Abstract
A crucial stage in the origin of life was the emergence of the first molecular entity that was able to replicate, transmit information, and evolve on the early Earth. The amyloid world hypothesis posits that in the pre-RNA era, information processing was based on catalytic amyloids. The self-assembly of short peptides into β-sheet amyloid conformers leads to extraordinary structural stability and novel multifunctionality that cannot be achieved by the corresponding nonaggregated peptides. The new functions include self-replication, catalytic activities, and information transfer. The environmentally sensitive template-assisted replication cycles generate a variety of amyloid polymorphs on which evolutive forces can act, and the fibrillar assemblies can serve as scaffolds for the amyloids themselves and for ribonucleotides proteins and lipids. The role of amyloid in the putative transition process from an amyloid world to an amyloid–RNA–protein world is not limited to scaffolding and protection: the interactions between amyloid, RNA, and protein are both complex and cooperative, and the amyloid assemblages can function as protometabolic entities catalyzing the formation of simple metabolite precursors. The emergence of a pristine amyloid-based in-put sensitive, chiroselective, and error correcting information-processing system, and the evolvement of mutualistic networks were, arguably, of essential importance in the dynamic processes that led to increased complexity, organization, compartmentalization, and, eventually, the origin of life.
Keywords: Origin-of-life theory, Amyloid world, RNA world, Prion, Molecular evolution, Primordial genetics
Introduction
It is generally believed that life on Earth passed through an RNA-world era in which RNA or an RNA-like polymer performed both informational and catalytic functions [1–4]. It is, however, unlikely that a functional ribonucleotide polymer could have existed under early Earth conditions [5–7]. The amyloid world hypothesis of the origin of life [8] posits that during that early period, about 4 billion years ago, peptide amyloids were the first molecular entities that were able to self-replicate, transmit information, and evolve. The model has gained momentum from recent empirical and theoretical studies clarifying the molecular mechanisms underlying amyloid formation, amyloid-related catalysis, and protein-encoded information processing. Here, I examine the model in light of recent advancements, develop the model further, and broaden the perspective to include an outline how a transition from a primitive β-sheet-based replicator world to a complex amyloid–ribonucleo-protein world might have occurred.
The amyloid replicator
Within the framework of origin-of-life research, the question of the mechanism of replication and information transfer is crucial. Given the stability and functionality problems connected with the RNA-world hypothesis [5–7, 9], the view that some other type of informational system probably preceded the RNA-based one has gained an increasing attention. According to the amyloid world hypothesis, the primordial information system was based on structurally stable catalytic and self-replicating β-sheet amyloid conformers [8, 10, 11]. The basis for the model is the conformational arrangement of the amyloid fold (Fig. 1). The cross-β sheet structure of amyloid, in addition to providing remarkable stability, can convey multifunctionality to peptides [12–26]. Even very short peptides may express diverse catalytic, replicative, and informational properties when adopting the amyloid conformation. This is in contrast to native peptides, which are easily denatured under harsh conditions, and whose functionality requires longer peptide sequences, the synthesis of which, again, would require an existing metabolic apparatus. Thus, under early Earth conditions, the amyloid fold would, obviously, have provided a substantial advantage for the survival and propagation of prebiotic peptides.
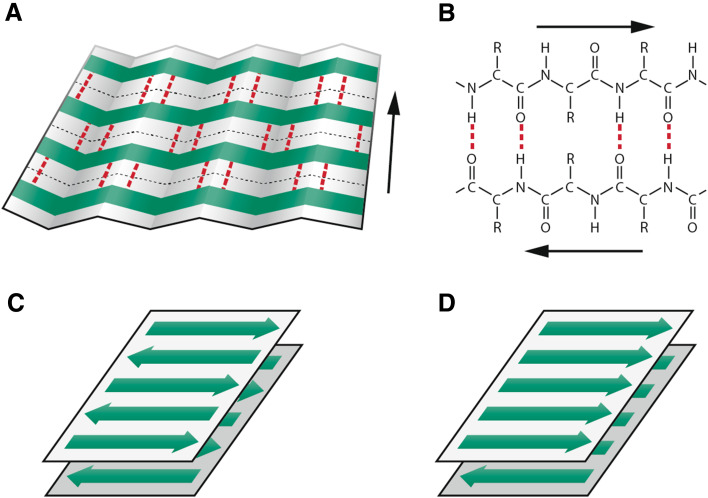
Fig. 1.
Schematic representation of the β-sheet structure of amyloid. a Section of a β-pleated sheet. The β-strands, which run perpendicular to the long axis of the fibril, are marked in green and interstrand hydrogen bonds in red. b Hydrogen-bonding pattern of two antiparallel β-strands. c Antiparallel bilayered β-sheet. d Parallel bilayered β-sheet. Typically, the repeating unit of the amyloid fibrils consists of two tightly packed layers of β-sheets with side chains within the bilayers forming a dry interdigitating zipper interface. The zippers differ in the organization of the β-strands within and between the β-sheets and in the stacking of the β-sheets enabling the formation of a diversity of structural variants. The cross β-structure gives rise to a characteristic X-ray diffraction pattern with a meridional reflection at 0.48 nm and an equatorial reflection at about 1.0 nm. These reflections correspond to the interstrand and intersheet spacings, respectively. The mature amyloid fibril is a highly ordered linear supramolecular structure forming long unbranched fibrils ranging from 5 to 12 nm in diameter. References are given in the text
Encrypting environmental information
Recent research has clarified many aspects of the molecular mechanisms underlying the amyloid-mediated information system. It has become evident that the coding element is the steric zipper structure of the amyloid motif and that recognition occurs by amino acid side chain complementarity [13, 18]. In the encryption process, environmental information is encoded in the three-dimensional structure of the amyloid conformer [27, 28]. The steric information can then be transferred to “daughter” molecular entities through the template-assisted conformational replication cycles generating replicas of the spatially altered amyloid conformer [11]. Fragmentation and the formation of new seeds characterize the primary replication process; the importance of fragment recycling in the prebiotic evolutive processes has been emphasized [29]. The oligomeric phase of the replication process appears to be essential for the inscription of environmental information.
