [13]. Rapid clearance of 125I–oxLDL from the blood and accumulation in the liver were also reported in rats [10] and in guinea pigs [14]. Sequential planar imaging in humans after i.v. injection of technetium-99 m-labeled oxLDL (99mTc-oxLDL), followed by visual or qualitative observation, revealed that the labeled oxLDL mostly accumulated ine the liver. This observation suggested that high accumulation in the liver is the major cause of rapid clearance of oxLDL from the blood [15]. 125I–oxLDL is not an ideal radio-ligand for in vivo imaging, due to the low energy of the gamma rays (30.5–35.3 keV) emitted from the 125I isotope; this is particularly true in larger objects, which exhibit greater attenuation. Iodine-123 (123I), which emits higher-energy gamma rays (159 keV), has been extensively utilized for clinical scans. With this isotope, application of suitable reconstruction procedures, including appropriate corrections for attenuation and scatter, enables quantitative imaging [16, 17].
In this study, we developed a technique for synthesizing 123I–labeled oxLDL (123I–oxLDL), and then evaluated the adequacy of the synthesized ligands by comparing the uptake rate of 123I–oxLDL to that of fluorescent oxLDL (DiI-oxLDL) in LOX-1-transfected CHO cells. We then quantitatively monitored the in vivo kinetics of 123I–oxLDL after i.v. injection, and assessed the biodistribution of 123I–oxLDL.
Materials and Methods
Subjects
Human LOX-1-transfected Chinese hamster ovary (hLOX-1-CHO) cells were used to evaluate the affinity of synthesized 123I–oxLDL and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate-labeled oxLDL (DiI-oxLDL). Detailed techniques for the preparation of the cells were described in a previous report [18]. Twenty seven C57BL/6 mice (CLEA Japan Inc., Tokyo, Japan), 10 weeks old at the time of the experiment, were used for in vivo imaging and ex vivo assessments. All mice were male, and average body weight was 22.7 ± 1.4 g. The mice were habituated for 2 weeks before the experiment in a controlled facility at 26 °C and 40% humidity, and then provided with water and chow ad libitum.
Preparation of Labeled oxLDL
OxLDL was prepared as described previously [19]. In brief, native albumin-free low-density lipoprotein (LDL) was isolated by sequential ultracentrifugation from plasma obtained from healthy human male volunteers. The LDL was oxidized by exposure to 7.5 μM CuSO4 for 16 h at 37 °C at a protein concentration of 3 mg/ml in phosphate-buffered saline (PBS). The protein content of oxLDL was measured using a BCA protein assay kit (PIERCE, Rockford, IL, USA).
The oxLDL was labeled with 123I as described previously [20] with some modifications. Iodine monochloride (ICl) (6.5 μl; 2600 nmol; Wako Pure Chemical Industries, Osaka, Japan) was added to a mixture of oxLDL (12.5 μl, 7 mg/ml; 0.05 nmol), to which was added [123I]NaI (111 MBq, 0.01 nmol in 0.04 M NaOH) from Fujifilm RI Pharma (Tokyo, Japan) in 1 M glycine/NaOH buffer (pH 10, 25 μl). The reaction proceeded for 10 min at 4 °C, and then the mixture was separated by high-performance liquid chromatography (HPLC) (L-2130; Hitachi, Tokyo, Japan) on a size-exclusion column (YMC-Pack Diol-200 (20 × 300 mm); YMC, Kyoto, Japan) eluted with buffer containing 150 mM NaCl and 0.24 mM EDTA at pH 7.4 (LDL buffer) at room temperature. At a flow rate of 1.0 ml/min, the retention time of 123I–oxLDL was 7.5 min. The eluate containing 123I–oxLDL (3 ml) was collected and concentrated to a volume of ~60 μl by centrifugation in an Amicon Ultra-4® (50 kDa cutoff; Millipore, Billerica, MA, USA). The number of oxLDL molecules and radiochemical purity of 123I–oxLDL were determined by size-exclusion HPLC, as described above (flow rate: 1.0 ml/min). The degree of oxidation, as indicated by relative electrophoretic mobility (REM) and thiobarbituric acid-reactive substances (TBARS assay kits; ZeptoMetrix Corp, Buffalo, NY), were compared between before and after labeling with the 123I using the ICl method. REM was calculated as the electrophoretic mobility to native-LDL ratio on agarose gels (Titan Gel Lipoprotein Kit 3045; Helena Laboratories, Beaumont, TX).
OxLDL was also labeled with DiI (Invitrogen, Carlsbad, CA, USA) as described previously [19]. Briefly, lipoprotein-deficient serum (LPDS; Sigma-Aldrich, St Louis, MO, USA) was added to yield a 1:5 ratio of oxLDL protein to LPDS protein. A stock solution of the fluorescent probe DiI was prepared by dissolving 30 mg DiI in 1 ml of dimethyl sulfoxide (DMSO; Wako Pure Chemical Industries). The stock DiI was added to the oxLDL-LPDS mixture to give a final concentration of 300 μg DiI/mg oxLDL. A fluorescent dye DiI (Ex: 549 nm, Em: 565 nm), which has lipophilic long-chain dialkylcarbocyanines in its structure, was used to stain the lipid protein components in oxLDL [21]. After this mixture was incubated for 18 h at 37 °C, the labeled oxLDL was isolated by ultracentrifugation, dialyzed against LDL buffer, and filter sterilized.
Cellular Studies
Uptake of labeled oxLDL to hLOX-1-CHO cell was evaluated for both 123I–oxLDL and DiI-oxLDL. Cells were washed twice with Ham’s F-12/10 mM HEPES, and were subsequently incubated for 2 h at 37 °C with 300 μl of Ham’s F-12/10 mM HEPES containing various concentrations of oxLDL (0.03, 0.1, 0.3, 1.0, 3.0, and 10 μg of protein/ml), prepared by successive dilution of 123I–oxLDL or DiI-oxLDL. Incubation was terminated by aspiration of the buffer and washing of the cell layers with PBS (500 μl × 2). The cells containing 123I–oxLDL were lysed with 0.2 M NaOH, and the radioactivity of the samples was measured using an automatic gamma counter (1480 WIZARD; PerkinElmer-Wallac, MA, USA). Protein concentrations of the sample cells were determined using the BCA protein assay kit. The cellular uptake of 123I–oxLDL was calculated as counts per minute (cpm) divided by the amount of total cellular protein. Cells incubated with DiI-oxLDL were fixed, counterstained with 4′,6-diamidino-2-phenylindole (DAPI; D9542, Sigma-Aldrich), and analyzed on an IN Cell Analyzer 1000 system (GE Healthcare, Waukesha, WI, USA). The total number of cell nuclei was counted in DAPI images. The cellular uptake of DiI-oxLDL was calculated as mean fluorescence intensity (MFI) divided by cell number.
