Abstract
Radiomics utilizes high-dimensional imaging data to discover the association with diagnostic, prognostic, predictive endpoint or radiogenomics. It is an emerging field of study that potentially depicts the intratumoral heterogeneity from quantitative and classified high-throughput data. The radiomics approach has an analytic pipeline where the imaging features are extracted, processed and analyzed. At this point, special data handling is essential because it faces issues of a high-dimensional biomarker compared to a single biomarker approach. This article describes the potential role of radiomics in oncologic studies, the basic analytic pipeline and special data handling with high-dimensional data to facilitate the radiomics approach as a tool for personalized medicine in oncology.
Keywords: Radiomics, High-dimensional, Imaging, Modeling, Neuro-oncology, Magnetic resonance
Introduction
Radiomics is an emerging field of study in which that quantitative, high-throughput data are extracted, processed and analyzed to discover the associations with meaningful information. The meaningful, relevant information is directed to pathologic or genomic data or to various clinical endpoints. The suffix -omics originally described a collective characterization and quantification of pools of biologic information, such as genomics, proteomics or metabolomics. The -omics concept in radiomics readily applies as radiomics utilizes millions of voxels from multi-section tomographic or volumetric imaging data from a single patient, which may comprehensively represent biologic information regarding a disease.
This collective data mining in radiomics has two distinctive benefits compared to the traditional radiologic data mining. First, a radiomics approach provides semiautomatic or automatic extraction of radiologic features and can provide robust information compared to subjective reading by readers or qualitative analysis. Second, high-dimensional radiomic data provide insight into intra-regional heterogeneity by identifying sub-regions and reflecting the spatial complexity of a disease. Therefore, radiomics is an especially promising tool for personalized medicine in oncology, which requires a clear understanding of tumoral heterogeneity in individual patients.
Recent advances in image processing and the availability of digital imaging data expand the boundaries of the radiomic approach beyond a research tool, and currently a number of studies test the important medical hypothesis of diagnosis, assessment of prognosis and prediction of treatment response. In this rapidly developing field of study, the goal of this article is to introduce the basics of radiomics workflow and practical considerations in data analysis to researchers who are about to engage high-dimensional data mining. Several review papers in this field are currently available, and each has its own strength and focus. This review highlights the potential role of the radiomics approach in oncologic imaging, basic radiomics workflow, illustrating examples of the promise in neuro-oncology research, and future consideration as a new imaging biomarker in oncology in conjunction with the imaging biomarker roadmap for cancer studies. Most of our examples are from neuro-oncologic articles, although these are representative of illustrative examples of the promise in general oncology research.
Radiomics Pipeline
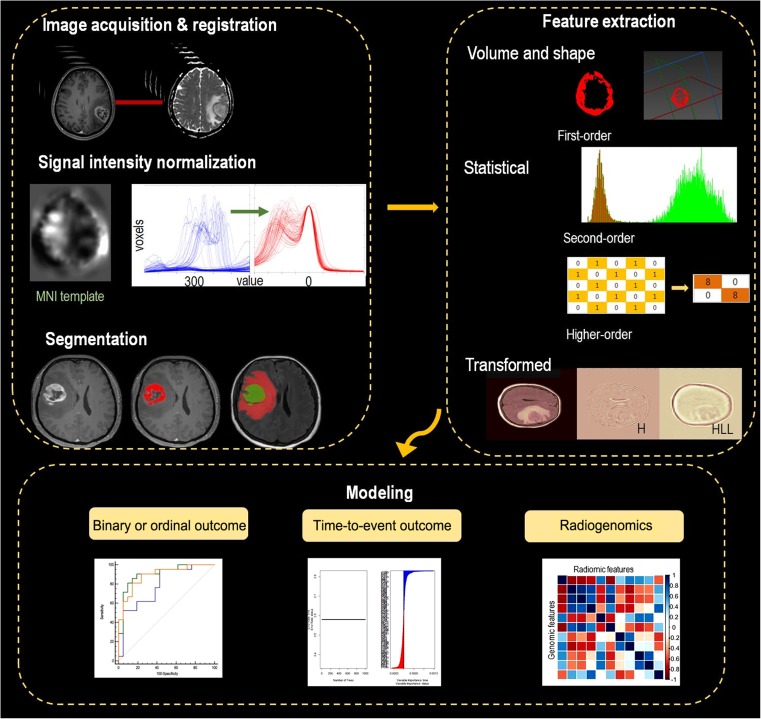
The workflow in the radiomics is summarized in Fig. 1. Followed by imaging acquisition, pre-processing including registration and signal intensity normalization is performed for each imaging datum. Radiomics features are extracted using low- and high-order statistics by computational methods. To reduce false positives from high-dimensional radiomics data, feature selection and dimension reduction are followed. Choosing a statistical model for radiomics to find associations with patient data greatly depends on the outcomes. A binary classification method will be used if it is a binary endpoint; survival modeling such as Cox proportional hazards regression will be used if it is a time-to-event outcome; multiple comparisons will be used if there are genomic data with multiple outcomes. This statistical model encompasses considerations of generalizability that create a model in a given data set as a training set and needs another data set to test the model, i.e., a so-called ‘validation’ or ‘test’ set.