A schematic representation of the replication cycles is shown in Fig. 2. Importantly, two different nucleation mechanisms exist. A primary one, that typically involves a short seed, and a secondary, fibril surface-catalyzed step, which occurs when a small, but critical amyloid concentration has been achieved [30–33]. The fibril-catalyzed step is characterized by an initial fast docking phase followed by a slow structural rearrangement locking phase [34]. From a prebiotic perspective, the demonstrations of template-assisted ligation of fibrillogenic peptides from two shorter building blocks [23–25] and of amyloid-directed synthesis of its constituent peptides from amino acids [26] are important.
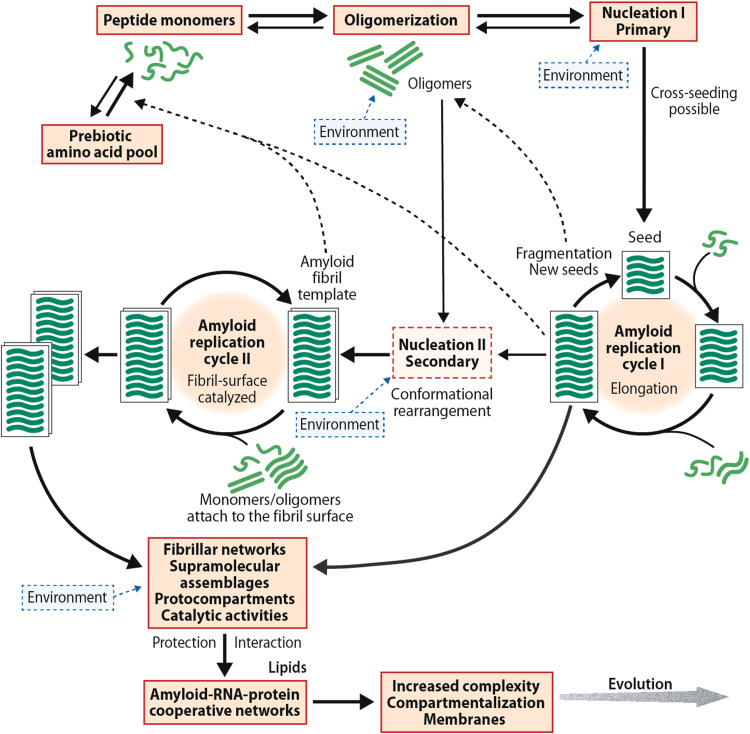
Fig. 2.
Schematic representation of the amyloid model of the origin of life. The figure outlines the proposed pathway of one type of peptide monomer from a prebiotic mixture of various protopeptides. The nucleation-dependent replication system is in-put sensitive, chiroselective, and error correcting. An initial slow nucleation process is followed by a fast polymerization phase where peptide monomers are added the growing end of the protofilament. Fragmentation generates new seeds that can initiate repeated replication cycles. The same peptide monomer can give rise to different amyloid structures and molecular rearrangements are possible. Specific conformational changes can be replicated in the fibril/protofibril-catalyzed cycle II. Amyloid is also able to direct the synthesis of its own constituent peptides. The β-sheet conformers and ribonucleotides interact dynamically and cooperatively, and the amyloid-based supramolecular fibrillar assemblies can function as a primitive metabolic apparatus catalyzing the formation metabolite precursors. The model does not exclude the possibility an extraterrestrial origin of the primordial amino acids or a contribution of extraterrestrial amino acids to the terrestrial prebiotic amino acid pool. References are given in the text
Adaptive and enantioselective amyloids
A distinctive feature of amyloid formation is that the same peptide monomer can generate functionally and structurally different amyloid conformers of which one or several can propagate and make new copies of itself/themselves [35, 36]. This ability is important with respect to the evolvability of the system: evolution requires variation. The replicating system can adapt to even small changes in the external milieu. The environmentally induced fine-tuned changes in the amyloid architecture can then be replicated and the pool of the fittest variants can expand. In this model, the environmentally less suitable conformers are degraded and the released monomers/oligomers are reutilized in the replication cycles [11].
The amyloid polymorphs differ from each other in the architecture of the β-strands within and between the β-sheets and in the stacking of the β-plates, as well as in filament length. The number of protofilaments per fibril and the degree of twisting of the fibril can also vary. The β-sheets are tightly packed without water molecules between them; hydrogen bonds, van der Waals forces, and electrostatic polarization hold the zippers together [13, 18, 37]. Typically, amyloid is found as a homochiral structure which has been explained by a poorer spatial fit of heterochiral structures [38, 39]. The high enantioselectivity of the molecular recognition of β-sheets is likely to be relevant to the question of the origin of biological homochirality [40, 41]. In a prebiotic setting even a minute enantiomeric precursor imbalance [42, 43], if present, could be amplified in the template-directed chiroselective β-sheet self-replication cycles, explaining both the amplification and the chiral transmission steps of terrestrial homochirality.
Catalytic amyloids
The catalytic activity of the peptide-based β-sheet assemblies is an important aspect of the amyloid world model. Amyloids not only catalyze their own formation, but they are able to catalyze other chemical reactions too. Rufo et al. [16] showed that small, 7-residue amyloid-forming peptides form efficient catalysts of ester hydrolysis. Other studies have demonstrated amyloid-related aldolase [17, 19], ATP-ase [44], and carbonic anhydrase [22] activities, as well as copper-mediated oxygen activation [45]. The catalytic functions are fibril/protofibril-dependent: the corresponding nonaggregated peptides are catalytically inactive. Another feature is the metal dependence of several amyloid catalysts; metal ions both stabilize the fibrillar structure and shape ligand geometry. In a recent study, Lee et al. [46] determined the structure of a metalloamyloid esterase catalyst by solid-state NMR. The peptide formed parallel β-sheets that assembled into stacked bilayers with alternating hydrophobic and polar interphases. The hydrophobic interphase was stabilized by apolar side chains, whereas the polar interphase contained zinc-binding histidines. In another recent study, Omosun et al. [19] examined the catalytic activities of peptides assembling into well-defined amyloid nanotubes. They found that the density and proximity of the extended arrays of chiroselective catalytic sites accomplished template-assisted polymerization of new polymers. The depth of the co-linear cross-β grooves of a heptapeptide assembling in antiparallel in-register β-strands was shown to be important for the retro-aldol activity; the shallower grooves limited substrate binding and diminished catalytic activity. Though the total efficiency of the catalytic activities of the cross-β assemblies is, in most cases, only moderate and the catalytic repertoire limited, the amyloid supramolecular network can, in a way, be regarded as a primitive metabolic apparatus with protoenzymatic activities.
