Abstract
Precision medicine offers better treatment options and improved survival for cancer patients based on individual variability. As the success of precision medicine depends on robust biomarkers, the requirement for improved imaging biomarkers that reflect tumor biology has grown exponentially. Radiomics, the field of study in which high-throughput data are generated and large amounts of advanced quantitative features are extracted from medical images, has shown great potential as a source of quantitative biomarkers in the field of oncology. Radiomics provides quantitative information about the morphology, texture, and intratumoral heterogeneity of the tumor itself as well as features related to pulmonary function. Hence, radiomics data can be used to build descriptive and predictive clinical models that relate imaging characteristics to tumor biology phenotypes. In this review, we describe the workflow of CT radiomics, types of CT radiomics, and its clinical application in thoracic oncology.
Keywords: Computed tomography, Lung cancer, Image processing, Biomarkers
Introduction: Role of Radiomics in Precision Medicine
Tumors are biologically complex and show phenotypic and genomic heterogeneity between different tumors and even within an individual tumor. In other words, although tumors have the same histopathological cell type, they can show vast variations in imaging features including vascularity, contrast enhancement, and necrosis. In parallel, such variations have also been reported in the genetic profile of cancers. Such genetic variation of cancers has become of great interest because patient-centered chemotherapy based on patient-specific tumor cell mutation, an approach called precision medicine, has recently been introduced and shows excellent results. Thus, during the past decade, large database studies have transferred the concept of cancer diagnosis from traditional histopathological cell type to a new classification based on molecular genetic data [1–3]. However, cancer treatment based on these results typically fails due to the amazing ability of tumor cells to acquire subclonal mutations during tumor evolution. Therefore, the key factor leading to successful precision medicine lies in a clear understanding of each patient’s tumoral heterogeneity and individual situation [4]. In other words, robust biomarkers are required to obtain a better understanding of the evolving biology of cancer.
During the last decade, dramatic advancements in high-throughput computing and automated pipeline systems have been introduced. Such advancements, especially in computed tomography (CT), have made it possible to extract innumerable quantitative features from medical CT images, a discipline known as radiomics. Thus, by extracting radiomics features, a great deal of information hidden within the layers of conventional CT images can be revealed for clinical use. Although radiomics can be applied to various conditions, its potential has been most promising in the field of oncology. Multiple studies using a radiomics approach have shown that quantitative features offer better characterization of the tumor, more precise prognosis assessment, and improved prediction of drug resistance [5–7]. Texture analysis has also been shown to be a highly significant independent predictor of survival in patients with non-small cell lung cancer [8]. Intratumor heterogeneity, which is near ubiquitous in malignant tumor, is a key challenge in cancer medicine. Genetic heterogeneity of a malignant tumor leads to regional difference in stromal architecture or function of individual tumors and imaging can quantify the adverse spatial feature and functional heterogeneity through measurement of quantitative features [9, 10]. In other words, quantitative tumor characteristics observable at medical imaging reflect the molecular, cellular, and tissue components, which might ultimately advance our understanding of the evolving biology of the whole tumor. In this article, we review the methodology of CT radiomics and discuss its application in thoracic oncology.
Methodology of CT Radiomics in Oncology
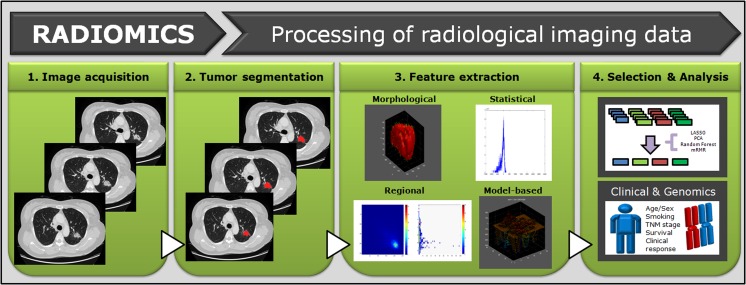
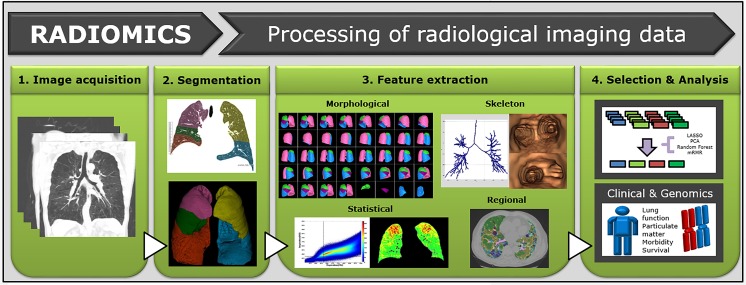
Radiomics is a quantitative, noninvasive method of revealing information embedded within conventional CT images performed clinically for diagnosis and preoperative planning. Radiomics data can be used to build descriptive and predictive clinical models relating imaging characteristics to tumor biology phenotypes. Although conceptually simple, each step of radiomics has its own challenges and applications. Furthermore, clinical translation of CT radiomics is a complex undertaking that requires the coordinated efforts of the radiologist, computer scientist, and oncologist (Figs. 1 and 2).