Animal Studies
Twenty seven mice were studied by dividing into four groups as follows: (a) consistency of 123I–oxLDL autoradiography (ARG) with the fluorescence technique, (b) whole-body distribution after i.v. injection of 123I–oxLDL, (c) assessment of the clearance of 123I–oxLDL from the blood, and (d) assessment of stability of 123I–oxLDL in the serum. All mice were fasted for 12 h before the experiments began.
Experiment (a)
Four mice were assigned to compare uptake of 123I–oxLDL and DiI-oxLDL in the spleen and liver. In this study, conscious mice were injected via the tail vein with a mixture of DiI-oxLDL (1.10 ± 0.05 MBq, 15 μg protein/50 μl) and 123I–oxLDL (15 μg protein/50 μl), so that the total amount of oxLDL was 30 μg/body (approximately 1.5 mg protein/kg). Animals were sacrificed by intraperitoneal administration of an overdose of pentobarbital (50 mg/ml, 150 μl) 10 min after injection. Animals were perfused with saline and perfusion-fixed with 4% paraformaldehyde phosphate buffer solution (PFA) (Wako Pure Chemical). Spleen and liver were excised and immersed in 4% PFA at 4 °C overnight. Two consecutive slices (100 μm thickness) were obtained from each organ using a microslicer (DTK-Zero 1; DSK, Kyoto, Japan). Nuclei in sliced organ samples were counterstained with DAPI. The bright-field and fluorescence images of sliced organs were acquired with a fluorescence microscope (AZ100; Nikon, Tokyo, Japan) affixed to a CCD camera (DS-Fi1-U2; Nikon). The slices were then placed in contact with an imaging plate (BAS IP MS-2025, Fujifilm, Tokyo, Japan) for 24 h, and autoradiograms were acquired using an image plate reader (BAS 5000, Fujifilm).
Experiment (b)
Sixteen mice were used to investigate the whole-body organ distribution of radioactivity after i.v. injection of 123I–oxLDL. Six mice among 16 were assigned to in vivo imaging prior to the ex vivo biodistribution study. Anesthesia was induced with 2% isoflurane, and subsequently maintained by intramuscular injection of ketamine (100 mg/kg)-xylazine (10 mg/kg) cocktail. Animals were then placed on a custom-designed holder in the supine position, in whicih equally spaced marker holes of 8 μl volume at 25 mm separation may be filled with 0.1 MBq [123I]NaI or gadolinium-diethylenetriaminepentaacetic acid (Gd-DTPA) solution, and were referred to register the planar 123I–oxLDL images to the MR images.
MR images were first obtained using a clinical 3 Tesla MRI scanner (Signa HD 3.0 T; GE Healthcare) fitted with an in-house eight-channel receive-only RF coil (diameter, 50 mm) to assess the whole-body T1-weighted images [22]. After moving the animals to a clinical SPECT system (GCA-7200A; TOSHIBA, Tokyo, Japan) fitted with a low-energy high-resolution (LEHR) parallel-beam collimator, an 123I–oxLDL sample of 32.5 ± 12.6 MBq with 30 ± 2.5 μg protein/60 μl was injected into the tail vein by bolus (<1.0 s) via a 26-gauge cannula (Safelet-Cath; Nipro, Osaka, Japan). Images were corrected for inhomogeneous detector sensitivity, and also for spatial distortion. Planar images of radioactivity distribution in those animals were initiated 6 s before 123I–oxLDL injection. The sequential planar imaging scan consisted of 246 frames of 1 s and 10 frames of 36 s, ending 600 s after 123I–oxLDL administration. Those six animals were sacrificed immediately after the end of the planer imaging, and the entire liver, spleen, heart, lung, kidney, bladder, stomach, gut, and thyroid were excised. The whole tail was also taken, and the rest of the body was also excised as “carcass”. Total radioactivity and weights were assessed for each tissue sample and also for the blood. Weights were measured using an electronic balance (GR-60; A&D, Tokyo, Japan), and the radioactivity was measured using an automatic gamma counter.
The ex vivo biodistribution study was also carried out for another ten animals, in which animals were sacrificed at 2 min (n = 5) and at 5 min (n = 5) post i.v. 123I–oxLDL, and tissue and blood samples were taken according to the same procedures as described for animals sacrificed at 10 min post injection of 123I–oxLDL. Radioactivity and weights were measured according to the sample for animals sacrificed at 10 min post injection of 123I–oxLDL.
Experiment (c)
In four mice, HPLC analysis was carried out on both 123I–oxLDL and serum samples. 123I–oxLDL (4.0 ± 1.2 MBq; 30 μg protein/60 μl) was injected into conscious mice, and a blood sample of approximately 800 μl was collected from the right ventricle 10 min post-injection. The blood sample was transferred to a serum separator tube (CAPIJECT CJ-AS; Terumo, Tokyo, Japan), and the tube was centrifuged for 15 min at 4 °C (1500 × g). The supernatant was collected as serum. Approximately 400 μl of serum was diluted with LDL buffer and applied to the size-exclusion HPLC mentioned above (flow rate: 1.0 ml/min). Areas under the curves (AUCs) were assessed for the two peaks corresponding to 123I–oxLDL and123I−.
Experiment (d)
Serum was obtained from the blood of the remaining three mice by centrifugation at 3000 × g for 10 min. A 20-μl solution of 123I–oxLDL (1.0 MBq) was added to the serum (400 μl). After incubation for 2 h at 37 °C, aliquots of the samples were drawn and the radioactivity was analyzed using size-exclusion HPLC at a flow rate of 1.0 ml/min.
Data Analysis
Cellular uptake of 123I–oxLDL (radioactivity per cell protein) and DiI-oxLDL (MFI per cell nuclei counts) was measured. The uptake values of both hLOX-1-CHO and control CHO cells were normalized against the value measured with 10 μg protein/ml of each labeled oxLDL. The dose-dependent uptakes of hLOX-1-CHO and control (wild type) CHO cells were compared between 123I–oxLDL and DiI-oxLDL.
The 123I–oxLDL autoradiograms images were co-registered to the DiI-oxLDL fluorescent images for both liver and spleen slices. The hepatic lobes were classified into two structures: around the central vein, and near the interlobular areas. The spleens were divided into three structures: white pulp, red pulp, and marginal zone. Three regions of interest (ROIs) (φ125 μm) were manually placed on each of these structures in two sections. Intensity values for each ROI were normalized against the average intensity at each section, and intensity values in ARG were compared with those of the fluorescence images. This image analysis was carried out using Fiji [23].
