Abstract
Climate change profoundly influences species distributions. These effects are evident in poleward latitudinal range shifts for many taxa, and upward altitudinal range shifts for alpine species, that resulted from increased annual global temperatures since the Last Glacial Maximum (LGM, ca. 22,000 BP). For the latter, the ultimate consequence of upward shifts may be extinction as species in the highest alpine ecosystems can migrate no further, a phenomenon often characterized as “nowhere to go”. To predict responses to climate change of the alpine plants on the Qinghai-Tibetan Plateau (QTP), we used ecological niche modelling (ENM) to estimate the range shifts of 14 Rhodiola species, beginning with the Last Interglacial (ca. 120,000–140,000 BP) through to 2050. Distributions of Rhodiola species appear to be shaped by temperature-related variables. The southeastern QTP, and especially the Hengduan Mountains, were the origin and center of distribution for Rhodiola, and also served as refugia during the LGM. Under future climate scenario in 2050, Rhodiola species might have to migrate upward and northward, but many species would expand their ranges contra the prediction of the “nowhere to go” hypothesis, caused by the appearance of additional potential habitat concomitant with the reduction of permafrost with climate warming.
Introduction
Climate change has profoundly impacted the distributions of species across the globe1. Rising temperatures of the last few decades have shifted the latitudinal and altitudinal ranges of many species2. This is particularly concerning for alpine species, for which there may not be sufficient suitable alpine habitats at higher altitudes to facilitate their migration. This phenomenon has been termed the “nowhere to go” hypothesis3,4. Chen et al.5 conducted a meta-analysis of over 1,000 species and found that the median rate of increased altitudinal range shifting was 11 meters per decade, while latitudinal range shifts showed a median rate of 16.9 kilometers poleward per decade (animals only). Lenoir et al.6 investigated 171 plant species in European montane regions, and found that mean elevation of ranges had moved upward by 29 meters per decade, coincident with rapidly rising temperatures occurring after 1986. These altitudinal shifts were faster for species adapted to montane regions than for more broadly-distributed species and for herbaceous plants compared to woody plants6,7. These studies collectively suggest that, under future climate warming scenarios, montane species, especially herbaceous plants, may face the aforementioned “nowhere to go” predicament. However, other researchers predict that global climate warming may drive snowlines upward, exposing new areas of suitable habitat that could facilitate alpine species’ upward migration; thus the notion of “nowhere to go” may be an oversimplification of the future of alpine taxa8. For example, Holzinger et al.9 reported that species richness of vascular plants in montane communities increased by 11% per decade under climatic warming. Clearly more research is required to fully explore the effects of global warming on alpine plants.
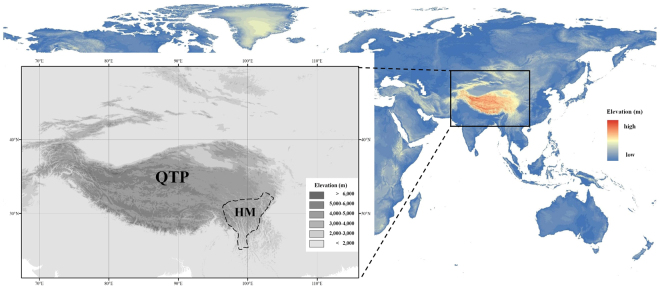
The Qinghai-Tibetan Plateau (QTP) is the highest alpine ecosystem in the world, and perhaps one of the most sensitive to climate change. The temperature increase in the QTP has been faster than the mean temperature increases in the Northern Hemisphere as well as in other regions at the same latitude10,11. The QTP and environs (like Hengduan Mountains (HM), Fig. 1) are also a key global biodiversity hotspot containing one of the richest alpine flora in the world12. Recent phylogeography studies of QTP plants reveal that Quaternary glaciations, especially the Last Glacial Maximum (LGM, ca. 22,000 years before present (BP)), have profoundly influenced the distributions of these species13,14. These studies rarely discuss how QTP plants will respond to future climate change, a major focus of our paper.
Figure 1.
Location and elevation of Rhodiola distributions within the Qinghai-Tibetan Plateau (QTP) and the Hengduan Mountains (HM). Elevation data were downloaded from WorldClim Dataset (www.worldclim.org/bioclim). The map was processed by ArcGIS ver 10.2 (ESRI, Redlands, California, USA) (http://www.esri.com/).
Rhodiola is a perennial herb genus comprising nearly 70 species, which are primarily distributed in the QTP and neighboring mountains15. Molecular phylogenetic analyses suggest that Rhodiola originated in the QTP about 21.0 Mya and rapidly diversified beginning 12.1 Mya coincident with the uplift of the QTP15. The genus subsequently expanded into adjacent regions, with a handful of species (e.g. R. rosea) dispersing to other parts of the globe16. The evolutionary history and contemporary geographic ranges suggest that Rhodiola species are adapted to low temperatures, making them an ideal model for investigating the response of montane herbaceous species to climate change. To date, the genealogical patterns of three Rhodiola species (R. alsia, R. dumulosa and R. kirilowii) of the QTP have been investigated17–19 and these studies suggested that contemporary Rhodiola distributions have been highly influenced by the LGM temperature where they expanded from refugia in the southeastern QTP and HM17–19. A limitation of these studies is that they do not utilize ecological niche modelling (ENM) with their molecular data, and thus exhibit only a cursory understanding of how changing environments might have shaped contemporary distributions and phylogeographic patterns.
ENM is based on the concept of the ecological niche, which associates a set of environmental variables to the ability of a species to persist in an environment, and is related to the fitness of a species20. These environmental variables (e.g. climate, soil moisture and nutrients, etc.) constrain the distribution and abundance of a species21,22. Using occurrence data of a particular species, ENM can be used to project its ecological niche back into palaeoclimatic scenarios, and also into future climate scenarios, predicting potential distributional patterns under the assumptions of niche conservatism23–25.
In this study, framing our research questions using a molecular phylogeny of Rhodiola15, we focus on 14 species with sufficient occurrence records for ENM analysis (Table 1). These species belong to different Rhodiola clades that vary in their distributions and breeding systems (Table 1)15, allowing us to test for the effects of evolutionary history, ecology niche differentiation, and reproductive strategies on plant responses to climate change. The specific aims of our study are to: 1) test which climatic factors underlie the distributions and range shifts of Rhodiola species in different periods (Last inter-glacial (LIG) ca. 120,000–140,000 years BP, LGM ca. 22,000 years BP, current, and future (2050), and 2) test the predictions of the “nowhere to go” hypothesis under future climate change scenarios.
Table 1.
Ecological information for the 14 Rhodiola species14 and occurrence data used for ENM.
