Abstract
The molecular clock plays key roles in daily physiological functions, development and cancer. Period 2 (PER2) is a repressive element, which inhibits transcription activated by positive clock elements, resulting in diurnal cycling of genes. However, there are gaps in our understanding of the role of the clock in normal development outside of its time-keeping function. Here, we show that PER2 has a noncircadian function that is crucial to mammalian mammary gland development. Virgin Per2-deficient mice, Per2−/−, have underdeveloped glands, containing fewer bifurcations and terminal ducts than glands of wild-type mice. Using a transplantation model, we show that these changes are intrinsic to the gland and further identify changes in cell fate commitment. Per2−/− mouse mammary glands have a dual luminal/basal phenotypic character in cells of the ductal epithelium. We identified colocalization of E-cadherin and keratin 14 in luminal cells. Similar results were demonstrated using MCF10A and shPER2 MCF10A human cell lines. Collectively this study reveals a crucial noncircadian function of PER2 in mammalian mammary gland development, validates the Per2−/− model, and describes a potential role for PER2 in breast cancer.
KEY WORDS: Mammary gland development, Molecular clock, PER2
Summary: The molecular clock is associated with crucial biological processes, but little is known about its role in development. Here, we show that PER2 plays a key role in mammary gland development.
INTRODUCTION
Biological clocks play a key role in how an organism adapts to daily and annual changes in the environment by regulating rhythmic fluctuations in metabolism, hormone and neurotransmitter release, and sensory capabilities and behaviors, including sleep (Okamura et al., 2002; Reppert and Weaver, 2001). In vertebrates, these physiological responses are controlled by the suprachiasmatic nucleus (SCN) of the anterior hypothalamus. In addition, peripheral tissues, including the liver, heart, kidney and mammary gland, contain functional endogenous clocks (Metz et al., 2006; Yamazaki et al., 2000; Zylka et al., 1998). These peripheral clocks, which regulate numerous physiological processes including proliferation and apoptosis, are similar to the central clock and are influenced by the SCN via a combination of neural and hormonal signals (Mohawk et al., 2012). The molecular basis of both the central and peripheral clocks is the interaction of positive and negative feedback loops governed by clock proteins. Recent studies have implicated several members of the Per-Arnt-Sim (PAS) family of proteins as central molecular components of the biological clock in invertebrates and vertebrates including mammals. Central to this mechanism, the positive elements, CLOCK and BMAL1 (MOP3; ARNTL1 – Mouse Genome Informatics), heterodimerize via their PAS domains and activate transcription of target genes that contain an E-box in their promoter regions. CLOCK/BMAL1 dimers also stimulate transcription of the negative elements of the molecular clock, including the Period (PER) genes, PER1 and PER2, which contain PAS domains, and the cryptochromes, CRY1 and CRY2, which do not. PER/CRY heterodimers function as repressors by blocking CLOCK/BMAL1-dependent transactivation. As PER and CRY proteins are degraded, this repression is released and the process begins again with a periodicity of ∼24 h. This transcriptional network has been shown to control tissue-specific and global gene expression in the SCN and peripheral tissues (Rey et al., 2011).
Clock genes have been shown to have differential tissue expression, suggesting that the molecular clock might have alternative biological activities. This is best observed in testes and thymus, where PER1 expression is high and cyclic expression of circadian clock genes does not occur (Alvarez et al., 2003; Morse et al., 2003). Indeed, we have demonstrated that PER1 and PER2 are differentially regulated during mouse mammary gland development. PER2 is highly expressed in virgin mammary glands and undifferentiated mammary epithelial cells (MECs), whereas PER1 expression is highest in lactating mouse mammary glands and differentiated HC11 cells (Metz et al., 2006). Furthermore, it has been shown that Clock mutant mice have a depressed mammary clock and a lactation defect, despite normal virgin development (Dolatshad et al., 2006; Hoshino et al., 2006). Based on these findings, it has been proposed that the circadian clock is developmentally regulated and is suspended in differentiating tissues to allow a ‘developmental clock’ to function independent of normal time-keeping functions. Recently, a role for PER proteins in maintaining progenitor stem cell division has been reported, as has a role for Clock in maintaining stem cell function in the mammary gland (Moriya et al., 2007; Tsinkalovsky et al., 2006, 2005; Yang et al., 2017). In hematopoietic and neural stem cells, PER1 was shown to be highly expressed and arrhythmic, whereas PER2 exhibited a robust circadian pattern of expression (Borgs et al., 2009). Furthermore, downregulation of PER2 in neural stem cells led to increased cell proliferation, suggesting that PER2 plays an important role in timing and regulating steps of cell lineage commitment and cell fate.
Mouse mammary gland development is dependent upon complex interactions between the stromal and epithelial compartments that drive cell division, migration, apoptosis and differentiation. These processes are regulated through functional gene expression, controlled by transcription factor cascades, and denote key events in regulating the differentiation potential of MECs. The mammary gland grows at the same rate as the organism until puberty, when high levels of circulating ovarian hormones initiate a branching morphogenesis program of ductal elongation and differentiation driven by the terminal end bud (TEB) (Daniel and Smith, 1999). Mammary ducts are composed of two epithelial cell types, luminal and myoepithelial. Similar to the hematopoietic system, a differentiation hierarchy of mammary stem cells (MaSCs) has been identified in the adult mammary gland that gives rise to the luminal and myoepithelial lineages (Visvader, 2009). The luminal lineage can be further subdivided into ductal cells that line the ducts and alveolar cells that expand in response to lactogenic hormones to form alveolar units (Shackleton et al., 2006; Stingl et al., 2006). Studies comparing the molecular signatures of MECs to breast cancer subtypes suggest that the mammary gland stem cell hierarchy is responsible for the inter- and intratumoral heterogeneity among breast cancers, which has been previously reviewed (Sreekumar et al., 2015).
Given that Per2 is differentially expressed in the developing mammary gland and a role for Clock has been identified in mammary gland function, we set out to understand the role of the repressive arm of the circadian clock in mammary gland development and function. Here, we identify a pathway regulating mammary epithelial subpopulations, which contributes to a better understanding of mammary gland development and breast cancer heterogeneity.