In the in vivo dynamic imaging study, ROIs were carefully placed on MR coronal images co-registered to sequential planar images of 123I–oxLDL to generate TACs for liver, heart, lungs, kidney, head, and abdomen. During ROI selection, care was taken to avoid the vena cava or the vasculature and bladder. TACs were calculated from sequential planar images using the AMIDE software [24], and temporally smoothed at later stages to reduce the statistical noise. Average TACs from six mice were plotted based on the moving average. Significant differences were detected at each time point using the LabVIEW 8.6 software (National Instruments, TX USA).
In the ex vivo biodistribution study, percent injected dose (%ID) was calculated as the fractional uptake of radioactivity in each organ divided by the total injected radioactivity, and the dose uptake ratio (DUR) was calculated as radioactivity per unit mass of each organ divided by the injected dose over whole body weight, yielding a dimensionless index. These parameters were calculated for all nine organs from all six mice, as well as from carcass, blood, and tail samples. Assuming the total blood volume of 72 ml per body weight of kg [25], the %ID and DUR were estimated from the measured radioactivity concentration of the sampled blood.
Fractions and calibrated radioactivity were assessed for the two peaks corresponding to 123I–oxLDL and 123I−, based on the amount of radioactivity injected into the HPLC and the AUC for each peak. Radioactivity was compared between the injection sample of 123I–oxLDL and the serum sample taken 10 min post-injection.
Summarized data are presented as means ± standard deviation. Statistical analysis was performed by Mann-Whitney U test, or ANOVA followed by Bonferroni correction, with significance accepted at P < 0.05. Statistical analyses were performed in R [26] with the EZR graphical interface [27].
Results
The radiochemical yield and purity of 123I–oxLDL were 30.0 ± 12.8% and 90.3 ± 2.5%, respectively. The REM of oxLDL and 123I–oxLDL were 2.11 ± 0.28 and 2.14 ± 0.22, respectively. The TBARS of oxLDL and 123I–oxLDL were 23.9 ± 3.0 nmol/mg and 23.8 ± 3.9 nmol/mg, respectively. The amount of radioactivity administered to mice in the dynamic planar studies was 32.5 ± 12.6 MBq, and the dose of oxLDL was 30 ± 2.5 μg of protein. The specific radioactivity of 123I–oxLDL (2500 kDa) [28] was calculated to be 2728 ± 1098 GBq/μmol.
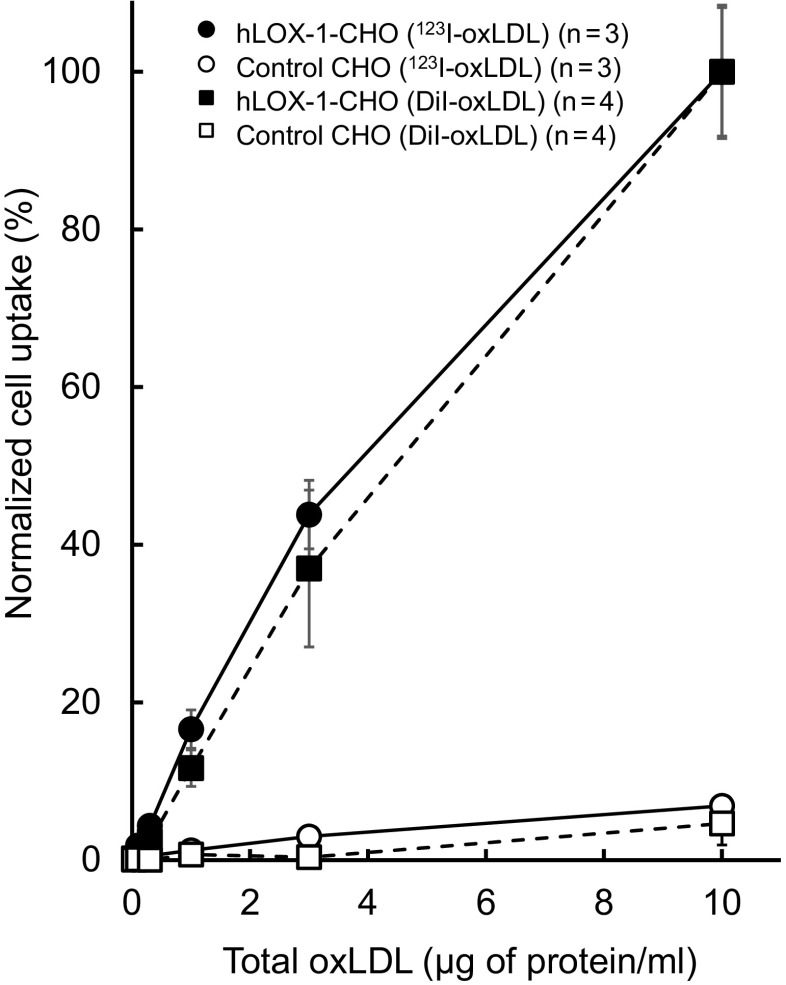
Figure 1 shows the cellular uptake of oxLDL, measured using 123I–oxLDL and DiI-oxLDL as tracers, in hLOX-1-CHO and control CHO cells. Note that the plotted values were normalized against the peak values for each of the ligands. Both 123I–oxLDL and DiI-oxLDL exhibited significantly higher uptake in hLOX-1-CHO cells than in control CHO cells. In the hLOX-1-CHO cells, a dose-dependent increase was seen in both 123I–oxLDL and DiI-oxLDL. We observed no difference between 123I–oxLDL and DiI-oxLDL at any oxLDL concentrations.
Fig. 1.
Accumulation of 123I–oxLDL and DiI-oxLDL in hLOX-1 CHO and control CHO cells. Cells were incubated with 123I–oxLDL and DiI-oxLDL (0.03–10 μg protein/ml) for 2 h at 37 °C. Each uptake value was normalized against the 10 μg/ml uptake value for hLOX-1-CHO cells
Fig. 2 shows a comparison of the relative distribution of 123I–oxLDL with that of DiI-oxLDL in liver and spleen slices. Fluorescence microscopy revealed high uptake of DiI-oxLDL in the interlobular area of the liver (panel 4 in Fig. 2a) and the marginal zone of the spleen (panel 4 in Fig. 2b). The distribution of 123I–oxLDL was in good agreement with that of DiI-oxLDL in both liver (panel 2 vs panel 3 in Fig. 2a) and spleen (panel 2 vs panel 3 in Fig. 2b) sections. The oxLDL uptake in the interlobular area was higher than that of the central vein area for both 123I–oxLDL and DiI-oxLDL. In the spleen, the highest and lowest accumulation was observed in the marginal zone and white pulp, respectively. Quantitative analysis confirmed that the intensity values differed significantly among the three ROIs for DiI-oxLDL and 123I–oxLDL (Table 1).