| Species | Habitat | Distribution | Elevation (m) | Breeding system | Clade in genus-level phylogeny | Records |
|---|---|---|---|---|---|---|
| R. alsia | Rhododendron forests, rocky slopes | Eastern QTP and Hengduan Mountains (GQTP) | 2450–5300 | Dioecism | Clade2 | 71 |
| R. bupleuroides | Thickets, grassy places, rock crevices on slopes | Eastern and southern QTP and Hengduan Mountains (GQTP) | 2300–5920 | Dioecism | Clade2 | 200 |
| R. chryanthemifolia | Grasslands, rocks, rock crevices | Eastern and southern QTP and Hengduan Mountains (GQTP) | 2500–5140 | Monoecism | Clade1 | 71 |
| R. crenulata | Thickets, grassland slopes, schist on mountain slopes, rocky places, rock crevices | Eastern and southern QTP and Hengduan Mountains (GQTP) | 2700–5850 | Dioecism | Clade2 | 96 |
| R. dumulosa | Rocky slopes | From eastern and southern QTP to northeastern China (Gwide) | 1570–5700 | Monoecism | Clade2 | 93 |
| R. fastigiata | Rocky slopes | Southern and eastern QTP, Hengduan Mountains (GQTP) | 2460–5600 | Dioecism | Clade2 | 161 |
| R. forrestii | Slopes | Western Sichuan, northwestern Yunnan (GHM) | 1600–4800 | Dioecism | Clade2 | 41 |
| R. henryi | Rocky slopes | Xichuan, Hubei, Gansu, Shanxi, Henan (Gplain) | 500–4200 | Dioecism | Clade2 | 125 |
| R. himalensis | Slopes, forests, scrub | Eastern and southern QTP and Hengduan Mountains (GQTP) | 2600–5300 | Dioecism | Clade2 | 92 |
| R. kirilowii | Forest margins, grassy slopes, often in partial shade | Eastern and southern QTP, northeastern China and Xinjiang (Gwide) | 1500–5300 | Dioecism | Clade2 | 174 |
| R. quadrifida | Alpine regions, stony slopes, rocks | QTP and the neighboring mountains (GQTP) | 2500–5200 | Dioecism | Clade2 | 78 |
| R. sacra | Grassland slopes, rock crevices on slopes | Southern QTP and Hengduan Mountains (GQTP) | 2700–5330 | Monoecism | Clade1 | 69 |
| R. wallichiana | Forests, rocky slopes | Southern and eastern QTP, Hengduan Mountains (Gwide) | 2500–5100 | Monoecism | Clade1 | 39 |
| R. yunnanensis | Forests on slopes | Hengduan Mountains and the neighboring plateau and plain (GHM) | 1400–4600 | Dioecism | Clade2 | 134 |
Results
Model performance
We used three modelling approaches to predict the distributions of Rhodiola species: (1) MEAN ENSEMBLE: using each Global Climate Model (GCM, one GCM for LIG and current, 4 GCMs for LGM, and 17GCMs for 2050, respectively) to do ensemble ENM, and then calculated the mean and standard deviation of all predictions; (2) MMM ENSEMBLE: using the multi-model mean (MMM) of all the GCMs to do ensemble ENM; (3) The maximum entropy algorithm, Maxent: using the MMM of all the GCMs to do Maxent modelling. The Area Under the Receiver Operator Curve (AUC) of ensemble models (MEAN ENSEMBLE and MMM ENSEMBLE) ranged from 0.968 to 0.995 for individual species with our designated groups (clade, distribution and breeding system) (Supplementary Table S1) showing high performance. Maxent also showed good performance in modelling the distributions of Rhodiola species: the AUC values ranged from 0.925 to 0.993 and True Skill Statistic (TSS) values from 0.902 to 0.993, both statistics being significantly higher than would be expected at random (all p < 0.0001, Wilcoxon rank sum test) (Supplementary Table S2). Similarly, all training omission (OR) values in Maxent were significantly lower than random predictions (all p < 0.0001, Wilcoxon rank sum test). Comparisons of model AUC values for the genus as a whole (WHOLE, 14 Rhodiola species), for each species group, and for constituent species of each species group revealed no significant differences (for all, p > 0.05, Wilcoxon rank sum test), indicating that all three modelling approaches performed well irrespective of the grouping.
Climate variables
The most important bioclimatic variables associated with the distribution of Rhodiola species were temperature-related (Isothermality, Mean Temperature of Wettest Quarter, and Mean Temperature of Driest Quarter) (Supplementary Table S3). Isothermality (BIO3) was the most important factor for predicting the distribution of R. bupleuroides, R. crenulata, R. fastigiata, R. sacra, Gclade1 (Rhodiola species found primarily in the first major clade in the genus-level phylogeny tree), and GQTP (species mainly confined to the QTP); whereas Mean Temperature of Wettest Quarter (BIO8) was the most important variable for the distributions of R. alsia, R. dumulosa, R. himalensi, R. kirilowii, R. quadrifida and Gwide (species with relatively wide distributions). Mean Temperature of Driest Quarter (BIO9) was the top performing variable for predicting the distributions of R. chryanthemifolia, R. forrestii, R. henryi, R. wallichiana, R. yunanensis, WHOLE, Gclade2 (species from the second major clade in the Rhodiola phylogeny), GHM (species mainly distributed within the HM), Gplain (comprising species not found in the QTP and HM), Gmon (comprising monoecious species), and Gdio (comprising dioecious species).
The principal components analysis (PCA) of temperature variables generated two temperature axis (Temp1 and Temp2) that explained 55.6% and 32.1% (87.7% cumulatively) of the total variation in the temperature dataset. The PCA of precipitation variables produced two precipitation axis (Prec1 and Prec2) that explained 62.3% and 25.6% (87.8% cumulatively) of total variation in the precipitation dataset. BIO8 (Mean temperature of wettest quarter) and BIO9 (Mean temperature of the driest quarter) both had the largest loadings (contributions to the summed variance) for Temp 1 and were both positive. Mean Diurnal Range (BIO2) and Isothermality (BIO3) showed the highest loadings for Temp2 (Supplementary Table S4). Three precipitation variables, Precipitation of Driest Month (BIO14), Precipitation of Coldest Quarter (BIO19), and Precipitation Seasonality (BIO15) loaded highly on Prec1, with the former two loading positively and the last negatively, whereas both Precipitation of Wettest Month (BIO13) and Precipitation of Warmest Quarter (BIO18) had positive loadings on Prec2 (Supplementary Table S5).
The canonical discriminant analysis (CDA) using Temp1, Temp2, Prec1 and Prec2 produced three discriminant functions, the first explained 76.9% among-group climatic variance, the second 20.1% (Fig. 2). The third explained the remaining 3% of the total variance, suggesting that it could be neglected in subsequent analysis. Of the four variables, Temp2 contributed the most to the first discriminant function, while Temp1 contributed most to the second (Supplementary Table S6).
Figure 2.
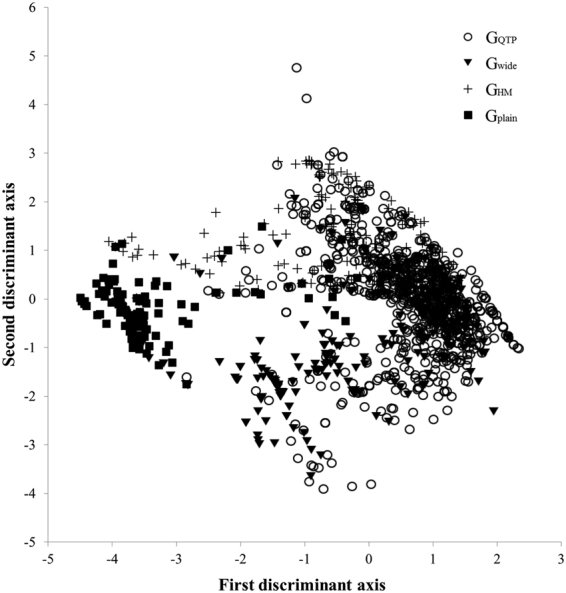
Canonical discriminant analysis (CDA) ordination of the four Rhodiola distribution groups: GQTP (○); Gwide (▽); GHM (+); Gplain (□).
The Wilks’ Lambda value for each CDA axis was significant at α = 0.05, suggesting that at least some of the 14 focal species occupied geographic ranges that were climatically distinguishable. The first discriminant function best separated Gplain and GQTP from other groups (Supplementary Fig. S1). Specifically, Gplain had the lowest first discriminant function corresponding to lower Mean Diurnal Range (BIO2) and Isothermality (BIO3), whereas GQTP had the highest first discriminant function relating to highest Mean Diurnal Range (BIO2) and Isothermality (BIO3). For Gplain and GHM the second discriminant function distinctly separated these groups from others. GHM individuals tended to score highest on the second discriminant axis, while Gplain individuals tended to have intermediate scores, where these two groups had higher values for Mean Temperature of Wettest Quarter (BIO8) and Mean Temperature of Driest Quarter (BIO9).
