Abstract
The radiological accident in Goiania in 1987 caused a trail of human contamination, animal, plant and environmental by a radionuclide. Exposure to ionizing radiation results in different types of DNA lesions. The mutagenic effects of ionizing radiation on the germline are special concern because they can endures for several generations, leading to an increase in the rate of mutations in children of irradiated parents. Thus, to evaluate the biological mechanisms of ionizing radiation in somatic and germline cells, with consequent determination of the rate mutations, is extremely important for the estimation of genetic risks. Recently it was established that Chromosomal Microarray Analysis is an important tool for detecting wide spectra of gains or losses in the human genome. Here we present the results of the effect of accidental exposure to low doses of ionizing radiation on the formation of CNVs in the progeny of a human population accidentally exposed to Caesium-137 during the radiological accident in Goiânia, Brazil.
Introduction
Unexpected events led to a serious radiological accident involving Caesium-137 in September of 1987 in Goiânia, Brazil. Consequently, people and the environment were contaminated with radionuclide and exposed to ionizing radiation (IR) during a two-week period in which the accident had not been discovered yet. A total of 249 people was exposed to IR with absorbed doses ranging from near zero up to 7 Gy. For an extensive review of the accident, the readers are referred to da Cruz et al. & IAEA1,2. Despite being disastrous, the accident provided a unique opportunity to study the biological effects of ionizing radiation-induced mutations in humans.
The genetic health of people accidentally exposed to Caesium-137 has been continuously biomonitored in Goiânia since the accident. To date, most of the genetic biomarkers of exposure available were examined for the cohort of families accidently exposed to IR. Currently, reports are accessible for chromosomal aberrations, glycophorin A mutations micronucleus assay, HPRT test, somatic chromosomal translocation using FISH germline mutation rate in STRs, assessment of BCL2/J(H) translocations, and date for serological autoimmune markers2–9.
At a cellular level, exposure of biomolecules to IR results in molecular lesions. When human DNA is exposed to IR, immediate damage is mostly of a result of either double strand breaks (DSBs) or single strand breaks (SSBs). The damage is enzymatically repaired, resulting in either the removal of damaged bases or the re-joining of strand breaks. However, incorrect repair of the DNA damage leads to mutations that contribute to adverse stochastic effects. The nature of cells and tissues defines their radiosensitivity, which is the highest in highly mitotic or undifferentiated cells. Thus, germline cells are highly sensitive to IR exposure, and mutations in germ cells could increase the mutation rate in the offspring of irradiated parents10,11.
Lately, chromosomal microarray analysis (CMA) has been recognized as a powerful tool to detect gains and losses of the human genome and has been recommended as the new standard test to accurately detect chromosomal variations of different types, sizes, and genomic locations at higher resolution than karyotyping and conventional fluorescent in situ hybridization (FISH)12,13. CMA is able to detect both losses and gains of copy number variants (CNV) based on the distribution of millions of SNP markers across the whole genome14. A CNV is defined as gain or loss of a DNA segment ≥50 pb broadly scattered through a given genome15–17. On the other hand, some authors only apply the definition of CNV to DNA segments ≥1 kb18.
The identification of CNVs was facilitated by the advent of new technologies that allowed high-resolution genomic analysis by oligonucleotide microarrays (14). Investigating CNVs revealed their crucial role in population diversity, both as the reason for genetic variability and the etiological causes of human diseases. The frequency with which CNVs are formed suggests a high rate of de novo mutations that can be transmitted by Mendelian over generations19–21.
The mechanisms subjacent to the formation of CNVs have gained much attention from researchers. It has been postulated that both DNA replication and homologous recombination are important events in the origin of CNVs15,21,22. Due to their nature, CNVs could be useful biomarkers of exposure to assess potential health risks related to environmental mutagens. The frequency of de novo CNVs increased in cells exposed in vitro to ionizing radiation23. Thus, the rate of CNV formation could be useful to discriminate both somatic as well as germline mutation in humans. Moreover, the identification of inherent biomarkers of radiation exposure is a priority in radiation research in the genome era24. The usefulness of the rate of de novo CNV formation as a biological marker for human exposure to IR remains mostly unknown and must be investigated25.
In general, studies of CNV have reported findings ≥500 kb. Some population studies have been generated to evaluate the role of larger CNVs. Itsara et al.26, estimated the frequency of large CNVs in the general population using the data of about 2,500 people, focusing on hotspots prone to recurrent mutations. The authors found CNVs ≥500 kb and ≥1 Mb in up to 1–2% of individuals. To date, studies describing the germline mutation rate and paternal age at conception for small CNVs have not been reported. The current study reports the results of germline mutation rates using CNVs from the progeny born to parents accidentally exposed to very low doses of IR during the radiological accident with Caesium-137 in Goiania (Brazil) in 1987. In this context, the authors aimed to demonstrate the potential of CNVs germline mutations to be retrospectively used as biomarkers of exposure to IR in human populations.
Results
The mean parental ages at conception were 31.4 and 32.3 for fathers, and 26.4 and 29.6 for mothers, in cases and controls, respectively. The exposed population incurred individual doses of ≤0.2 Gy. The average ages of the F1 generations were 14.5 and 12.3 for cases and controls, respectively. Tables 1 and 2 summarize some of the data in the current study.
Table 1.
Summary of data from case and control groups for both parental and F1 generation included in the study of induced germline mutation in the offspring of people accidently exposed to low doses of ionizing radiation from Caesium-137 in Goiânia (Brazil).
| Groups | Family | Number of CNVloss | Number of CNVgain | Paternal Age | Maternal Age | F1’s Age | F1’s Sex |
|---|---|---|---|---|---|---|---|
| Control | CMA001-1F | 39 | 1 | 40 | 36 | 9 | Female |
| CMA002-1F | 34 | 1 | 22 | 21 | 9 | Male | |
| CMA003-1F | 20 | 1 | 26 | 26 | 25 | Female | |
| CMA004-1F | 32 | 1 | 30 | 31 | 10 | Female | |
| CMA005-1F | 29 | 2 | 42 | 31 | 5 | Female | |
| CMA006-1F | 24 | 3 | 37 | 35 | 6 | Male | |
| CMA007-1F | 27 | 3 | 47 | 36 | 9 | Male | |
| F09-3F | 19 | 2 | 19 | 21 | 25 | Male | |
| Total | — | 224 | 14 | — | — | — | — |
| Case | F04-2F | 18 | 3 | 27a | 27 | 20 | Male |
| F06-2F | 26 | 3 | 35a | 26 | 9 | Male | |
| F07-1F | 26 | 5 | 54 | 24b | 19 | Male | |
| F07-4F | 30 | 2 | 56 | 26b | 17 | Female | |
| F08-2F | 13 | 5 | 18 | 20b | 8 | Male | |
| F09-2F | 9 | 1 | 24c | 26c | 20 | Female | |
| F10-2F | 16 | 1 | 21 | 24b | 2 | Female | |
| F12-2F | 43 | 3 | 31 | 30b | 3 | Male | |
| F15-2F | 26 | 0 | 18a | 27 | 16 | Male | |
| F18-2F | 36 | 11 | 47a | 30 | 18 | Male | |
| F21-2F | 18 | 2 | 38 | 27b | 20 | Female | |
| F22-2F | 42 | 2 | 29 | 31b | 20 | Female | |
| F22-3F | 45 | 2 | 32 | 34b | 17 | Female | |
| F22-4F | 25 | 3 | 33 | 35b | 16 | Male | |
| FAD-24F | 29 | 4 | 21a | 19 | 12 | Female | |
| FAD-25F | 20 | 4 | 18a | 16 | 15 | Female | |
| Total | — | 422 | 51 | — | — | — | — |
CNVloss: genomic losses; CNVgain: genomic gains. aExposed father; bexposed mother; and cboth parents exposed.