Fig. 2.
Typical examples of 123I–oxLDL autoradiogram and DiI-oxLDL fluorescence images of the liver (Fig. 2a) and spleen (Fig. 2b). Bright field (1), DiI fluorescence (2), and ARG (3) images are shown. Enlarged fluorescence images merged with nuclei image of hepatic lobule and spleen slices are also shown (4). In the liver slice, a star represents the central vein, and triangles the interlobular area, including the interlobular artery and vein of the hepatic lobule. In the spleen slice, circles represent red pulp, rectangles the white pulp, and diamonds the marginal zone. Circular regions of interest (ROIs) of 125 μm diameter were placed on each of those regions
Table 1.
Structural differences in accumulation of labeled oxLDL in the liver and spleen
| Organ | Structure | Fluorescence | ARG |
|---|---|---|---|
| Liver | Around central vein area | 0.89 ± 0.01 | 0.98 ± 0.05 |
| Near interlobular area | 1.38 ± 0.10*,a | 1.10 ± 0.03*,a | |
| Spleen | White pupl | 0.42 ± 0.07 | 0.80 ± 0.04 |
| Red pulp | 0.96 ± 0.10**,b | 1.00 ± 0.03****,b | |
| Marginal zone | 2.10 ± 0.31****,b ****,c | 1.11 ± 0.03****,b **,c |
Both fluorescence and ARG are reported in arbitrary units. Data represent means ± SD (n = 4 in each group)
*: P < 0.05, **: P < 0.01, ****: P < 0.0001
a vs. near the central vein area; b vs. white pulp; c vs. red pulp
Dynamic planar images acquired following i.v. injection of 123I–oxLDL are shown in Fig. 3. These images show that the injected radioactivity moved from the right ventricle (at 2 s) to the left ventricle (at 3 s), and then to the lungs (2–4 s). Re-circulated radioactivity arriving at the heart could also be seen around 7–8 s. Radioactivity then gradually accumulated in the liver ~100 s post-injection. Ultimately, the radioactivity accumulated gradually throughout the body, which could not be seen in early phase images.
Fig. 3.
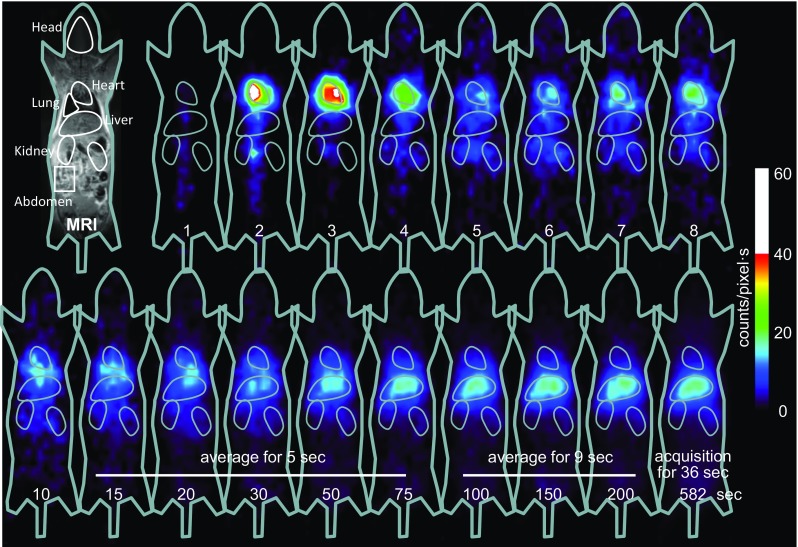
Representative dynamic planar images of a mouse acquired after i.v. injection of 123I–oxLDL. Accumulation of radioactivity in the heart, followed by the liver, is apparent. A small but significant distribution of radioactivity can also be seen in the whole body, particularly at later stages
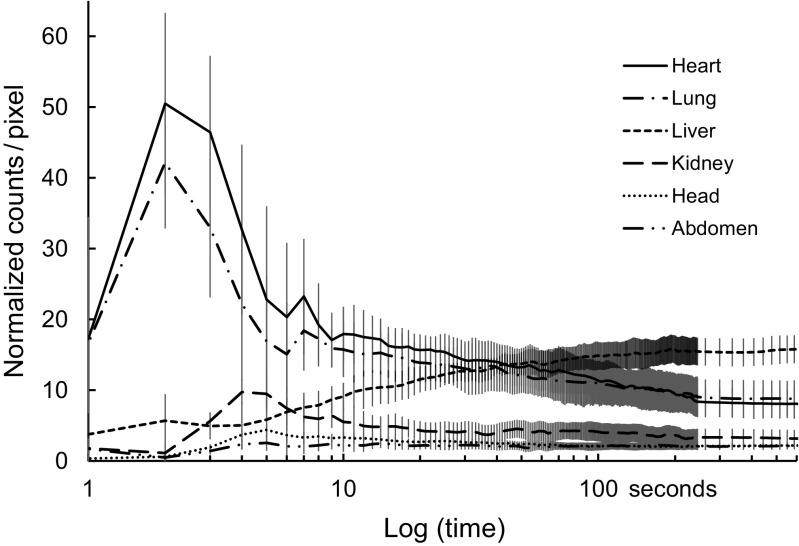
Figure 4 shows TACs averaged over six mice for each of the six ROIs. The heart region exhibited an initial peak at 2 s and a second at 7 s post-injection; radioactivity in the heart region subsequently decreased to a plateau after 200 s. TAC for the lung was similar to that of the heart. The kidney region exhibited an initial peak at 4 s post-injection, and then the radioactivity decreased to a constant level after approximately 30 s. In the liver, TAC increased and reached a plateau at ~200 s post-injection. Marked accumulation of radioactivity was not observed in the head or abdomen regions.
Fig. 4.