Fig. 1.
The radiomics pipeline. Part I includes image acquisition, registration and segmentation. Signal intensity normalization is conducted for conventional MR imaging with signal intensity of arbitrary units. Part II includes feature extraction. Part III includes modeling according to the outcomes, with special consideration for high-dimensional data
Steps in the Radiomics Pipeline and Practical Considerations
Imaging Acquisition and Preprocessing
Any digitalized imaging data are utilized in radiomics and include simple radiographs, computed tomography, magnetic resonance imaging and positron emission tomography (PET). Although radiomics is regarded as a ‘post-processing’ computational analytic method, it is not. Acquisition of raw data is as important as other imaging biomarkers because both spatial resolution (voxel size) and gray-level resolution (contrast) can affect the calculation of radiomics features, which are determined by imaging acquisition techniques and parameters. Previous CT studies showed that different imaging acquisition parameters change the radiologic features [1, 2]. Therefore, standardization of image acquisition is essential to maintain the integrity of the radiomics analysis. However, in actual clinical practice, acquisitions and scanning protocols vary among patients and medical institutions.
Preprocessing of imaging data is, therefore, performed to standardize heterogeneous imaging data before feature extraction. A solution for heterogeneous ‘spatial resolution’ is resampling voxels into isotropic pixels or voxels after co-registration of multi-spectral imaging or different sequences in the same imaging modality. A solution for heterogeneous ‘contrast’ is intensity normalization. In MRI, the two most routinely centered modalities in glioma are fluid-attenuated inversion recovery (FLAIR) and T1-post contrast enhancement sequences. Signal intensity normalization was used to reduce variance in the T1-based signal intensity of the brain. This preserves ranks among tissues and matches the intensity of tissues without disturbing the natural balance of tissue intensities [3]. Statistical packages, such as the hybrid white-stripe method [4] for intensity normalization using the ANTsR and WhiteStripe packages [3, 5] in R, are readily available for this method.
Consideration of standardized imaging acquisition and preprocessing is used for the reproducibility of radiomics features. A homogeneous imaging protocol strengthens standardization of imaging, although it degrades the generalizability of the radiomics features, which will be discussed later in this article.
Defining the Region of Interest
Segmentation of the region-of-interest (ROI) is often performed semi-automatically, first by computer-aided or software-based edge detection followed by manual curation. User-friendly software programs aid this procedure: 3D-Slicer (http://www.slicer.org) [6], (MITK, www.mitk.org German Cancer Research Center, Heidelberg, Germany) [4]; etc. Segmentation is probably the most critical component in radiomics because actual radiomics features are generated from the segmented ROIs. This is especially challenging in most tumors, including gliomas, because they have indistinct margins. Also, as there is no defined ground truth, consistency (reproducibility) of segmentation becomes an important issue. Although the semiautomated method is the current state-of-the-art method in cancer imaging studies, fully automated methods are consistently sought to improve reproducibility. Pattern recognition, advanced machine learning or the deep learning technique explores this distinct research topic to improve robustness and optimize the segmentation of tumors.
Radiomic Feature Extraction
Two types of features can be extracted from the segmented volume, “semantic” and “agnostic” features, using the radiomics approach [7]. Semantic features are well-known descriptors for the radiologists and include the size, shape, location, vascularity speculation or necrosis. Visually Accessible Rembrandt Images (VASARI) features on MRI (VASARI ResearchProject. https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project) are representative semantic imaging descriptors for gliomas and showed relevance in genomic data [8] and patient survival prediction [9]. On the other hand, agnostic features are mathematically extracted, quantitative imaging descriptors. Using computational methods, morphologic features and low- and high-order statistics features can be extracted. Table 1 summarizes the agnostic quantitative radiomic features.
Morphologic features
Table 1.
Quantitative radiomic features
| Methods | Simple methods | Statistical methods | Transformed methods | ||
|---|---|---|---|---|---|
| Type | Morphologic features | First-order features | Second-order features | Higher-order, model-based features | |
| Examples | Volume | Histogram parameters | Gray-level matrix | Fractal dimensions | Wavelet transformation |
| Shape | Minkowski functionals | Laplacian transforms | |||
Rather than semantic feature extraction from individual readers, shape and physical characteristics can be extracted mathematically. Commonly used volume and shape features include the surface area, volume, surface-to-volume ratio, compactness and sphericity, all of which are used to describe the 3D geometric properties of a tumor.