The prebiotic relevance of the β-sheet networks and assemblies was recently highlighted by Tena-Solsona et al. [17] who provided evidence for emergent catalytic behavior of self-assembled low-molecular-weight peptide aggregates, and by Nanda et al. [21] who demonstrated error correction within replication networks through the emergence of short polymers exhibiting selective autocatalytic properties. Very recently, Rout et al. [26] showed that an amyloid can direct the sequence-, regio-, and stereo-selective condensation of amino acid synthesis, and that the templating reaction is stable over a wide range of pH (5.6–8.6) and temperature (25–90 °C). This is of particular interest from the perspective of early molecular evolution as it demonstrates that an amyloid formed from short peptides can direct the synthesis of its own constituent peptides under plausible prebiotic Earth conditions.
Emergence of cooperative networks
The amyloid model fulfils key criteria of a valid origin-of-life theory, i.e., the requirements of replication, information transfer, and variation. By repeated replication cycles, the amyloid conformers can generate a variety of polymorphic fibrillar networks (Fig. 3) and structures such as nanotubes, nanospheres, and hydrogels [47]. The fibrillar assemblies can act as scaffolds for the amyloids themselves and for RNA, protein, and lipids [48–55]. They could also, under harsh prebiotic conditions, provide protection for nucleobases and natively folded peptides/proteins. Notably, extant organisms utilize amyloid structures for protection against environmental hazards [56–58] and for driving compartment formation [59].
Fig. 3.
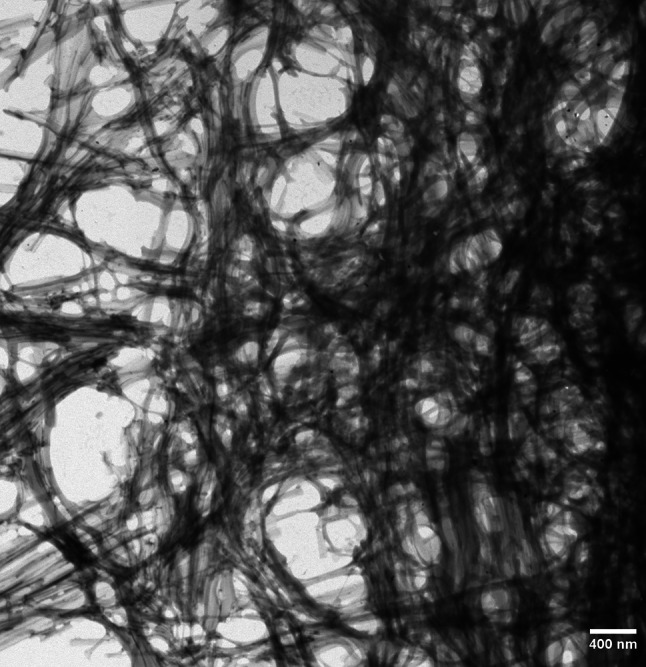
Electron micrograph of a polymorphic fibrillar amyloid network self-assembled from a prebiotically relevant 9-mer peptide (EGGSVVAAD) in aqueous environment. Experimental conditions were as described in Ref. [10]
The role of amyloid in the putative transition of the pristine amyloid world to an amyloid–RNA–protein world is not limited to scaffolding and protection; the interactions of amyloid with ribonucleotides and protein are both cooperative and dynamic. Fibrillar amyloid can act as an auto-catalyzing surface [34, 60–62] and, in addition to promoting its own formation, can, in a similar manner to that of clay and other mineral surfaces [63, 64], bind nucleic acids and enhance their polymerization, which, in turn, can promote amyloid production [15, 65–68]. Moreover, the amyloid–nucleic acid complexes may enhance nucleic acid hybridization [65]. With respect to compartmentalization and membrane formation, amyloid–lipid interactions are of key importance [50, 52, 55, 69, 70]. Amyloidogenesis is promoted by lipid interphases that lead to accelerated fibrillar network formation and the recruitment of both nucleic acids and lipids generating feed-back loops. The complex interactions between amyloids and ribonucleotides are also reflected in current biology: amyloid-forming proteins are overrepresented among the factors that modulate the transcription, translation, and storage of RNA [71], and amyloid-like aggregation has been implicated in the formation of both RNA granules [72] and P-bodies [73]. In certain instances, protein’s amyloidogenic properties and RNA-modulating activity are associated with the occurrence of glutamine/asparagine-rich sequence motifs.
The transition from an amyloid world to an amyloid–RNA–protein world is likely to have evolved over a long period of time. However, the amyloid model [8, 11, 74] is also compatible with a gradually coevolving RNA–protein world [75, 76]. The scaffolding properties, protection, and protocompartments provided by amyloid supramolecular structures in combination with the cooperative interactions and interdependence between amyloid, ribonucleotides, and protein are likely to have constituted a driving force in prebiotic molecular evolution.