Time-activity curves (TACs) averaged over six mice for each of the six ROIs
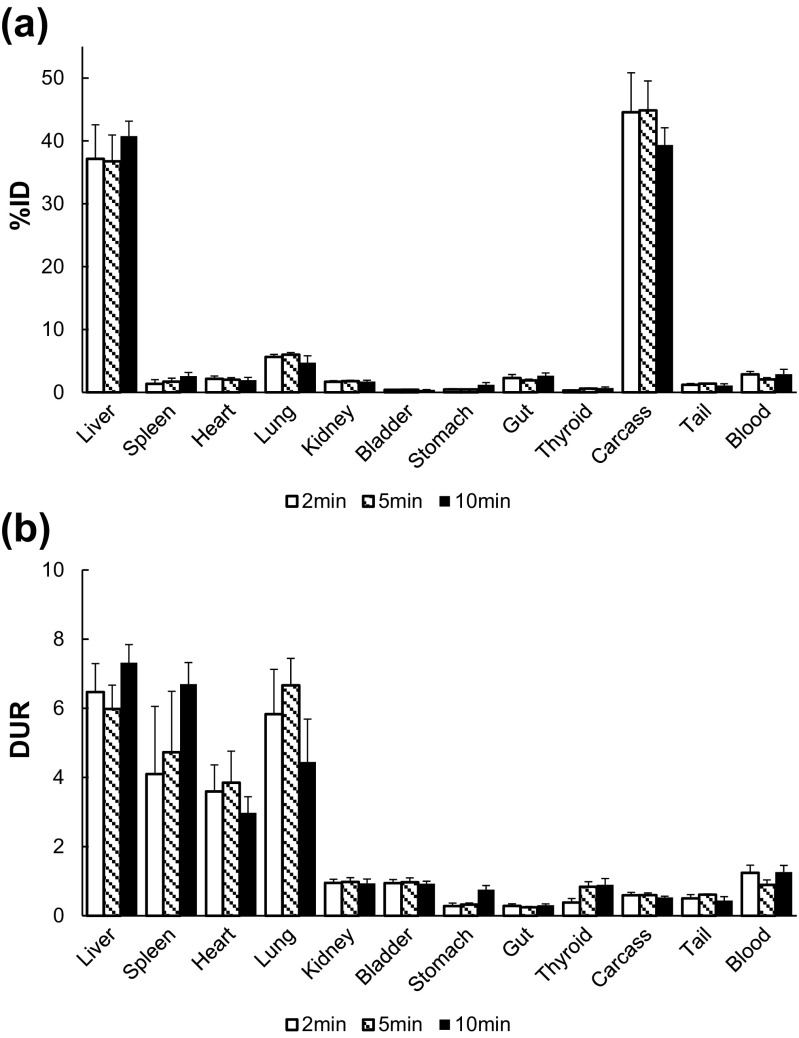
Figure 5 shows %ID and DUR of mice at 2, 5, and 10 min after injection of 123I–oxLDL. The DUR of 123I–oxLDL in the liver, spleen, heart, and lungs were higher than that in the blood (Fig. 5a). The summed %ID over these organs was 50.0 ± 2.3%, and the remaining half of the radioactivity was mainly in the carcass. Accumulation of radioactivity in the entire carcass was 39.4 ± 2.7% ID, almost equal to that of the liver 40.8 ± 2.4% ID (Fig. 5b). DUR and %ID in the kidney, bladder, and thyroid were low: 0.94 ± 0.13, 0.93 ± 0.07, and 0.89 ± 0.18 for DUR, and 1.71 ± 0.22, 0.41 ± 0.04, and 0.69 ± 0.18 for %ID, respectively. None of samples showed significant changes among different time points for both %ID and DUR.
Fig. 5.
Dose-uptake ratio (DUR: a) and percent injected dose (%ID: b) assessed by ex vivo biodistribution analysis for 123I–oxLDL
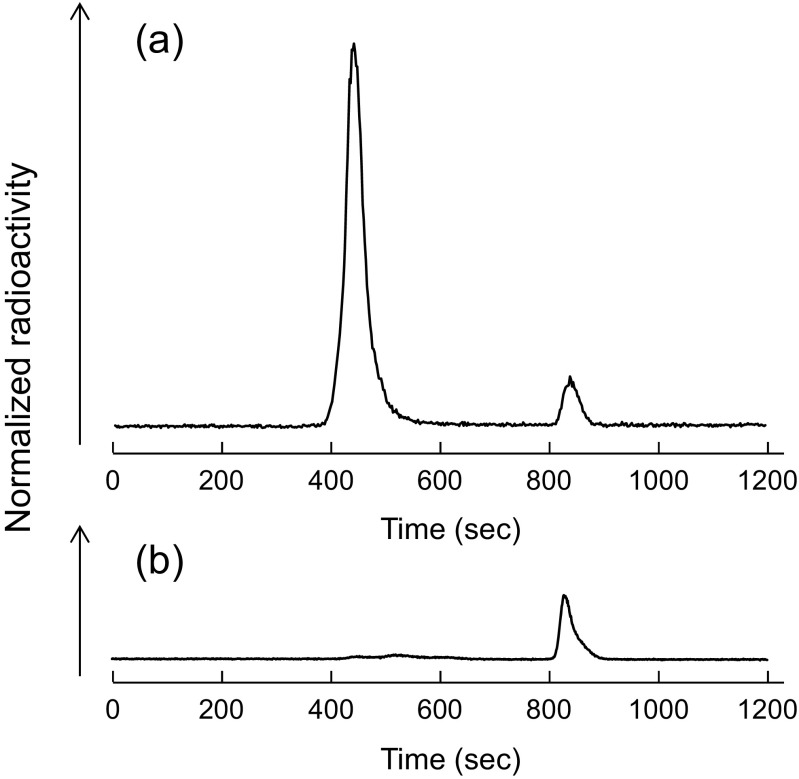
Figure 6 shows the results of size-exclusion radiochromatographic analysis of 123I–oxLDL in the serum. We observed two peaks in the radiochromatogram of 123I–oxLDL, corresponding to 123I–oxLDL and 123I− at retention times of 450 s and 840 s, respectively (Fig. 6a). Most of the 123I–oxLDL fraction had disappeared from the serum by 10 min post-injection (Fig. 6b). A considerable reduction of 123I–oxLDL was apparent in the serum sampled 10 min post-injection. The amounts of radioactivity injected into the mice were 4.0 ± 1.8 MBq and 0.4 ± 0.4 MBq for 123I–oxLDL and 123I−, respectively, and the amounts in the serum were 0.04 ± 0.01 MBq and 0.3 ± 0.16 MBq, respectively. Radioactivity in the form of 123I–oxLDL disappeared from the serum by 10 min post-injection, whereas the amount of 123I− was also reduced in all cases.
Fig. 6.