-
(2)
Statistical methods
The detailed description of statistical radiomic features refers to previous literature [7, 10]. Briefly, the first-order features were derived from the intensity histogram using first-order statistics including the intensity range, energy, entropy, kurtosis, maximum, mean, median, uniformity and variance. A histogram method is a commonly used tool for imaging parameters displaying the range and frequency of pixel values. Histogram parameters of imaging have been shown to be useful in aiding the differential diagnosis [11] and for monitoring the treatment response in glioblastomas [12]. Although histogram analysis facilitates the objective assessment of the entire volume of interest, it has the limitation that spatial information regarding each voxel is lost and information related to the relative positions of the various gray levels is not available.
On the other hand, texture features can retain spatial information among pixels. Texture can be assessed using statistical methods, i.e., histogram and gray-level dependence matrices, model-based methods, i.e., fractal models, or transformed-based methods, i.e., Fourier and the wavelet transform. Gray-level dependence matrices are the gray level co-occurrence matrix (GLCM) and the gray level run length matrix (GLRLM). When using GLCM, various textural features are extracted, and GLRLM characterizes coarse textures as having many pixels in a constant gray level run and fine textures as having few pixels in such a run. Both GLCM and GLRLM can be constructed from 3D analysis of a volume of interest from matrices. For 3D data, all 13 directions are considered with 26-voxel connectivity for a given location, after which the varying distances of 1, 2 and 3 voxels in 13 directions can be computed.
-
(3)
Transform-based methods
Wavelet transformation can be jointly used together with the morphologic features and higher-order statistics. It is applied with a single-level directional discrete wavelet transform of a high- and low-pass filter [13]. For example, eight wavelet-decomposition images were generated from each MRI imaging sequence input, i.e., HHH, HHL, HLH, HLL, LHH, LHL, LLH and LLL images. The above texture features were then applied to the wavelet-transformed images for texture features multiplied by eight images and thus yielding high-dimensional data.
Notably, statistical-based texture analysis has made the most significant contribution in testing medical hypotheses [14]. A recent pixel-wise correlation study showed the correlation between texture analyses and histopathology [15] and the potential of texture analysis in biologic usage.
Feature Selection and Classifier Modeling Strategies According to Outcomes
By extracting a large amount of through-put data, radiomic analysis faces issues of a high-dimensional biomarkers compared to existing single-parameter analysis. Table 2 summarizes the methods of radiomic analysis according to the outcomes and considerations for high-dimensional data.
For binary outcomes
Table 2.
Feature selection and classifier modeling strategies according to outcomes
| Outcome | Diagnosis | Prognosis/predictive | Radiogenomics |
|---|---|---|---|
| Outcome type | Binary | Time-to-event | Single, binary or multiple |
| Example of study hypothesis | ‘Radiomic features distinguish glioblastoma from primary central nervous lymphoma’ | ‘Radiomic features in glioblastoma have added prognostic value to clinical data’ | ‘Radiomic features in glioblastoma have a significant correlation with specific genes’ |
| Traditional statistical methods | Classification | Cox proportional hazard regression | False discovery rate |
| Considerations for high-dimensional data | Machine-learning- based feature selection Dimension reduction | Statistical methods: Partial least squares Cox-regression under lasso penalization Machine learning: Random survival forest Principle component analysis |
Machine learning: Dimension reduction Permutation test to correct multiple testing issues for multiple outcomes |
| Performance measurement | Receiver-operating characteristics curve | C-index Integrated Brier score |
Correlation coefficients |
Because radiomics has highly redundant feature space [16], it is important to reduce highly correlated features in the selected feature subset as correlated features can cause collinearity [17]. The accuracy test and diagnostic performance are most commonly used to measure the performance of a model for binary outcomes, such as the area under the curve (AUC) in receiver-operating curve analysis. However, before testing the performance, appropriate classification methods are required to be used to reduce the potential risk of over-fitting or false discovery [18].
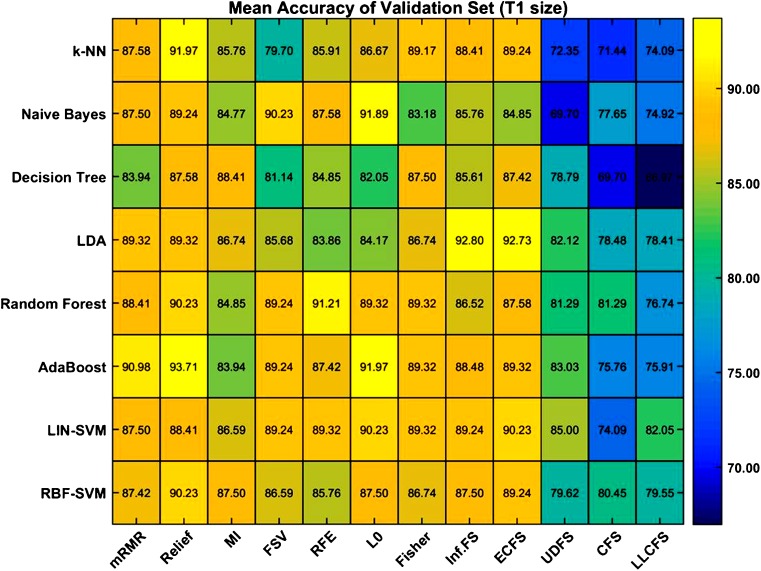
Various machine learning methods can be used for feature selection and classification for binary outcomes, and the best combination of different feature selection and classification methods can be computed. The combination depends on the researchers, and usage is based on commonly adopted methods [16, 19, 20]. Figure 2 demonstrates accuracy results for binary outcomes using different combinations of feature-selection methods and classification methods. The 12 methods for feature selection were correlation-based and local learning-based clustering feature selection, eigenvector centrality, feature selection via concave minimization, generalized Fisher score, infinite feature selection, minimum redundancy maximum relevance, mutual information, relief, recursive feature elimination, unsupervised discriminative feature selection and zero-norm minimization. The classification methods included adaptive boosting, decision tree, k-nearest neighbor, linear kernel support vector machine, linear discriminant analysis, naïve Bayes classifier, random forest and radial basis function support vector machine. This shows that the combination of feature selection and classification method affects the performance in binary outcomes.