Range shifts
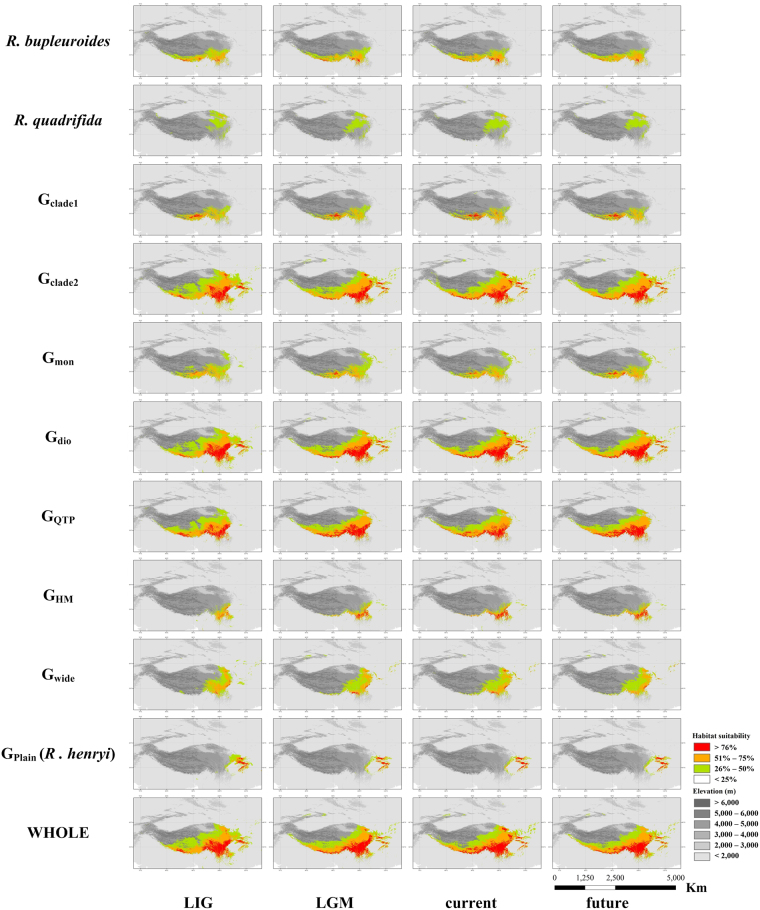
The predictions generated using MEAN ENSEMBLE, MMM ENSEMBLE, and Maxent all showed that potential distribution of Rhodiola varies among species and groups under four climate scenarios: LIG, LGM, current and future (2050 RCP8.5) (Table 2; Fig. 3; Supplementary Figs S2–S4). MEAN ENSEMBLE and MMM ENSEMBLE predicted similar patterns of range shifting (Supplementary Tables S7, S8), which both appear to give more conservative predictions than Maxent. For example, Maxent, on average, predicted relatively larger range shifts and showed some differences in tendency for range shifting to occur (e.g. from current to 2050 for R. henryi; see Table 2).
Table 2.
Predicted suitable area (km2) of Rhodiola species and species groups for each time period using MEAN ENSEMBLE, MMM ENSEMBLE and Maxent.
| MEAN ENSEMBLE | MMM ENSEMBLE | Maxent | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LIG | LGM | current | 2050 | LIG | LGM | current | 2050 | LIG | LGM | current | future | |
| R. alsia | 25105 | 29754 | 29189 | 29261 | 25105 | 26271 | 29189 | 30754 | 256304 | 995152 | 1127870 | 1126001 |
| R. bupleuroides | 35105 | 34687 | 34381 | 34554 | 35105 | 34239 | 34381 | 34873 | 416941 | 537477 | 737503 | 1151287 |
| R. chryanthemifolia | 18938 | 16815 | 18313 | 17072 | 18938 | 15054 | 18313 | 18076 | 80522 | 304446 | 445371 | 875260 |
| R. crenulata | 27447 | 24662 | 20730 | 25781 | 27447 | 15037 | 20730 | 27666 | 58188 | 1060539 | 906215 | 984226 |
| R. dumulosa | 12272 | 9319 | 9594 | 8183 | 12272 | 7012 | 9594 | 10080 | 164429 | 392825 | 323833 | 606667 |
| R. fastigiata | 28974 | 28098 | 27107 | 28843 | 28974 | 27224 | 27107 | 33222 | 86127 | 561174 | 745397 | 1031277 |
| R. forrestii | 6068 | 7336 | 8082 | 8113 | 6068 | 7891 | 8082 | 8556 | 50141 | 488778 | 377406 | 647803 |
| R. henryi | 16634 | 11019 | 10386 | 11119 | 16634 | 10924 | 10386 | 12826 | 173804 | 344487 | 262348 | 34270 |
| R. himalensis | 28047 | 30835 | 31790 | 30171 | 28047 | 26956 | 31790 | 33109 | 798050 | 748932 | 1067350 | 1270866 |
| R. kirilowii | 11038 | 9301 | 9308 | 8666 | 11038 | 7885 | 9308 | 9241 | 803288 | 1015632 | 1212586 | 1352739 |
| R. quadrifida | 19668 | 19659 | 23196 | 20056 | 19668 | 11754 | 23196 | 22277 | 1920453 | 2749254 | 2377665 | 1800716 |
| R. sacra | 9116 | 7972 | 8152 | 8260 | 9116 | 7548 | 8152 | 9260 | 25017 | 100974 | 257310 | 580758 |
| R. wallichiana | 5719 | 5744 | 7169 | 6680 | 5719 | 5660 | 7169 | 7700 | 765199 | 1249080 | 1305554 | 1642499 |
| R. yunnanensis | 17391 | 19182 | 18540 | 18587 | 17391 | 17754 | 18540 | 18777 | 257468 | 446269 | 407214 | 419536 |
| Gclade1 | 31596 | 28961 | 27869 | 27446 | 31596 | 28556 | 27869 | 28701 | 351642 | 308435 | 554068 | 1099216 |
| Gclade2 | 115702 | 101676 | 100741 | 103019 | 115702 | 101137 | 100741 | 115453 | 1234504 | 1442743 | 1871221 | 2032588 |
| Gmon | 43140 | 42180 | 36810 | 38612 | 43140 | 39934 | 36810 | 40825 | 1704898 | 685087 | 1079352 | 1813421 |
| Gdio | 110781 | 96342 | 95643 | 96534 | 110781 | 96477 | 95643 | 113463 | 1046104 | 1409270 | 1758647 | 1922653 |
| GQTP | 80186 | 81275 | 79792 | 82687 | 80186 | 80406 | 79792 | 83951 | 834374 | 970820 | 1434682 | 1731064 |
| GHM | 15606 | 18180 | 18577 | 18794 | 15606 | 19096 | 18577 | 20879 | 123193 | 460259 | 395055 | 381566 |
| Gwide | 42492 | 42441 | 42872 | 42687 | 42492 | 42164 | 42872 | 44300 | 209539 | 466811 | 400191 | 420343 |
| Gplain | 16634 | 11019 | 10386 | 11119 | 16634 | 10924 | 10386 | 12826 | 173804 | 344487 | 262348 | 34270 |
| WHOLE | 116609 | 105768 | 107735 | 104059 | 116609 | 98201 | 107735 | 102625 | 1190909 | 1372133 | 1801155 | 1979687 |
Figure 3.
MEAN ENSEMBLE predicted maps for R. bupleuroides, R. quadrifida, and species group for the Last inter-glacial (LIG ca. 120,000–140,000 years BP), the Last glacial maximum (LGM ca. 21,000 years BP), current and future (2050). ENM predicted results were processed by ArcGIS ver 10.2 (ESRI, Redlands, California, USA) (http://www.esri.com/), and then integrated using Microsoft Office Visio 2013 (http://office.microsoft.com/visio/).
Between the LIG and LGM, the predictions of MEAN ENSEMBLE and MMM ENSEMBLE indicated no significant range shifts for any species or species group (the values of ΔS1 were near to zero), whereas the results from Maxent showed that most species would have expanded their respective ranges (Table 2; Supplementary Table S9), migrating to lower elevations (Table 3; Supplementary Table S10) and longitudes, but showing increases in the mean latitudes of their distributions (Table 4; Supplementary Tables S11 and S12). Between the LGM and current time points, the combined results of the three modelling approaches showed that most focal Rhodiola species expanded their ranges; an exception, R. henryi, was predicted to have undergone range contraction over this same period (Fig. 3; Supplementary Table S9; Supplementary Figs S2–S4).
Table 3.
Predicted mean elevation (m) of Rhodiola species and species groups for each time period using MEAN ENSEMBLE, MMM ENSEMBLE and Maxent.