The prion connection
Prions are self-propagating infectious protein-based agents that can cause severe neurodegenerative disease. The prion hypothesis postulates that a misfolded form of the prion protein is the causative and transmissible agent of prion disorders [77]. Most, if not all, of the extraordinary characteristics of prions, such as protease-resistance, thermal stability, transmissibility, and strain specificity, are closely related to the amyloid fold [12, 14, 35, 78, 79]. The conversion of the prion protein into the infectious form involves a conformational rearrangement of the protein that generates an amyloid structure, and the propagation of the prions occurs via a template-seeded replication mechanism very similar to that of amyloids in general [12, 78, 80]. In addition to disease-related prions, a number of functional prions that exploit the amyloid fold for evolutionarily selected biological processes have been identified [81]. These processes include polyamine regulation [82], signal transduction [83, 84], regulation of gametogenesis [85], hormone storage [86], epigenetic inheritance [87], and memory persistence [88].
The amyloidogenic regions of prions are evolutionarily conserved, and in many instances represented by short, low-complexity sequences enriched in glutamine/asparagine domains [89, 90]. Importantly, a marked homology exists between the amino acid sequences of peptides preferentially produced in the salt-induced peptide formation reaction simulating early Earth conditions and the sequences of known prions [91]. Extant prions, and their amyloid-based functional entities, may represent a relic of the pristine β-sheet-based information system, and the amyloid fold may represent the first functional protein fold [8, 74, 91–93].
Genetics before genetics
Information transfer on the early Earth for about 4000 million years ago occurred, according to the amyloid hypothesis, by means of a β-sheet peptide-based prion-like amyloid system in which environmentally derived information encrypted in the β-sheet zipper structure was transmitted by a templated conformational self-replication mechanism to “daughter” amyloid entities [8, 11, 74]. Recognition was mediated by amino acid side chain complementarity and coding by the β-sheet zipper structure [13, 18, 94]. The proposed system is characterized by both robustness and variability: the replication cycles are able to produce optimized stable molecular variants for evolutive forces to act on. From a primordial pool of random uncoded short protopeptides, the adaptive template-directed chiroselective and error correcting replication cycles generated amyloids that represented the first “coded” peptide polymers. Direct chemical interaction between amino acids/peptides and ribonucleotides in the primordial environment was probably important the evolution of the genetic code [95, 96].
The amyloid model emphasizes the importance of the cooperative interactions between amyloid and ribonucleotides and of the transition from a primitive β-sheet world to a more complex amyloid–RNA–protein world, a transition that was necessary from both an evolutionary and informational point of view: The information content of the β-sheet system, though potentially large, is very limited when compared to the virtually unlimited information content of a nucleic acid-based genetic system [13]. The β-system allows, on the other hand, for more rapid responses to environmental changes which would likely have been an advantage during early molecular evolution.
The amyloid hypothesis versus peptide/protein-first hypotheses
Though based on amino acids and peptides, there are fundamental differences between the amyloid world hypothesis [8, 11] and the peptide-/protein-/GADV-first hypotheses of the origin of life [97–100]. The key point is conformation: the differences are based on the particular folding patterns of the molecular entities involved. The amyloid fold is structurally different from all the other protein folds and it is functionally unique. It has an extraordinarily stable molecular structure and conveys functionality even to short (e.g., 3- to 9-mer) peptides. This is in contrast to nonaggregated peptides that are likely to decompose under harsh conditions, and functionality requires longer peptide lengths. In a prebiotic setting, short functional (aggregated) peptides with a rigid structure would, obviously, have had a selective evolutionary advantage over less stable, natively folded longer peptides. Importantly, template-directed reactions in β-sheet-driven replication networks can perform error correction and lead to the enrichment of a functional polymer within prebiotically relevant mixtures [21].
Amyloid fibril formation by prebiotically relevant peptides
It is well documented that under conditions simulating early Earth conditions, amino acids and short peptides are readily formed [101–105]. A high content of hydrophobic amino acids and the presence of alternating hydrophobic and hydrophilic residues tend to increase the β-sheet forming potential of peptide mixtures [106–111]. In a recent study, Greenwald et al. [112] specifically addressed the question of whether amyloid fibers can result from a condensation of amino acids under prebiotically plausible conditions. The study showed that fibrillar amyloid spontaneously formed under such conditions from a mixture of glycine, alanine, aspartate, and valine, all representing prebiotic “consensus” amino acids. An earlier study [10] had shown that a nonapeptide composed of six of the most abundantly produced amino acids in experiments simulating early Earth conditions [113], and also present in carbonaceous meteorites [114], generated polymorphic amyloid networks in an aqueous solution at temperatures likely to have existed on the primitive Earth. Moreover, intriguingly, a marked amino acid sequence homology has been observed between experimentally produced prebiotic peptides and extant amyloid-forming prions [91]. These findings are in line with the view that amyloids could have been formed and existed under early Earth conditions.
Toward life
A crucial stage in the origin of life was the emergence of the first informational molecular entity that was able to self-organize and evolve on the primordial Earth. The amyloid model, a hybrid replication-metabolism model of the origin of life, posits that, in the pre-RNA era, information storage and transfer was based on peptide-based catalytic amyloids. The template-assisted conformational replication cycles generate a variety of amyloid polymorphs on which evolutive forces can act. Amyloid, RNA, and protein interact dynamically and cooperatively, and the amyloid assemblages can function as primitive metabolic entities catalyzing the formation of simple metabolite precursors.
In conclusion, the emergence of an amyloid-based pristine in-put sensitive, chiroselective, and error correcting information-processing system, and the evolvement of mutualistic networks were, arguably, of essential importance in the dynamic processes that led to increased complexity, organization, compartmentalization, and, eventually, the origin of life.
Acknowledgements
I thank Fang Zhao for taking the electron micrograph in Fig. 3.