Size-exclusion HPLC analyses of 123I–oxLDL. (a) HPLC of the injected sample of 123I–oxLDL, (b) HPLC of serum prepared at 10 min post-injection of 123I–oxLDL. The main peak (retention time: 450 s) corresponds to 123I–oxLDL, and the second peak (retention time: 840 s) to 123I−. Only the peak for 123I− can be seen in the serum sample
123I–oxLDL in murine serum as evaluated in vitro using size-exclusion HPLC analysis was 94.5 ± 2.3% (n = 3) in the beginning, and was 88.8 ± 3.5% at 2 h after the incubation. The degradation was approximately 94% even after 2 h of incubation at 37 °C, which is greater than a typical value of 90% considered as a stability reference.
Discussion
OxLDL was labeled with 123I efficiently by the direct method [20], with a radiochemical yield of 30% and purity of 90%. The degree of oxidation, as indicated by REM and TBARS, did differ before and after labeling of oxLDL by 123I using the ICl method. Notably, the specific radioactivity of 2700 GBq/μmol was considerably higher than that of previously reported oxLDLs labeled with other radioisotopes, such as 125I–oxLDL (6–7 GBq/μmol) [10, 13, 14], 99mTc-labeled oxLDL (300–900 GBq/μmol) [15], and fluorine-18-labeled oxLDL (200–500 GBq/μmol) [29]. The high specific radioactivity of 123I–oxLDL is simply attributed to that of the prepared 123I–NaI. In previous 125I–oxLDL studies, much effort was not carried out to maximize the specific radioactivity because the high specific radioactivity was not essential in those experiments. The approximately 400-fold higher specific radioactivity of 123I–oxLDL relative to 125I–oxLDL is partly attributed to the shorter half-life of 123I (13.4 h) relative to 125I (59.4 days), and would be beneficial when one intends to target low-density binding distributions for in vivo imaging particularly in man.
Oxidizing agents are not used during the labeling process of DiI-oxLDL, which is widely used in cellular and in vivo studies [19, 30, 31]. Uptake of 123I–oxLDL by hLOX-1-CHO cells increased in a dose-dependent manner, and was approximately 15 times higher than that in control CHO cells. Notably, the uptake of 123I–oxLDL was very similar to that of DiI-oxLDL, as demonstrated in Fig. 1. This suggested that the binding properties of 123I–oxLDL are comparable to those of DiI-oxLDL, suggesting that the oxidation procedures used during the synthesis of 123I–oxLDL are unlikely to have altered the binding properties of oxLDL. In fact, the slice comparison shown in Fig. 2 and Table 1 indicated that distributions were consistent between 123I–oxLDL ARG and DiI-oxLDL fluorescence images of the liver and spleen. 123I–oxLDL uptake in the liver was higher in the interlobular area, which includes the interlobular artery and vein, but lower in the vicinity of the central vein. In the interlobular area, uptake is mediated by scavenger receptors on Kupffer cells. Evidence for selective expression of a receptor for oxLDL has been obtained in liver Kupffer cells isolated from rat [10], human [11], and rabbit [12]. The highest uptake of oxLDL in the spleen were observed at the marginal zone, followed by red pulp, for both types of labeled oxLDL. The marginal zone is an important transition area, and the most distinctive cell type in this region is the marginal-zone macrophage, which expresses the type I scavenger receptor MRCO [32]. The reduced contrast among regions in the 123I–oxLDL autoradiogram in comparison with the DiI-oxLDL fluorescence images shown in Table 1 is attributed to the poorer spatial resolution of the 123I–oxLDL autoradiogram. The ROI analysis was likely influenced by the spillover effect from the surrounding tissue areas. Because the in vivo receptor binding characteristics of oxLDL were not modified during 123I–labeling with the ICl method, we used this compound as an in vivo imaging tracer.
On the basis of previous ex vivo biodistribution studies [10, 13, 14] and human dynamic planar scintigraphy [15], oxLDL is thought to be rapidly cleared from blood due to high liver uptake. Our whole-body dynamic planar imaging in mice also clearly visualized the decrease in radioactivity in the heart, concomitant with accumulation of radioactivity in the liver, under anesthetized conditions (Fig. 3). The liver contained the highest level of radioactivity, which reached a plateau after 200 s (Fig. 4). The TAC of the heart exhibited an initial peak at 2 s and a second at 7 s post-injection; the interval between these peaks was 5 s, corresponding to the whole-body recirculation time in mice [33]. In the biodistribution study, we found that %ID was approximately 40% in the liver, and 20% in other specifically excised organs (Fig. 5b) at 10 min post injection of 123I–oxLDL. In addition, radioactivity equal to that in the liver was distributed in the rest of the body (carcass). This distribution was almost completed in 2 min after i.v. injection of 123I–oxLDL, which is compatible with the results of the in vivo imaging study (Figs. 3 and 4). Radioactivity of collected blood was 2.91 ± 0.76% ID, indicating that total %ID of blood was ~9.2% ID, calculated based on the assumption that the total blood volume of mice is 72 ml/kg [25]. Therefore, the total radioactivity of residual blood in the mouse body, including organs, was calculated to be approximately 6.3% ID; thus, 123I–oxLDL was incorporated and retained in particular organs or tissues within 10 min. The carcass includes brain, skeletal muscle, fat, genitalium, vasculature, and skin. The planar image and TAC revealed no specific accumulation of radioactivity in the brain or bone (Figs. 3 and 4). Thus, the radioactivity of the carcass was very low, but widely distributed throughout the whole body. Therefore, we predict that vessels or muscle fiber in skeletal muscle and/or fat might contribute to oxLDL kinetics. Further studies should seek to identify the accumulation sites and mechanism that regulate the 123I–oxLDL in the blood.
According to TACs and the ex vivo biodistribution study, the radioactivity accumulation per unit mass of organ was high in both spleen and lung (Figs. 4 and 5a) in spite of the lower total radioactivity. This would be due to higher expression levels of scavenger receptors on the macrophage-like cells which are widely distributed in these tissues [34]. This is also consistent with recent findings of increased accumulation of a translocator protein targeted radio-ligands [35]. In addition, abundant microvessels surrounding the pulmonary alveoli might lead to the uptake of 123I–oxLDL in the lung.