| MEAN ENSEMBLE | MMM ENSEMBLE | Maxent | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LIG | LGM | current | 2050 | LIG | LGM | current | 2050 | LIG | LGM | current | 2050 | |
| R. alsia | 4174 | 4083 | 4107 | 4110 | 4174 | 4147 | 4107 | 4160 | 3698 | 3726 | 4243 | 4492 |
| R. bupleuroides | 4155 | 4158 | 4160 | 4203 | 4155 | 4136 | 4160 | 4181 | 2893 | 3222 | 4158 | 4500 |
| R. chryanthemifolia | 3941 | 3970 | 3971 | 3960 | 3941 | 3923 | 3971 | 3950 | 3150 | 2951 | 3872 | 4445 |
| R. crenulata | 4449 | 4536 | 4570 | 4582 | 4449 | 4617 | 4570 | 4564 | 4136 | 4034 | 4467 | 4638 |
| R. dumulosa | 3367 | 3267 | 3220 | 3080 | 3367 | 3206 | 3220 | 3194 | 3318 | 3125 | 3904 | 4546 |
| R. fastigiata | 4086 | 4053 | 4069 | 4093 | 4086 | 4063 | 4069 | 4140 | 3603 | 3540 | 4175 | 4570 |
| R. forrestii | 3155 | 3176 | 3176 | 3199 | 3155 | 3169 | 3176 | 3296 | 2638 | 2438 | 3310 | 4222 |
| R. henryi | 1494 | 1727 | 1737 | 1712 | 1494 | 1703 | 1737 | 1614 | 1049 | 1555 | 1902 | 3557 |
| R. himalensis | 4274 | 4193 | 4208 | 4209 | 4274 | 4205 | 4208 | 4229 | 4131 | 3885 | 4382 | 4611 |
| R. kirilowii | 3311 | 3201 | 3188 | 3146 | 3311 | 3100 | 3188 | 3111 | 4192 | 2714 | 3818 | 4311 |
| R. quadrifida | 4106 | 4200 | 4201 | 4119 | 4106 | 4181 | 4201 | 4164 | 4002 | 4166 | 4555 | 4757 |
| R. sacra | 4403 | 4310 | 4310 | 4320 | 4403 | 4309 | 4310 | 4319 | 3090 | 3734 | 4142 | 4561 |
| R. wallichiana | 4092 | 4119 | 4089 | 4088 | 4092 | 4133 | 4089 | 4051 | 2685 | 2962 | 4135 | 4357 |
| R. yunnanensis | 3109 | 3097 | 3099 | 3146 | 3109 | 3131 | 3099 | 3107 | 2571 | 2324 | 3076 | 3994 |
| Gclade1 | 4110 | 4104 | 4063 | 4096 | 4110 | 4078 | 4063 | 4129 | 2766 | 3159 | 4063 | 4469 |
| Gclade2 | 3644 | 3760 | 3764 | 3766 | 3644 | 3768 | 3764 | 3798 | 3661 | 2819 | 3855 | 4364 |
| Gmon | 3991 | 3882 | 3926 | 3882 | 3991 | 3905 | 3926 | 3867 | 3955 | 2733 | 3927 | 4478 |
| Gdio | 3692 | 3795 | 3788 | 3790 | 3692 | 3779 | 3788 | 3528 | 3460 | 2796 | 3883 | 4376 |
| GQTP | 4249 | 4282 | 4237 | 4290 | 4249 | 4268 | 4237 | 4259 | 4101 | 3645 | 4291 | 4527 |
| GHM | 3010 | 3063 | 3061 | 3107 | 3010 | 3054 | 3061 | 3129 | 2622 | 2357 | 3106 | 4016 |
| Gwide | 3702 | 3762 | 3765 | 3749 | 3702 | 3779 | 3765 | 3785 | 4359 | 2702 | 3836 | 4343 |
| Gplain | 1494 | 1727 | 1737 | 1712 | 1494 | 1703 | 1737 | 1614 | 1049 | 1555 | 1902 | 3557 |
| WHOLE | 3723 | 3794 | 3780 | 3797 | 3723 | 3757 | 3780 | 3685 | 3698 | 2833 | 3885 | 4366 |
Table 4.
Mean longitude/latitude (degree) of Rhodiola species and species groups for each time period using MEAN ENSEMBLE, MMM ENSEMBLE and Maxent.
| MEAN ENSEMBLE | MMM ENSEMBLE | Maxent | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LIG | LGM | current | 2050 | LIG | LGM | current | 2050 | LIG | LGM | current | 2050 | |
| R. alsia | 98.5/30.5 | 98.7/31.1 | 98.7/31.1 | 98.6/30.9 | 98.5/30.5 | 98.7/31.0 | 98.7/31.1 | 98.3/31.1 | 99.3/28.9 | 96.1/31.1 | 95.7/31.2 | 95.0/31.7 |
| R. bupleuroides | 93.8/28.9 | 93.7/29.0 | 93.2/28.9 | 93.4/28.8 | 93.8/28.9 | 93.7/29.1 | 93.2/28.9 | 93.2/28.8 | 95.7/31.9 | 95.3/28.6 | 93.5/29.6 | 92.9/30.6 |
| R. chryanthemifolia | 96.0/28.6 | 95.6/28.6 | 95.7/28.5 | 95.9/28.5 | 96.0/28.6 | 95.5/28.5 | 95.7/28.5 | 94.8/28.5 | 100.3/28.1 | 97.6/27.9 | 95.5/29.1 | 93.7/30.2 |
| R. crenulata | 94.5/29.5 | 93.5/29.4 | 93.4/29.2 | 93.4/29.3 | 94.5/29.5 | 92.0/29.1 | 93.4/29.2 | 93.5/29.5 | 99.4/28.0 | 93.9/29.7 | 93.2/30.0 | 93.1/30.5 |
| R. dumulosa | 101.2/33.7 | 101.2/34.6 | 101.3/34.5 | 102.1/35.0 | 101.2/33.7 | 101.7/34.4 | 101.3/34.5 | 101.4/34.5 | 98.9/31.4 | 100.4/31.9 | 96.6/32.9 | 92.6/33.1 |
| R. fastigiata | 96.7/29.2 | 97.1/29.4 | 96.5/29.2 | 96.4/29.2 | 96.7/29.2 | 96.2/29.3 | 96.5/29.2 | 95.2/29.2 | 100.3/27.9 | 98.1/29.2 | 95.5/29.9 | 93.6/30.7 |
| R. forrestii | 100.5/27.4 | 99.8/27.5 | 99.2/27.4 | 99.5/27.5 | 100.5/27.4 | 99.1/27.5 | 99.2/27.4 | 99.1/27.7 | 97.9/27.3 | 98.0/27.2 | 97.2/28.5 | 95.7/29.9 |
| R. henryi | 106.6/31.6 | 106.9/31.3 | 106.7/31.3 | 106.9/31.3 | 106.6/31.6 | 106.7/31.9 | 106.7/31.3 | 106.7/31.8 | 108.6/28.5 | 105.7/29.9 | 105.3/31.9 | 84.1/35.8 |
| R. himalensis | 96.0/30.4 | 97.2/30.7 | 96.8/30.6 | 96.9/30.5 | 96.0/30.4 | 97.2/30.5 | 96.8/30.6 | 96.6/30.5 | 95.9/29.8 | 97.1/30.3 | 94.8/30.8 | 93.5/31.4 |
| R. kirilowii | 100.9/34.1 | 101.9/34.4 | 101.6/34.6 | 101.9/34.6 | 100.9/34.1 | 102.5/34.6 | 101.6/34.6 | 101.7/34.7 | 95.5/30.4 | 102.3/32.7 | 97.2/32.4 | 94.1/33.2 |
| R. quadrifida | 98.5/34.4 | 98.6/33.6 | 98.3/33.7 | 98.6/34.3 | 98.5/34.4 | 99.2/33.6 | 98.3/33.7 | 98.2/34.1 | 93.0/33.3 | 90.3/34.4 | 90.5/33.4 | 90.3/33.4 |
| R. sacra | 91.3/28.9 | 91.6/29.0 | 91.7/28.9 | 91.5/28.9 | 91.3/28.9 | 91.4/28.9 | 91.7/28.9 | 92.0/28.8 | 101.6/29.7 | 94.4/29.3 | 93.3/29.1 | 92.4/29.8 |
| R. wallichiana | 90.1/28.5 | 91.4/28.4 | 93.2/28.3 | 93.2/28.3 | 90.1/28.5 | 90.3/28.5 | 93.2/28.3 | 93.9/28.4 | 95.1/32.5 | 90.0/32.0 | 91.3/30.9 | 91.1/31.9 |
| R. yunnanensis | 99.9/28.1 | 99.5/28.3 | 99.5/28.1 | 99.3/28.1 | 99.9/28.1 | 99.2/28.2 | 99.5/28.1 | 98.9/28.0 | 99.3/27.2 | 100.9/28.0 | 99.5/28.7 | 97.0/29.7 |
| Gclade1 | 95.0/29.1 | 95.0/29.1 | 94.3/28.9 | 94.6/28.8 | 95.0/29.1 | 94.3/29.0 | 94.3/28.9 | 94.4/28.8 | 95.8/32.6 | 97.0/28.4 | 94.3/29.5 | 93.0/30.8 |
| Gclade2 | 96.6/31.2 | 95.9/31.0 | 96.0/31.1 | 96.0/31.1 | 96.6/31.2 | 95.8/31.0 | 96.0/31.1 | 95.5/31.2 | 96.8/29.6 | 100.6/30.8 | 95.9/31.6 | 93.1/32.5 |
| Gmon | 95.7/29.9 | 96.7/30.2 | 95.9/29.9 | 96.0/29.9 | 95.7/29.9 | 95.8/30.1 | 95.9/29.9 | 95.8/30.0 | 95.5/31.4 | 101.8/31.6 | 96.4/31.1 | 93.1/32.3 |
| Gdio | 96.4/31.0 | 95.8/30.8 | 96.1/30.9 | 96.1/30.9 | 96.4/31.0 | 95.8/30.8 | 96.1/30.9 | 96.4/31.5 | 97.7/29.3 | 100.5/30.5 | 95.6/31.3 | 93.5/32.1 |
| GQTP | 93.7/30.8 | 93.5/30.7 | 94.1/30.8 | 93.7/30.8 | 93.7/30.8 | 93.4/30.7 | 94.1/30.8 | 93.7/30.7 | 96.2/29.9 | 97.1/30.1 | 93.7/30.9 | 93.1/31.8 |
| GHM | 99.8/27.8 | 99.7/28.3 | 100.5/28.1 | 99.6/28.2 | 99.8/27.8 | 100.1/28.2 | 100.5/28.1 | 98.8/28.2 | 98.8/27.2 | 100.2/27.9 | 99.5/28.6 | 97.1/29.7 |
| Gwide | 99.7/31.8 | 99.1/31.7 | 99.2/31.5 | 99.4/31.7 | 99.7/31.8 | 99.1/31.1 | 99.2/31.5 | 98.8/31.0 | 93.7/30.9 | 104.0/33.2 | 98.0/32.5 | 94.1/33.3 |
| Gplain | 106.6/31.6 | 106.9/31.3 | 106.7/31.3 | 106.9/31.3 | 106.6/31.6 | 106.7/31.9 | 106.7/31.3 | 106.7/31.8 | 108.6/28.5 | 105.7/29.9 | 105.3/31.9 | 84.1/35.8 |
| WHOLE | 96.2/31.1 | 95.6/31.1 | 96.1/31.4 | 95.9/31.1 | 96.2/31.1 | 95.9/31.1 | 96.1/31.4 | 95.9/31.1 | 96.6/29.7 | 100.6/30.8 | 95.8/31.5 | 93.5/32.3 |
Between the current to future scenarios, a total of 8, 11 and 10 Rhodiola species are predicted to expand their ranges in MEAN ENSEMBLE, MMM ENSEMBLE, and Maxent, with the remaining species predicted to exhibit range contractions; this was especially evident for R. quadrifida for which all three modelling approaches predicted range contraction. In addition, we found that ranges were projected to shift more extensively (two or three orders of magnitude) between the current and future scenarios compared to between the LGM to current time points (Supplementary Tables S9–S12). This implies that accelerated climate warming will have marked impacts on future species’ distributions.