References
- 1.Gilbert W. The RNA world. Nature. 1986;319:618. doi: 10.1038/319618a0. [DOI] [Google Scholar]
- 2.Cech TR. A model for the RNA-catalyzed reaction. Proc Natl Acad Sci USA. 1986;83:4360–4363. doi: 10.1073/pnas.83.12.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 4.Higgs PG, Lehman N. The RNA world. Molecular cooperation at the origin of life. Nat Rev Genet. 2015;16:7–17. doi: 10.1038/nrg3841. [DOI] [PubMed] [Google Scholar]
- 5.Yarus M (2011) Life from an RNA world: the ancestor within. Harvard University Press, Cambridge, pp 1–194. ISBN 978-0-674-06071-5
- 6.Robertson MP, Joyce GF. The origins of the RNA world. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernhardt HS. The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others) Biol Direct. 2012;7:23–37. doi: 10.1186/1745-6150-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maury CPJ. Self-propagating β-sheet polypeptide structures as prebiotic informational molecular entities: the amyloid world. Orig Life Evol Biosph. 2009;39:141–150. doi: 10.1007/s11084-009-9165-6. [DOI] [PubMed] [Google Scholar]
- 9.Ikehara K. Evolutionary steps in the emergence of life deduced from the bottom-up approach and GADV hypothesis (top-down approach) Life. 2016;6:E6. doi: 10.3390/life6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maury CPJ, Liljeström M, Zhao F. Was the first molecular replicator on the primitive Earth an informational amyloid? EGGSVVAAD, a prebiotically plausible peptide, spontaneously forms amyloid assemblies. J Biol Res. 2012;18:332–335. [Google Scholar]
- 11.Maury CPJ. Origin of life. Primordial genetics: information transfer in a pre-RNA world based on self-replicating beta-sheet amyloid conformers. J Theor Biol. 2015;382:292–297. doi: 10.1016/j.jtbi.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 13.Wiltzius JJW, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickner RB, Edskes HJ, Bateman DA, Kelly AC, Gorkowsky A, Dayani Y, Zhou A. Amyloid-diseases of yeast: prions are proteins acting as genes. Essays Biochem. 2014;56:193–205. doi: 10.1042/bse0560193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carny O, Gazit E. Creating a prebiotic sanctuary: self assembling supramolecular peptide structures bind and stabilize RNA. Orig Life Evol Biosph. 2011;41:121–132. doi: 10.1007/s11084-010-9219-9. [DOI] [PubMed] [Google Scholar]
- 16.Rufo CM, Moroz YS, Moroz OV, Stör J, Smith TA, Hu X, DeGrado WF, Korendovych IV. Short peptides self-assemble to produce catalytic amyloids. Nat Chem. 2014;6:303–309. doi: 10.1038/nchem.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tena-Solsona M, Nanda J, Diaz-Oltra S, Chotera A, Ashkenasy G, Escuder B. Emergent catalytic behavior of self-assembled low molecular weight peptide-based aggregates and hydrogels. Chem Eur J. 2016;22:6687–6694. doi: 10.1002/chem.201600344. [DOI] [PubMed] [Google Scholar]
- 18.Riek R, Eisenberg DS. The activities of amyloids from a structural perspective. Nature. 2016;539:227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 19.Omosun TO, Hsieh M-C, Childers WS, Das D, Mehta AK, Anthony NR, Pan T, Grover MA, Berland KM, Lynn DG. Catalytic diversity in self-propagating peptide assemblies. Nat Chem. 2017;9:805–809. doi: 10.1038/nchem.2738. [DOI] [PubMed] [Google Scholar]
- 20.Ivnitski D, Amit M, Rubinov B, Cohen-Luria R, Ashkenasy N, Ashkenasy G. Introducing charge transfer functionality into prebiotically relevant peptide fibrils. Chem Commun (Camb) 2014;50:6733–6736. doi: 10.1039/c4cc00717d. [DOI] [PubMed] [Google Scholar]
- 21.Nanda J, Rubinov B, Ivnitski D, Mukherjee R, Shtelman E, Motro Y, Miller Y, Wagner N, Cohen-Luria R, Ashkenasy G. Emergence of native peptide sequences in prebiotic replication networks. Nat Commun. 2017;8:434. doi: 10.1038/s41467-017-00463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Garawi ZS, McIntosh BA, Neill-Hall D, Hatimy AA, Bagely MC, Serpell LC. The amyloid architecture provides a scaffold fold for enzyme-like catalysis. Nanoscale. 2017;9:10773–10783. doi: 10.1039/C7NR02675G. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi Y, Mihara H. Construction of a chemically and conformationally self-replicating system of amyloid-like fibrils. Bioorg Med Chem. 2004;12:693–699. doi: 10.1016/j.bmc.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Rubinov B, Wagner N, Rapaport H, Ashkenasy G. Self-replicating amphiphilic beta-sheet peptides. Angew Chem Int Ed. 2009;48:6683–6686. doi: 10.1002/anie.200902790. [DOI] [PubMed] [Google Scholar]
- 25.Bourbo V, Matmor M, Shtelman E, Rubinov B, Ashkenasy N, Ashkenasy G. Self-assembly and self-replication of short amphiphilic beta-sheet peptides. Orig Life Evol Biosph. 2011;41:563–567. doi: 10.1007/s11084-011-9257-y. [DOI] [PubMed] [Google Scholar]
- 26.Rout SK, Friedmann MP, Riek R, Greenwald J. A prebiotic templated-directed peptide synthesis based on amyloids. Nat Commun. 2018;9:234–242. doi: 10.1038/s41467-017-02742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westergard L, True HL. Extracellular environment modulates the formation and propagation of particular amyloid structures. Mol Microbiol. 2014;92:698–715. doi: 10.1111/mmi.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhak G, Lee J, Kim TH, Lee S, Lee D, Palk SR. Molecular inscription of environmental information into protein suprastructures: temperature effects on unit assembly of alfa-synuclein oligomers into polymorphic amyloid fibrils. Biochem J. 2014;464:259–269. doi: 10.1042/BJ20140723. [DOI] [PubMed] [Google Scholar]