The rapid clearance of 123I–oxLDL could also be observed in HPLC analysis of serum sampled at 10 min post-i.v. injection of 123I–oxLDL. In the serum analysis, the dominant peak (t = 840 s) agreed with the retention time of 123I−, and no other peaks were observed (Fig. 6). It should be noted that free 123I− was not completely removed from the injection solution, despite purification by ultrafiltration. However, the data presented in this analysis supports that 123I–oxLDL is mostly bound to binding sites in the body rather than the catabolism of 123I–oxLDL or isolation of 123I− from 123I–oxLDL. If the majority of 123I–oxLDL generates 123I− in the serum, the amount of radioactivity should become much larger than that reflected by the HPLC peak corresponding to 123I− in the serum. Moreover, low DUR and %ID values of thyroid also provided support for the high stability of 123I–oxLDL. Indeed, in vitro assay indicated that more than 90% of this probe remained stable after 2 h incubation in the murine serum.
Accumulation of oxLDL in organs is attributed to endocytosis by scavenger receptors. Widespread distribution of oxLDL in the carcass could therefore be thought associated with retention via LOX-1 expressed in vascular endothelial cells [19] or CD36 expressed in fat and skeletal muscle [36, 37].
The main limitation of this study was low spatial resolution of the clinical SPECT system; however, by applying high-resolution in vivo imaging techniques to various animal models, we might detect differences in the biodistributions of oxLDL, and thus obtain novel insights into the kinetics of oxLDL. Elucidation of the involvement of scavenger receptors in lesion site-specific uptake of 123I–oxLDL might reveal the pathophysiological significance of these receptors.
Conclusion
123I–oxLDL was synthesized, and was confirmed to have sufficient yield and purity as well as high specific radioactivity. 123I–oxLDL in the blood is rapidly cleared, attributed to the uptake not only by the liver but also by tissues throughout the whole body. The amount of 123I–oxLDL in the carcass was similar to that of the liver. Further studies need to identify the tissues, receptors, and mechanisms that regulate oxLDL in the blood.
Acknowledgements
The authors would like to thank Dr. Kyoko Shioya, DVM from Laboratory of Animal Experiment and Medicine Management, National Cerebral and Cardiovascular Center, Osaka, Japan, for her assistance and advise on animal care and experimental procedures.
Compliance with Ethical Standards
Conflict of Interest
Atushi Nakano, Hidekazu Kawashima, Yoshinori Miyake, Tsutomu Zeniya, Kazuhiro Koshino, Takashi Temma, Tetsuya Fukuda, Yoshiko Fujita, Akemi Kakino, Shigehiko Kanaya, and Tatsuya Sawamura declare that they have no conflict of interest. Hidehiro Iida received research grants from Chugai Yakuhin, Japan, Nihon Medi Physics, Japan and Molecular Imaging Labo, Japan. Akihide Yamamoto is paid by Molecular Imaging lab, Japan. This study was supported by the Budget for Nuclear Research of MEXT (Ministry of Education, Culture, Sports, Science and Technology Japan), a Grant for Translational Research from MHLW (Ministry of Health, Labor and Welfare, Japan), a Grant for Strategic Japanese-Finnish Research Cooperative Program on “Application of Medical ICT Devices” from Japan Agency for Medical Research and Development (AMED), Japan, and JSPS KAKENHI Grants (Number: 24,601,021 and 15 K01309).
Ethical Approval
The animal experiments in this study were conducted in accordance with guidelines for animal research on Human Care and Use of Laboratory Animals (Rockville, National Institute of Health/Office for Protection from Research Risks, 1996). The study protocol was approved by the Sub-committee for Laboratory Animal Welfare, National Cerebral and Cardiovascular Center Research Institute, Osaka, Japan.
Informed Consent
The present study included only animal data, thus our institute approved that the requirement to obtain informed consent was waived.
Footnotes
Atushi Nakano and Hidekazu Kawashima contributed equally to this work.
References
- 1.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340(2):115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 2.Tabas I, Williams KJ, Boren J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation. 2007;116(16):1832–1844. doi: 10.1161/CIRCULATIONAHA.106.676890. [DOI] [PubMed] [Google Scholar]
- 3.Glass CK, Witztum JL. Atherosclerosis. The road ahead. Cell. 2001;104(4):503–516. doi: 10.1016/S0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 4.Kopprasch S, Pietzsch J, Ansurudeen I, et al. Prediabetic and diabetic in vivo modification of circulating low-density lipoprotein attenuates its stimulatory effect on adrenal aldosterone and cortisol secretion. J Endocrinol. 2009;200(1):45–52. doi: 10.1677/JOE-08-0293. [DOI] [PubMed] [Google Scholar]
- 5.Portelinha A, Belo L, Cerdeira AS, et al. Lipid levels including oxidized LDL in women with history of preeclampsia. Hypertens Pregnancy. 2010;29(1):93–100. doi: 10.3109/10641950902968593. [DOI] [PubMed] [Google Scholar]
- 6.Dai L, Zhang Z, Winyard PG, et al. A modified form of low-density lipoprotein with increased electronegative charge is present in rheumatoid arthritis synovial fluid. Free Radic Biol Med. 1997;22(4):705–710. doi: 10.1016/S0891-5849(96)00389-9. [DOI] [PubMed] [Google Scholar]
- 7.Meisinger C, Baumert J, Khuseyinova N, Loewel H, Koenig W. Plasma oxidized low-density lipoprotein, a strong predictor for acute coronary heart disease events in apparently healthy, middle-aged men from the general population. Circulation. 2005;112(5):651–657. doi: 10.1161/CIRCULATIONAHA.104.529297. [DOI] [PubMed] [Google Scholar]
- 8.Inoue N, Okamura T, Kokubo Y, et al. LOX index, a novel predictive biochemical marker for coronary heart disease and stroke. Clin Chem. 2010;56(4):550–558. doi: 10.1373/clinchem.2009.140707. [DOI] [PubMed] [Google Scholar]
- 9.Holvoet P, Lee DH, Steffes M, Gross M, Jacobs DR., Jr Association between circulating oxidized low-density lipoprotein and incidence of the metabolic syndrome. JAMA. 2008;299(19):2287–2293. doi: 10.1001/jama.299.19.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Berkel TJ, De Rijke YB, Kruijt JK. Different fate in vivo of oxidatively modified low density lipoprotein and acetylated low density lipoprotein in rats. Recognition by various scavenger receptors on Kupffer and endothelial liver cells. J Biol Chem. 1991;266(4):2282–2289. [PubMed] [Google Scholar]