Extrapolating from our findings for the future climate warming scenario, the mean upward rate of elevational shifting for Rhodiola species would be ca. 0.273 (±0.217) m/year, 0.495 (±0.521) m/year and 7.07 (±5.42) m/year estimated using MEAN ENSEMBLE, MMM ENSEMBLE and Maxent, respectively. The three modelling approaches predicted that the mean northward rate of range movement should be ca. 0.288 (±0.288) km/year, 0.384 (±0.192) km/year and 1.24 (±1.19) km/year for (estimated by 1 degree equals 111 × cos30 km, at about 30° latitude). Maxent had relatively higher predictions than the other approaches. In contrast, some species displayed southward movement, including R. yunnanensis, R. alsia, R. himalensis (Supplementary Table S12).
From the LGM to current and future scenarios, the differences in mean range shifting between sister species versus between species groups were significant for predictions from MEAN ENSEMBLE, MMM ENSEMBLE (except for GQTP and Gwide), and Maxent (except for GQTP and Gwide, and Gclade1 and Gclade2) (χ2 tests, Supplementary Table S13).
Discussion
The reliability of model projections
Assertions of historical or future range shifts of species largely depend on the reliability of climatic data used and the ENM modelling process. In order to consider the range of plausible climates and of possible predictions by different models26, we used the multi-model mean of all the GCMs to do ENM (MMM ENSEMBLE), and also applied each GCM to do ENM (MEAN ENSEMBLE), and then calculated the mean and standard deviations for all predictions. The predictions of MEAN ENSEMBLE for the LGM and 2050 scenarios overlapped those of MMM ENSEMBLE, implying that modelling with the MMM of the GCMs was reasonable27,28. We also found that Maxent29,30 models for Rhodiola outperformed other sub-model algorithms and predictions generally agreed with those from ensemble models, although Maxent consistently predicted more significant range shifts under future climate warming scenarios. Oppel et al.31 also reported that Maxent predicted larger areas than ensemble models, the cause of the difference between Maxent and ensemble models calls for further studies.
Our ENM results for three Rhodiola species for which there are molecular phylogeographic information (R. kirilowii, R. dumulosa and R. alsia) were consistent with published studies, suggesting that the southeastern QTP and HM were the center from which Rhodiola lineages spread to occupy broader geographic ranges from the LGM to the present day (Fig. 3; Supplementary Figs S2–S4)17–19. Consistency of these published results with our own findings, increases confidence in our ENM interpretations.
Upward and northward movement
From the LIG to the LGM, concomitant with climate cooling, most Rhodiola species expanded their geographic ranges, consistent with what happened with R. integrifolia in North America32,33. A similar pattern was also detected in other QTP plants including Taxus wallichiana and Picea likiangensis34,35. Thus, as would be expected of cold-adapted species responding to climate warming5,6,36, we found that Rhodiola species have generally shifted upward in altitude and also northward since the LGM (Fig. 3; Supplementary Tables S10 and S12; Supplementary Figs S2–S4). Although our estimates of upward rate of movement varied somewhat using different modelling approaches, the mean elevation upward movement of Rhodiola (0.273~7.07 m/year) is similar to estimates of upward movement rate for the tree line in the Baima Mountains of the HM (0.84 m/year, between 1923 to 2003)37 and montane herbaceous plants in Europe (8 m/year estimated from Fig. 4 in Lenoir et al.6). We have no estimates for rate of latitudinal movement in plants, although the northward rate of range shifting of Rhodiola (0.288~1.24 km/year) is comparable to that of animals (1.69 km/year)5.
Not “nowhere to go”
Contrary to the expectations under the “nowhere to go” hypothesis3,4, most Rhodiola species broadened their geographic ranges rather than contracting with future climate warming (Fig. 3; Supplementary Table S9; Supplementary Figs S2–S4). DeChaine et al.32 and Forester et al.33 found that R. integrifolia exhibited a similar pattern of range expansion in North America. Other QTP plants, such as Hippophae gyantsensis, H. rhamnoides ssp. yunnanensis, H. neurocarpa, Eriophyton wallichii, Thalictrum squamiferum, Paraquilegia microphylla and Allium przewalskianum also tend towards range expansion with climate warming38–40. Such patterns may be explained by a number of factors. First, these species have relatively small contemporary geographical ranges. For instance, Rhodiola is mainly distributed in the eastern and southern regions of the QTP while the more extensive areas available above 4000 m above sea level (asl) in the northern and western parts of the QTP remain uncolonized. Second, warmer temperatures and higher precipitation under future warming conditions could transform unsuitable regions, such as those currently with permafrost, into suitable habitats for Rhodiola at higher latitudes and elevations. Moreover, increased vegetation coverage could also play a role in expansion of Rhodiola species ranges. Cannone et al.41 reported that vegetation coverage, particularly that of shrubs, increased dramatically in the alpine belt of Italian Central Alps from 1952 to 2003, coincident with higher precipitation and diminution of permafrost. The extent of shrub coverage has also increased in the Arctic over the past 50 years42. Rhodiola species typically co-occur with shrubs15, and thus we predict that areas suitable for colonization by Rhodiola will be increasingly available as shrub vegetation expands in the QTP and environs. Overall then, our results provide evidence that vascular plant species richness in montane communities will increase with climate warming9.
In contrast to the patterns evident for its congeners, all predictions generated from MEAN ENSEMBLE, MMM ENSEMBLE and Maxent showed that R. quadrifida would have a more restricted geographic range under future climate warming (Fig. 3; Supplementary Table S9; Supplementary Figs S2–S4). This implies that, for higher elevation montane species like R. quadrifida, there may be insufficient suitable alpine habitat to facilitate future migration. Thus, for this species at least, projections of future distributions in areas above 4000 m asl on the QTP are consistent with predictions of the “nowhere to go” hypothesis3,4.