- 29.Valdya N, Walker SI, Lehman N. Recycling of informational units leads to selection of replicators in a prebiotic soup. Chem Biol. 2013;20:241–252. doi: 10.1016/j.chembiol.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Jeong JS, Ansaloni A, Mezzenga R, Lashuel HA, Dietler G. Novel mechanistic insight into the molecular basis of amyloid polymorphism and secondary nucleation during amyloid formation. J Mol Biol. 2013;425:1765–1781. doi: 10.1016/j.jmb.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Saric A, Chebaro YC, Knowles TP, Frenkel D. Crucial role of nonspecific interactions in amyloid nucleation. Proc Natl Acad Sci USA. 2014;111:17869–17874. doi: 10.1073/pnas.1410159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaels TC, Lazell HW, Arosio P, Knowles TP. Dynamics of protein aggregation and oligomer formation governed by secondary nucleation. J Chem Phys. 2015;143:054901. doi: 10.1063/1.4927655. [DOI] [PubMed] [Google Scholar]
- 33.Castello F, Paredes JM, Ruedas-Rama MJ, Martin M, Roldan M, Casares S, Orte A. Two-step amyloid aggregation: sequential lag phase intermediates. Sci Rep. 2017;7:40065. doi: 10.1038/srep40065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwierz N, Frost CV, Geissler PL, Zacharias M. From Abeta filament to fibril: molecular mechanism of surface activated secondary nucleation from all-atom MD simulations. J Phys Chem B. 2017;121:671–682. doi: 10.1021/acs.jpcb.6b10189. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka M, Chien P, Naber M, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 36.Tycko R. Physical and structural basis for polymorphism in amyloid fibrils. Prot Sci. 2014;23:1528–1539. doi: 10.1002/pro.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J, Kahng B, Uwang W. Thermodynamic selection of steric zipper pattern of the amyloid cross-beta spine. PLoS Comput Biol. 2009;5:e1000492. doi: 10.1371/journal.pcbi.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung DM, Nowick JS. Enantioselective molecular recognition between β-sheets. J Am Chem Soc. 2004;126:3062–3063. doi: 10.1021/ja031632z. [DOI] [PubMed] [Google Scholar]
- 39.Wadai H, Yamaguchi K-I, Takahashi S, Kanno T, Kawai T, Naiki H, Goto Y. Sterospecific amyloid-like fibril formation by a peptide fragment of beta-2-microglobulin. Biochemistry. 2005;44:157–164. doi: 10.1021/bi0485880. [DOI] [PubMed] [Google Scholar]
- 40.Weissbuch I, Illos RA, Bolbach G, Lahav M. Racemic beta-sheets as templates of relevance to the origin of homochirality of peptides: lessons from crystal chemistry. Acc Chem Res. 2009;42:1128–1140. doi: 10.1021/ar900033k. [DOI] [PubMed] [Google Scholar]
- 41.Wagner N, Rubinov B, Ashkenasy G. Beta-sheet-induced chirogenesis in polymerization of oligopeptides. Chem Phys Chem. 2011;12:2771–2780. doi: 10.1002/cphc.201100292. [DOI] [PubMed] [Google Scholar]
- 42.Bada JL. Origins of homochirality. Nature. 1995;374:594–595. doi: 10.1038/374594a0. [DOI] [PubMed] [Google Scholar]
- 43.Hein JE, Blackmond DG. On the origin of single chirality of amino acids and sugars in biogenesis. Acc Chem Res. 2012;45:2045–2054. doi: 10.1021/ar200316n. [DOI] [PubMed] [Google Scholar]
- 44.Monasterio O, Nova E, Diaz-Espinoza R. Development of a novel catalytic amyloid displaying a metal-dependent ATPase-like activity. Biochem Biophys Res Commun. 2017;482:1194–1200. doi: 10.1016/j.bbrc.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Sternisha A, Makhlynets O. Catalytic amyloid fibrils that bind copper to activate oxygen. Methods Mol Biol. 2017;1596:59–68. doi: 10.1007/978-1-4939-6940-1_4. [DOI] [PubMed] [Google Scholar]
- 46.Lee M, Wang T, Makhlynets OV, Wu Y, Polizzi NF, Wu H, Gosavi PM, Stöhr J, Korendovych IV, DeGrtado WF, Hong M. Zinc-binding structure of a catalytic amyloid from solid-state NMR. Proc Natl Acad Sci USA. 2017;114:6191–6196. doi: 10.1073/pnas.1706179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamley IW. Peptide fibrillization. Angew Chem Int Ed Engl. 2007;46:8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- 48.Kodama H, Matsumura S, Yamashita T, Mihara H. Construction of a protein array on amyloid-like fibrils using co-assembly of designed peptides. Chem Commun. 2004;24:2876–2877. doi: 10.1039/b409641j. [DOI] [PubMed] [Google Scholar]
- 49.Nandi PK, Nicole JC. Nucleic acid and prion interaction produces spherical amyloids which can function in vivo as coats of spongiform encephalopathy agent. J Mol Biol. 2004;344:827–837. doi: 10.1016/j.jmb.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 50.Domanov YA, Kinnunen PK. Islet amyloid polypeptide forms rigid lipid-protein amyloid fibrils on supported phospholipid bilayers. J Mol Biol. 2008;376:42–54. doi: 10.1016/j.jmb.2007.11.077. [DOI] [PubMed] [Google Scholar]
- 51.Liu P, Ni R, Mehta AK, Childres WS, Lakdawala A, Pingali SW, Thyjagarajan P, Lynn DG. Nucleobase-directed amyloid nanotube assembly. J Am Chem Soc. 2008;130:16867–16869. doi: 10.1021/ja807425h. [DOI] [PubMed] [Google Scholar]