RESULTS
Per2 regulates branching morphogenesis
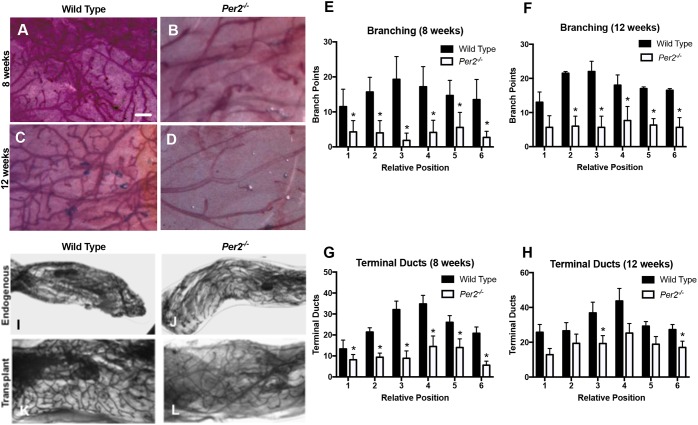
We and others have reported that circadian clock expression changes with development (Alvarez et al., 2003; Metz et al., 2006; Xiao et al., 2003). Per1 and Per2 are differentially regulated in mouse MECs. Per2 is highly expressed in luminal epithelial cells in the virgin gland, whereas Per1 and Bmal1/Clock are expressed at higher levels during lactation, suggesting that they play different roles in mammary gland development (Metz et al., 2006). To address whether PER2 contributes to mammary gland development, we analyzed glands from 8- and 12-week-old wild-type (WT) and Per2−/− mice by whole-mount staining (Fig. S1). The results showed dramatic morphological differences in Per2−/− mammary glands relative to WT glands (Fig. 1A-D). Per2−/− mammary glands displayed fewer bifurcations (Fig. 1E-H) as well as a lack of distal migration of the ducts. To determine whether these defects were the result of systemic effects of circadian rhythm disruption, or were intrinsic to the gland, mammary epithelia from WT and Per2−/− mice were transplanted contralaterally into the cleared fat pads of 21-day-old syngeneic mice and analyzed after 8 weeks of outgrowth. A similar phenotype for Per2−/− was observed in the transplanted mutant glands (Fig. 1I-L), suggesting that these defects are caused by processes intrinsic to the mammary gland and not systemic factors. These results show, for the first time, that Per2 is required for normal mouse mammary gland development.
Fig. 1.
Disruption of branching morphogenesis in Per2−/− virgin mice. (A-D) Animal development stage is indicated along the left edge of panels. Representative images of normal branching and development of terminal ducts in a WT animal are shown (A,C). Representative images of Per2−/− mammary glands showing fewer bifurcations than in WT mammary glands (B,D). (E-H) The number of branch points (E,F) and terminal ducts (G,H) at the relative positions differed significantly between the WT and Per2−/− mice (*P<0.05; error bars indicate s.e.m.; n=3). (I-L) Mammary epithelia from WT and Per2−/− mice were transplanted contralaterally into the cleared fat pads of 21-day-old syngeneic mice and analyzed after 8 weeks of outgrowth. A phenotype consistent with Per2−/− mice was observed in the transplanted mutant glands. Per2−/−: n=4; WT: n=3. Scale bar: 500 μm.
Loss of Per2 disrupts cell fate
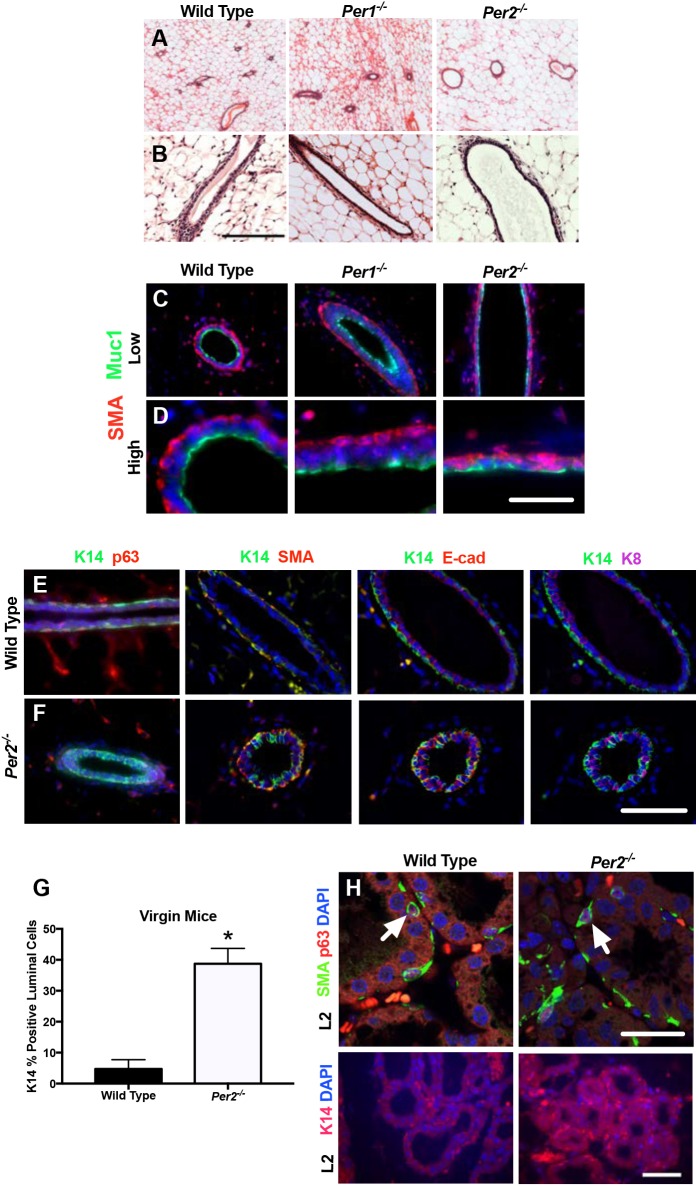
To determine whether the developmental phenotype is Per2 dependent, we compared mammary glands from virgin WT, Per1−/− and Per2−/− mice. Histological analysis showed that, compared with WT and Per1−/− ducts, the ducts in the Per2−/− glands were larger in size (Fig. 2A,B). Because Per2−/− mammary glands show decreased branching morphogenesis compared with WT mice, we wanted to determine differences in the epithelial character of luminal cells. To determine epithelial cell organization, we stained virgin glands of all three genotypes for the myoepithelial and luminal polarity markers, smooth muscle actin (SMA; ACTA2) and mucin 1 (MUC1), respectively. Per1−/− glands showed proper basal localization of SMA and presence of MUC1 on the apical surface, which was consistent with WT glands (Fig. 2C,D). The Per2−/− ducts revealed colocalization of SMA and MUC1 (Fig. 2I,L), as well as epithelium lacking distinct luminal and basal layers, but still maintaining cell polarity. We then compared the expression and colocalization of the basal marker keratin 14 (K14; KRT14) with that of basal markers p63 (Trp63) and SMA, and luminal markers keratin 8 (K8; KRT8) and E-cadherin (E-cad; CDH1), in virgin WT and Per2−/− mammary glands. Co-staining for K14 and all markers shows aberrant localization of the basal marker K14 to luminal cells in Per2−/−, indicating defects in cell fate of Per2−/− cells (Fig. 2E,F). We were surprised to find that the luminal epithelial cell layer contained K14-positive cells in Per2−/− mice. This is in contrast to staining in WT ducts, which showed that myoepithelial cells displayed K14 expression colocalized with SMA expression, and luminal epithelial E-cadherin staining that did not colocalize with K14 staining (Fig. 2E,F). Furthermore, we show that a subset of luminal epithelial cells in Per2−/− mice expressed K14, which colocalized with E-cadherin and K8 (Fig. 2F,G). These data indicate that PER2 contributes to cellular fate during mammary gland development and plays a role in lineage commitment.
Fig. 2.
Altered expression of luminal and basal markers in Per2−/− mammary glands. (A,B) H&E staining of mammary gland sections from adult WT, Per1−/− and Per2−/− mice. (C-F) IF staining for myoepithelial and luminal cell markers, SMA and MUC1 (C,D), K14/p63, K14/SMA, K14/E-cad and K14/K8 (E,F), showing inappropriate localization of myoepithelial and luminal markers. (G) Quantification of K14-positive cells in WT and Per2−/− virgin mice (*P<0.05; error bars indicate s.e.m.). (H) IF staining of mammary gland sections in WT and Per2−/− mice at lactation day 2 (L2), showing colocalization (arrows) of SMA/p63 expression (upper panels) and normal K14 expression (lower panels). Per2−/−: n=4; WT: n=3. Scale bars: 50 μm.