-
(2)
For time-to-event outcomes
Fig. 2.
The heatmap demonstrates that diagnostic accuracy can be changed depending on which feature-selection method and classification method is applied. Color scale: expressed from yellow (accuracy, 100%) to blue (accuracy, 65%)
The high-dimensionality of radiomics data also poses challenges in studying the survival data. Univariate analysis using the Cox-regression model was often initially used to select features from multivariable data, although it is often not sufficient to deal with high-dimensional data. Statistical methods of a regularized Cox model [21] or a penalized Cox model [22] can be applied. Machine learning methods have gained considerable interest, for example, boosting methods [23, 24] and random survival forests methods [25–27] have been proposed to overcome hurdles of sparse meaningful data.
-
(3)
For radiogenomics
Dealing with genomic data presents issues of multiple testing problems, and the false discovery rate (FDR) [28] is a commonly used method to correct the p value. Briefly, FDR attempts to adjust the p value to each test or to reduce the p value threshold from 5% to a more reasonable value. The q values refer to the adjusted p values found using the FDR approach and give a more accurate indication of the level of false positives for a given cut-off value. Also, specific data handling is required to erase noise from the genomic data.
Generalizability Issue of Radiomics
Radio-informatic analysis is based on modeling, and the performance of this model needs to be tested in a validation set to gain generalizability. Although complex statistical and machine learning methods are often used to address the challenges presented by a large data set, there is a risk of over-fitting the data in overly parameterized models [29]. Therefore, the purpose of prediction or prognostic modeling is to provide valid information for new patients and necessitates the validation in the modeling process.
Internal validation utilizes the resampling technique within the given data set, such as cross-validation and boot-strapping methods in the original sample. However, the results are from the derivative cohort and have less general applicability compared to a separate cohort. A separate cohort is more desirable to support generalizability by including temporally different cohorts (temporal validation), cohorts from other places (geographic validation) or both temporally and geographically different cohorts (fully external validation). This issue complicates radiomics models because the external validation cohort has different scan parameters and may fundamentally affect radiomics features. Although the standardization of image acquisition is central to the integrity of the entire radiomics pipeline [30], it is very difficult to achieve when the external validation is performed using the different image acquisition in an outside hospital. Nontheless, the generalizability is the purpose of the radiomics approach, and it is a still difficult problem to be solved in the radiomics research.
Examples of Radiomics Research in Neuro-Oncology
Radiomics and radiogenomics research for cancer studies have expanded dramatically over the past decade. In the subsequent section, we will demonstrate examples from neuro-oncology, which is one of the most active fields in radio-informatic research utilizing various quantitative imaging biomarkers and advanced imaging techniques. Table 3 summarizes the currently published articles dealing with radiomics in the field of neuro-oncology.
Table 3.