Table 2.
Descriptive data from case and control groups for both parental and F1 generations included in the study of germline mutation induction detected in the progeny of people accidentally exposed to ionizing radiation from Caesium-137.
| Generation | Variables | Controls | Cases | |
|---|---|---|---|---|
| Parental | n | 16 | 24 | |
| Age Range | 16 to 56 | 19 to 47 | ||
| Average Age at Conception (±SD) | Fathers | 32.3 (10.1) | 31.4 (12.3) | |
| Mothers | 29.6 (6.3) | 26.4 (5.1) | ||
| Dose (Gy) | 0 | 0.2 | ||
| F1 | n | 8 | 16 | |
| Age Range | 5 to 25 | 2 to 20 | ||
| Average Age (±DP) | 12.3 (8.0) | 14.5 (6.0) | ||
| Sex Ratio (M/F) | 4/4 | 9/7 | ||
| Mean (SD) | 1.5 × 10−5 (±9.7 × 10−6) |
2.2 × 10−5 (±1.23 × 10−5) | ||
| Mean (SD) | 6.2 × 10−6 (±6.4 × 10−6) |
1.0 × 10−5 (±1.3 × 10−5) | ||
| Mean (SD) | 2.2 × 10−5 (±1.1 × 10−5) |
3.2 × 10−5 (±1.9 × 10−5) | ||
SD: Standard Deviation; M: Males; F: Females; ger: Generation; β: Burden.
No significant differences between cases and controls in the number of mutational events, either gains or losses, were observed. The differences arose when the data were transformed to the proportion of the gained or lost genome as a function of the total size of CNVs (Equation 1), where genomic losses and gains were higher in the offspring of exposed parents, corresponding to an increase of 1.5x for losses and 1.6x for gains when compared to the control group. In this context, the burden of the germline mutation rate in CNVs (MRCNVβ) increased about 50% in the genome of children whose parents were exposed to low doses of IR when compared to the offspring born to parents with no history of IR exposure from the same population (Goiânia).
With respect to the total number of CNVs, we identified 473 and 238 CNVs for case and control groups, respectively. Distribution and characterization of CNVs between the groups can be found in Table 3. In the current study, the largest CNV size was 393 kb. All observed CNVs in the current study were included in the statistical analysis. Genomic losses ranged from 1 kb to 147 kb, while gains ranged from 1 kb to 393 kb. The total size of the gains and the losses in the genomes of the exposed group were 1,892 kb and 4,103 kb. However, in the control group, gains added up to 579 kb and losses up to 1,428 kb (Table 3). Using the tools intrinsic to ChAS® (Affymetrix, USA), it was possible to assign the parent of origin (POR) for 60% (428/711) of all called CNVs, distributed as follow: 133 and 285 CNVs for control and cases, respectively. With respect to POR, 71 CNVs we transmitted by fathers as well as 81 transmitted by mothers to their offspring, considering the transmission by the exposed parents in the group of cases cohort.
Table 3.
Distribution of CNVs between cases and controls observed in the study of the impact of accidental exposure to low doses of ionizing radiation on the frequency of germline mutations in the progeny of people exposed during the Goiânia radiological accident with Caesium-137. ΣTCNV: Total size of CNVs (kb); na: not applicable.
| Group | CNV | Gain | Loss | Total |
|---|---|---|---|---|
| Case | Interval (kb) | 1 to 393 | 1 to147 | 1 to 393 |
| n | 51 | 422 | 473 | |
| Average of CNV Size (kb) | 37.1 (±58.8) |
9.7 (±13.8) |
na | |
| ΣTCNV (kb) | 1.892 | 4.103 | 5.995 | |
| Control | Interval (kb) | 1 a 140 | 1 a 85 | 1 a 140 |
| n | 14 | 224 | 238 | |
| Average of CNV Size (kb) | 41.4 (±48.2) |
6.4 (±8.5) |
na | |
| ΣTCNV (kb) | 579 | 1.428 | 2.007 |
Figure 1 shows the differences in the distribution of MRCNV between case and control groups. The Mann-Whitney U test indicated the difference for genomic losses was statistically significant (p < 10−3). However, no statistical significance for genomic gains was observed between the studied groups (p = 0.76). The results of Two-way ANOVA followed by the Tukey’s Test a posteriori were performed to compare the contribution of parental exposure and the parent of origin (POR) to the burden of the de novo CNV in their progenies showed significant differences among parental exposure (F = 61,03; p < 0,0001). On the other hand, there was no significant difference (F = 0,00; p = 1,00) between the origin of the CNVS and POR (Fig. 2).
Figure 1.
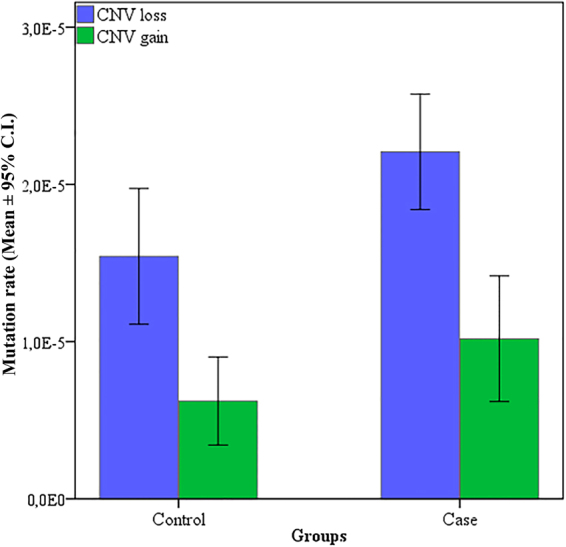
Average of germline mutation rates in CNVs from the progeny of controls and cases who were accidentally exposed to low doses of ionizing radiation from Caesium-137 in Goiânia (Brazil). CNVloss: genomic loss; CNVgain: genomic gain.
Figure 2.