Current MaSC hierarchy models indicate that early alveolar progenitor cells of the luminal lineage give rise to the alveolar secretory cells during pregnancy (Visvader and Stingl, 2014). It is believed that the existing myoepithelial cell layer surrounding the luminal duct expands around the alveolus and provides secretory properties via SMA, which confer the ability to secrete milk. Analysis of lactating Per2−/− mice shows that the mammary gland proliferates and fills the fat pad in response to lactogenic hormones and is capable of supporting pup growth (Fig. S2). As previously discussed (Fig. 2), Per2−/− virgin mice have a disrupted cell lineage commitment, indicated by delocalization of K14. Interestingly, in early lactation (L2), we observed that the virgin phenotype has recovered proper localization of basal markers, showing K14- and p63/SMA-positive myoepithelial cells in Per2−/− mice (Fig. 2H). These results suggest that luminal progenitors are present in the Per2−/− virgin mice and are capable of conferring the ability for these mice to establish the fully differentiated cell types necessary for lactation. Furthermore, these findings indicate that there is a defect in the ductal progenitors that are important in ductal growth and branching morphogenesis during pubertal development.
Characterization of hormone receptor status in Per2−/− mice
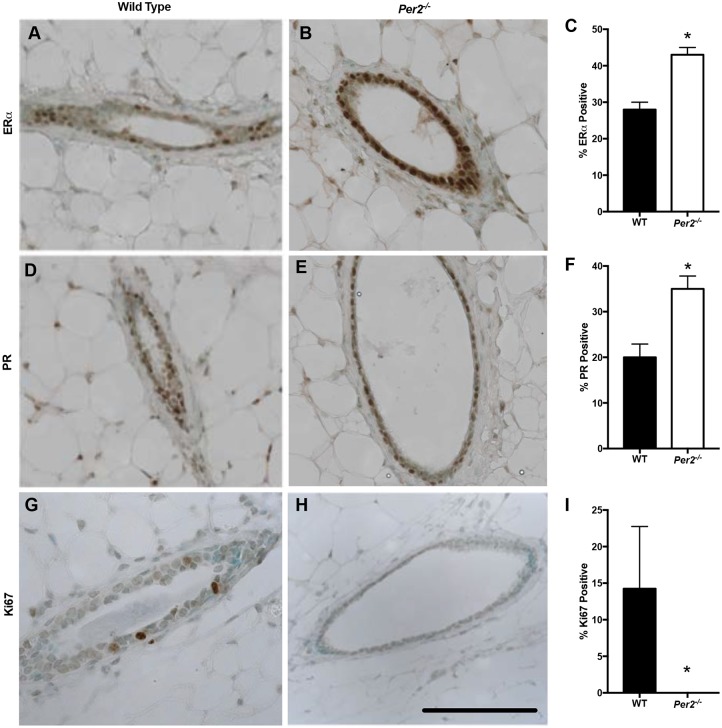
Having observed a cellular defect inhibiting branching morphogenesis and ductal growth, we characterized the hormone receptor status of Per2−/− mice. Both estrogen receptor α (ERα; ESR1) and progesterone receptor (PR; PGR) have been shown to play important roles in mammary gland development and branching morphogenesis (Brisken et al., 1998; Korach et al., 1996). A recent study has demonstrated that Per2 inhibits stabilization of ERα expression, providing a link between the molecular clock and estrogen signaling (Gery et al., 2007). To determine whether ERα and PR levels are affected by loss of Per2 in mouse mammary glands, we stained 10-week-old virgin WT and Per2−/− mammary gland sections for ERα and PR. The results show that Per2−/− mice had a significant increase in the number of ERα- (Fig. 3A-C) and PR-positive cells (Fig. 3D-F) compared with WT mice. As ERα and PR are involved in the regulation of mammary epithelial proliferation, we determined whether the MECs of Per2−/− and WT mice differed in proliferation status. We stained virgin WT and Per2−/− glands for Ki67 (Mki67), which is a marker indicative of active proliferation (Bruno and Darzynkiewicz, 1992). WT glands harbored ∼15% of MECs which were actively dividing, compared with almost no Ki67-positive cells in Per2−/− glands – a significant decrease (Fig. 3G-I). These results indicate that although Per2−/− mice have increased ERα- and PR-positive cells, they do not demonstrate active proliferation, indicated by the lack of Ki67 staining.
Fig. 3.
Altered hormone receptor status in Per2−/− virgin mice. (A-F) IHC staining indicated a significant increase in the number of ERα-positive (A-C) and PR-positive (D-F) cells in virgin Per2−/− mice compared with WT mice. (G-I) Staining for Ki67 revealed ∼15% Ki67-positive cells in WT mice, but almost no Ki67-positive cells in 10-week-old virgin Per2−/− mice. *P<0.05; error bars indicate s.e.m. Per2−/−: n=4; WT: n=3. Scale bar: 50 μm.
Loss of PER2, but not PER1, disrupts cell fate in MCF10A cells
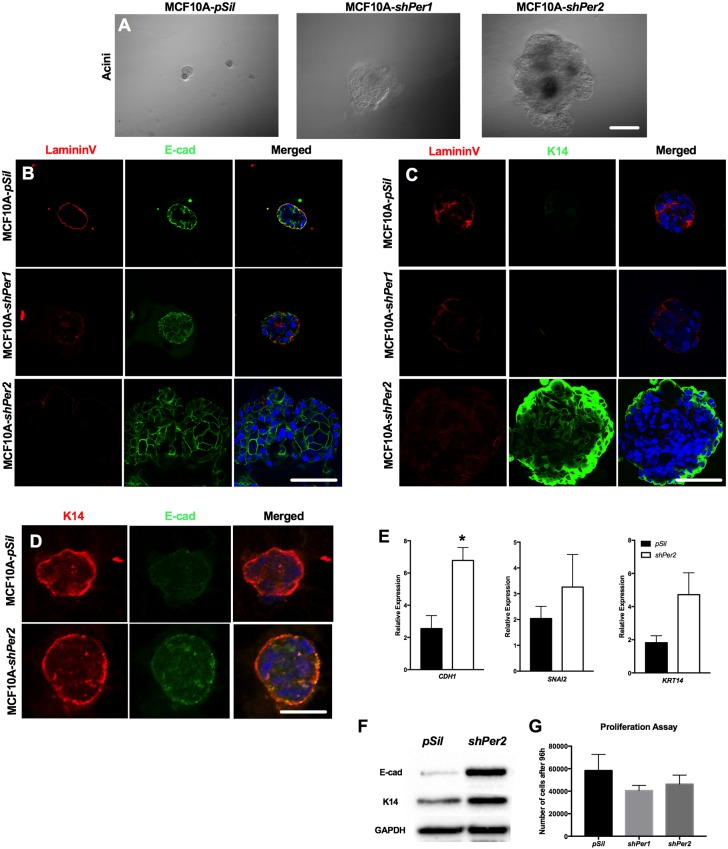
To determine whether the dual luminal/basal phenotype observed in Per2−/− mammary glands could be reproduced in human MECs, we analyzed MCF10A cells with and without a short hairpin construct to suppress PER1 and PER2 expression in 3D culture, as we previously described (Schmitt et al., 2017). The MCF10A cell line was chosen because it is characterized as a normal breast epithelial cell line that is under circadian control (Xiang et al., 2012). Normal MCF10A cells and MCF10A-shPER1 formed hollow spherical structures, indicating proper acini formation, whereas MCF10A-shPER2 cells developed large, multilayered and filled spherical structures, inconsistent with proper development (Fig. 4A). Furthermore, staining for the basal marker lamininV, and luminal marker E-cadherin, revealed an increase in E-cadherin staining in the filled-in lumen of the PER2 knockdown acini compared with control and PER1 knockdown acini (Fig. 4B). In addition, we observed an increase in the amount of K14-positive cells in PER2 knockdown acini (Fig. 4C), which we also observed in Per2−/− mouse mammary glands (Fig. 2F). Consistent with the mouse mammary gland (Fig. 2F), we also observed colocalization of E-cadherin and K14 in MCF10A-shPER2 acini (Fig. 4D). Gene expression analysis of luminal and basal markers showed that CDH1, SNAI2 and KRT14 levels were higher in the PER2 knockdown cells (Fig. 4E), and were consistent with increased protein levels of K14 and E-cadherin (Fig. 4F). Moreover, although MCF10A-shPER2 acini grew larger in size (Fig. 4A-C), these cells tended to proliferate more slowly than control cells in monolayer (Fig. 4G). These results indicate that PER2 plays a similar role in both mouse and human MECs and regulates cell fate commitment, with a trend towards a bipotent cell type.