Fig. 1.
Overview of radiomics in lung cancer. Whole tumors are segmented by drawing regions of interest that traced tumor edge, and quantitative features are extracted within defined tumor contours on CT images. Relationships among the radiomics features, clinical data, and genomic data are analyzed
Fig. 2.
Schematic diagram of quantitative analysis of diffuse lung disease in lung cancer patients. Whole lung and lobes are segmented, and quantitative features are extracted using histogram or texture-based method. Relationships among the quantitative features, clinical data, and genomic data are analyzed
Steps
-
Image acquision
Image acquisition is the first step in the practice of radiomics. One major challenge in this step is the wide variation in image acquisition parameters including radiation dose, scanning protocol, reconstruction algorithm, and slice thickness used in routine clinical practice. Yan et al. successfully identified several features that remained stable even at different PET image reconstruction settings [11], of which peak standardized uptake value (SUVpeak), SUVmean, multiple texture features, and entropy were the most robust. However, comparison of radiomics features extracted from different methods of image acquisition needs further investigation.
-
2)
Segmentation
The next step is to define the region of interest (ROI) that contains the whole tumor or subregions within the tumor, a process called tumor segmentation. This is generally not a problem for solid tumors with definite tumor margins. However, when tumors have indistinct borders, e.g., peripheral ground glass opacity (GGO) in invasive lung adenocarcinoma, identification of the tumor margin becomes a much more complex task [12].
In addition, particular consideration should be paid to whole lung and lobe segmentation, which provides the advantage of predicting postoperative residual lung function, morbidity, and mortality. For lobe segmentation, the first step is to segment the lung region including the lung parenchyma, airways, and vessels by applying an airway threshold. Next, the major airways and vessels are removed to separate the left and right lungs. Fissure detection is essential for accurate lobe segmentation and is based on image intensity computed from the local neighborhoods around each voxel and anatomical information, such as the airways and vasculature [13]. Segmentation of the airways can be performed by manual, semiautomatic, or automatic methods. Manual segmentation is extremely time consuming. Region growing and wave propagation are common methods of airway segmentation based on threshold (cutoff) pixel values in Hounsfield units (HU). A morphology-based method is also used for segmentation of airways [14]. Some additional algorithms have been developed to improve this basic segmentation process. The luminal segmentation is condensed to the centerline that runs exactly through the center of the airway [15]. Airway branches are identified by detection of the divergence of each point on the skeleton, and the airway is labeled or classified by the identified airway branches [16].
-
3)
Feature extraction
After accurate tumor segmentation, a nearly limitless supply of radiomics features can be extracted from the identified tumor ROI. The advantage of including radiomics features in the field of oncology is quite clear: quantitative features will allow better tumor characterization and can objectively reveal valuable patterns reflecting the tumor biology that are hard to detect with the human eye. Furthermore, extracted radiomics features are constantly being refined and developed [17–19]. We will discuss major types of currently available radiomics features in detail later in this article.
-
4)
Feature selection
Having extracted a massive amount of radiomics features, the next step is to capture the true clinical value of such features. Although the full potential of extracted features has yet to be realized, they have been shown to have associations with cancer detection, diagnosis, prognosis assessment, and even monitoring of treatment response [5, 7, 17, 20]. Commonly use methods are least absolute shrinkage and selection operator, principle component analysis, and random forest. The challenge in this step is the considerable variability in predictive performance that has been reported for the different methods of feature selection and classification [21]. Therefore, the goal is to select the most useful radiomics features for clinical translation in the field of oncology.
Types of Radiomics Features
We present five major classes of radiomics feature: (a) morphological, (b) statistical, (c) regional, (d) model-based, and (e) skeleton features [22]. Morphological features provide detailed information about the shape and volume of a tumor. Features calculated by statistical methods can be further classified into first-order statistical (histogram) features and higher-order statistical (texture) features. Regional features can allow quantification beyond the immediate neighborhood and represent intratumor clonal heterogeneity by subregional clustering. Model-based features are extracted using mathematical approaches, such as the fractal model. Skeleton features provide information about the alteration, shape, thickness, and narrowing of airways.
In the subsequent subsections, we briefly summarize the details of each class of feature.
-
Morphological features
Morphological features are used to define the physical characteristics of a tumor. For example, the roundness of a tumor can be quantified using features, such as spherical disproportion, sphericity, and discrete compactness. Surface area can be calculated by triangulation, which is a technique of generating a net of triangles that completely covers the tumor surface. In terms of spiculation, a larger surface-to-volume ratio demonstrates a more spiculated and irregular tumor, while a lower surface-to-volume ratio demonstrates a smoother and rounder tumor. Another morphological feature of interest is tumor mass, a parameter that integrates volume and density. A wide spectrum of lung adenocarcinomas, the most common histologic type of lung cancer, manifest as sub-solid nodules including a GGO portion. Tumor mass measurement enables the detection of GGO growth earlier than traditional measurements [23, 24].