Factors underlying species’ distribution
Species’ distributions are shaped by both abiotic (e.g. climatic and edaphic) and biotic factors (e.g. evolutionary history and interspecific competition)43,44. ENM is tacitly based on the assumption of niche conservation and is often used to estimate the influence of climate and other abiotic factors on species distributions; however, it can also be used to predict the effects of evolutionary history on species ranges (i.e. examining the evolutionary niche sensu Mao & Wang45 and Aguirre-Gutiérrez et al.46). Sister species and closely related species existing in sympatry should have distinct niches for them to coexist, and thus are expected to respond differently to climate change45,46. In our study we did detect significantly different patterns of range shifting between sister species and related species within particular clades (Supplementary Table S13). This suggests that there is an effect of evolutionary history, consistent with the view that species diversity of Rhodiola resulted from rapid radiation into different environments after their origination in the QTP16. In addition, this assertion is consonant with our CDA results that indicated differences in ecological niches among Rhodiola species (Fig. 2; Supplementary Fig. S1).
Our ENM showed that, in comparison to precipitation, temperature-related variables are more important influencing factors to Rhodiola species distributions, implying that cold-adapted plants exhibit sensitivity to climate warming. Results from our PCA and CAD analyses suggested that divergence in ecological niches accounted for the different distribution patterns among Rhodiola species (Fig. 2). Niche breadths of Gplain species were distinct from high elevation plants with the former showing broader distributions than the latter. Generally low elevation Rhodiola taxa need relatively stable higher temperatures and more homogeneous precipitation throughout the year, whereas higher elevation species are adapted to relatively low temperature, larger diel and seasonal temperature differences and more heterogeneous, and sporadic precipitation. Among the high elevation taxa, the ranges of those within GHM had higher temperatures and summer precipitation levels than taxa within GQTP and Gwide, with temperature appearing to be more important. The geographic ranges for species within our Gwide group exhibited higher temperature stability than those in GQTP (Fig. 2).
Model performance was higher for groups than for the genus in its entirety (WHOLE) (Supplementary Tables S1 and S2), implying that classifications based on distribution patterns may yield more insights about habitat suitability and climate requirements. It was especially notable that the responses to climate change varied between lower versus higher elevation species. For instance, the suitable area available for low elevation Rhodiola species (Gplain) increased from the LIG to the LGM, and then decreased from the LGM to the current and future scenarios. This was in contrast to higher elevation taxa (e.g. GQTP) which were predicted to continue to increase from the LIG using Maxent (Supplementary Table S9; Supplementary Fig. S4). These findings together imply that even closely-related species might diverge in their responses to climate change because of niche differentiation45,46.
Theoretically, if not self-incompatible, hermaphroditic plants can produce seeds by selfing, restricting pollen mediated gene flow. In contrast, dioecious plants are obligatorily outbreeding and thus have a relatively wider range of pollen mediated gene flow patterns47,48. Such differences might explain the range of responses to environmental change47,48. MEAN and MMM ENEMBLE predicted that the Rhodiola species with divergent breeding systems exhibited differential responses to climate change consistent with the above expectation. However, the results of Maxent implied that the Rhodiola species with different breeding systems or no close evolutionary affinities respond similarly to climate change40. Species from a biological community will necessarily share adaptations to similar climatic conditions and thus by definition have similar climatic niche requirements45,46. Therefore, our study shows that ENM based on communities or functional groups may better reflect the impact of climate on constituent plant taxa within particular regions49.
Conclusion
Our study revealed that Rhodiola species were primarily confined to the HM during the LIG, and then expanded into the southeastern QTP during LGM. From the LGM to the present day our results thus indicate that most species expanded to inhabit much broader geographic ranges on the QTP and adjacent regions concomitant with climate warming. These results together imply that high elevation regions in the southeastern QTP and HM were the center of origin and radiation for Rhodiola, and the location of refugia. Our analyses suggested that many Rhodiola species will not show diminished range sizes under future climate warning as would be predicted by the “nowhere to go” scenario hypothesis. This in part could be because diminution of permafrost at higher altitudes could create large swaths of new, suitable habitat. However, distributions of Rhodiola species were projected to show both latitudinal and elevational shifts. Finally, our analyses indicated that species with similar ecological niche requirements will respond similarly to climate change.
Methods
Species sampling
We selected Rhodiola species for ENM based on the following criteria: (1) phylogeographic data availability, (2) variation in breeding system (dioecious or monoecious), (3) distinction in evolutionary history (sister group vs. different clades in phylogeny tree), (4) dissimilarity in spatial distribution patterns (dispersed or clumped over the QTP), and (5) availability of at least 39 occurrence records. Based on these criteria, a total of 14 species were selected for ENM analysis (Table 1; Supplementary Fig. S5). These species derived from the two main clades at the genus-level phylogeny (Supplementary Fig. S6)16 (Gclade1 comprised three species, Gclade2 comprised 11 species). Among these 14 targeted species, we included three pairs of sister species (R. alsia and R. fastigiata, R. forrestii and R. yunnanensis, R. henryi and R. quadrifida) that, despite overlapping ranges, show some allopatry15. The distributions of the 14 species can be classified into four groups: 1) GQTP = species for which the bulk of their distributions occur on the QTP (R. alsia, R. bupleuroides, R. chryanthemifolia, R. crenulata, R. fastigiata, R. himalensis, R. quadrifida, R. sacra and R. wallichiana), 2) GHM = species that are mainly distributed within the HM (R. forrestii, R. yunnanensis), 3) Gwide = species widely distributed from the QTP to northern China (R. dumulosa and R. kirilowii), and 4) Gplain, = not distributed on the QTP or other high elevation (<3000 m) regions (R. henryi) (see Table 1; Supplementary Fig. S6). These species can also be classified based on breeding system: dioecy (Gdio, with 10 species) and monoecy (Gmon, with 4 species) (Table 1; Supplementary Fig. S6).
Occurrence records were derived from our field expeditions, on-line herbarium databases (Global Biodiversity Information Facility Data Portal, GBIF: www.gbif.org; Chinese Virtual Herbarium Data Portal, CVH: www.cvh.org.cn) and published research (see Supplementary Material1). We removed duplicate records and records with obvious errors in their geographic coordinates and thinned records that occurred within the same 1 km pixel so that only one occurrence remained. After pruning and verifying our data, we were left with 1,444 records for ENM spanning 39 to 200 unique locations (Table 1; Supplementary Fig. S6; see Supplementary Material1).
Climate variables
We obtained bioclimatic data to test our predictions from the WorldClim Dataset (www.worldclim.org/bioclim) (Supplementary Table S14). We downloaded 19 bioclimatic variables for the LIG (ca. 120,000–140,000 years BP)50, LGM (ca. 22,000 years BP), current conditions (average from years 1950 to 2000) and 2050 RCP8.5 (average from 2041 to 2060) from WorldClim (all 30 arc sec resolution, original 2.5 minute resolution of LGM were resampled to a spatial resolution of 30 arc sec by ArcGIS). Future climate data were accessed from Phase 5 of the Coupled Model Intercomparison Project (CIMP5)51, where we selected scenario 2050 RCP8.5, the highest emission scenario, so that we could forecast the warmest future environments for alpine species2. For LGM and 2050 which include multi-GCMs, the 19 bioclimatic variables were averaged for 4 GCMs for the LGM (Supplementary Table S7) and 17 GCMs for 2050 (Supplementary Table S8)27,28,52, standard deviations of multi-GCMs were also calculated (Supplementary Fig. S7)26.
To mitigate issues of multicollinearity in our models, we selected bioclimatic variables with lower correlations bioclimatic variables by using the variance inflation factor (VIF) statistics using Biodiversity R package in R version 3.1.225. We retained bioclimatic variables with VIF values lower than 10 for all species records in each run, until all remaining VIF values were less than 10, as VIF values higher than 10 indicate strong collinearity53,54. Our final dataset included nine bioclimatic variables with VIF values lower than 10 (4 temperature-related variables (BIO2, BIO3, BIO8 and BIO9) and 5 precipitation variables (BIO13, BIO14 BIO15 BIO18 and BIO19)) (Supplementary Table S15).
Modelling process
We used three methods in our modelling process: (1) MEAN ENSEMBLE (using each GCM to do ensemble ENM, and then calculating the mean and standard deviation of all predictions); (2) MMM ENSEMBLE (using the multi-model mean (MMM) of all GCMs to do ensemble ENM); (3) Maxent (using the MMM of all GCMs to do Maxent modelling). For each method, we did ENM for individual species, clade, distribution, and breeding system groups defined above (Gclade1, Gclade2, GQTP, GHM, Gwide, Gplain, Gdio, Gmon) and finally for all 14 species combined (WHOLE).