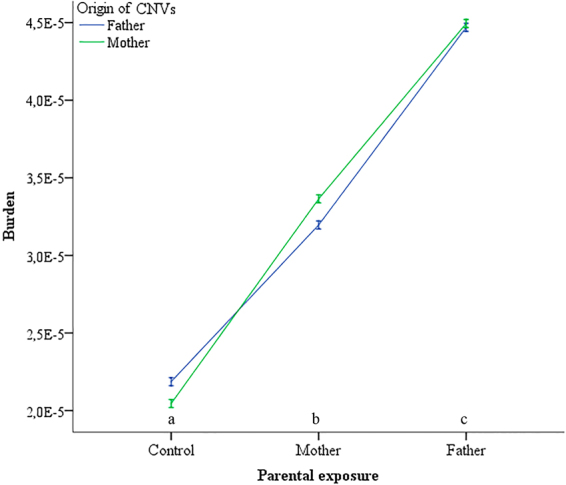
Burden of CNVs as a function of parental exposure and origin of CNVs for cases and control groups in the study of the effect of low absorbed doses of IR on the rate of germline mutations. Letters indicate significant differences among parental exposure at a significance level of 5%.
The binomial test showed the genome was affected differently between cases and controls, with respect to genomic losses (χ2 = 39.06; p < 0.00001) and gains (χ2 = 284.8; p < 0.00001). A similar statistical significance was observed even if the unequal cohort size was taken into account for genomic losses (χ2 = 378.99; p < 0.00001) and gains (χ2 = 5.7; p = 0.02).
Discriminant function analysis was used to determine which variables discriminated between exposed and control groups. In the current study, the best three predictors to discriminate the groups were number of paternal meiosis, mean parental age at conception, and number of markers in a CNV. The differences were statistically significant for both genomic gains (p = 0.02; r2 = 0.17) and losses ((p < 0.001; r2 = 0.08). The scores of linear discrimination are depicted in Fig. 3.
Figure 3.
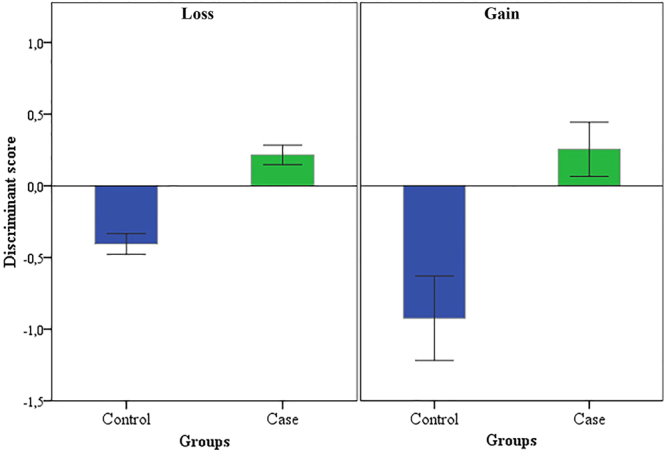
Discriminant function analysis of predictors: number of paternal meiosis, mean parental age at conception, and number of marker in a CNV on the germline mutation rate of genomic losses and gains for cases and controls observed in the progeny of a population exposed to low doses of ionizing radiation and a control population from Goiânia (Brazil).
The Mann-Whitney U was used to test the differences in the distribution of CNV size, number of markers and number of genes within the CNVs (Fig. 4). There were no statistical differences for the tested variables between the studied groups, in the current study.
Figure 4.
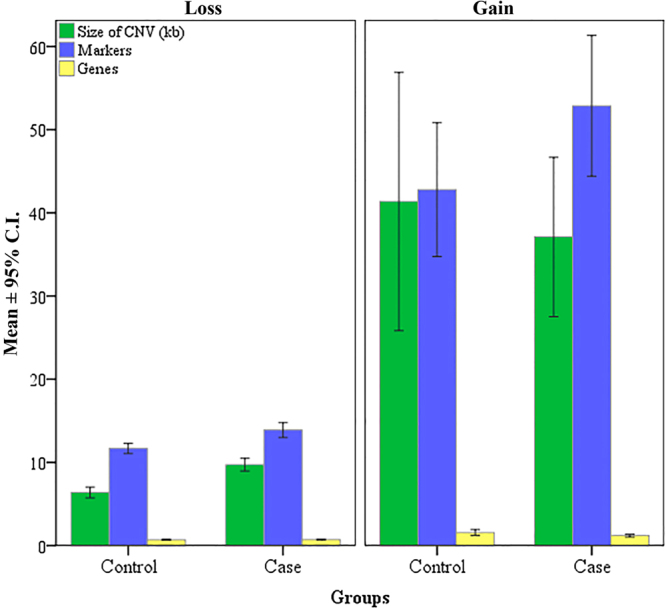
Distribution of CNV size, number of markers and number of genes in the CNVs for both control and exposed groups to low doses of ionizing radiation of Caesium-137.
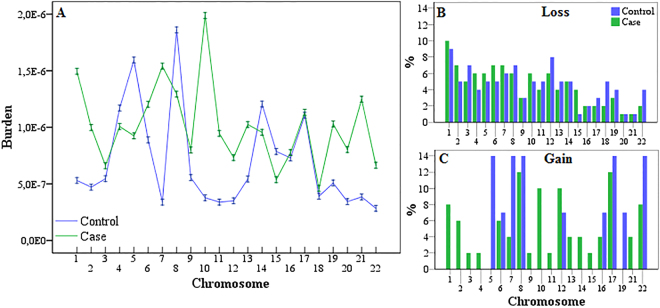
In order to compare the burden of de novo CNVs (Fig. 5A) and the proportion of chromosomal genome affected by the CNVs (Fig. 5B,C) per chromosome for both losses and gains, we applied the Mann-Whitney U test. Statistically significant differences between case and control groups were observed for chromosomes 1, 2, 7, 10, 11, 12, 19, and 20 (Table 4).
Figure 5.
Results of the Mann- Whitney U test of the burden of CNVs (A) and the total of CNVs per chromosome for genomic losses (B) and genomic gains (C) between case and control groups in the study of the induction of germline mutation due to parental exposure to ionizing radiation of Caesium-137.
Table 4.