Laplacian of Gaussian is a spatial filtering technique that enhances the marginal features from surrounding regions. This technique enables quantitative analysis regarding tumor margin characterization, which can reflect the relationship between tumor and surrounding tissue and thus the tumor microenvironment.
-
2)
Statistical features
-
First-order histogram features
The basis of first-order statistics is a histogram, which is a simple plot of tumor pixel attenuation along one axis versus the frequency of pixels at each attenuation value along the other axis. Thus, a histogram displays the range and frequency of pixel values within the defined lesion ROI. Multiple features including mean, median, standard deviation, kurtosis, skewness, energy, entropy, uniformity, and variance can be calculated from this histogram, and most features are reported to be reproducible [25].
Constructing a histogram from conventional CT images is easy, and histogram analysis yields multiple quantitative features; therefore, histogram-based features have been used widely in the field of oncology. Quantitative features from the histogram demonstrate information from the voxel level, which can reflect subtle changes in lung cancers. However, a major limitation of histogram-based features is the loss of spatial information about each voxel.
-
b.
Higher-order texture features
In contrast to histogram features, higher-order texture features denote spatial information about each voxel. A gray level co-occurrence matrix (GLCM) is constructed using the number, distance, and angle of a combination of gray levels in the image. From the GLCM, features of cluster, correlation, contrast, energy, and entropy can be extracted. A gray level run length matrix (GLRL) characterizes continuous voxels with the same gray level in any direction. From the GLRL, features such as long run emphasis, short run emphasis, run length non-uniformity, gray level non-uniformity, and run percentage can be extracted. The neighborhood gray-tone difference matrix (NGTDM) uses the intensity values of a neighborhood instead of one voxel to represent how similar or dissimilar voxel intensities are within a neighborhood. Features of busyness, complexity, and texture strength can be extracted from the NGTDM. There is a large body of literature on texture analysis showing an association with tumor stage, metastasis, treatment response, survival, and molecular genetic profiles in lung cancer [8, 26–30].
-
3)
Regional features
As mentioned above, a great deal of heterogeneity exists even within a single tumor. Intratumoral heterogeneity is important because certain subregions can initiate cancer cell transformation leading to tumor progression. Intratumoral heterogeneity can be exhibited by mapping the spatial distribution of similar gray level intensities within a tumor, namely regional features. In other words, regional features demonstrate the number of subregions and how often certain subregions occur within a tumor. Methods of subregional partitioning include data-driven segmentation and the use of threshold values [6, 10, 31]. Data-driven segmentation groups voxels with similar intensity into clusters, and threshold values are also used to group voxels into clusters.
-
4)
Model-based features
Fractal alteration characterizes the shape complexity of an object over a range of scales. In other words, fractal dimension is a mathematical calculation that reflects the intrinsic shape of an object. In this context, morphological complexity and spatial heterogeneity of tumors can be quantified and assigned a numerical value.
Advantages of fractal dimension are that it is relatively stable, less susceptible to noise than other features, and can be used for longitudinal assessment in a single patient [32]. Another feature of interest is the recently developed fractal signature dissimilarity method, which has been suggested as a novel image texture analysis technique [33]. In that study, the fractal signature dissimilarity method was used to quantitatively assess contrast agent uptake heterogeneity dynamics, indicating a potential role in monitoring the early response to anti-angiogenesis treatment [33].
-
5)
Skeleton features
Skeletonization, referred to as medial axis extraction, is widely used in computerized shape analysis [16]. Quantitative measurement of airways follows segmentation to accurately find the location of the inner airway and is then computed, allowing segmentation of perpendicular plans across the targeted bronchi [14, 34]. The full-width-at-half-maximum (FWHM) method, based on the difference between the two extreme values at which the HU value is equal to half of its maximum, is mostly used to find the inner and outer pixels of the airway wall and calculate the airway wall dimension [15, 34]. The luminal area, wall area (WA%), is automatically extracted and has been used for quantification of airway wall thickening and airway narrowing [34]. Bifurcation angle and airway luminal circularity are used to identify the alteration of airway skeletal structure and heterogeneous airway luminal shape [35].