Fig. 4.
MCF10A (pSIL, shPER1, shPER2) human MECs in 3D culture. (A) MCF10A normal (pSIL) and shPER1 cells formed hollow spherical structures, indicating proper acini formation. MCF10A-shPER2 cells developed large, multilayered and filled in spherical structures, inconsistent with proper development. (B) Staining of a luminal marker (E-cad) indicates proper development in pSIL cells and shPER1 cells, yet an increase in the amount of E-cad fluorescence in shPER2 cells. (C) Staining of basal markers (lamininV and K14) indicates proper development in pSIL cells and shPER1 cells, yet improper localization in shPER2 cells. (D) K14 and E-cad staining showing colocalization in shPER2 acini. (E) qPCR analysis of luminal and basal markers indicated that CDH1, SNAI2 and KRT14 levels relative to β-actin were higher in the PER2 knockdown cells than in normal cells. (F) KRT14 and E-cad protein levels were also higher in the PER2 knockdown cells than in normal cells. (G) Proliferation assay results of MCF10A pSIL, shPER1 and shPER2 cell lines following 96 h under normal culture conditions. *P<0.05; error bars indicate s.e.m. Scale bars: 50 μm.
Loss of Per2 alters luminal and basal mammary cell populations
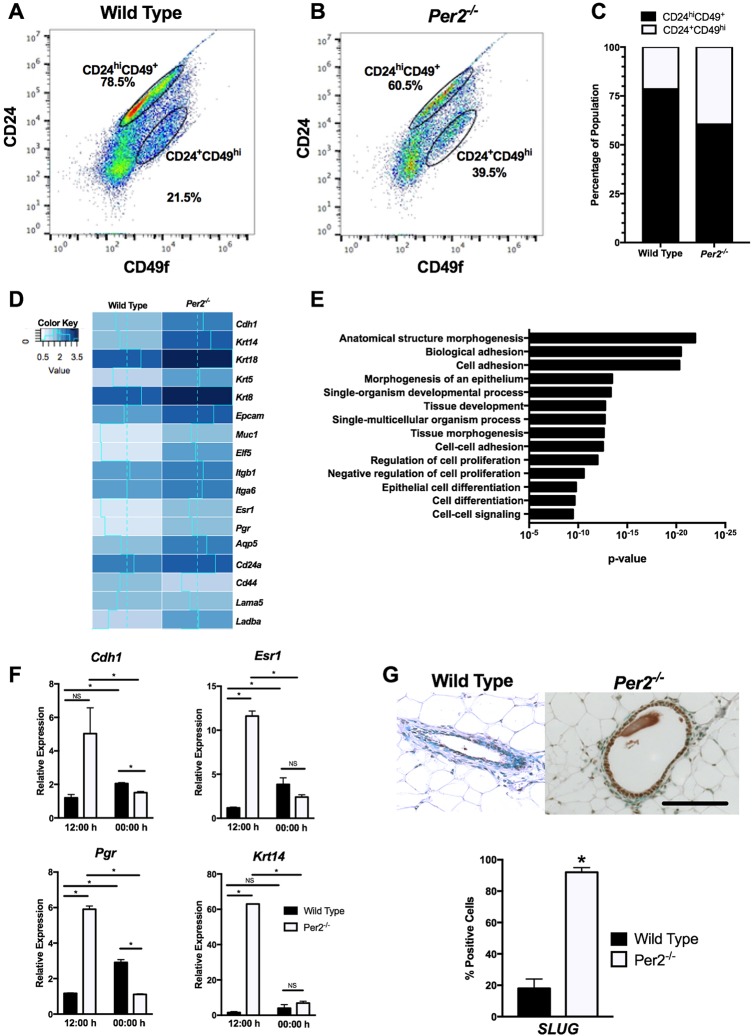
To determine whether MECs isolated from Per2−/− mice do indeed possess the bipotent phenotype observed in our immunofluorescence (IF) studies, we used fluorescence-activated cell sorting (FACS) to sort epithelial cells using the cell surface markers CD24 and CD49f (ITGA6). WT (n=10) and Per2−/− (n=13) MEC preparations from pooled mammary glands were lineage depleted (Lin−), labeled with both CD24(PE) and CD49f(FITC), and sorted (Fig. 5A-C). Per2−/− MECs showed a reduction in Lin− Cd24hi Cd49f+ luminal cells (18.5%) when compared with WT cells. Furthermore, Per2−/− mice were enriched for myoepithelial/basal-like cells, indicated by an increase (18.0%) in Lin− Cd24+ Cd49fhi cells. Together, these results suggest that PER2 regulates cell lineage commitment and, in its absence, increases the basal-like cell populations of the mouse mammary gland.
Fig. 5.
Cell lineage and RNA-Seq analysis of WT and Per2−/− MECs. (A-C) Per2−/− MECs showed a 19.55% reduction in luminal progenitors (CD24hi) and 6.63% increase in myoepithelial/basal-like cells (CD49fhi) when compared with WT cells. (D) RNA-Seq results of WT and Per2−/− mice, showing that genes associated with epithelial and basal phenotypes, including Krt14, Krtr5, Krt18 and Krt8, were upregulated in Per2−/− MECs. (E) GOrilla was used to identify biological pathways differentially regulated between the WT and Per2−/− mice. (F) qPCR analysis of luminal and basal markers confirmed that Chd1, Esr1, Pgr and Krt14 levels were higher relative to 18 s in Per2−/− MECs compared with WT MECs at 12:00 h. At 00:00 h, Chd1 and Pgr levels were lower in Per2−/− MECs compared with WT MECs, but no differences in the levels of Esr1 and Krt14 were observed. (G) SLUG staining showed a significant increase in SLUG in Per2−/− mouse mammary tissues compared with WT tissues. *P<0.05; error bars indicate s.e.m. Per2−/−: n=13; WT: n=10. Scale bar: 50 μm.