- 52.Relini A, Cavalleri O, Rolandi R, Gliozzi A. The two-fold aspect of the interplay of amyloidogenic proteins with lipid membranes. Chem Phys Lipids. 2009;158:1–9. doi: 10.1016/j.chemphyslip.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Silva JL, Cordeiro Y. The “Jekyll and Hyde” actions of nucleic acids on the prion-like aggregation of proteins. J Biol Chem. 2016;291:15482–15490. doi: 10.1074/jbc.R116.733428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silva JL, Lima LM, Foguel D, Cordeiro Y. Intriguing nucleic acid-binding features of mammalian prion protein. Trends Biochem Sci. 2008;33:132–140. doi: 10.1016/j.tibs.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Lindberg DJ, Wesen E, Björkeroth J, Esbjörner EK. Lipid membranes catalyse the fibril formation of the amyloid-beta (1–42) peptide through lipid-fibril interactions that reinforce secondary pathways. Biochim Biophys Acta. 2017;1859:1921–1929. doi: 10.1016/j.bbamem.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 56.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iconomidou VA, Hamodrakas SJ. Natural protective amyloids. Curr Protein Pept Sci. 2008;9:291–309. doi: 10.2174/138920308784534041. [DOI] [PubMed] [Google Scholar]
- 58.Tagialegna A, Lasa I, Valle J. Amyloid structures as biofilm matrix scaffolds. J Bacteriol. 2016;198:2579–2588. doi: 10.1128/JB.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boke E, Ruer M, Wuhr M, Coughlin M, Lemaltre R, Gygi SP, Alberti SS, Drechsel D, Hyman AA, Mitchison TJ. Amyloid-like self-assembly of a cellular compartment. Cell. 2016;166:637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kellermayer MS, Karsai A, Benke M, Soos K, Penke B. Stepwise dynamics of epitaxially growing single amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:141–144. doi: 10.1073/pnas.0704305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammarström P, Ali MM, Mishra R, Salagic B, Svensson S, Tengvall P, Lundström I. An auto-catalytic surface for conformational replication of amyloid fibrils-genesis of an amyloid world? Orig Life Evol Biosph. 2011;41:373–383. doi: 10.1007/s11084-010-9230-1. [DOI] [PubMed] [Google Scholar]
- 62.Rubio MA, Schlamadinger DE, White EM, Miranker AD. Peptide amyloid surface display. Biochemistry. 2014;54:987–993. doi: 10.1021/bi5011442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joyce GF. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- 64.Huang W, Ferris JP. One step, regio-selective synthesis of up to 50-mers RNA oligomers by montmorrionite catalysis. J Am Chem Soc. 2006;128:8914–8919. doi: 10.1021/ja061782k. [DOI] [PubMed] [Google Scholar]
- 65.Braun S, Humphreys C, Fraser E, Brancale A, Bochtler M, Dale TC. Amyloid associated nucleic acid hybridization. PLoS One. 2011;6:e19125. doi: 10.1371/journal.pone.0019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu C, Zhang Y. Nucleic acid-mediated protein aggregation and assembly. Adv Protein Chem Struct Biol. 2011;84:1–40. doi: 10.1016/B978-0-12-386483-3.00005-7. [DOI] [PubMed] [Google Scholar]
- 67.Alred EJ, Nguyen M, Martin M, Hansmann U. Molecular dynamic simulations of early steps in RNA-mediated conversion of prions. Prot Sci. 2017;26:1524–1534. doi: 10.1002/pro.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macedo B, Cordeiro Y. Unraveling prion protein interactions with aptamers and other PrP-binding nucleic acids. Int J Mol Sci. 2017;18:1023. doi: 10.3390/ijms18051023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahalka AK, Maury CPJ, Kinnunen PK. 1-palmoyl-2-(9′-oxononaoyl)-9-sn glycero-3-phosphocholine, an oxidized phospholipid, accelerates Finnish gelsolin amyloidosis in vitro. Biochemistry. 2011;50:4877–4889. doi: 10.1021/bi200195s. [DOI] [PubMed] [Google Scholar]
- 70.Deamer D. The role of lipid membranes in life’s origin. Life. 2017;7:5. doi: 10.3390/life7010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nizhnikov AA, Antonets KS, Bondarev SA, Inge-Vechtomov SG, Derkatch IL. Prions, amyloids, and RNA: pieces of a puzzle. Prion. 2016;10:182–206. doi: 10.1080/19336896.2016.1181253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reijns MAM, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maury CPJ. Self-replicating protein conformations and information transfer: the adaptive β-sheet model. Biosci Hypotheses. 2008;1:82–89. doi: 10.1016/j.bihy.2008.02.010. [DOI] [Google Scholar]
- 75.Caetano-Anolles G, Seufferheld MJ. The coevolutionary roots of biochemistry and cellular organization challenge the RNA world paradigm. J Mol Microbiol Biotechnol. 2013;23:152–177. doi: 10.1159/000346551. [DOI] [PubMed] [Google Scholar]
- 76.Carter CW., Jr What RNA world? Why a peptide/RNA partnership merits renewed experimental attention. Life. 2015;5:294–320. doi: 10.3390/life5010294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones E, Surewicz WK. Fibril conformation as the basis of species- and strain- dependent seeding specificity of mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 79.Wickner RB, Shewmaker F, Edskes H, Kryndushkin D, Nemeecek J, McGlinchey R, Bateman D, Winchester CL. Prion amyloid structure explains templating: how proteins can be genes. FEMS Yeast Res. 2010;10:980–991. doi: 10.1111/j.1567-1364.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collins SR, Douglas A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maury CPJ. The emerging concept of functional amyloid. J Int Med. 2009;265:329–334. doi: 10.1111/j.1365-2796.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- 82.Namy O, Galopier A, Martini C, Matsufuij S, Fabret C, Roussel JP. Epigenetic control of polyamines by the prion [PSI+] Nat Cell Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- 83.Li J, McQuade T, Siemer AB, Napetschinig J, Moriwaki K, Hsiao Y-S, Darnko E, Moquin D, Walz T, McDermott A, Ka-Ming Chan F, Wu H. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai X, Chen J, Xu H, Jiang Q-X, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KP, Simon R, Schubert D, Eisenberg D, Rivier J, Sawchenko P, Vale VV, Riek R. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 88.Khan MR, Li L, Perez-Sanchez C, Saraf A, Florens L, Slaughter BD, Unruch JR, Si K. Amyloidogenic oligomerization transforms Drosophila Orb2 from a translation repressor to an activator. Cell. 2015;163:1468–1483. doi: 10.1016/j.cell.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Epinosa Angarica VP, Ventura S, Sancho J. Discovering putative prion sequences in complete proteomes using probalistic representations of Q/N-rich domains. BMC Genomics. 2013;14:316. doi: 10.1186/1471-2164-14-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rode BM, Flader W, Sotriffer C, Righi A. Are prions a relic of an early stage of peptide evolution. Peptides. 1999;20:1513–1516. doi: 10.1016/S0196-9781(99)00163-1. [DOI] [PubMed] [Google Scholar]