Clinical Application of CT Radiomics in Oncology
Radiomics Approach to Lung Cancer
In 2011, the International Association for the Study of Lung Cancer (IASLC), the American Thoracic Society (ATS), and the European Respiratory Society (ERS) introduced a new classification for lung adenocarcinomas. A vast volume of literature has covered sub-solid nodules, namely nodules with a GGO component, which correlate with the spectrum of lung adenocarcinoma. CT findings of early-stage lung adenocarcinomas and their precursors are usually pure GGO nodules or part-solid nodules. Thus, the imaging spectrum of GGO reflects the evolving process of adenocarcinoma from preinvasive lesions caused by the accumulation of gene mutations. However, discrimination between the invasive and non-invasive proportions is challenging in GGO lesions due to limited visual perception and subjective analysis of conventional CT scans [36, 37]. Multiple investigations have shown that quantitative radiomics features of GGO lesions can help find small pathologically invasive components that are hard to visually perceive at the medical imaging voxel level [7, 18, 38]. Entropy or a high attenuation value, such as the 75th percentile CT attenuation value from histograms, has been reported as a significant discrimination factor for invasive adenocarcinomas [7]. Furthermore, the 97.5th percentile CT attenuation value and the slope of CT attenuation values have been suggested as predictors for future CT attenuation changes and the growth rate of pure GGO lesions [39]. Therefore, it is not surprising that lung cancer-specific (GGO-related) radiomics features can provide additional information about tumor invasiveness and progression from other indolent or non-invasive lesions and can even predict tumor growth.
In addition, regional features have shown great potential in depicting the spatial heterogeneity of cancers. By grouping similar voxels together, multiple subregions that respond differently to therapy or result in tumor progression can be revealed. In fact, in one recent study, researchers were able to identify clinically relevant high-risk subregions in lung cancer using intratumor partitioning of 18F-FDG PET and CT images [31].
Tissue stiffness is a widely accepted biomechanical property of fibrotic tumors and affects tumor growth, invasion, metastasis, and treatment. Analysis of tissue displacement after disruption of the confining structure showed that solid stress depends on both cancer cell type and the microenvironment; solid stress increased with tumor size, and mechanical confinement by the tumor surroundings substantially contributes to intratumoral solid stress [40].
Radiomics features have also shown favorable results when linked to underlying genomic alterations. Features of tumor size, edge shape, and sharpness showed the highest prognostic significance to predict metagenes in patients with non-small cell lung cancer [30]. Another study using semantic features and clinical variables was able to predict patients with ALK rearrangements [41]. Finally, Yoon et al. combined radiomics features and clinical information to successfully predict oncogenic fusion genes in lung cancer [42].
Prediction of Postoperative Lung Function or Postoperative Morbidity
Prediction of postoperative lung function plays a key role in the preoperative evaluation of lung cancer patients with impaired lung function in order to identify an increased risk of postoperative complication and mortality [43]. Currently, postoperative lung function is predicted using spirometry, including forced expiratory volume in 1 s and diffusing capacity for carbon monoxide, and radionuclide lung scanning [44]. An accurate prediction of postoperative pulmonary function is considered for conditions with inhomogeneous effective pulmonary function such as pulmonary emphysema or interstitial lung disease (ILD) [45]. Quantitative CT can be used to calculate the volume of regional and total functional lung, enabling the normal functional volume to be distinguished from the non-functional volume resulting from emphysema, tumor, and atelectasis using histogram-based lung densitometry, separately in the resected lobe and the remaining lung regions [46, 47]. The effectiveness of quantitative CT for predicting postoperative lung function was first proposed by Wu et al. [48]. Further studies have demonstrated the role of quantitative CT in predicting postoperative lung function, and its prediction seems to correlate well with perfusion scintigraphy and pulmonary function tests [43, 49–51]. A recent study showed that volumetry from inspiration/expiration CT could be useful for prediction of postoperative lung function [45]. Although lung lobectomy results in permanent loss of functional lung, patients with lung cancer and chronic obstructive pulmonary disease (COPD) who undergo cancer resection can have minimal loss or improvement in postoperative lung function, a phenomenon known as the lung volume reduction effect [52]. A combined evaluation using spirometry and quantitative CT could characterize the respiratory dynamics and might be used as a predictor of the volume reduction effect [47]. In addition, dual-energy CT (DECT) provides images presenting the lung perfusion at a specific time point. By extraction and quantification of the iodine concentration, DECT provides the ratio of lobar perfusion of the lung, which allows accurate prediction of postoperative lung function [46, 53]. Choe et al. reported that a modified method incorporating postoperative lung volume change using DECT can be considered a comparable method for predicting postoperative lung function [54].
Furthermore, the frequencies of postoperative complications and mortality are higher in patients with COPD and ILD [52, 55]. Quantitative CT in combination with spirometeric measurements contribute to improved prediction of cardiopulmonary complications after lobectomy for lung cancer [56]. In one study, the authors reported that lung density lower than −787.5 HU and volume of emphysema greater than 5.41% increase the risk for developing postoperative pulmonary morbidity [57]. In addition, the severity of lung fibrosis on preoperative CT images was an independent predictive factor of postoperative mortality in lung cancer patients with combined pulmonary fibrosis and emphysema [58]. The severity of lung fibrosis can be quantitatively analyzed for extent and pattern on CT using histogram-based quantification, texture-based quantification, and deep learning. Several studies showed that automated quantification of radiologic patterns of ILD, including normal, GGO, reticular opacity, honeycombing, emphysema, and consolidation, could predict lung function, disease severity, and progression [59–63]. Therefore, quantitative analysis of ILD and COPD can be used to predict mortality and morbidity after treatment in lung cancer patients.