To investigate gene expression patterns of interest in Per2−/− mice compared with their WT counterparts, we used pooled MEC preparations for RNA sequencing (RNA-Seq). Loss of Per2 altered the expression of 2257 genes greater than twofold. Unsurprisingly, many genes associated with epithelial and basal phenotypes were upregulated in Per2−/− MECs, including Krt14, Krt5, Krt18 and Krt8 (Fig. 5D). Other notable genes that specifically relate to epithelial cell fate and were overexpressed included Muc1, aquaporin 5 (Aqp5), Cdh1 and epithelial cell adhesion molecule (Epcam) (Fig. 5D). RNA-Seq results also indicated that Pgr and Esr1 were differentially expressed in the Per2−/− mammary glands. Differential expression of Cdh1, Krt14, Esr1 and Pgr in Per2−/− mice at 12:00 h was confirmed using quantitative PCR (qPCR) (Fig. 5F). We observed that Cdh1, Krt14, Esr1 and Pgr mRNA levels were upregulated in Per2−/− MECs at 12:00 h, consistent with RNA-Seq analysis results. We also compared the expression of these markers in a time-resolved manner. Per2−/− MECs expressed these mRNAs in lower quantities at 00:00 h than at 12:00 h, whereas WT MECs had higher expression of these genes at 00:00 h than at 12:00 h. Although the mRNA levels of these genes differed between 12:00 h and 00:00 h, we observed no changes in staining for these markers in virgin mammary glands harvested at 12:00 h and at 00:00 h (data not shown). These results further support the previously described staining (Figs 2 and 3) results. Pathway analysis of differentially expressed genes identified enriched biological processes relating to cell adhesion, differentiation and tissue morphogenesis (Fig. 5E). Collectively, these data confirm our findings that in the absence of Per2, MECs take on a dual phenotypic character, exhibiting both luminal and basal epithelial characteristics, further supporting a role for PER2 in regulating cell fate commitment.
SLUG (SNAI2) is an important transcription factor known to regulate cell fate determination and epithelial-to-mesenchymal (EMT) transition and has been shown to be suppressed by PER2 in MCF10A cells (Bolos et al., 2003; Guo et al., 2012; Hwang-Verslues et al., 2013). To confirm SLUG upregulation in Per2−/− mice, we analyzed WT and Per2−/− mammary sections for SLUG protein levels. Indeed, levels of SLUG protein were significantly elevated in Per2−/− mouse mammary tissues (Fig. 5G). We did not observe any change in the expression levels of the related transcription factors SNAIL (SNAI1) and TWIST (TWIST1) (data not shown).
DISCUSSION
Per2−/− mice exhibit a dual basal/luminal epithelial phenotype that clearly plays a role in cell fate determination. Per2 mutant mammary glands exhibit an underdeveloped ductal system that lacks extensive branching and cell differentiation. Interestingly, these phenotypes are similar Gata3 KO and Elf5 KO mammary glands, in which altered cell populations disrupt various aspects of mammary development (Asselin-Labat et al., 2007; Chakrabarti et al., 2012; Choi et al., 2009). Furthermore, Per2−/− mammary glands have some similarity to Elf5 KO mice, which have a dual luminal/basal phenotype, indicated by Krt8 and Krt14 expression (Chakrabarti et al., 2012; Choi et al., 2009). These similarities further strengthen our hypothesis that PER2 plays a role in mammary cell differentiation.
Previous studies demonstrated that Clock mutant mice are unable to support pups owing to a lactation defect (Dolatshad et al., 2006; Hoshino et al., 2006). This lactation phenotype was later attributed to deficiencies in MaSC maintenance, in which genes crucial to MaSC status were under circadian control via Clock (Yang et al., 2017). The phenotypes of Per2 and Clock mutant mice are similar in that they both affect regulation and maintenance of cellular populations that underlie mammary morphogenesis. It is not surprising, however, that they affect the gland in different ways and at different developmental time points. A previous study found that PER2, PER1 and CLOCK have little overlap amongst their transcription targets, and thus suggests that these differential targets are crucial to functions outside of time-keeping duties (Koike et al., 2012). MaSC hierarchy models suggest that early alveolar progenitor cells of the luminal lineage give rise to the alveolar secretory cells during pregnancy, as previously discussed. We observed that Per2−/− virgin mice have a disrupted altered cell identity in their luminal cells, indicated by altered localization of K14, that recovers during late pregnancy and throughout lactation. These results suggest that, during alveolar expansion, a more differentiated cell population can be generated from a luminal progenitor present in the affected virgin gland, despite altered cell identity or defects in the ductal progenitors that are important in growth and branching morphogenesis during pubertal development in virgin Per2−/− mice.
Furthermore, it was shown that, during pubertal development, ERα is a key regulator of ductal growth and branching (Mallepell et al., 2006). Mallepell et al. demonstrated that ERα signals in a paracrine fashion, driving ductal outgrowth. We have observed that the majority of MECs from Per2−/− virgin mice are ERα positive, yet these glands fail to develop. It is unclear whether the cause of this is the lack of ERα-negative MECs capable of receiving positive signal or some other mechanism. Gery et al. found that ERα stabilization is inhibited by PER2 in MCF-7 cells, but, conversely, PER2 expression is upregulated through ERα binding of estrogen receptor response elements located in the PER2 promoter (Gery et al., 2007). Further work is needed to elucidate the mechanisms and interactions involving PER2 and ERα.
SLUG is a member of the SNAIL/SLUG family of transcription factors and plays important roles in cell fate determination and cancer progression. In many tissues, SLUG regulates EMT to ensure proper migration during development (Savagner, 2001; Tucker, 2004). During breast cancer progression, increased SLUG leads to deregulation of E-cadherin expression, and contributes to cell mobility, metastasis and poor patient outcome (Bolos et al., 2003; Martin et al., 2005). Our results contradict those of a previous study, which found that knockdown of PER2 in MCF10A cells results in upregulation of SLUG and decreased E-cadherin levels (Hwang-Verslues et al., 2013). Instead, we found that both SLUG and E-cadherin levels increased with loss of PER2 in the mouse mammary gland, as well as in MCF10A cells. This raises an interesting question of the association of Per2 and Slug. Previous studies have shown that Slug is expressed in basal MaSCs and early luminal epithelial cells during initial ductal morphogenesis, requiring co-expression of Sox9, which was not observed in Per2−/− mammary glands (data not shown) (Guo et al., 2012; Nassour et al., 2012). The contrasting results presented in this study and others suggest that SLUG might not be directly involved in the observed phenotype, but merely an additional marker of bipotency.
Previously, it was established that p53 regulates Per2 expression by binding a p53 response element, which overlaps the E-box in the Per2 promoter (Miki et al., 2013). Miki et al. reported that this transcriptional repression of Per2 occurs because p53 binding inhibits CLOCK/BMAL1 binding of the E-box. It has also been shown that PER2 interacts with p53 to stabilize p53 by inhibiting degradation and keeping the cell poised for a p53 response, and also by modulating expression of p53 target genes (Gotoh et al., 2014). With these studies in mind, it becomes apparent that p53 or its targets might play a potential role in the PER2-deficient phenotype observed here. Purvis et al. found that modulating p53 expression resulted in induction of downstream targets related to terminal fates (Purvis et al., 2012). Interestingly, overexpression of WT p53 in the mouse was shown to inhibit ductal outgrowth and dramatically impact development of the gland (Gatza et al., 2008). These studies suggest a potential role for p53 in contributing to the PER2 phenotype we observe in our model.