Published radiomics research in neuro-oncology
| Study | No. of patients | Patients | Purpose | Outcome type | Feature type | No. of features | Validation set |
|---|---|---|---|---|---|---|---|
| Zacharaki et al. (2009) [31] | 102 | Gliomas, Metasta-ses, meningi-oma | Classification | Diagnostic | Agnostic | 161 | No |
| Qin et al. (2017) [32] | 66 | Gliomas | Grade | Diagnostic | Agnostic | 114 | No |
| Yu et al. (2017) [33] | 140 (110 + 30) | Grade II | IDH mutation estimation | Diagnostic | Agnostic | 671 | Yes |
| Lopez et al. (2017) [34] | 17 | GBM | Metabolic tumor volumes | Diagnostic | Semantic, agnostic | 48 | No |
| Wiestler et al. (2016) [35] | 37 | Gliomas | Grade | Diagnostic | Agnostic | 116 | No |
| Zhou et al. (2017) [36] | 165 | Grade II and III | IDH mutation 1p/19q status Grade, progression |
Diagnostic Prognostic |
Semantic, agnostic | 3360 | No |
| Lee et al.(2015) [37] | 65 | GBM | EGFR mutation, Survival | Diagnostic Prognostic |
Agnostic | 36 | No |
| Gevaert et al. (2014) [8] | 55 | GBM | Molecular subtype, survival | Diagnostic Prognostic |
Agnostic | 79 | No |
| Macyszyn et al. (2016) [38] | 134 (105 + 29) | GBM | Molecular subtype, survival | Diagnostic Prognostic |
Agnostic | 60 | Yes |
| Kickingereder et al. (2016) [39] | 119 (79 + 40) | GBM | Survival | Prognostic | Agnostic | 12,190 | Yes |
| Ingrisch et al. (2017) [40] | 66 | GBM | Survival | Prognostic | Agnostic | 208 | No |
| Rao et al. (2016) [41] | 92 | GBM | Survival | Prognostic | Semantic | 9 | No |
| Prasanna et al. (2016) [42] | 66 | GBM | Survival | Prognostic | Agnostic | 402 | No |
| McGarry et al. (2016) [43] | 81 | GBM | Survival | Prognostic | Agnostic | 81 | No |
| Kickingereder et al. (2016) [44] | 172 (112+ 60) | GBM | Treatment response to anti-angiogenic agent | Predictive | Agnostic | 4842 | Yes |
| Lohmann et al. (2017) [45] | 23 | GBM | Pseudo-progression | Predictive | Agnostic | 34 | No |
| Kickingereder et al. (2016) [46] | 152 | GBM | Genetic driven molecular characteristic | Radiogenomics | Agnostic | 31 | No |
| Hu et al. (2017) [47] | 13, 48 samples | GBM | 6 Driver genes | Radiogenomics | Agnostic | 256 | No |
| Gutman et al. (2015) [48] | 66 | GBM | 7 Genes | Radiogenomics | Agnostic | 11 | No |
| Jamshidi et al. (2014) [49] | 23 | GBM | 34 Gene loci | Radiogenomics | Semantic | 6 | No |
GBM = glioblastoma. Number (No.) of patients indicates the patients in the entire study, with the number of derivation (training) set + validation (test) set expressed in parentheses. Semantic features were used with Visually Accessible Rembrandt Images (VASARI). Agnostic features refer to quantitative radiomics features obtained using statistical or model-based methods
Radiomics as a Diagnostic Biomarker
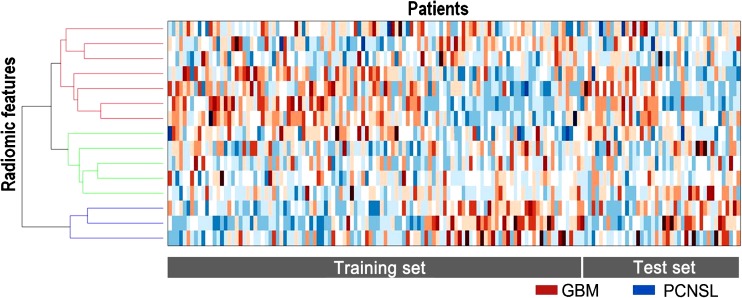
The diagnostic biomarker is a popular topic for the radiomics approach [8, 31–38]. Texture features have long been investigated as a diagnostic marker before the term ‘radiomics’ was established. Texture analyses successfully distinguished gliomas from metastases [31] with an accuracy of 85%, sensitivity of 87% and specificity of 79% in a previous study. The radiomics approach can help glioma grading and determine tumor aggressiveness. In a study of 66 patients with pathologically proven gliomas, texture analysis of T1, T1 CE and ADC showed improved accuracy of glioma grading [32]. Conventional T1, FLAIR and T1 with contrast enhancement (T1 CE) are the most commonly used techniques in radiomics, although investigations now attempt to apply radiomics to advanced imaging such as the apparent diffusion coefficient (ADC). In our pilot study, ADC radiomics was performed for differentiating atypical primary central nervous system lymphoma from glioblastoma, and showed better diagnostic performance (AUC, 0.98) compared to a single but strong parameter of ADC (AUC, 0.78) or CBV (0.90). Figure 3 demonstrates the distinction between two disease entities using the radiomics model.
Fig. 3.
The heatmap is a representation for radiomics analysis, with each row expressing one z-score normalized feature, while each column represents a single patient. GBM = glioblastoma, PCNSL = primary central nervous system lymphoma
The binary outcome of the presence or absence of a molecular biomarker was tested in low-grade glioma characterization [36]. In 165 patients with grade II and grade III gliomas, a radiomic model was built to predict isocitrate dehydrogenase 1 (IDH1) mutation and the 1p/19 co-deletion status as well as the histologic grade. AUC reached 0.86 for IDH1 mutation and 0.96 for the 1p/19q co-deletion status. Another study used 671 features for non-invasive diagnosis of IDH1 mutation in 110 patients with grade II gliomas and showed an AUC of 0.86. These results imply the utility of the radiomics approach in the differential diagnosis, glioma grading and non-invasive diagnosis of molecular markers in neuro-oncology. The results will be strengthened when they are tested with external validation cohorts.