Conclusion and Future Aspects
Although radiomics is still in its infancy, the approaches used to date are very promising sources of robust imaging biomarkers for predicting molecular genetic subtypes related to patient prognosis, optimizing treatment such as selecting the appropriate chemotherapeutic agent, and predicting treatment response. Nevertheless, the hurdle of reproducibility of radiomics features remains. Although radiomics features were found to be mostly unstable in earlier studies [64–66], there are continuous improvements in their standardization. In addition, most studies have extracted radiomics features from a single imaging modality, and extraction of radiomics features using multispectral analysis across different modalities has substantial potential. By combining anatomic, functional, and metabolic imaging, a radiomics approach can provide valuable information for phenotyping tumor biology in correlation with tumor diagnosis, classification, and treatment response prediction.
Another issue is sharing of data cross multiple institutions. Data sharing is a critical point in the field of radiomics which must be overcome. Answers to this may be large centralized data repositories or federated approaches. For tumor response and prognosis, any imaging biomarker must be reliable and meaningful. Thus, by incorporating different image protocols and reconstructions, data sharing among institutes can help to create a predictive and prognostic model with high accuracy. Furthermore, as intensity inhomogeneity may significantly affect the extracted radiomics features, special consideration is required when applying radiomics to magnetic resonance images [67, 68].
In conclusion, the role of medical imaging in human cancer is now larger than ever, and analysis of radiologic data, namely radiomics, has enormous potential to further enrich the knowledge obtained from medical images. Therefore, we anticipate that radiomics will have an essential position in the development and implementation of precision medicine in the field of oncology in the foreseeable future.
Compliance with Ethical Standards
Conflict of Interest
Geewon Lee, So Hyeon Bak, and Ho Yun Lee declare that they have no conflict of interest. This research was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare (HI17C0086) and by National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIP; Ministry of Science, ICT, & Future Planning) (No. NRF-2016R1A2B4013046 and NRF-2017M2A2A7A02018568).
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Requirement to obtain informed consent was waived.
Footnotes
Geewon Lee and So Hyeon Bak contributed equally to this work.
References
- 1.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. [DOI] [PMC free article] [PubMed]
- 2.The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. [DOI] [PMC free article] [PubMed]
- 3.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366:489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 5.Chong Y, Kim JH, Lee HY, Ahn YC, Lee KS, Ahn MJ, et al. Quantitative CT variables enabling response prediction in neoadjuvant therapy with EGFR-TKIs: are they different from those in neoadjuvant concurrent chemoradiotherapy? PLoS One. 2014;9:e88598. doi: 10.1371/journal.pone.0088598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Divine MR, Katiyar P, Kohlhofer U, Quintanilla-Martinez L, Pichler BJ, Disselhorst JA. A population-based Gaussian mixture model incorporating 18F-FDG PET and diffusion-weighted MRI quantifies tumor tissue classes. J Nucl Med. 2016;57:473–479. doi: 10.2967/jnumed.115.163972. [DOI] [PubMed] [Google Scholar]
- 7.Son JY, Lee HY, Kim JH, Han J, Jeong JY, Lee KS, et al. Quantitative CT analysis of pulmonary ground-glass opacity nodules for distinguishing invasive adenocarcinoma from non-invasive or minimally invasive adenocarcinoma: the added value of using iodine mapping. Eur Radiol. 2016;26:43–54. doi: 10.1007/s00330-015-3816-y. [DOI] [PubMed] [Google Scholar]
- 8.Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur Radiol. 2012;22:796–802. doi: 10.1007/s00330-011-2319-8. [DOI] [PubMed] [Google Scholar]
- 9.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 10.O'Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21:249–257. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan J, Chu-Shern JL, Loi HY, Khor LK, Sinha AK, Quek ST, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med. 2015;56:1667–1673. doi: 10.2967/jnumed.115.156927. [DOI] [PubMed] [Google Scholar]
