ABSTRACT
Plants, with cells fixed in place by rigid walls, often utilize spatial and temporally distinct cell division programs to organize and maintain organs. This leads to the question of how developmental regulators interact with the cell cycle machinery to link cell division events with particular developmental trajectories. In Arabidopsis leaves, the development of stomata, two-celled epidermal valves that mediate plant-atmosphere gas exchange, relies on a series of oriented stem cell-like asymmetric divisions followed by a single symmetric division. The stomatal lineage is embedded in a tissue in which other cells transition from proliferation to postmitotic differentiation earlier, necessitating stomatal lineage-specific factors to prolong competence to divide. We show that the D-type cyclin, CYCD7;1, is specifically expressed just prior to the symmetric guard cell-forming division, and that it is limiting for this division. Further, we find that CYCD7;1 is capable of promoting divisions in multiple contexts, likely through RBR1-dependent promotion of the G1/S transition, but that CYCD7;1 is regulated at the transcriptional level by cell type-specific transcription factors that confine its expression to the appropriate developmental window.
KEY WORDS: Stomatal development, Cell cycle, Cyclin, Cell division, Differentiation, Guard cell
Highlighted Article: Stomatal transcription factors define GMC-specific expression of the D-type cyclin CYCD7;1, which promotes the G1/S transition in partnership with RBR1, enabling a direct link between cell cycle control and developmental fate decisions in Arabidopsis leaves.
INTRODUCTION
Development of multicellular organisms requires the coordination and control of cell proliferation with differentiation programs to generate distinct cell types, tissues and organs. Different cell lineages are specified by sets of developmental regulators and display various cell proliferation dynamics, suggesting that the cell cycle machinery might not always be comprised of the same components or controlled in the same way. In Arabidopsis, the mature leaf epidermis contains pavement cells, trichomes and stomata, three different functional cell types with their own developmental trajectories. In the past decade, genetic analyses of these cell types have enabled the discovery of various connections between cell cycle and development. For example, trichome precursors are specified early and patterned via lateral inhibition networks (Schellmann et al., 2002), and their maturation requires a shift from mitotic to endoreplication programs (Bramsiepe et al., 2010). Pavement cells also endoreplicate as they acquire their lobed morphologies (Katagiri et al., 2016). Stomata, pivotal for gas exchange between the plant and the environment, are derived from protodermal cells in a process that requires them to first become self-renewing and multipotent, but then to navigate an ordered set of divisions and differentiation programs to create the mature stoma (Matos and Bergmann, 2014).
Key transcriptional regulators of the stomatal lineage – the stage-specific, basic-helix loop-helix (bHLH) transcription factors SPEECHLESS (SPCH), MUTE and FAMA, and their broadly expressed heterodimer partners, SCRM/ICE1 and SCRM2 (Kanaoka et al., 2008) – each have roles in cell division and cell fate (Fig. 1A). SPCH drives asymmetric cell divisions that initiate the lineage, creating meristemoids (M) that can undergo continued self-renewing divisions. Plants lacking SPCH have no stomatal lineage. MUTE is essential to terminate the asymmetric self-renewing divisions and to induce the differentiation of meristemoids into guard mother cells (GMCs) (MacAlister et al., 2007; Pillitteri et al., 2007); loss of MUTE results in excess meristemoids at the expense of GMCs (MacAlister et al., 2007; Pillitteri and Torii, 2007). FAMA is required for the establishment of guard cells (GCs) but also to restrict GMCs to a single division. fama mutants exhibit numerous rounds of symmetric and parallel GMC divisions without acquisition of terminal GC identities (Matos et al., 2014; Ohashi-Ito and Bergmann, 2006). Plants bearing mutations in two R2R3 MYB transcription factor genes, FOUR LIPS (FLP) and MYB88, also exhibit fama-like GMC overproliferation phenotypes (Lai et al., 2005; Xie et al., 2010).
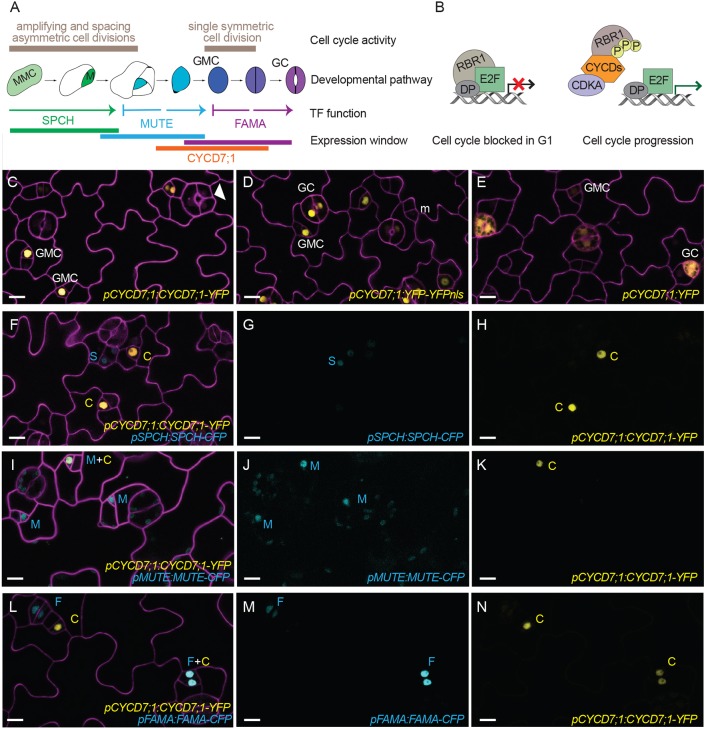
Fig. 1.