For the MEAN ENSEMBLE and MMM ENSEMBLE approaches, we undertook a three-step process in the BiodiversityR package25 to prepare for consensus mapping for each species and species group. The first step involved calibration of niche modelling algorithms, applying the ‘ensemble.test.splits’ function where we calibrated 17 ENM sub-models and defined a 4-fold cross-validation using 75% of the data to evaluate the remaining 25% data. AUC values for sub-models were used to determine their weights for ensemble models (Supplementary Table S1)25. For the second step we applied the “ensemble.test” function with sub-model weights >0.05 retained for ensemble modelling (Supplementary Table S16) – this used 10 internal test runs and 4-fold cross-validations predicting final weights for the sub-models. The last step was “ensemble.raster” function, which generated our consensus mapping. Upon completion of our MEAN ENSEMBLE models, we calculated the mean and standard deviation of all predictions.
AUC values of sub-models from the ensemble ENM indicated that Maxent had the highest performance among all the sub-models. Thus, we used Maxent to predict range shifts for Rhodiola species and species groups. We randomly selected 25% of the locations as test data and used the remaining 75% as the training dataset. This was the same approach used for ensemble modelling; we used default values for other parameters30.
Estimating range shifts
We created a python script (see Supplementary Material2) to calculate the predicted suitable area, mean elevation, and mean center of the predicted distribution in ArcGIS for each model based on 25% habitat suitability. The predicted suitable area was calculated using rectangular projections. We calculated the mean elevation of each suitable area in meters, and found the mean center of the suitable area by calculating the mean longitude and latitude (for python script see Supplementary Material2).
The following equations were used to estimate the range shifts between different time periods:
| 1 |
| 2 |
where for (1) ΔS is the range of distribution shift; St represents the suitable area at time ‘t’; tn represents time for different periods (four periods in total: LIG, LGM, current, and future); ΔT equals the years from the periods tn to tn+1, ΔT1 ≈ 98,000 (from LIG to LGM), ΔT2 ≈ 22,000 (LGM to current), ΔT3 ≈ 75 (current to future) and for (2) u equals the variation rate of elevation (ualt) or longitude and latitude of distribution center (ulon and ulat) and Vt equals the mean elevation or mean center at time ‘t’.
Data analysis
Wilcoxon rank sum tests were used to evaluate whether models differed in AUC values for models with all clades, breeding and distribution groups, individual species, and genus49.
Jackknife tests were used to measure variable importance29, with higher gain indicating a variable that contributes highly to the species’ spatial distribution. For a better understanding of the contributions of temperature and precipitation variables to the distribution of Rhodiola species, we first performed separate Principal Components Analysis (PCA) on the four temperature variables and five precipitation variables using records for all species (WHOLE)49. These ordinations produced two major temperature axes (Temp1 and Temp2) and two major precipitation axes (Prec1 and Prec2). To further reduce the dimensionality of the PCA data, we then used the scores of PC axes to do canonical discriminants analysis (CDA) (considering both temperature variables and precipitation variables) for GQTP, GHM, Gplain and Gwide (Fig. 2)49. Mean discriminant scores of species groups were plotted against the bioclimatic variables with the highest contributions to evaluate the relationship between known geographical distributions and climate variables (Supplementary Fig. S1). Wilks’ Lambda values were used to test the significance of our CDA results.
To test the influence of evolutionary history on range shifts induced by climate warming, we did χ2 tests on ΔS, ualt, ulon and ulat during the LGM to current and current to future between each sister species pair and between two clades (Gclade1 and Gclade2). Additionally, χ2 tests were used to compare Gmon and Gdio, as well as between groups pairs from GQTP, GHM, Gplain and Gwide to examine the effects of breeding systems and ecological niche differentiation on the response to climate change. These analyses were performed using SPSS Statistics ver. 22.
Electronic supplementary material
Acknowledgements
This study was supported by the National Key Basic Research Program of China (2014CB954100), the Applied Fundamental Research Foundation of Yunnan Province (2014GA003), and the National Natural Science Foundation of China (31400327).
Author Contributions
Zhiping Song conceived the idea and proposed the research. Jianling You and Xiaoping Qin collected the data and performed the analysis. Sailesh Ranjitkar guided the modelling work. Mingcheng Wang wrote the python scripts. Jianling You wrote the first draft of the MS and all other co-authors (Xiaoping Qin, Sailesh Ranjitkar, Stephen C. Lougheed, Mingcheng Wang, Wen Zhou, Dongxin Ouyang, Yin Zhou, Jianchu Xu, Wenju Zhang, Yuguo Wang, Ji Yang and Zhiping Song) contributed substantially to its editing. Stephen C. Lougheed, provided insightful comments on the analysis and conclusions.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24360-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hoffmann AA, Sgro CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- 2.IPCC authors. In Climate Change: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (eds Stocker, T. F. et al.) Ch. 12, 1055 (Cambridge University Press, NY, USA, 2013).
- 3.Loarie SR, et al. The velocity of climate change. Nature. 2009;462:1052–1055. doi: 10.1038/nature08649. [DOI] [PubMed] [Google Scholar]
- 4.Nogués-Bravo D, Araujo MB, Errea MP, Martinez-Rica JP. Exposure of global mountain systems to climate warming during the 21st Century. Global environmental change. 2007;17:420–428. doi: 10.1016/j.gloenvcha.2006.11.007. [DOI] [Google Scholar]
- 5.Chen IC, Hill JK, Ohlemueller R, Roy DB, Thomas CD. Rapid range shifts of species associated with high levels of climate warming. Science. 2011;333:1024–1026. doi: 10.1126/science.1206432. [DOI] [PubMed] [Google Scholar]
- 6.Lenoir J, Gegout JC, Marquet PA, de Ruffray P, Brisse H. A significant upward shift in plant species optimum elevation during the 20th century. Science. 2008;320:1768–1771. doi: 10.1126/science.1156831. [DOI] [PubMed] [Google Scholar]
- 7.Dainese M, et al. Human disturbance and upward expansion of plants in a warming climate. Nature Climate Change. 2017;7:577–580. doi: 10.1038/nclimate3337. [DOI] [Google Scholar]
- 8.Gaur UN, Raturi GP, Bhatt AB. Quantitative response of vegetation in Glacial Moraine of Central Himalaya. Environmentalist. 2003;23:237–247. doi: 10.1023/B:ENVR.0000017378.55926.a7. [DOI] [Google Scholar]
- 9.Holzinger B, Hülber K, Camenisch M, Grabherr G. Changes in plant species richness over the last century in the eastern Swiss Alps: Elevational gradient, bedrock effects and migration rates. Plant Ecology. 2008;195:179–196. doi: 10.1007/s11258-007-9314-9. [DOI] [Google Scholar]
- 10.Liu XD, Chen BD. Climatic warming in the Tibetan Plateau during recent decades. International Journal of Climatology. 2000;20:1729–1742. doi: 10.1002/1097-0088(20001130)20:14<1729::AID-JOC556>3.0.CO;2-Y. [DOI] [Google Scholar]
- 11.Liu X, Zhang M. Contemporary climatic change over the Qinghai-Xizang plateau and its response to the green-house effect. Chinese Geographical Science. 1998;8:289–298. doi: 10.1007/s11769-997-0034-9. [DOI] [Google Scholar]
- 12.Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 13.Yang F, Li Y, Ding X, Wang X. Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with the Quaternary climate change. Molecular Ecology. 2008;17:5135–5145. doi: 10.1111/j.1365-294X.2008.03976.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, et al. Phylogeographic structure of Hippophae tibetana (Elaeagnaceae) highlights the highest microrefugia and the rapid uplift of the Qinghai-Tibetan Plateau. Molecular Ecology. 2010;19:2964–2979. doi: 10.1111/j.1365-294X.2010.04729.x. [DOI] [PubMed] [Google Scholar]