Rate of germline mutation per chromosome in the offspring of parents exposed to ionizing radiation of Caesium-137 and the control group.
| Chromosome | Control | Case | P value | ||
|---|---|---|---|---|---|
| Average | SD | Average | SD | ||
| 1 | 5.3E-07 | 8.3E-07 | 1.5E-06 | 2.5E-06 | 0.04 |
| 2 | 4.7E-07 | 4.8E-07 | 1.0E-06 | 1.1E-06 | 0.04 |
| 3 | 5.5E-07 | 5.3E-07 | 6.6E-07 | 7.3E-07 | 0.51 |
| 4 | 1.2E-06 | 1.3E-06 | 1.0E-06 | 9.7E-07 | 0.98 |
| 5 | 1.6E-06 | 1.5E-06 | 9.2E-07 | 8.7E-07 | 0.11 |
| 6 | 8.9E-07 | 2.0E-06 | 1.2E-06 | 1.5E-06 | 0.07 |
| 7 | 3.4E-07 | 3.2E-07 | 1.5E-06 | 2.6E-06 | 0.003 |
| 8 | 1.9E-06 | 3.7E-06 | 1.3E-06 | 2.4E-06 | 0.81 |
| 9 | 5.6E-07 | 4.9E-07 | 8.0E-07 | 9.0E-07 | 0.63 |
| 10 | 3.8E-07 | 3.3E-07 | 2.0E-06 | 6.2E-06 | 0.007 |
| 11 | 3.4E-07 | 4.9E-07 | 9.5E-07 | 2.0E-06 | 0.02 |
| 12 | 3.5E-07 | 3.5E-07 | 7.3E-07 | 7.7E-07 | 0.03 |
| 13 | 5.4E-07 | 5.7E-07 | 1.0E-06 | 1.2E-06 | 0.06 |
| 14 | 1.2E-06 | 2.2E-06 | 9.6E-07 | 8.4E-07 | 0.27 |
| 15 | 7.9E-07 | 6.2E-07 | 5.4E-07 | 4.7E-07 | 0.35 |
| 16 | 7.3E-07 | 9.3E-07 | 7.8E-07 | 8.5E-07 | 0.44 |
| 17 | 1.1E-06 | 9.4E-07 | 1.1E-06 | 1.4E-06 | 0.47 |
| 18 | 3.9E-07 | 4.1E-07 | 4.6E-07 | 2.7E-07 | 0.23 |
| 19 | 5.1E-07 | 5.4E-07 | 1.0E-06 | 6.3E-07 | 0.04 |
| 20 | 3.4E-07 | 8.5E-08 | 8.0E-07 | 4.8E-07 | 0.04 |
| 21 | 3.9E-07 | 6.4E-08 | 1.2E-06 | 1.4E-06 | 0.99 |
| 22 | 2.8E-07 | 2.3E-07 | 6.6E-07 | 1.1E-06 | 0.39 |
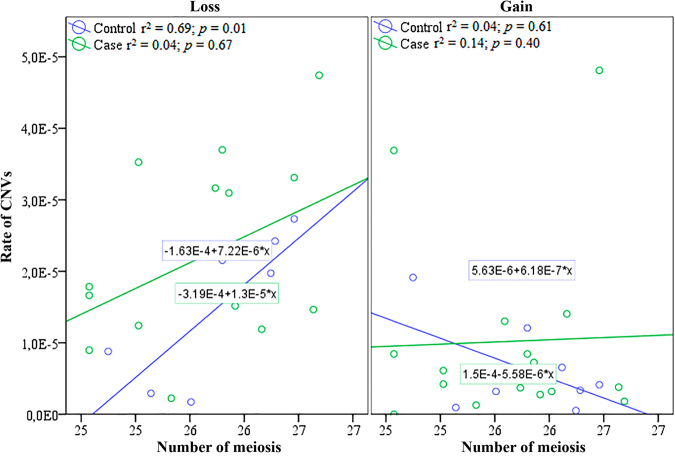
Spearman’s correlation test was applied to the number of paternal meiosis at the time of conception and the burden of CNV germline mutations in the progenies of case control population (Fig. 6). For the exposed pupation, the test was performed using only the fathers exposed to IR from 137Cs. Positive and statistically significant association was observed for genomic losses in the control group. However due to the paternal exposure, the significance was lost for genomic gains in the exposed population. Microduplications were not statistically significant for neither case nor control populations.
Figure 6.
Spearman’s correlation test applied to the number of paternal meiosis at the time of conception and the burden of CNV germline mutation in F1 of controls and cases, corresponding to children born to fathers accidentally exposed to low doses of ionizing radiation of Caesium-137 in Goiânia (Brazil).
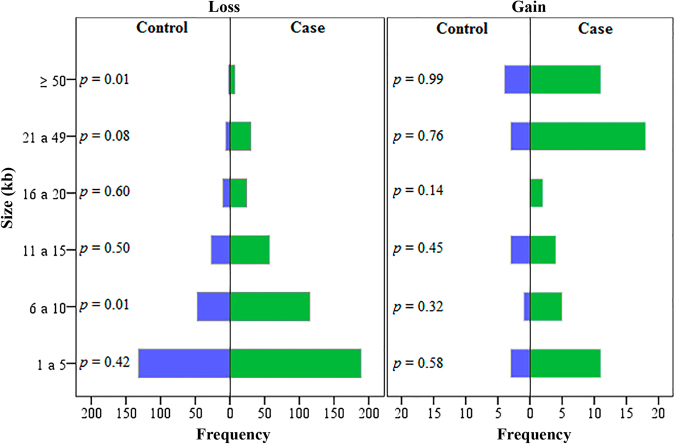
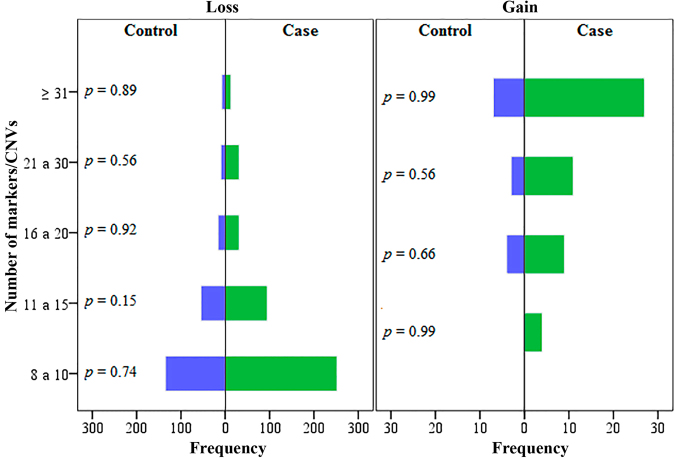
In order to test for the differences of the frequency of CNV size (kb) and the number of markers/CNV for both genomic losses and gains, the χ2 post-hoc test was used. CNV sizes for genomic losses ranging from 6 to 10 kb and ≥50 kb from cases were statistically different from the control group. On the other hand, no differences were observed for the genomic gains with respect to their size (Fig. 7). Similarly, there were no statically significant differences for the number of markers/CNV between cases and controls (Fig. 8).
Figure 7.
Frequency distribution of different classes of CNV sizes (kb) between controls and exposed groups for both genomic losses and gains, in the study of the effect of ionizing radiation in the induction of germline mutations CNV.
Figure 8.
The χ2 post-hoc test showed no statistical differences in the frequency distribution of the classes of CNV according to the number of markers between cases and controls in the study of the effect of ionizing radiation in the induction of germline mutations CNV.