- 12.Rios Velazquez E, Aerts HJ, Gu Y, Goldgof DB, De Ruysscher D, Dekker A, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists' delineations and with the surgical specimen. Radiother Oncol. 2012;105:167–173. doi: 10.1016/j.radonc.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doel T, Gavaghan DJ, Grau V. Review of automatic pulmonary lobe segmentation methods from CT. Comput Med Imaging Graph. 2015;40:13–29. doi: 10.1016/j.compmedimag.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Tschirren J, Hoffman EA, McLennan G, Sonka M. Segmentation and quantitative analysis of intrathoracic airway trees from computed tomography images. Proc Am Thorac Soc. 2005;2:484–487. doi: 10.1513/pats.200507-078DS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley-Zaporozhan J, Kauczor HU. Imaging of airways: chronic obstructive pulmonary disease. Radiol Clin N Am. 2009;47:331–342. doi: 10.1016/j.rcl.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Gu S, Wang Z, Siegfried JM, Wilson D, Bigbee WL, Pu J. Automated lobe-based airway labeling. Int J Biomed Imaging. 2012;2012:382806. doi: 10.1155/2012/382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko JP, Suh J, Ibidapo O, Escalon JG, Li J, Pass H, et al. Lung Adenocarcinoma: correlation of quantitative CT findings with pathologic findings. Radiology. 2016;280:931–939. doi: 10.1148/radiol.2016142975. [DOI] [PubMed] [Google Scholar]
- 19.Park J, Kobayashi Y, Urayama KY, Yamaura H, Yatabe Y, Hida T. Imaging characteristics of driver mutations in EGFR, KRAS, and ALK among treatment-naive patients with advanced lung Adenocarcinoma. PLoS One. 2016;11:e0161081. doi: 10.1371/journal.pone.0161081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messiou C, Orton M, Ang JE, Collins DJ, Morgan VA, Mears D, et al. Advanced solid tumors treated with cediranib: comparison of dynamic contrast-enhanced MR imaging and CT as markers of vascular activity. Radiology. 2012;265:426–436. doi: 10.1148/radiol.12112565. [DOI] [PubMed] [Google Scholar]
- 21.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ. Machine learning methods for quantitative Radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee G, Lee HY, Park H, Schiebler ML, van Beek EJ, Ohno Y, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: state of the art. Eur J Radiol. 2017;86:297–307. doi: 10.1016/j.ejrad.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 23.de Hoop B, Gietema H, van de Vorst S, Murphy K, van Klaveren RJ, Prokop M. Pulmonary ground-glass nodules: increase in mass as an early indicator of growth. Radiology. 2010;255:199–206. doi: 10.1148/radiol.09090571. [DOI] [PubMed] [Google Scholar]
- 24.Lee HY, Jeong JY, Lee KS, Kim HJ, Han J, Kim BT, et al. Solitary pulmonary nodular lung adenocarcinoma: correlation of histopathologic scoring and patient survival with imaging biomarkers. Radiology. 2012;264:884–893. doi: 10.1148/radiol.12111793. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, Zhang L, Fave XJ, Fried DV, Stingo FC, Ng CS, et al. Uncertainty analysis of quantitative imaging features extracted from contrast-enhanced CT in lung tumors. Comput Med Imaging Graph. 2016;48:1–8. doi: 10.1016/j.compmedimag.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Kadi OS, Watson D. Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans Biomed Eng. 2008;55:1822–1830. doi: 10.1109/TBME.2008.919735. [DOI] [PubMed] [Google Scholar]
- 27.Cook GJ, Yip C, Siddique M, Goh V, Chicklore S, Roy A, et al. Are pretreatment 18F-FDG PET tumor textural features in non-small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. 2013;54:19–26. doi: 10.2967/jnumed.112.107375. [DOI] [PubMed] [Google Scholar]
- 28.Fried DV, Tucker SL, Zhou S, Liao Z, Mawlawi O, Ibbott G, et al. Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2014;90:834–842. doi: 10.1016/j.ijrobp.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganeshan B, Abaleke S, Young RC, Chatwin CR, Miles KA. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010;10:137–143. doi: 10.1102/1470-7330.2010.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gevaert O, Xu J, Hoang CD, Leung AN, Xu Y, Quon A, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264:387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Gensheimer MF, Dong X, Rubin DL, Napel S, Diehn M, et al. Robust intratumor partitioning to identify high-risk subregions in lung cancer: a pilot study. Int J Radiat Oncol Biol Phys. 2016;95:1504–12. [DOI] [PMC free article] [PubMed]