Although the PER2-deficient phenotype appears to be more bipotent in nature and not a classical EMT phenotype, and there are potential interactions of ERα and p53 with PER2, these recent studies open the door to a multitude of possibilities involving other factors that could play a crucial role in mammary development. Altogether, this study suggests that PER2 is required for proper mouse mammary gland development and thus has a crucial role outside of its circadian function. These results further strengthen our hypothesis that Per2-deficient MECs are held in a pre-differentiated state, which is a precursor to both luminal and myoepithelial mammary cell types. Further mechanistic and lineage tracing studies are required to unravel the role of PER2 in the developing mammary gland.
MATERIALS AND METHODS
Animals
Female WT, Per1−/− and Per2ldc mice on a C57Bl/6J background were generously provided by Dr David Weaver (University of Massachusetts Medical School, Worcester, MA, USA). Establishment, characterization and behavioral analysis of these mice have been described previously (Bae et al., 2001). Briefly, the mPer2-targeting construct deleted exon 5 and part of exon 6, encompassing part of the PAS domain, resulting in a null allele, and we refer to mice with this allele as Per2−/−. Animals for this study were maintained at Texas A&M Institute for Genomic Medicine (TIGM) under a standard 12 h light:12 h dark cycle (LD 12:12; lights on at 06:00 h), with access to food and water ad libitum. All protocols for this study were reviewed and approved by the Texas A&M University Institutional Animal Care and Use Committee and complied with relevant regulatory standards.
MCF10A cell culture
Human MCF10A-pSIL, MCF10A-shPER1 and MCF10A-shPER2 MECs were generated as previously described (Schmitt et al., 2017). Cells were cultured in growth medium consisting of Dulbecco's modified Eagle medium (DMEM)/F12 with 5% donor horse serum, epidermal growth factor, hydrocortisone, insulin and penicillin/streptomycin as previously described (Debnath et al., 2003). Acini growth assays and IF were performed as previously described (Gustafson et al., 2009). Briefly, 5000 cells were plated on Matrigel Matrix (Corning) in eight-well chamber slides (Lab-Tek) and allowed to grow for 15 days to form acini. Following day 15, cells were fixed with 2% paraformaldehyde and stained by IF. For cell proliferation assays, 10,000 cells were plated in triplicate on six-well plates and counted in triplicate at 96 h.
qPCR
RNA from mouse MECs and human MCF10A cells was isolated using High Pure RNA and High Pure RNA Tissue Isolation Kits (Roche Diagnostics). Following RNA isolation, cDNA was synthesized from 1 µg RNA using an iScript cDNA Synthesis Kit (Bio-Rad). qPCR analysis reactions consisted of 6 µl 2× SYBR GoTaq qPCR master mix (Promega), 1 µl primer mix (10 nM) and 3 µl cDNA input, and reactions were processed on a CFX384 Real Time System (Bio-Rad). All qPCR analyses were performed using the ΔΔcT method to indicate the fold change in target genes relative to β-actin in the MCF10A cell line and 18 s (Rn18s) in the mouse studies as previously described (Livak and Schmittgen, 2001). Primer sequences can be found in Table S1.
Whole-mount, immunohistochemical and IF staining
Mouse mammary glands from C57Bl/6J and Per2−/− mice were harvested from 8-week-old (Per2−/− n=4, WT n =3), 10-week-old (Per2−/− n=4, WT n=4) and 12-week-old (Per2−/− n=4, WT n=3) estrous-staged virgin female mice at 12:00 h, and whole-mount stained with Carmine Alum. Tissues were fixed for 2 h in 4% paraformaldehyde at 4°C then moved to 70% ethanol until staining. To begin staining, tissues were rinsed in PBS and stained in Carmine Alum solution (Carmine C-1022 and Aluminum Potassium Sulfate A-7167, Sigma-Aldrich) overnight at room temperature. The following morning, tissues were washed in a series of ethanol dilutions, cleared in xylene overnight, and stored in methyl salicylate at room temperature. To analyze branching morphogenesis, whole-mount mammary gland images were divided into six equal parts (or relative positions), and branch junction points and terminal ducts were counted in each relative position section.
For immunohistochemical (IHC) and IF staining, mammary glands from 8-, 10- and 12-week-old diestrus-staged WT, Per1−/− and Per2−/− mice were fixed in 4% paraformaldehyde solution. Serial mammary sections were cut at 8 μm and used for either IHC or IF staining, as previously described (Scribner et al., 2011). A standard IF and IHC protocol was followed for all sections used in this study. Briefly, sections were deparaffinized for 30 min at 60°C followed by 3-5 min washes in xylene, 100% ethanol, 95% ethanol, 70% ethanol and 1× PBS. Antigen retrieval was performed by boiling sections in 10 mM sodium citrate for 5 min. All sections were then washed in 1× PBS for 5 min, followed by a 6 min incubation in 3% hydrogen peroxide (IHC only). Sections were then blocked for 60 min in PBS with Tween (PBS-T) containing 10% horse serum. All sections were incubated overnight at 4°C in primary antibodies (Table S2) and washed in PBS-T for 10 min the following day. For IHC, sections were incubated for 60 min in secondary antibodies, washed in PBS-T for 5 min, incubated for 30 min with ABC (Vector Laboratories) and then incubated with antibody-dependent DAB (Vector Laboratories) for a timed period. Sections were counterstained with Methyl Green and dehydrated with 95% ethanol, 100% ethanol and xylene, and then coverslips were mounted with Permount mounting medium (Electron Microscopy Sciences). For IF, sections were incubated in secondary antibodies for 45 min, washed in PBS-T for 5 min, washed in PBS for 5 min and incubated with DAPI for 5 min. Sections were then washed two times in Milli-Q water and cover slips were mounted with ProLong Gold (Invitrogen). For histological sections requiring a mouse primary antibody, the 60 min block in PBS-T was modified. Briefly, slides were incubated in MOM blocking reagent (Vector Laboratories) for 60 min, followed by a 5 min wash in PBS. Slides were then incubated for 5 min in MOM diluent before proceeding to primary antibody incubation overnight.
Mammary transplants
Mouse mammary epithelium was isolated from the number 4 inguinal mammary gland of 12-week-old WT and Per2−/− mice, transplanted contralaterally into the cleared mammary fat pads of 21-day-old syngeneic mice and analyzed after 8 weeks of outgrowth, as previously described (Laffin et al., 2008).
FACS
Mouse mammary glands were harvested from 12-week-old diestrus-staged virgin WT (n=10) and Per2−/− (n=13) mice at 12:00 h, followed by a primary MEC isolation. Glands were homogenized on a 10 cm plate using scalpels, and digested in DMEM/F12 containing 10% FBS and 2 mg/ml collagenase A for 30 min at 37°C. Samples were then treated with DNase I and pulse spins were performed to remove red blood cells. Remaining cells were re-suspended in trypsin and incubated for 20 min at 37°C to generate a single cell suspension. Epithelial cells were then isolated using the EasyStep Mouse Epithelial Cell Enrichment Kit (StemCell Technologies), following the manufacturer’s protocol. Primary MECs were labeled with CD24(PE)- and CD49f(FITC)-conjugated antibodies to identify myoepithelial and luminal lineages. Cells were sorted using an Accuri C6 (BD Biosciences) and analyzed using FlowJo v10 (BD) analysis software with the auto-compensation setting.