- 11.Kamps JA, Kruijt JK, Kuiper J, van Berkel TJ. Characterization of the interaction of acetylated LDL and oxidatively modified LDL with human liver parenchymal and Kupffer cells in culture. Arterioscler Thromb. 1992;12(9):1079–1087. doi: 10.1161/01.ATV.12.9.1079. [DOI] [PubMed] [Google Scholar]
- 12.Ueda Y, Arai H, Kawashima A, et al. Different expression of modified low density lipoprotein receptors in rabbit peritoneal macrophages and Kupffer cells. Atherosclerosis. 1993;101(1):25–35. doi: 10.1016/0021-9150(93)90098-F. [DOI] [PubMed] [Google Scholar]
- 13.Ling W, Lougheed M, Suzuki H, Buchan A, Kodama T, Steinbrecher UP. Oxidized or acetylated low density lipoproteins are rapidly cleared by the liver in mice with disruption of the scavenger receptor class a type I/II gene. J Clin Invest. 1997;100(2):244–252. doi: 10.1172/JCI119528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinbrecher UP, Witztum JL, Parthasarathy S, Steinberg D. Decrease in reactive amino groups during oxidation or endothelial cell modification of LDL. Correlation with changes in receptor-mediated catabolism. Arteriosclerosis. 1987;7(2):135–143. doi: 10.1161/01.ATV.7.2.135. [DOI] [PubMed] [Google Scholar]
- 15.Iuliano L, Signore A, Vallabajosula S, et al. Preparation and biodistribution of 99m technetium labelled oxidized LDL in man. Atherosclerosis. 1996;126(1):131–141. doi: 10.1016/0021-9150(96)05888-1. [DOI] [PubMed] [Google Scholar]
- 16.Iida H, Nakagawara J, Hayashida K, et al. Multicenter evaluation of a standardized protocol for rest and acetazolamide cerebral blood flow assessment using a quantitative SPECT reconstruction program and split-dose 123I-iodoamphetamine. J Nucl Med. 2010;51(10):1624–1631. doi: 10.2967/jnumed.110.078352. [DOI] [PubMed] [Google Scholar]
- 17.Iida H, Narita Y, Kado H, et al. Effects of scatter and attenuation correction on quantitative assessment of regional cerebral blood flow with SPECT. J Nucl Med. 1998;39(1):181–189. [PubMed] [Google Scholar]
- 18.Fujita Y, Kakino A, Nishimichi N, et al. Oxidized LDL receptor LOX-1 binds to C-reactive protein and mediates its vascular effects. Clin Chem. 2009;55(2):285–294. doi: 10.1373/clinchem.2008.119750. [DOI] [PubMed] [Google Scholar]
- 19.Sawamura T, Kume N, Aoyama T, et al. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386(6620):73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- 20.Atsma DE, Kempen HJ, Nieuwenhuizen W, van’t Hooft FM. Pauwels EK. Partial characterization of low density lipoprotein preparations isolated from fresh and frozen plasma after radiolabeling by seven different methods. J Lipid Res. 1991;32(1):173–181. [PubMed] [Google Scholar]
- 21.Gullapalli RR, Demirel MC, Butler PJ. Molecular dynamics simulations of DiI-C18(3) in a DPPC lipid bilayer. Phys Chem Chem Phys. 2008;10(24):3548–3560. doi: 10.1039/b716979e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto A, Sato H, Enmi J, et al. Use of a clinical MRI scanner for preclinical research on rats. Radiol Phys Technol. 2009;2(1):13–21. doi: 10.1007/s12194-008-0038-x. [DOI] [PubMed] [Google Scholar]
- 23.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loening AM, Gambhir SS. AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging. 2003;2(3):131–137. doi: 10.1162/153535003322556877. [DOI] [PubMed] [Google Scholar]
- 25.Diehl KH, Hull R, Morton D, et al. A good practice guide to the administration of substances and removal of blood, including routes and volumes. J Appl Toxicol. 2001;21(1):15–23. doi: 10.1002/jat.727. [DOI] [PubMed] [Google Scholar]
- 26.R Core Team. R: A language and environment for statistical computing, version 3.2.2. R Foundation for Statistical Computing, Vienna, Austria; 2014. http://www.R-project.org/.
- 27.Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48(3):452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobal G, Resch U, Sinzinger H. Modification of low-density lipoprotein by different radioiodination methods. Nucl Med Biol. 2004;31(3):381–388. doi: 10.1016/j.nucmedbio.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Pietzsch J, Bergmann R, Wuest F, Pawelke B, Hultsch C, van den Hoff J. Catabolism of native and oxidized low density lipoproteins: in vivo insights from small animal positron emission tomography studies. Amino Acids. 2005;29(4):389–404. doi: 10.1007/s00726-005-0203-z. [DOI] [PubMed] [Google Scholar]
- 30.Lougheed M, Moore ED, Scriven DR, Steinbrecher UP. Uptake of oxidized LDL by macrophages differs from that of acetyl LDL and leads to expansion of an acidic endolysosomal compartment. Arterioscler Thromb Vasc Biol. 1999;19(8):1881–1890. doi: 10.1161/01.ATV.19.8.1881. [DOI] [PubMed] [Google Scholar]
- 31.Nakano A, Inoue N, Sato Y, et al. LOX-1 mediates vascular lipid retention under hypertensive state. J Hypertens. 2010;28(6):1273–1280. doi: 10.1097/HJH.0b013e32833835d4. [DOI] [PubMed] [Google Scholar]
- 32.Mebius RE, Kraal G. Structure and function of the spleen. Nat Rev Immunol. 2005;5(8):606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 33.Kreissl MC, Wu HM, Stout DB, et al. Noninvasive measurement of cardiovascular function in mice with high-temporal-resolution small-animal PET. J Nucl Med. 2006;47(6):974–980. [PMC free article] [PubMed] [Google Scholar]
- 34.Palanisamy GS, Kirk NM, Ackart DF, et al. Uptake and accumulation of oxidized low-density lipoprotein during mycobacterium tuberculosis infection in guinea pigs. PLoS One. 2012;7(3):e34148. doi: 10.1371/journal.pone.0034148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luoto P, Laitinen I, Suilamo S, Nagren K, Roivainen A. Human dosimetry of carbon-11 labeled N-butan-2-yl-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide extrapolated from whole-body distribution kinetics and radiometabolism in rats. Mol Imaging Biol. 2010;12(4):435–442. doi: 10.1007/s11307-009-0293-1. [DOI] [PubMed] [Google Scholar]
- 36.Yamada Y, Doi T, Hamakubo T, Kodama T. Scavenger receptor family proteins: roles for atherosclerosis, host defence and disorders of the central nervous system. Cell Mol Life Sci. 1998;54(7):628–640. doi: 10.1007/s000180050191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy JE, Tedbury PR, Homer-Vanniasinkam S, Walker JH, Ponnambalam S. Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis. 2005;182(1):1–15. doi: 10.1016/j.atherosclerosis.2005.03.036. [DOI] [PubMed] [Google Scholar]