- 32.Lennon FE, Cianci GC, Cipriani NA, Hensing TA, Zhang HJ, Chen CT, et al. Lung cancer-a fractal viewpoint. Nat Rev Clin Oncol. 2015;12:664–675. doi: 10.1038/nrclinonc.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Subashi E, Yin FF, Chang Z. Dynamic fractal signature dissimilarity analysis for therapeutic response assessment using dynamic contrast-enhanced MRI. Med Phys. 2016;43:1335–1347. doi: 10.1118/1.4941739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dournes G, Laurent F. Airway remodelling in asthma and COPD: findings, similarities, and differences using quantitative CT. Pulm Med. 2012;2012:670414. doi: 10.1155/2012/670414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi S, Hoffman EA, Wenzel SE, Castro M, Fain S, Jarjour N, et al. Quantitative computed tomographic imaging-based clustering differentiates asthmatic subgroups with distinctive clinical phenotypes. J Allergy Clin Immunol. 2017;140:690-700.e8. [DOI] [PMC free article] [PubMed]
- 36.Eguchi T, Yoshizawa A, Kawakami S, Kumeda H, Umesaki T, Agatsuma H, et al. Tumor size and computed tomography attenuation of pulmonary pure ground-glass nodules are useful for predicting pathological invasiveness. PLoS One. 2014;9:e97867. doi: 10.1371/journal.pone.0097867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HY, Choi YL, Lee KS, Han J, Zo JI, Shim YM, et al. Pure ground-glass opacity neoplastic lung nodules: histopathology, imaging, and management. AJR Am J Roentgenol. 2014;202:W224–W233. doi: 10.2214/AJR.13.11819. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda K, Awai K, Mori T, Kawanaka K, Yamashita Y, Nomori H. Differential diagnosis of ground-glass opacity nodules: CT number analysis by three-dimensional computerized quantification. Chest. 2007;132:984–990. doi: 10.1378/chest.07-0793. [DOI] [PubMed] [Google Scholar]
- 39.Bak SH, Lee HY, Kim JH, Um SW, Kwon OJ, Han J, et al. Quantitative CT scanning analysis of pure ground-glass opacity nodules predicts further CT scanning change. Chest. 2016;149:180–191. doi: 10.1378/chest.15-0034. [DOI] [PubMed] [Google Scholar]
- 40.Nia HT, Liu H, Seano G, Datta M, Jones D, Rahbari N, et al. Solid stress and elastic energy as measures of tumour mechanopathology. Nat Biomed Eng. 2016;1:1–11. doi: 10.1038/s41551-016-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong CJ, Lee HY, Han J, Jeong JY, Lee KS, Choi YL, et al. Role of imaging biomarkers in predicting anaplastic lymphoma kinase-positive lung adenocarcinoma. Clin Nucl Med. 2015;40:e34–e39. doi: 10.1097/RLU.0000000000000581. [DOI] [PubMed] [Google Scholar]
- 42.Yoon HJ, Sohn I, Cho JH, Lee HY, Kim JH, Choi YL, et al. Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung Adenocarcinoma using a Radiomics approach. Medicine (Baltimore) 2015;94:e1753. doi: 10.1097/MD.0000000000001753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papageorgiou CV, Antoniou D, Kaltsakas G, Koulouris NG. Role of quantitative CT in predicting postoperative FEV1 and chronic dyspnea in patients undergoing lung resection. Multidiscip Respir Med. 2010;5:188–193. doi: 10.1186/2049-6958-5-3-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poonyagariyagorn H, Mazzone PJ. Lung cancer: preoperative pulmonary evaluation of the lung resection candidate. Semin Respir Crit Care Med. 2008;29:271–284. doi: 10.1055/s-2008-1076747. [DOI] [PubMed] [Google Scholar]
- 45.Yabuuchi H, Kawanami S, Kamitani T, Yonezawa M, Yamasaki Y, Yamanouchi T, et al. Prediction of post-operative pulmonary function after lobectomy for primary lung cancer: a comparison among counting method, effective lobar volume, and lobar collapsibility using inspiratory/expiratory CT. Eur J Radiol. 2016;85:1956–1962. doi: 10.1016/j.ejrad.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Chae EJ, Kim N, Seo JB, Park JY, Song JW, Lee HJ, et al. Prediction of postoperative lung function in patients undergoing lung resection: dual-energy perfusion computed tomography versus perfusion scintigraphy. Investig Radiol. 2013;48:622–627. doi: 10.1097/RLI.0b013e318289fa55. [DOI] [PubMed] [Google Scholar]
- 47.Ueda K, Murakami J, Sano F, Hayashi M, Kobayashi T, Kunihiro Y, et al. Assessment of volume reduction effect after lung lobectomy for cancer. J Surg Res. 2015;197:176–182. doi: 10.1016/j.jss.2015.03.064. [DOI] [PubMed] [Google Scholar]
- 48.Wu MT, Chang JM, Chiang AA, Lu JY, Hsu HK, Hsu WH, et al. Use of quantitative CT to predict postoperative lung function in patients with lung cancer. Radiology. 1994;191:257–262. doi: 10.1148/radiology.191.1.8134584. [DOI] [PubMed] [Google Scholar]
- 49.Moloney F, McWilliams S, Crush L, Laughlin PD, Kenneddy M, Henry M, et al. CT densitometry as a predictor of pulmonary function in lung cancer patients. Open Respir Med J. 2012;6:139–144. doi: 10.2174/1874306401206010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohno Y, Koyama H, Nogami M, Takenaka D, Onishi Y, Matsumoto K, et al. State-of-the-art radiological techniques improve the assessment of postoperative lung function in patients with non-small cell lung cancer. Eur J Radiol. 2011;77:97–104. doi: 10.1016/j.ejrad.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Wu MT, Pan HB, Chiang AA, Hsu HK, Chang HC, Peng NJ, et al. Prediction of postoperative lung function in patients with lung cancer: comparison of quantitative CT with perfusion scintigraphy. AJR Am J Roentgenol. 2002;178:667–672. doi: 10.2214/ajr.178.3.1780667. [DOI] [PubMed] [Google Scholar]