Radiomics as a Prognostic Biomarker
Survival research for patients with glioblastoma has been conducted in several aspects using both semantic and agnostic features [8, 39–43, 50]. A comprehensive data set of the Cancer Imaging Archive (http://cancerimagingarchive.net/) with clinical, imaging and genomic information is widely available, and early radiomics studies show that sematic features by expert radiologists were useful in predicting survival in patients with glioblastoma [8, 50].
A representative quantitative analysis was conducted in 119 glioblastoma patients with 12,190 features [39] in which the radiomics approach stratified a low- or high-risk group and showed better performance than the clinical (age and Karnofsky performance score) and radiologic parameters of CBV and ADC. Peri-enhancing tumoral lesions also showed prognostic value in glioblastomas [42] in which a subset of ten radiomic ‘peritumoral’ features was found to be predictive of survival compared to features from an enhancing tumor portion, necrotic regions and known clinical factors.
Radiomics Approach for Other Surrogate Endpoints
Radiomics showed potential for use as a predictive marker for treatment response in glioblastoma patients treated with an antiangiogenic agent [44]. The study included 172 patients with recurrent glioblastoma, extracted 4842 quantitative radiomic features and generated a prediction model for stratifying the low- and high-risk response group for bevacizumab treatment.
Pseudoprogression can become a surrogate endpoint for radiomics studies, and preliminary results (not shown in Table 3) reported that radiomic features from either MRI [51] or PET [45] can become predictive for discriminating pseudoprogression from true progression. These results showed that the radiomics approach may become helpful to address challenging diagnoses in neuro-oncology.
Radiogenomics
Both semantic and agnostic features have been studied for radiogenomic analysis [46–49] to find the association between radiomics features and known gene loci. The set of genes can vary from a single gene to multiple gene loci, and a significant correlation is determined using Pearson’s correlation with the adjusted p value. Radiogenomic analysis found an association between semantic features with gene mutation in glioblastoma, i.e., the contrast-to-necrosis ratio with KLK3 and RUNX3, association of SVZ involvement with Ras oncogene family members, such as RAP2A, and the metabolic enzyme TYMS, and the association of vasogenic edema with the oncogene FOXP1 and PIK3IP1 [49]. A more recent study using 48 samples identified significant imaging correlations for six driver genes, EGFR, PDGFRA, PTEN, CDKN2A, RB1 and TP53, with various agnostic features among 256 imaging phenotypes [47]. This research field has yet to be validated with external validation data, and further investigation with large cohort numbers is particularly required in this field of research.
Outlook—Radiomics as a New Imaging Biomarker in Cancer Studies
Imaging data can non-invasively assess tissue characteristics using the radiomics approach and therefore have potential to be routinely used for diagnostic and predictive purposes in cancer studies. Biomarkers that can be reliably used to test medical hypotheses cross the first gap and become a useful ‘medical research tool;’ if the biomarker crosses the second gap, then it becomes a ‘clinical decision-making tool’ [52]. Radiomics has currently become a useful tool for research studies. However, as it is a new imaging biomarker, further validation and qualification are needed for it to become a clinical decision-making tool in health care. Also, a potential limitation in image analysis is the heterogeneous imaging acquisition parameters being used in clinical practice and affecting the results. A previous CT study [1] showed that radiomic features are reproducible over a wide range of imaging settings, unless smooth and sharp reconstruction algorithms are used. MR-based radiomic features may be more vulnerable to changes in acquisition parameters, wherein the margin and signal-to-noise ratio can be easily varied across imaging protocols. Parametric maps such as ADC maps or normalized cerebral blood volume may be robust across the different acquisition schemes, but this issue needs to be further studied. Consequently, it is important to determine how different imaging acquisition parameters affect the computed values of the radiomic features.
Conclusion
Radiomics mines high-throughput quantitative imaging features to depict tissue characteristics noninvasively and to find relationships with meaningful clinical and genetic information. In neuro-oncologic research, radiomics can be studied as either a diagnostic, prognostic, predictive biomarker or radiogenomics. Selection of a proper model needs to be based on the outcome types. Also, validation of the model with independent data set is critical to generalize the results from radiomic analysis. When it obtains generalizability across heterogeneous imaging acquisition protocols, it can become a clinical decision-making tool in clinical practice.
Acknowledgements
This study was supported by a grant from the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (1720030).
Compliance with Ethical Standards
Conflict of Interest
Ji Eun Park and Ho Sung Kim declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed Consent
The institutional review board of our institute approved this retrospective study, and the requirement to obtain informed consent was waived.