RNA-Seq
Mammary glands were harvested from 12-week-old diestrus-staged mice at 12:00 h, glands were pooled (Per2−/− n=4, WT n=3) and MEC preparation performed, as described above. RNA was isolated from MECs using a High Pure RNA Isolation Kit (Roche), and RNA-Seq libraries were made with a TrueSeq RNA preparation kit (Illumina) and polyA selection step. The indexed libraries were pooled and sequenced (150-bp, paired-end sequencing) on a HiSeq 2500 (Illumina). The Texas A&M AgriLife Genomics and Bioinformatics Core generated the RNA-Seq libraries and performed the RNA-Seq reactions. Raw sequence data were processed using bioinformatic tools provided by the Texas A&M Institute for Genome Sciences and Society. FASTQ reads were de-multiplexed and assessed for quality using FastQC. Samples were then aligned to mouse reference genome assembly (mm10, ENSEMBL 74) using HISAT2 (version 2.0.5) (Kim et al., 2015). Aligned sequences were processed with cuffdiff to identify differentially expressed genes (Trapnell et al., 2013). GOrilla (http://cbl-gorilla.cs.technion.ac.il) was used to identify biological pathways differentially regulated between the Per2−/− and WT mice (Eden et al., 2007, 2009). Briefly, a fold change sorted gene list with cuffdiff no test status genes removed was inputted into GOrilla.
Statistical analysis
All samples for qPCR were run in biological and technical triplicate. Image analysis and cell counts were generated from at least three replicates. Student's t-tests were performed with significance indicated by P≤0.05 using JMP PRO12 (SAS). Where appropriate, data are represented by the mean±s.e.m.
Supplementary Material
Acknowledgements
The authors thank the Texas A&M Institute for Genome Sciences and Society for providing sequence analysis hardware, software and support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: W.W.P.; Methodology: C.M.M., E.E.S., R.P.M., D.E., W.W.P.; Validation: C.M.M., E.E.S.; Formal analysis: C.M.M., E.E.S., M.R.; Investigation: E.E.S., T.R.S., J.E., R.P.M., M.R., W.W.P.; Data curation: C.M.M., E.E.S.; Writing - original draft: C.M.M., E.E.S.; Writing - review & editing: C.M.M., E.E.S., J.E., M.R., W.W.P.; Visualization: W.W.P.; Supervision: W.W.P.; Project administration: W.W.P.; Funding acquisition: W.W.P.
Funding
This work was supported by the National Institutes of Environmental Health Sciences [R01-ES02344 and P30-ES023512 to W.W.P.]. E.E.S. was supported by a National Institutes of Health institutional training grant [T32 ES026568]. Deposited in PMC for release after 12 months.
Data availability
RNA-Seq data from this study have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through accession number GSE109522.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.157966.supplemental
References
- Alvarez J. D., Chen D., Storer E. and Sehgal A. (2003). Non-cyclic and developmental stage-specific expression of circadian clock proteins during murine spermatogenesis. Biol. Reprod. 69, 81-91. 10.1095/biolreprod.102.011833 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M.-L., Sutherland K. D., Barker H., Thomas R., Shackleton M., Forrest N. C., Hartley L., Robb L., Grosveld F. G., van der Wees J. et al. (2007). Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201-209. 10.1038/ncb1530 [DOI] [PubMed] [Google Scholar]
- Bae K., Jin X., Maywood E. S., Hastings M. H., Reppert S. M. and Weaver D. R. (2001). Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525-536. 10.1016/S0896-6273(01)00302-6 [DOI] [PubMed] [Google Scholar]
- Bolos V., Peinado H., Pérez-Moreno M. A., Fraga M. F., Esteller M. and Cano A. (2003). The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116, 499-511. 10.1242/jcs.00224 [DOI] [PubMed] [Google Scholar]
- Borgs L., Beukelaers P., Vandenbosch R., Nguyen L., Moonen G., Maquet P., Albrecht U., Belachew S. and Malgrange B. (2009). Period 2 regulates neural stem/progenitor cell proliferation in the adult hippocampus. BMC Neurosci. 10, 30 10.1186/1471-2202-10-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C., Park S., Vass T., Lydon J. P., O'Malley B. W. and Weinberg R. A. (1998). A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc. Natl. Acad. Sci. USA 95, 5076-5081. 10.1073/pnas.95.9.5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno S. and Darzynkiewicz Z. (1992). Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 25, 31-40. 10.1111/j.1365-2184.1992.tb01435.x [DOI] [PubMed] [Google Scholar]
- Chakrabarti R., Wei Y., Romano R.-A., DeCoste C., Kang Y. and Sinha S. (2012). Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells 30, 1496-1508. 10.1002/stem.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. S., Chakrabarti R., Escamilla-Hernandez R. and Sinha S. (2009). Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev. Biol. 329, 227-241. 10.1016/j.ydbio.2009.02.032 [DOI] [PubMed] [Google Scholar]
- Daniel C. W. and Smith G. H. (1999). The mammary gland: a model for development. J. Mammary Gland Biol. Neoplasia 4, 3-8. 10.1023/A:1018796301609 [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S. K. and Brugge J. S. (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256-268. 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Dolatshad H., Campbell E. A., O'Hara L., Maywood E. S., Hastings M. H. and Johnson M. H. (2006). Developmental and reproductive performance in circadian mutant mice. Hum. Reprod. 21, 68-79. 10.1093/humrep/dei313 [DOI] [PubMed] [Google Scholar]
- Eden E., Lipson D., Yogev S. and Yakhini Z. (2007). Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 3, e39 10.1371/journal.pcbi.0030039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D. and Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatza C. E., Dumble M., Kittrell F., Edwards D. G., Dearth R. K., Lee A. V., Xu J., Medina D. and Donehower L. A. (2008). Altered mammary gland development in the p53+/m mouse, a model of accelerated aging. Dev. Biol. 313, 130-141. 10.1016/j.ydbio.2007.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S., Virk R. K., Chumakov K., Yu A. and Koeffler H. P. (2007). The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene 26, 7916-7920. 10.1038/sj.onc.1210585 [DOI] [PubMed] [Google Scholar]
- Gotoh T., Vila-Caballer M., Santos C. S., Liu J., Yang J. and Finkielstein C. V. (2014). The circadian factor Period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol. Biol. Cell 25, 3081-3093. 10.1091/mbc.E14-05-0993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Keckesova Z., Donaher J. L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zürrer-Härdi U., Bell G. et al. (2012). Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015-1028. 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson T. L., Wellberg E., Laffin B., Schilling L., Metz R. P., Zahnow C. A. and Porter W. W. (2009). Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBPbeta. Oncogene 28, 1561-1568. 10.1038/onc.2008.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K., Wakatsuki Y., Iigo M. and Shibata S. (2006). Circadian Clock mutation in dams disrupts nursing behavior and growth of pups. Endocrinology 147, 1916-1923. 10.1210/en.2005-1343 [DOI] [PubMed] [Google Scholar]