- 52.Dai J, Yang P, Cox A, Jiang G. Lung cancer and chronic obstructive pulmonary disease: from a clinical perspective. Oncotarget. 2017;8:18513–18524. doi: 10.18632/oncotarget.14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lapointe A, Bahig H, Blais D, Bouchard H, Filion E, Carrier JF, et al. Assessing lung function using contrast-enhanced dual energy computed tomography for potential applications in radiation therapy. Med Phys. 2017. 10.1002/mp.12475. [DOI] [PubMed]
- 54.Choe J, Lee SM, Chae EJ, Lee SM, Kim YH, Kim N, et al. Evaluation of postoperative lung volume and perfusion changes by dual-energy computed tomography in patients with lung cancer. Eur J Radiol. 2017;90:166–173. doi: 10.1016/j.ejrad.2017.02.040. [DOI] [PubMed] [Google Scholar]
- 55.Chiyo M, Sekine Y, Iwata T, Tatsumi K, Yasufuku K, Iyoda A, et al. Impact of interstitial lung disease on surgical morbidity and mortality for lung cancer: analyses of short-term and long-term outcomes. J Thorac Cardiovasc Surg. 2003;126:1141–1146. doi: 10.1016/S0022-5223(03)00791-8. [DOI] [PubMed] [Google Scholar]
- 56.Ueda K, Kaneda Y, Sudoh M, Mitsutaka J, Tanaka N, Suga K, et al. Role of quantitative CT in predicting hypoxemia and complications after lung lobectomy for cancer, with special reference to area of emphysema. Chest. 2005;128:3500–3506. doi: 10.1378/chest.128.5.3500. [DOI] [PubMed] [Google Scholar]
- 57.Kaplan T, Atac GK, Gunal N, Kocer B, Alhan A, Cubuk S, et al. Quantative computerized tomography assessment of lung density as a predictor of postoperative pulmonary morbidity in patients with lung cancer. J Thorac Dis. 2015;7:1391–1397. doi: 10.3978/j.issn.2072-1439.2015.07.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mimae T, Suzuki K, Tsuboi M, Ikeda N, Takamochi K, Aokage K, et al. Severity of lung fibrosis affects early surgical outcomes of lung cancer among patients with combined pulmonary fibrosis and emphysema. Medicine (Baltimore) 2016;95:e4314. doi: 10.1097/MD.0000000000004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humphries SM, Yagihashi K, Huckleberry J, Rho BH, Schroeder JD, Strand M, et al. Idiopathic pulmonary fibrosis: data-driven textural analysis of extent of fibrosis at baseline and 15-month follow-up. Radiology. 2017;285:270-278. [DOI] [PMC free article] [PubMed]
- 60.Maldonado F, Moua T, Rajagopalan S, Karwoski RA, Raghunath S, Decker PA, et al. Automated quantification of radiological patterns predicts survival in idiopathic pulmonary fibrosis. Eur Respir J. 2014;43:204–212. doi: 10.1183/09031936.00071812. [DOI] [PubMed] [Google Scholar]
- 61.Moon JW, Bae JP, Lee HY, Kim N, Chung MP, Park HY, et al. Perfusion- and pattern-based quantitative CT indexes using contrast-enhanced dual-energy computed tomography in diffuse interstitial lung disease: relationships with physiologic impairment and prediction of prognosis. Eur Radiol. 2016;26:1368–1377. doi: 10.1007/s00330-015-3946-2. [DOI] [PubMed] [Google Scholar]
- 62.Park HJ, Lee SM, Song JW, Lee SM, Oh SY, Kim N, et al. Texture-based automated quantitative assessment of regional patterns on initial CT in patients with idiopathic pulmonary fibrosis: relationship to decline in forced vital capacity. AJR Am J Roentgenol. 2016;207:976–983. doi: 10.2214/AJR.16.16054. [DOI] [PubMed] [Google Scholar]
- 63.Yoon RG, Seo JB, Kim N, Lee HJ, Lee SM, Lee YK, et al. Quantitative assessment of change in regional disease patterns on serial HRCT of fibrotic interstitial pneumonia with texture-based automated quantification system. Eur Radiol. 2013;23:692–701. doi: 10.1007/s00330-012-2634-8. [DOI] [PubMed] [Google Scholar]
- 64.Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, Hawkins S, et al. Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol. 2014;7:72–87. doi: 10.1593/tlo.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014;27:805–823. doi: 10.1007/s10278-014-9716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med. 2012;53:693–700. doi: 10.2967/jnumed.111.099127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antunes J, Viswanath S, Rusu M, Valls L, Hoimes C, Avril N, et al. Radiomics analysis on FLT-PET/MRI for characterization of early treatment response in renal cell carcinoma: a proof-of-concept study. Transl Oncol. 2016;9:155–162. doi: 10.1016/j.tranon.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vovk U, Pernus F, Likar B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging. 2007;26:405–421. doi: 10.1109/TMI.2006.891486. [DOI] [PubMed] [Google Scholar]