- 92.Chernoff YO. Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquistion? Curr Opin Chem Biol. 2004;8:665–671. doi: 10.1016/j.cbpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 93.Greenwald J, Riek R. On the possible amyloid origin of protein folds. J Mol Biol. 2012;421:417–426. doi: 10.1016/j.jmb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 94.Eisenberg DS, Sawaya MR. Structural studies of amyloid proteins at the molecular level. Annu Rev Biochem. 2017;86:69–95. doi: 10.1146/annurev-biochem-061516-045104. [DOI] [PubMed] [Google Scholar]
- 95.Yarus M, Widmann JJ, Knight R. RNA-amino acid binding: a stereochemical era for the genetic code. J Mol Evol. 2009;69:406–429. doi: 10.1007/s00239-009-9270-1. [DOI] [PubMed] [Google Scholar]
- 96.Beier A, Zagrovic B, Polyansky A. On the contribution of protein spatial organization to the physicochemical interconnection between proteins and their cognate mRNAs. Life. 2014;4:788–799. doi: 10.3390/life4040788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rode BM. Peptides and the origin of life. Peptides. 1999;20:773–786. doi: 10.1016/S0196-9781(99)00062-5. [DOI] [PubMed] [Google Scholar]
- 98.Woolfson A. Life without genes. London: Flamingo; 2000. [Google Scholar]
- 99.Ikehara K. Possible steps to the emergence of life: the GADV-protein world. Chem Res. 2005;5:107–118. doi: 10.1002/tcr.20037. [DOI] [PubMed] [Google Scholar]
- 100.Kurland CG. The RNA dreamtime: modern cells feature proteins that might have supported a prebiotic peptide world, but nothing indicates that RNA world ever was. BioEssays. 2010;32:866–871. doi: 10.1002/bies.201000058. [DOI] [PubMed] [Google Scholar]
- 101.Miller SL. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 102.Brack A. From interstellar amino acids to prebiotic catalytic peptides: a review. Chem Biodivers. 2007;4:665–679. doi: 10.1002/cbdv.200790057. [DOI] [PubMed] [Google Scholar]
- 103.Li F, Fitz D, Fraser DG, Rode BM. Methionine peptide formation under primordial earth conditions. J Inorg Biochem. 2008;102:1212–1217. doi: 10.1016/j.jinorgbio.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 104.Bada JL. New insights into prebiotic chemistry from Stanley Miller’s spark discharge experiments. Chem Soc Rev. 2013;42:2186–2196. doi: 10.1039/c3cs35433d. [DOI] [PubMed] [Google Scholar]
- 105.Parker ET, Zhou M, Burton AS, Glavin DP, Dworkin JP, Krishnamurthy R, Fernandez FM, Bada JL. A plausible simultaneous synthesis of amino acids and simple peptides on the primordial Earth. Angew Chem Int Ed Engl. 2014;53:8132–8136. doi: 10.1002/anie.201403683. [DOI] [PubMed] [Google Scholar]
- 106.Brack A, Orgel LE. Beta structures of alternating polypeptides and their possible prebiotic significance. Nature. 1975;256:383–387. doi: 10.1038/256383a0. [DOI] [PubMed] [Google Scholar]
- 107.de la Chapelle A, Tolvanen R, Boysen G, SantavyJ Bleeker-Wagemaker L, Maury CPJ, Kere J. Gelsolin-derived familial amyloidosis caused by asparagine or tyrosine substitution for aspartic acid at residue 187. Nat Genet. 1992;2:157–160. doi: 10.1038/ng1092-157. [DOI] [PubMed] [Google Scholar]
- 108.Maury CPJ, Nurmiaho-Lassila E-L. Creation of amyloid fibrils from mutant Asn-187 gelsolin peptides. Biochem Biophys Res Commun. 1992;183:227–231. doi: 10.1016/0006-291X(92)91632-Z. [DOI] [PubMed] [Google Scholar]
- 109.Maury CPJ, Nurmiaho-Lassila EL, Rossi H. Amyloid fibril formation in gelsolin-derived amyloidosis. Definition of the amyloidogenic region and evidence of accelerated amyloid formation of mutant Asn-187 and Tyr-187 gelsolin peptides. Lab Investig. 1994;70:558–564. [PubMed] [Google Scholar]
- 110.West MW, Wang W, Patterson J, Mancias JD, Beasley JR, Hecht MH. De novo amyloid proteins designed combinatorial libraries. Proc Natl Acad Sci USA. 1999;96:11211–11216. doi: 10.1073/pnas.96.20.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dyson HJ, Wright PE, Scheraga HA. The role of hydrophobic interactions in the initiation and propagation of protein folding. Proc Natl Acad Sci USA. 2006;103:13057–13061. doi: 10.1073/pnas.0605504103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Greenwald J, Friedmann MP, Riek R. Amyloid aggregates arise from amino acid condensations under prebiotic conditions. Angew Chem Int Ed. 2016;55:11609–11613. doi: 10.1002/anie.201605321. [DOI] [PubMed] [Google Scholar]
- 113.Van Gulik P, Massar S, Gilis D, Buhrman H, Rooman M. The first peptides: the evolutionary transition between prebiotic amino acids and early proteins. J Theor Biol. 2009;261:531–539. doi: 10.1016/j.jtbi.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 114.Botta O, Glavin DP, Kminek G, Bada JL. Relative aminoacid concentrations as a signature for parent body processes in carbonaceous chondrites. Orig Life Evol Biosph. 2002;32:143–163. doi: 10.1023/A:1016019425995. [DOI] [PubMed] [Google Scholar]