- Hwang-Verslues W. W., Chang P.-H., Jeng Y.-M., Kuo W.-H., Chiang P.-H., Chang Y.-C., Hsieh T.-H., Su F.-Y., Lin L.-C., Abbondante S. et al. (2013). Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl. Acad. Sci. USA 110, 12331-12336. 10.1073/pnas.1222684110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B. and Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K. and Takahashi J. S. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349-354. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korach K. S., Couse J. F., Curtis S. W., Washburn T. F., Lindzey J., Kimbro K. S., Eddy E. M., Migliaccio S., Snedeker S. M., Lubahn D. B. et al. (1996). Estrogen receptor gene disruption: molecular characterization and experimental and clinical phenotypes. Recent Prog. Horm. Res. 51, 159-186; discussion 186-158. [PubMed] [Google Scholar]
- Laffin B., Wellberg E., Kwak H.-I., Burghardt R. C., Metz R. P., Gustafson T., Schedin P. and Porter W. W. (2008). Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol. Cell. Biol. 28, 1936-1946. 10.1128/MCB.01701-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mallepell S., Krust A., Chambon P. and Brisken C. (2006). Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc. Natl. Acad. Sci. USA 103, 2196-2201. 10.1073/pnas.0510974103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. A., Goyal A., Watkins G. and Jiang W. G. (2005). Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann. Surg. Oncol. 12, 488-496. 10.1245/ASO.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Metz R. P., Qu X., Laffin B., Earnest D. and Porter W. W. (2006). Circadian clock and cell cycle gene expression in mouse mammary epithelial cells and in the developing mouse mammary gland. Dev. Dyn. 235, 263-271. 10.1002/dvdy.20605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Matsumoto T., Zhao Z. and Lee C. C. (2013). p53 regulates Period2 expression and the circadian clock. Nat. Commun. 4, 2444 10.1038/ncomms3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk J. A., Green C. B. and Takahashi J. S. (2012). Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445-462. 10.1146/annurev-neuro-060909-153128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya T., Hiraishi K., Horie N., Mitome M. and Shinohara K. (2007). Correlative association between circadian expression of mousePer2 gene and the proliferation of the neural stem cells. Neuroscience 146, 494-498. 10.1016/j.neuroscience.2007.02.018 [DOI] [PubMed] [Google Scholar]
- Morse D., Cermakian N., Brancorsini S., Parvinen M. and Sassone-Corsi P. (2003). No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol. Endocrinol. 17, 141-151. 10.1210/me.2002-0184 [DOI] [PubMed] [Google Scholar]
- Nassour M., Idoux-Gillet Y., Selmi A., Côme C., Faraldo M.-L., Deugnier M.-A. and Savagner P. (2012). Slug controls stem/progenitor cell growth dynamics during mammary gland morphogenesis. PLoS ONE 7, e53498 10.1371/journal.pone.0053498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H., Yamaguchi S. and Yagita K. (2002). Molecular machinery of the circadian clock in mammals. Cell Tissue Res. 309, 47-56. 10.1007/s00441-002-0572-5 [DOI] [PubMed] [Google Scholar]
- Purvis J. E., Karhohs K. W., Mock C., Batchelor E., Loewer A. and Lahav G. (2012). p53 dynamics control cell fate. Science 336, 1440-1444. 10.1126/science.1218351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert S. M. and Weaver D. R. (2001). Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647-676. 10.1146/annurev.physiol.63.1.647 [DOI] [PubMed] [Google Scholar]
- Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M. and Naef F. (2011). Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 9, e1000595 10.1371/journal.pbio.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner P. (2001). Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. BioEssays 23, 912-923. 10.1002/bies.1132 [DOI] [PubMed] [Google Scholar]
- Schmitt E. E., Barhoumi R., Metz R. P. and Porter W. W. (2017). Circadian regulation of Benzo[a]Pyrene metabolism and dna adduct formation in breast cells and the mouse mammary gland. Mol. Pharmacol. 91, 178-188. 10.1124/mol.116.106740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scribner K. C., Wellberg E. A., Metz R. P. and Porter W. W. (2011). Singleminded-2s (Sim2s) promotes delayed involution of the mouse mammary gland through suppression of Stat3 and NFkappaB. Mol. Endocrinol. 25, 635-644. 10.1210/me.2010-0423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K. J., Stingl J., Smyth G. K., Asselin-Labat M.-L., Wu L., Lindeman G. J. and Visvader J. E. (2006). Generation of a functional mammary gland from a single stem cell. Nature 439, 84-88. 10.1038/nature04372 [DOI] [PubMed] [Google Scholar]
- Sreekumar A., Roarty K. and Rosen J. M. (2015). The mammary stem cell hierarchy: a looking glass into heterogeneous breast cancer landscapes. Endocr Relat. Cancer 22, T161-T176. 10.1530/ERC-15-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H. I. and Eaves C. J. (2006). Purification and unique properties of mammary epithelial stem cells. Nature 439, 993-997. 10.1038/nature04496 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L. and Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46-53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinkalovsky O., Rosenlund B., Laerum O. D. and Eiken H. G. (2005). Clock gene expression in purified mouse hematopoietic stem cells. Exp. Hematol. 33, 100-107. 10.1016/j.exphem.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Tsinkalovsky O., Filipski E., Rosenlund B., Sothern R. B., Eiken H. G., Wu M. W., Claustrat B., Bayer J., Lévi F. and Laerum O. D. (2006). Circadian expression of clock genes in purified hematopoietic stem cells is developmentally regulated in mouse bone marrow. Exp. Hematol. 34, 1249-1261. 10.1016/j.exphem.2006.05.008 [DOI] [PubMed] [Google Scholar]
- Tucker R. P. (2004). Neural crest cells: a model for invasive behavior. Int. J. Biochem. Cell Biol. 36, 173-177. 10.1016/S1357-2725(03)00243-7 [DOI] [PubMed] [Google Scholar]
- Visvader J. E. (2009). Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 23, 2563-2577. 10.1101/gad.1849509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J. E. and Stingl J. (2014). Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 28, 1143-1158. 10.1101/gad.242511.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S., Mao L., Duplessis T., Yuan L., Dauchy R., Dauchy E., Blask D. E., Frasch T. and Hill S. M. (2012). Oscillation of clock and clock controlled genes induced by serum shock in human breast epithelial and breast cancer cells: regulation by melatonin. Breast Cancer (Auckl) 6, 137-150. 10.4137/BCBCR.S9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Li C., Zhu N.-L., Borok Z. and Minoo P. (2003). Timeless in lung morphogenesis. Dev. Dyn. 228, 82-94. 10.1002/dvdy.10346 [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Numano R., Abe M., Hida A., Takahashi R., Ueda M., Block G. D., Sakaki Y., Menaker M. and Tei H. (2000). Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682-685. 10.1126/science.288.5466.682 [DOI] [PubMed] [Google Scholar]
- Yang N., Williams J., Pekovic-Vaughan V., Wang P., Olabi S., McConnell J., Gossan N., Hughes A., Cheung J., Streuli C. H. et al. (2017). Cellular mechano-environment regulates the mammary circadian clock. Nat. Commun. 8, 14287 10.1038/ncomms14287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka M. J., Shearman L. P., Levine J. D., Jin X., Weaver D. R. and Reppert S. M. (1998). Molecular analysis of mammalian timeless. Neuron 21, 1115-1122. 10.1016/S0896-6273(00)80628-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.