Discussion
The vast majority of studies about CNVs in humans are focused on DNA segments ≥500 kb27,28. To the best of our knowledge, the current study is the first to report the use of genomic gains and losses of small CNVs (≤500 kb) to estimate de novo human germline mutation rates in the offspring of a parental population accidentally exposed to ionizing radiation of caesium-137. In the current study, the number of mutational events weren’t statistically different between cases and controls. However, when, the data were transformed to the proportion of genomic gain or loss for an individual genome as a function of the total size of CNVs, the difference emerged. Thus, given the potential deleterious effect of acute whole-body external exposure to IR on the germ cell lines, the genome of children born to parents exposed to low doses rates of 137Cs gamma radiation exhibited about 3x more of global genomic loss and gain when compare to the offspring born from the control population (Table 3).
There is a considerable worldwide effort to estimate the mutation rates of CNVs in humans. Initial studies estimated CNV mutation rate at 2.5 × 10−6 per locus per generation29,30. Whole genome analysis for 400 parent-child trios detecting CNVs ≥100 kb reported a de novo mutation rate of 1.2 × 10−2 variants per generation27. A higher mutation rate of 3.6 × 10−2 CNVs per generation was observed in individuals with intellectual disabilities31. In the current study, CNV mutation rates for both gains and losses were defined as burden. The burden of CNV mutations in the germ cells was 2.2 × 10−5 and 3.2 × 10−5 for control and exposed populations, respectively, thus, increasing 1.5x the mutation burden in the offspring born to parents exposed to ionizing radiation when compared to unexposed controls. To date, there are no reports on the rate of CNV germline mutations due to population exposure to IR in humans. Nevertheless, our findings are in agreement with the CNV mutation rate reported by Campbell & Eichler30 in their extensive review of the properties and rates of germline mutations in humans. For advanced understanding on the genome-wide effects of ionizing radiation on mutation induction in the mammalian germline the authors suggest the work of Adewoye et al.11. With respect to the transmission of de novo CNVs based on parental exposure, we report the highest rate was observed for exposed fathers and mothers when compared to the controls. Moreover, the paternal contribution was higher than the maternal contribution to the burden of CNV in their offspring within the group of exposed parents.
It has been well established that mutation rates differ within in between families32,33. Reports on comparing de novo mutations in sibs of multisibling families in which one sib was conceived prior to a parent being exposed and the other after has ben helpful to understand how the environment interacts with the inherited genome. For both human and animal models exposed to radiation, most studies reported the post exposure offspring had higher significantly germline mutation rates when compared with pre-exposure offspring34. On the other hand, epidemiologic studies have failed to demonstrate a significantly increased risk of genetic defects or congenital malformations in children born to parents treated with radiotherapy35. Curiously, in the current study, for one family (F09) we had siblings conceived before (F09-3F) and after (F09-2F) both parents were accidentally exposed to IR. Interestingly, the child born after the parental exposure showed fewer CNV loses and gains (Table 1) despite being born 5 years after her brother. Moreover, this child has the smallest mutation rate in the entire cohort. Although, nothing could be concluded based on a single observation, our finding warrants the need for further evaluation of the potential effect of genome and environmental interactions. Thus, the discussion about the impact of human exposure to low doses of IR and its consequence on the germline mutation rate in the offspring conceived after parental exposure is still far from over.
The variables namely number of paternal meiosis, mean parental age and number of markers distributed in the CNVs, were the strongest predictors to contribute to the burden of CVN mutations in the offspring and allowed the discrimination between exposed and unexposed groups. Regarding the recurrence of CNVs, no hotspots in the chromosomes were identified, confirming that induced CVNs fit the random-effect model of radiation exposure. This observation was expected due to the random deposition of radiation energy on biological systems. Here, we report that the mutation rate of genomic losses was greater than gains and the effect of paternal age was better discriminated when analysing the frequency of losses. This observation is supported by previous reports about the increase of mutations frequencies and rates in relation to the advancing age of fathers36.
The common knowledge that most radiation-induced mutations are deletions is nothing new. Genetic analysis studies of specific loci reported that radiation exposure of mouse germ cells induces deletions of different sizes, more frequently observed when animals have incurred high absorbed doses of radiation37. However, a central question remains about the induction of small deletions following human germ cells exposure to low absorbed doses of ionizing ration38–41. Despite the vast amount of information accumulated and the technological advances in the field, the knowledge about the mechanism of DNA repair associated with DSBs and SSBs subjacent to the induction of large and small deletions by ionizing radiation in human models have been consolidated37,41–45.
NAHR and MMBIR are considered as responsible for generating deletions of varying sizes, which is consistent with independent data on genomic disorders associated with recurrent deletions of different lengths (mostly by NAHR) and other disorders associated with non-recurrent and complex rearrangements (mostly by MMBIR). However, with respect to NHEJ and MMEJ, the situation is different. Data on structural variation in the human genome suggest that a proportion of deletions of smaller sizes (≤100 kb) could result from NHEJ/MMEJ. On the other hand, mutations induced in mouse germ cells, following irradiation at G0/G1 phases, mostly derived from the repair of DSBs, predominantly via NHEJ, resulted in deletions of different sizes, some on the megabase scale. Although this discrepancy remains to be explained, our CNV dataset was largely comprised of structural variants without a flanking segmental duplication, 423/473 and 222/238 for case and control groups, respectively. The data favoured NHEJ and MMBIR as the two potential DNA repair mechanisms underlying the recovery of DSBs following human exposure to very low doses of ionizing radiation. Both mechanisms can mitotically generate genome rearrangements ranging from several megabases to a few hundred base pairs in size. Our findings were in agreement with37,46,47.
On the other hand, data from the present study indicated LCRs (SD) flanked 10.6% and 6.7% of the CNVs observed in the exposed and control groups, respectively. There was an increase of approximately 3 × in the fraction of induced CNVs harbouring SD among the children born to exposed parents, suggesting CNVs could also rise from NAHR, a potential mechanism for repairing DSBs. However, NAHR was not found to be the most relevant repair mechanism to fix DNA damage induced by low radiations doses in our cohort. Our findings are in agreement with48, who described the role of mutagenic agents in the formation DSB in the DNA, which in turn will lead to an increase in non-recurrent CNVs but not an increase in the occurrence rate of NAHR in the cells.
Arlt et al.15 used SNP array methodology to detected CNVs ranging in size from 2.7 kb to 34.2 Mb in fibroblasts exposed to IR. The authors reported that 1.5 to 3.0 Gy of ionizing radiation significantly induced somatic de novo CNVs in cultured human fibroblast cells with no difference in size distribution. In that study, the frequency of new CNVs increased in a dose-dependant manner and duplications were more frequently found. The results of the current study, reporting on germline mutations, were in disagreement with the aforementioned study as genomic losses were more frequent than genomic gais in both groups. On the other hand, our results were concordant with the report by11, who analyzed immediate and long-term effects of ionizing radiation on germline mutation in mice and found a significant increase in the frequency of CNVs in the offspring of exposed male mice. Animals incurred an absorbed dose of 0.45 Gy, generating 14 unique germline de novo CNV mutations, 12 were deletions and 2 were duplications.