References
- 1.Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428. doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One. 2016;11:e0166550. doi: 10.1371/journal.pone.0166550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014;6:9–19. doi: 10.1016/j.nicl.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nolden M, Zelzer S, Seitel A, Wald D, Muller M, Franz AM, et al. The medical imaging interaction toolkit: challenges and advances: 10 years of open-source development. Int J Comput Assist Radiol Surg. 2013;8:607–620. doi: 10.1007/s11548-013-0840-8. [DOI] [PubMed] [Google Scholar]
- 5.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain R, Poisson LM, Gutman D, Scarpace L, Hwang SN, Holder CA, et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology. 2014;272:484–493. doi: 10.1148/radiol.14131691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee G, Lee HY, Park H, Schiebler ML, van Beek EJR, Ohno Y, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: state of the art. Eur J Radiol. 2017;86:297–307. doi: 10.1016/j.ejrad.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Park JE, Kim HS, Park KJ, Choi CG, Kim SJ. Histogram analysis of amide proton transfer imaging to identify contrast-enhancing low-grade brain tumor that mimics high-grade tumor: increased accuracy of MR perfusion. Radiology. 2015;277:151–161. doi: 10.1148/radiol.2015142347. [DOI] [PubMed] [Google Scholar]
- 12.Baek HJ, Kim HS, Kim N, Choi YJ, Kim YJ. Percent change of perfusion skewness and kurtosis: a potential imaging biomarker for early treatment response in patients with newly diagnosed glioblastomas. Radiology. 2012;264:834–843. doi: 10.1148/radiol.12112120. [DOI] [PubMed] [Google Scholar]
- 13.Wang JZ. Wavelets and imaging informatics: a review of the literature. J Biomed Inform. 2001;34:129–141. doi: 10.1006/jbin.2001.1010. [DOI] [PubMed] [Google Scholar]
- 14.Alobaidli S, McQuaid S, South C, Prakash V, Evans P, Nisbet A. The role of texture analysis in imaging as an outcome predictor and potential tool in radiotherapy treatment planning. Br J Radiol. 2014;87:20140369. doi: 10.1259/bjr.20140369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin YC, Lin G, Hong JH, Lin YP, Chen FH, Ng SH, et al. Diffusion radiomics analysis of intratumoral heterogeneity in a murine prostate cancer model following radiotherapy: pixelwise correlation with histology. J Magn Reson Imaging. 2017;46(2):483–489. doi: 10.1002/jmri.25583. [DOI] [PubMed] [Google Scholar]
- 16.Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. 2016;6:71. doi: 10.3389/fonc.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography. 2013;36:27–46. doi: 10.1111/j.1600-0587.2012.07348.x. [DOI] [Google Scholar]
- 18.Friedman JH. On bias, variance, 0/1 - loss, and the curse-of-dimensionality. Data Min Knowl Disc. 1997;1:55–77. doi: 10.1023/A:1009778005914. [DOI] [Google Scholar]
- 19.Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016. 10.1038/npjbcancer.2016.12. [DOI] [PMC free article] [PubMed]
- 20.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinnott JA, Cai T. Inference for survival prediction under the regularized Cox model. Biostatistics. 2016;17:692–707. doi: 10.1093/biostatistics/kxw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du P, Ma SG, Liang H. Penalized variable selection procedure for Cox models with semiparametric relative risk. Ann Stat. 2010;38:2092–2117. doi: 10.1214/09-AOS780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hothorn T, Buhlmann P. Model-based boosting in high dimensions. Bioinformatics. 2006;22:2828–2829. doi: 10.1093/bioinformatics/btl462. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Luan Y. Boosting proportional hazards models using smoothing splines, with applications to high-dimensional microarray data. Bioinformatics. 2005;21:2403–2409. doi: 10.1093/bioinformatics/bti324. [DOI] [PubMed] [Google Scholar]
- 25.Mogensen UB, Ishwaran H, Gerds TA. Evaluating random forests for survival analysis using prediction error curves. J Stat Softw. 2012;50:1–23. doi: 10.18637/jss.v050.i11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishwaran H, Gerds TA, Kogalur UB, Moore RD, Gange SJ, Lau BM. Random survival forests for competing risks. Biostatistics. 2014;15:757–773. doi: 10.1093/biostatistics/kxu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishwaran H, Kogalur UB, Chen X, Minn AJ. Random survival forests for high-dimensional data. Stat Anal Data Mining. 2011;4:115–132. doi: 10.1002/sam.10103. [DOI] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 29.Kuo MD, Jamshidi N. Behind the numbers: decoding molecular phenotypes with radiogenomics—guiding principles and technical considerations. Radiology. 2014;270:320–325. doi: 10.1148/radiol.13132195. [DOI] [PubMed] [Google Scholar]
- 30.Narang S, Lehrer M, Yang D, Lee J, Rao A. Radiomics in glioblastoma: current status, challenges and potential opportunities. Transl Cancer Res. 2016;5:383–397. doi: 10.21037/tcr.2016.06.31. [DOI] [Google Scholar]