- 15.Wu CY, Raven PH. Flora of China 8. Beijing: Science Press; 2001. [Google Scholar]
- 16.Zhang J, Meng S, Allen GA, Wen J, Rao G. Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae) Molecular Phylogenetics and Evolution. 2014;77:147–158. doi: 10.1016/j.ympev.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Gao Q, et al. Intraspecific divergences of Rhodiola alsia (Crassulaceae) based on plastid DNA and internal transcribed spacer fragments. Botanical Journal of the Linnean Society. 2012;168:204–215. doi: 10.1111/j.1095-8339.2011.01193.x. [DOI] [Google Scholar]
- 18.Hou Y, Lou A. Phylogeographical patterns of an alpine plant, Rhodiola dumulosa (Crassulaceae), inferred from chloroplast DNA sequences. Journal of Heredity. 2014;105:101–110. doi: 10.1093/jhered/est072. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Meng S, Rao G. Phylogeography of Rhodiola kirilowii (Crassulaceae): A story of Miocene divergence and Quaternary expansion. PLoS ONE. 2014;9:e112923. doi: 10.1371/journal.pone.0112923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grinnell J. The niche-relationships of the California Thrasher. The Auk. 1917;34:427–433. doi: 10.2307/4072271. [DOI] [Google Scholar]
- 21.Hirzel AH, Le Lay G. Habitat suitability modelling and niche theory. Journal of Applied Ecology. 2008;45:1372–1381. doi: 10.1111/j.1365-2664.2008.01524.x. [DOI] [Google Scholar]
- 22.Sillero N. What does ecological modelling model? A proposed classification of ecological niche models based on their underlying methods. Ecological Modelling. 2011;222:1343–1346. doi: 10.1016/j.ecolmodel.2011.01.018. [DOI] [Google Scholar]
- 23.Elith J, et al. A statistical explanation of Maxent for ecologists. Diversity and Distributions. 2011;17:43–57. doi: 10.1111/j.1472-4642.2010.00725.x. [DOI] [Google Scholar]
- 24.Soberón J, Peterson AT. Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodiversity Informatics. 2005;2:1–10. doi: 10.17161/bi.v2i0.4. [DOI] [Google Scholar]
- 25.Kindt R. GUI for biodiversity, suitability and community ecology analysis. R package BiodiversityR version. 2014;2:4–4. [Google Scholar]
- 26.Harris RMB, et al. Climate projections for ecologists. Wiley Interdisciplinary Reviews: Climate Change. 2014;5:621–637. [Google Scholar]
- 27.Mora C, et al. The projected timing of climate departure from recent variability. Nature. 2013;502:183–187. doi: 10.1038/nature12540. [DOI] [PubMed] [Google Scholar]
- 28.Zomer RJ, Xu J, Wang M, Trabucco A, Li Z. Projected impact of climate change on the effectiveness of the existing protected area network for biodiversity conservation within Yunnan Province, China. Biological Conservation. 2015;184:335–345. doi: 10.1016/j.biocon.2015.01.031. [DOI] [Google Scholar]
- 29.Elith J, et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. doi: 10.1111/j.2006.0906-7590.04596.x. [DOI] [Google Scholar]
- 30.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. doi: 10.1016/j.ecolmodel.2005.03.026. [DOI] [Google Scholar]
- 31.Oppel S, et al. Comparison of five modelling techniques to predict the spatial distribution and abundance of seabirds. Biological Conservation. 2012;156:94–104. doi: 10.1016/j.biocon.2011.11.013. [DOI] [Google Scholar]
- 32.DeChaine EG, Forester BR, Schaefer H, Davis CC. Deep genetic divergence between disjunct refugia in the Arctic-Alpine King’s Crown, Rhodiola integrifolia (Crassulaceae) PLoS ONE. 2013;8:e79451. doi: 10.1371/journal.pone.0079451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forester BR, DeChaine EG, Bunn AG. Integrating ensemble species distribution modelling and statistical phylogeography to inform projections of climate change impacts on species distributions. Diversity and Distributions. 2013;19:1480–1495. doi: 10.1111/ddi.12098. [DOI] [Google Scholar]
- 34.Li L, et al. Pliocene intraspecific divergence and Plio-Pleistocene range expansions within Picea likiangensis (Lijiang spruce), a dominant forest tree of the Qinghai-Tibet Plateau. Molecular Ecology. 2013;22:5237–5255. doi: 10.1111/mec.12466. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, et al. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytologist. 2013;199:1093–1108. doi: 10.1111/nph.12336. [DOI] [PubMed] [Google Scholar]
- 36.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 37.Baker BB, Moseley RK. Changes in the Hengduan Mountains: Advancing treeline and retreating glaciers. Arctic, Antarctic, and Alpine Research. 2007;39:200–209. doi: 10.1657/1523-0430(2007)39[200:ATARGI]2.0.CO;2. [DOI] [Google Scholar]
- 38.Jia DR, et al. Diploid hybrid origin of Hippophae gyantsensis (Elaeagnaceae) in the western Qinghai-Tibet Plateau. Biological Journal of the Linnean Society. 2015;117:658–671. doi: 10.1111/bij.12707. [DOI] [Google Scholar]
- 39.Liang Q, Hu X, Wu G, Liu J. Cryptic and repeated “allopolyploid” speciation within Allium przewalskianum Regel. (Alliaceae) from the Qinghai-Tibet Plateau. Organisms Diversity & Evolution. 2015;15:265–276. doi: 10.1007/s13127-014-0196-0. [DOI] [Google Scholar]
- 40.Luo D, et al. Evolutionary history of the subnival flora of the Himalaya-Hengduan Mountains: first insights from comparative phylogeography of four perennial herbs. Journal of Biogeography. 2016;43:31–43. doi: 10.1111/jbi.12610. [DOI] [Google Scholar]
- 41.Cannone N, Sgorbati S, Guglielmin M. Unexpected impacts of climate change on alpine vegetation. Frontiers in Ecology and the Environment. 2007;5:360–364. doi: 10.1890/1540-9295(2007)5[360:UIOCCO]2.0.CO;2. [DOI] [Google Scholar]
- 42.Tape KEN, Sturm M, Racine C. The evidence for shrub expansion in northern Alaska and the Pan-Arctic. Global Change Biology. 2006;12:686–702. doi: 10.1111/j.1365-2486.2006.01128.x. [DOI] [Google Scholar]
- 43.Elith J, Leathwick JR. Species distribution models: ecological explanation and prediction across space and time. Annual Review of Ecology, Evolution, and Systematics. 2009;40:677. doi: 10.1146/annurev.ecolsys.110308.120159. [DOI] [Google Scholar]
- 44.Roux PCL, Virtanen R, Heikkinen RK, Luoto M. Biotic interactions affect the elevational ranges of high-latitude plant species. Ecography. 2012;35:1048–1056. doi: 10.1111/j.1600-0587.2012.07534.x. [DOI] [Google Scholar]
- 45.Mao J, Wang X. Distinct niche divergence characterizes the homoploid hybrid speciation of Pinus densata on the Tibetan Plateau. The American Naturalist. 2011;177:424–439. doi: 10.1086/658905. [DOI] [PubMed] [Google Scholar]
- 46.Aguirre-Gutiérrez J, Serna-Chavez HM, Villalobo-Arambula AR, Pérez de la Rosa JA, Raes N. Similar but not equivalent: ecological niche comparison across closely–related Mexican white pines. Diversity and Distributions. 2015;21:245–257. doi: 10.1111/ddi.12268. [DOI] [Google Scholar]
- 47.Grossenbacher D, Briscoe Runquist R, Goldberg EE, Brandvain Y. Geographic range size is predicted by plant mating system. Ecology letters. 2015;18:706–713. doi: 10.1111/ele.12449. [DOI] [PubMed] [Google Scholar]
- 48.Hargreaves AL, Eckert CG. Evolution of dispersal and mating systems along geographic gradients: implications for shifting ranges. Functional ecology. 2014;28:5–21. doi: 10.1111/1365-2435.12170. [DOI] [Google Scholar]
- 49.Riordan EC, Rundel PW. Modelling the distribution of a threatened habitat: The California sage scrub. Journal of Biogeography. 2009;36:2176–2188. doi: 10.1111/j.1365-2699.2009.02151.x. [DOI] [Google Scholar]
- 50.Otto-Bliesner BL, Marsha SJ, Overpeck JT, Miller GH, Hu AX. Simulating arctic climate warmth and icefield retreat in the last interglaciation. Science. 2006;311:1751–1753. doi: 10.1126/science.1120808. [DOI] [PubMed] [Google Scholar]
- 51.Overpeck JT, Meehl GA, Bony S, Easterling DR. Climate data challenges in the 21st century. Science. 2011;331:700–702. doi: 10.1126/science.1197869. [DOI] [PubMed] [Google Scholar]
- 52.Hijmans, R. J., Phillips, S., Leathwick, J. & Elith, J. dismo: Species distribution modeling. R package version 0.7–17 (2012).
- 53.Ranjitkar S, et al. Climate modelling for agroforestry species selection in Yunnan Province, China. Environmental Modelling & Software. 2016;75:263–272. doi: 10.1016/j.envsoft.2015.10.027. [DOI] [Google Scholar]
- 54.Hair JF, Black WC, Babin BJ, Anderson RE, Tatham RL. Multivariate data analysis 6. NJ: Pearson Prentice Hall Upper Saddle River; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.