Most germline mutations are paternally inherited due to the continuous nature of cell divisions in spermatogenesis throughout mitotic maintenance of spermatogonia in males as opposed to a few dozens cell divisions to form an oocyte in females49. Several studies have confirmed that the number of de novo mutations increases with paternal age. The linear relationship observed between paternal age and the number of mutations in the progeny is most often explained as a result of increased cell division, replication and DNA repair errors40,50–54. The current study also tested the hypothesis that males may contribute more to the burden of de novo CNVs in their offspring. We also found a positive relationship between the number of paternal meiosis at conception and de novo germline mutation rate in the progeny born to parents accidentally exposed to very low doses of ionising radiation in Goiânia (Brazil). We also confirmed that increased paternal age correlates positively to increased frequency of losses in the genomes of the offspring of both control and exposed groups. Among the cases, the extensive inter-individual variation observed in the rate of genomic losses obscured the effect of parental age. This finding may be explained by the individual dose of radiation absorbed by the fathers during the accident, which caused the induction of de novo CNVs in the offspring, confounding the effect of age of the exposed fathers on the observed mutation rate. In the current study, this finding in the control group is consistent with historical evidence that older men contribute more mutations to their children.
There has been a long-standing interest in the area of radiation protection and mutagenicity to declare ionizing radiation as a human germ cell mutagen. The current work was aimed to give some insights regarding the transmission of mutations through the germ line of the irradiated parents and we found it feasible to claim low doses of IR as a potent germline mutagen in humans.
Conclusions
High density SNP array technology proved useful to retrospectively estimate the germline mutation rate in CNV in the offspring of a population accidentally exposed to very low doses of ionizing radiation. In the context of radiological protection, our data suggest that even doses of IR below 0.2 Gy incurred during whole-body exposure at lower dose rates could prove harmful by increasing the de novo mutation rate in the offspring. Thus, understating the genetic and biological consequences of exposure to IR has remained a long-standing goal for human populations, especially in the germ cell lines as the accumulation of additional mutations in the offspring of irradiated parents adds to the genetic risk for that particular population.The transgenerational effect of parental exposure to ionizing radiation has been well documented in animal models, focused on relatively large radiation doses and the conclusions have been extrapolated to humans55. However, little data on germline mutations has been generated on humans exposed to low absorbed doses of ionizing radiation. The mutation rate of de novo CNV, especially genomic losses, was proven to be a good biomarker of parental exposure to retrospectively study human populations exposed to low absorbed doses of IR as reported here.
Materials and Methods
Sampling Groups
The exposed group was comprised of 12 families of which at least one parent was directly exposed to ionizing radiation (IR) of Caesium-137. This group included 40 people: 12 mothers, 12 fathers, and 16 children conceived after the accident. All those exposed people incurred absorbed doses of ≤0.2 Gy, mostly due to whole-body external exposure to gamma IR. For extensive review on dose estimates for the Goiânia population, please refer to IAEA’s report on the accident2. The cut-off at 0, 2 Gy was selected as the lowest absorbed dose detected by traditionally used genetic biomarkers, such as micronuclei and chromosome aberrations, as the biological effect of very low doses of IR has remained unclear and inconsistent in human populations. Moreover, discrepant information has been generated regarding the induction of germline mutations in the offspring of exposed parents. Additionally, ≤0.2 Gy included the smallest absorbed doses recorded for the Goiânia population based on biodosimetry using dicentric chromosomes. A group of people with no history of occupational, accidental or medical exposure to IR participated as a control population from Goiânia. The control group comprised of 8 trios (24 people), corresponding to a mother, a father, and one biological child from the Goiânia population who voluntarily agreed to participate in the study. For both cases and controls, biological paternity and maternity were confirmed to an index of 99.99%.
A total of 10 mL of peripheral blood was withdrawn from all participants who voluntarily agreed to take part in the study. DNA was isolated from each sample and stored at −20 °C. Participants answered a lifestyle questionnaire and signed an informed consent from according to the national ethical guidelines for research with humans. All participants, including mothers, fathers, and children (for minors consent was signed by the legal guardian) voluntarily signed the informed consent forms approved by the Ethics Committee on Human Research from the Pontifical Catholic University of Goiás (CEP-PUC Goiás), kept under the registration numbers CAAE-205000.073183/2004-45, and CAAE-49338615.2.0000.0037/2016. The study was carried out at Núcleo de Pesquisas Replicon from PUC Goiás having all methods and procedures carried out in accordance with relevant international guidelines and regulations. Moreover, the CEP-PUC Goiás also approved the experimental protocols.
DNA Extraction, Isolation, and Quantification
DNA extraction and isolation was carried out using DNA Illustra Blood GenomicPrep® Mini Kit (GE Healthcare, USA) following the instructions provided by the manufacturer. DNA concentration was spectrophotometrically estimated in a NanoVue® Plus (GE Healthcare Life Sciences, United Kingdom).
Chromosomal Microarray Analysis (CMA)
CMA was performed with the GeneChip® CytoScanHDTM (Affymetrix, USA) following the manufacturer’s recommendations without modifications. Capture and digitalization of fluorescent signals were done with the aid of the GeneChip® Command Console® software (AGCC®, Affymetrix, USA). Chips that met the quality control metrics, namely Median Absolute Pairwise Difference (MAPD) e SNP-QC, with parameters set at de MAPD ≤ 0.25 e de SNP-QC ≥ 15, had their data included in the analyses.
Chromosomal analyses were performed using ChAS® (Affymetrix, USA). Filters were set at 8 and 15 SNP markers for microdeletion and microduplication calling, respectively. A de novo CNV was considered if the mean distribution of markers was ≤2 kb, CNV’s size were set as ≥1 kb, and median log2 ratio ±0.40. Whenever a CNV was found with a chance to be imputed, based on the distance of its markers, that segment was manually cut to adjust the area around the markers, following the rules for marker counts and their mean distribution along the DNA segment. In the current study, only de novo CNVs were scored. Sex chromosomes were not included in the analysis. Using the mendelian error rate and the graph table from ChAS® (Affymetrix, USA), the parent of origin (POR) for each CNV was assessed.
Germline Mutation Rate Estimates (MRcNV)
The rate of de novo CNV was estimated taking into consideration the formation of CNV/locus/generation. The MRCNV was estimated applying the following equation 1 (6, 7):
| 1 |
where:
ΣTCNV: Total size o f CNVs (bp)
ε!: Number of meiosis (2)
a: Bialelic locus (2)
shg: Size of haploid genome (2.9 × 109), according to the genome build GRCh37/hg19.
Equation 1 was applied to both CNVp (loss) and CNVg (gain). The MRCNV corresponded to the total mutation rate, referred to in the text as CNV burden, which pertains to the sum of MRCVN of gain and MRCNV of losses in a given genome.
In order to estimate the number of paternal meiosis for both cases and controls, we followed Equation 2, based on the age of father at the time of child conception:
| 2 |
where:
No. ρ!: Number of paternal meiosis
PA: Paternal age (years)
m: Months/year
dp: Duration of pregnancy (months)
A: Average age for starting sperm production in humans [12 year-old] (months)
d/m: Total days in a month.
tρ!d: Average of the total number of meiosis per day in human males (~25 × 106).