- 31.Zacharaki EI, Wang S, Chawla S, Yoo DS, Wolf R, Melhem ER, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med. 2009;62:1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J-b Q, Liu Z, Zhang H, Shen C, Wang X-C, Tan Y, et al. Grading of gliomas by using radiomic features on multiple magnetic resonance imaging (MRI) sequences. Med Sci Monit. 2017;23:2168–2178. doi: 10.12659/MSM.901270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Shi Z, Lian Y, Li Z, Liu T, Gao Y, et al. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol. 2017;27:3509–3522. doi: 10.1007/s00330-016-4653-3. [DOI] [PubMed] [Google Scholar]
- 34.Lopez CJ, Nagornaya N, Parra NA, Kwon D, Ishkanian F, Markoe AM, et al. Association of radiomics and metabolic tumor volumes in radiation treatment of glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2017;97:586–595. doi: 10.1016/j.ijrobp.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiestler B, Kluge A, Lukas M, Gempt J, Ringel F, Schlegel J, et al. Multiparametric MRI-based differentiation of WHO grade II/III glioma and WHO grade IV glioblastoma. Sci Rep. 2016;6:35142. doi: 10.1038/srep35142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H, Vallieres M, Bai HX, Su C, Tang H, Oldridge D, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19(6):862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Narang S, Martinez JJ, Rao G, Rao A. Associating spatial diversity features of radiologically defined tumor habitats with epidermal growth factor receptor driver status and 12-month survival in glioblastoma: methods and preliminary investigation. J Med Imaging (Bellingham) 2015;2:041006. doi: 10.1117/1.JMI.2.4.041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macyszyn L, Akbari H, Pisapia JM, Da X, Attiah M, Pigrish V, et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-Oncology. 2015;18:417–425. doi: 10.1093/neuonc/nov127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kickingereder P, Burth S, Wick A, Gotz M, Eidel O, Schlemmer HP, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280:880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 40.Ingrisch M, Schneider MJ, Norenberg D, Negrao de Figueiredo G, Maier-Hein K, Suchorska B, et al. Radiomic analysis reveals prognostic information in T1-weighted baseline magnetic resonance imaging in patients with glioblastoma. Investig Radiol. 2017;52:360–366. doi: 10.1097/RLI.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 41.Rao A, Rao G, Gutman DA, Flanders AE, Hwang SN, Rubin DL, et al. A combinatorial radiographic phenotype may stratify patient survival and be associated with invasion and proliferation characteristics in glioblastoma. J Neurosurg. 2016;124:1008–1017. doi: 10.3171/2015.4.JNS142732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prasanna P, Patel J, Partovi S, Madabhushi A. Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. 2017;27(10):4188–4197. doi: 10.1007/s00330-016-4637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGarry SD, Hurrell SL, Kaczmarowski AL, Cochran EJ, Connelly J, Rand SD, et al. Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography. 2016;2:223–228. doi: 10.18383/j.tom.2016.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kickingereder P, Gotz M, Muschelli J, Wick A, Neuberger U, Shinohara RT, et al. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res. 2016;22:5765–5771. doi: 10.1158/1078-0432.CCR-16-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohmann P, Lerche C, Stoffels G, Filss CP, Stegmayr C, Neumaier B, et al. P09.26 FET PET radiomics - diagnosis of pseudoprogression in glioblastoma patients based on textural features. Neuro-Oncology. 2017;19:iii75–iiiii. doi: 10.1093/neuonc/nox036.282. [DOI] [Google Scholar]
- 46.Kickingereder P, Bonekamp D, Nowosielski M, Kratz A, Sill M, Burth S, et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology. 2016;281:907–918. doi: 10.1148/radiol.2016161382. [DOI] [PubMed] [Google Scholar]
- 47.Hu LS, Ning S, Eschbacher JM, Baxter LC, Gaw N, Ranjbar S, et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro-Oncology. 2017;19:128–137. doi: 10.1093/neuonc/now135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutman DA, Dunn WD, Jr, Grossmann P, Cooper LA, Holder CA, Ligon KL, et al. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology. 2015;57:1227–1237. doi: 10.1007/s00234-015-1576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamshidi N, Diehn M, Bredel M, Kuo MD. Illuminating radiogenomic characteristics of glioblastoma multiforme through integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology. 2014;270:1–2. doi: 10.1148/radiol.13130078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267:560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abrol S, Kotrotsou A, Hassan A, Elshafeey N, Hassan I, Idris T, et al. Radiomic analysis of pseudo-progression compared to true progression in glioblastoma patients: a large-scale multi-institutional study. J Clin Oncol. 2017;35:2015. 10.1200/JCO.2017.35.15_suppl.2015
- 52.O'Connor JPB, Aboagye EO, Adams JE, Aerts HJWL, Barrington SF, Beer AJ, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2016;advance online publication. [DOI] [PMC free article] [PubMed]