Segmental Duplication Flanking CNVs
The occurrence of segmental duplications (SD) around the annotated CNVs was carried out doing active search in the databank from UCSC Genome Browser (http://www.genome.ucsc.edu). In order to associate a CNV to a SD we considered all surrounding DNA sequences up to 100x the size of a particular CNV. SD flanking a CNV must have met the criteria of similarity ≥90%.
Statistical Analysis
The non-parametric test of Mann-Whitney U was applied to estimate the differences in mutation frequencies, size of CNVs, MRCNV/chromosome, number of markers and genes for losses, gains and burden, when comparing cases and controls. MRCNV did not follow a normal distribution according to the Shapiro-Wilk’s test. Spearman’s rank correlation coefficient was applied to analyse the effect of paternal meiosis at conception on the burden of CNV mutation. The binomial test was used to test the differences between cases and controls with respect to the frequencies of CNVloss and CNVgain, and the frequency of flanking SD in CNVs. Discriminant function analysis was used to predict the contribution of the variables (predictors): number of paternal meiosis, frequency of CNVloss and CNVgain, mean parental age at conception, number of markers in a CNV, number of genes in a CNV, and size of CNVs (kb) in discriminating cases and controls. In these cases, exposed and control groups were used as the categorical dependent variables. We took into consideration the 3 predictors that had the largest contribution to discriminate the groups in order to calculate the coefficient for the discriminating function. The post-hoc χ2 test was used to examine the differences in the frequency of CNVs classified in categories according to their size and number of SNP markers. Also, Chi square was used to test for the distribution in CNVs in chromosomes in order to validate potential hotspots for both genomic losses and genomic gains for case and control groups. In order to compare the contribution of the parental exposure and the parent of origin (POR) for the CNVs on the burden of mutations found in the current study, Two-way ANOVA followed by Tukey’s test a posteriori were performed. All statistical analyses were performed using SPSS® 23.0 at a level of significance of 5% (p < 0.05).
Data availability Statement
All relevant data are within the paper.
Acknowledgements
The authors wish to thank the voluntarily participation of the patients. This study was supported by FAPEG-GO (Fundação de Amparo à Pesquisa do Estado de Goiás), ExeGenS (Rede de Excelência em Genética e Genética Molecular Aplicada à Saúde Humana), Laboratório de Citogenética Humana e Genética molecular (LaGene/SES-GO), Núcleo de Pesquisas Replicon, Pontifícia Universidade Católica de Goiás (PUC-GO), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Capes (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). E.O.A.C., D.M.S., I.P.P., R.W.P. and A.D.C. wish to thank for their scholarships and fellowships. The author also express their gratitude to Mr. Sean M. Quail for his contribution in proofreading the draft. Moreover, we thank Dr. Nicole de Leeuw from the Radboud University Medical Center for her valuable assistance in assessing the parent of origin of a CNV using ChAS® (Affymetrix, USA). Lastly, we thank Mr. Hugo Leite Filho for assisting with bioinformatics and scripts writing. This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), with the number 407948/2016-3 and Fundação de Amparo à Pesquisa do Estado de Goiás. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
E.O.A.C., A.D.C., R.W.P., C.C.S., D.M.S. and I.P.P. designed the study. E.O.A.C., I.P.P. and L.G.O. performed the experiments. E.O.A.C., I.P.P., A.S.C., M.W.G., A.D.C. analyzed data and performed the statistical analysis. J.F.D.S. performed sample and data collection. E.O.A.C., I.P.P., A.D.C. and R.W.P. contributed to manuscript writing. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.da Cruz AD, et al. Radiation risk estimation in human populations: lessons from the radiological accident in Brazil. Mutat Res. 1997;373:207–214. doi: 10.1016/S0027-5107(96)00199-6. [DOI] [PubMed] [Google Scholar]
- 2.IAEA. Cytogenetic Dosimetry Applications in Preparedness for and Response to Radiation Emergencies. International Atomic Energy Agency, 1–247 (2011).
- 3.Straume T, et al. Novel biodosimetry methods applied to victims of the Goiânia accident. Health Phys. 1991;60:71–76. doi: 10.1097/00004032-199101000-00011. [DOI] [PubMed] [Google Scholar]
- 4.da Cruz AD, McArthur AG, Silva CC, Curado MP, Glickman BW. Human micronucleus counts are correlated with age, smoking, and cesium-137 dose in the Goiânia (Brazil) radiological accident. Mutat Res. 1994;313:57–68. doi: 10.1016/0165-1161(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 5.da Cruz AD, Curry J, Curado MP, Glickman BW. Monitoring hprt Mutant Frequency Over Time in T-Lymphocytes of People Accidentally Exposed to High Doses of Ionizing Radiation. Environ Mol Mutagen. 1996;27:165–175. doi: 10.1002/(SICI)1098-2280(1996)27:3<165::AID-EM1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 6.da Cruz AD, et al. Microsatellite mutations in the offspring of irradiated parents 19 years after the Cesium-137 accident. Mutat Res. 2008;652:175–179. doi: 10.1016/j.mrgentox.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Costa EOA, et al. The effect of low-dose exposure on germline microsatellite mutation rates in humans accidentally exposed to caesium-137 in Goiânia. Mutagenesis. 2008;26:651–655. doi: 10.1093/mutage/ger028. [DOI] [PubMed] [Google Scholar]
- 8.Nunes HF, et al. Assessment of BCL2/J(H) translocation in healthy individuals exposed to low-level radiation of 137CsCl in Goiânia, Goiás, Brazil. Genet Mol Res. 2013;12:28–36. doi: 10.4238/2013.January.16.6. [DOI] [PubMed] [Google Scholar]
- 9.Cruvinel JAA, et al. Avaliação de Auto-Anticorpos em Células HEP-2 nos Indivíduos Expostos ao 137Cs no Acidente Radioativo de Goiânia. Estudos (Goiânia Online) 2014;41:603–613. [Google Scholar]
- 10.Asakawa J, Kodaira M, Cullings HM, Katayama H, Nakamura N. The genetic risk in mice from radiation: an estimate of the mutation induction rate per genome. Radiat Res. 2013;179:293–303. doi: 10.1667/RR3095.1. [DOI] [PubMed] [Google Scholar]
- 11.Adewoye AB, Lindsay SJ, Dubrova YE, Hurles ME. The genome-wide effects of ionizing radiation on mutation induction in the mammalian germline. Nat Commun. 2015;26:1–8. doi: 10.1038/ncomms7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu XY, et al. Genomic imbalances in neonates with birth defects: high detection rates by using chromosomal microarray analysis. Pediatrics. 2008;122:1310–1318. doi: 10.1542/peds.2008-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong SW, et al. Peripheral blood gene expression signature differentiates children with autism from unaffected siblings. Neurogenetics. 2013;14:143–152. doi: 10.1007/s10048-013-0363-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller DT, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. Copy number variants are produced in response to low-dose ionizing radiation in cultured cells. Environ Mol Mutagen. 2014;55:103–113. doi: 10.1002/em.21840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:986–992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 18.Hu G, et al. Copy number variations in 119 Chinese children with idiopathic short stature identified by the custom genome-wide microarray. Mol Cytogenet. 2016;9:2–7. doi: 10.1186/s13039-016-0214-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dauber A, et al. Genome-wide association of copy-number variation reveals an association between short stature and the presence of low-frequency genomic deletions. Am J Hum Genet. 2011;89:751–759. doi: 10.1016/j.ajhg.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arlt MF, Wilson TE, Glover TW. Replication stress and mechanisms of CNV formation. Curr Opin Genet Dev. 2012;22:204–210. doi: 10.1016/j.gde.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci USA. 2011;108:17360–17365. doi: 10.1073/pnas.1109272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rana S, Kumar R, Sultana S, Sharma RK. Radiation-induced biomarkers for the detection and assessment of absorbed radiation doses. J Pharm Bioallied Sci. 2010;2:189–196. doi: 10.4103/0975-7406.68500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernot E, et al. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat Res. 2012;751:258–286. doi: 10.1016/j.mrrev.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Itsara A, et al. Population Analysis of Large Copy Number Variants and Hotspots of Human Genetic Disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itsara A, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ku CS, Vasiliou V, Cooper DN. A new era in the discovery of de novo mutations underlying human genetic disease. Hum Genomics. 2012;6:1–4. doi: 10.1186/1479-7364-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner DJ, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends Genet. 2013;29:575–584. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hehir-Kwa JY, et al. De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J. Med. Genet. 2011;48:776–778. doi: 10.1136/jmedgenet-2011-100147. [DOI] [PubMed] [Google Scholar]
- 32.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahbari R, et al. Timing, rates and spectra of human germline mutation. Nat Genet. 2016;48:126–133. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kryukov GV, et al. Genetic Effect of Chemotherapy Exposure in Children of Testicular Cancer Survivors. Clin Cancer Res. 2016;22:2183–2189. doi: 10.1158/1078-0432.CCR-15-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Signorello LB, et al. Stillbirth and neonatal death in relation to radiation exposure before conception: a retrospective cohort study. Lancet. 2010;376:624–630. doi: 10.1016/S0140-6736(10)60752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnaswamy S, Subramaniam K, Ramachandran P, Indran T, Abdul AJ. Delayed fathering and risk of mental disorders in adult offspring. Early Hum Dev. 2011;87:171–175. doi: 10.1016/j.earlhumdev.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Sankaranarayanan K, Taleei R, Rahmanian S, Nikjoo H. Ionizing radiation and genetic risks. XVII. Formation mechanisms underlying naturally occurring DNA deletions in the human genome and their potential relevance for bridging the gap between induced DNA double-strand breaks and deletions in irradiated germ cells. Mutat Res. 2013;753:114–130. doi: 10.1016/j.mrrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Sankaranarayanan K. Ionizing radiation and genetic risks. II. Nature of radiation-induced mutations in experimental mammalian in vivo systems. Mutat Res. 1991;258:51–73. doi: 10.1016/0165-1110(91)90028-T. [DOI] [PubMed] [Google Scholar]
- 39.Cattanach, B. M., Evans, E. P., Burtenshaw, M. D., Rasberry, C. Incidence and distribution of radiation-induced large deletions in the mouse. Proceedings of the 10th International Congress of Radiation Research. Radiation Research, 531–534 (1996).
- 40.Sankaranarayanan K. Ionizing radiation and genetic risks. X. The potential “disease phenotypes” of radiation-induced genetic damage in humans: perspectives from human molecular biology and radiation genetics. Mutat Res. 1999;429:45–83. doi: 10.1016/S0027-5107(99)00100-1. [DOI] [PubMed] [Google Scholar]
- 41.Sankaranarayanan K, Nikjoo H. Ionising radiation and genetic risks. XVI. A genome-based framework for risk estimation in the light of recent advances in genome research. Int J Radiat Biol. 2011;87:161–178. doi: 10.3109/09553002.2010.518214. [DOI] [PubMed] [Google Scholar]
- 42.Taleei R, Weinfeld M, Nikjoo H. A kinetic model of single-strand annealing for the repair of DNA double-strand breaks. Radiat Prot Dosimetry. 2011;143:191–195. doi: 10.1093/rpd/ncq535. [DOI] [PubMed] [Google Scholar]
- 43.Taleei R, Nikjoo H. Repair of the double-strand breaks induced by low energy electrons: a modelling approach. Int J Radiat Biol. 2012;88:948–953. doi: 10.3109/09553002.2012.695098. [DOI] [PubMed] [Google Scholar]
- 44.Taleei R, Nikjoo H. The non-homologous end-joining (NHEJ) pathway for the repair of DNA double-strand breaks: I. A mathematical model. Radiat Res. 2013;179:530–539. doi: 10.1667/RR3123.1. [DOI] [PubMed] [Google Scholar]
- 45.Taleei R, Girard PM, Sankaranarayanan K, Nikjoo H. The non-homologous end-joining (NHEJ) mathematical model for the repair of double-strand breaks: II. Application to damage induced by ultrasoft X rays and low-energy electrons. Radiat Res. 2013;179:540–548. doi: 10.1667/RR3124.1. [DOI] [PubMed] [Google Scholar]
- 46.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harper JV, Anderson JA, O’Neill P. Radiation induced DNA DSBs: Contribution from stalled replication forks? DNA Repair (Amst). 2010;9:907–913. doi: 10.1016/j.dnarep.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Conover HN, Argueso JL. Contrasting mechanisms of de novo copy number mutagenesis suggest the existence of different classes of environmental copy number mutagens. Environ Mol Mutagen. 2016;57:3–9. doi: 10.1002/em.21967. [DOI] [PubMed] [Google Scholar]
- 49.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat. Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 50.Thomas NS, et al. De novo apparently balanced translocations in man are predominantly paternal in origin and associated with a significant increase in paternal age. J Med Genet. 2010;47:112–115. doi: 10.1136/jmg.2009.069716. [DOI] [PubMed] [Google Scholar]
- 51.Flatscher-Bader T, et al. Increased de novo copy number variants in the offspring of older males. Transl Psychiatry. 2011;1:1–8. doi: 10.1038/tp.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buizer-Voskamp JE, et al. Increased paternal age and the influence on burden of genomic copy number variation in the general population. Hum Genet. 2013;132:443–450. doi: 10.1007/s00439-012-1261-4. [DOI] [PubMed] [Google Scholar]
- 55.Dubrova YE, Plumb M, Gutierrez B, Boulton E, Jeffreys AJ. Transgenerational mutation by radiation. Nature. 2000;405:37. doi: 10.1038/35011135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.