CYCD7;1 is expressed in GMCs prior to the last symmetric division of the stomatal lineage. (A) Schematic of stomatal development in Arabidopsis thaliana. Cell cycle activity, depicted in beige, along with cell fate transitions, transcription factor (TF) function and expression window of the master bHLH transcription factors SPCH (green), MUTE (blue), and FAMA (purple) and CYCD7;1 (orange) are shown. Meristemoid mother cells (MMC, light green) divide asymmetrically to enter the lineage. Meristemoids (green) can undergo amplifying and spacing asymmetric cell divisions until activity is terminated. Guard mother cells (GMC, blue) reenter the cell cycle only once to generate the pair of symmetric guard cells (GC, purple). (B) Plant RBR1/CYCD complexes driving the G1 to S transition and commitment to divide. RBR1 binds to E2F-DP transcription factors and blocks their ability to induce transcription of S phase genes. CYCDs interact with RBR1 through their LxCxE motif and facilitate phosphorylation of RBR1 by the CDKA;1/CYCD complex. Upon phosphorylation, RBR1 releases E2F transcription factors, which leads to expression of S phase genes for DNA replication. (C-E) Expression of the translational reporter pCYCD7;1:CYCD7;1-YFP, and the transcriptional reporters pCYCD7;1:YFP-YFPnls and pCYCD7;1:YFP (all yellow), in abaxial cotyledons. White arrowhead points at ectopic cell divisions. (F-N) Co-expression of pCYCD7;1:CYCD7;1-YFP (yellow, C) and pSPCH:SPCH-CFP (cyan, S), pMUTE:MUTE-CFP (cyan, M) and pFAMA:FAMA-CFP (cyan, F). Confocal images were taken at 5 dag. Cell outlines (magenta) are visualized with propidium iodide. All images are at the same magnification. Scale bars: 10 μm.
Presumably, among targets and partners of these transcription factors are cell cycle regulators that enable the diverse trajectories and division behaviors of epidermal cells. The components of the core cell cycle machinery are highly conserved among eukaryotes, although there has been a large expansion of genes in plants (Harashima et al., 2013; Inzé and De Veylder, 2006). The plant cell cycle is regulated by five main cyclin-dependent kinases (CDKs): CDKA;1, CDKB1;1, CDKB1;2, CDKB2;1 and CDKB2;2. CDKs require cyclins (CYCs) as binding partners for their kinase activity toward downstream phosphorylation targets. Plants genomes encode much larger families of cyclin genes than animals; for example, Arabidopsis encodes at least 32 cyclins (Vandepoele et al., 2002; Wang et al., 2004), and it has been speculated that this expansion allows plants to specifically regulate their postembryonic development (De Veylder et al., 2007; Harashima et al., 2013; Inzé and De Veylder, 2006). D-type cyclins as partners of CDKA;1 are crucial for the G1/S cell cycle transition and commitment to divide (Dewitte et al., 2007; Harbour and Dean, 2000; Riou-Khamlichi et al., 2000). Eight of 10 plant CYCDs have an RBR1-binding motif (LxCxE) (Kono et al., 2007; Menges et al., 2003). RBR1, the Arabidopsis homolog of the human tumor suppressor protein retinoblastoma, is crucial for the negative control of the cell cycle at G1/S transition (Desvoyes et al., 2006; Gutzat et al., 2012; Nowack et al., 2012; Uemukai et al., 2005; Zhao et al., 2012). Phosphorylation of RBR1 by CDKA;1/CYCD complexes inactivates its suppression of E2F transcription factors, allowing entry into S phase and commitment to divide (Fig. 1B) (Harashima et al., 2013; Nakagami et al., 2002; Nowack et al., 2012; Umen and Goodenough, 2001).
Here, we show how the cell cycle and cell fate transition from GMCs to GCs is regulated by the stomatal-lineage specific G1-S phase cell cycle regulator CYCD7;1. We demonstrate that CYCD7;1 activity is that of a typical D-type cyclin, but its expression window is narrowed by stomatal lineage specific transcription factors. By examining how CYCD7;1 works with the core cell cycle machinery and with stomatal regulators, and by revealing the phenotypes upon loss and gain of CYCD7;1 function, we link a core cell cycle regulator with a specific differentiation process and show how a formative division is initiated, but also restricted, to allow ‘one and only one division’ in GMCs to create a physiologically functional valve structure from its two identical daughters.
RESULTS
CYCD7;1 is expressed prior to the last symmetric division in the stomatal lineage
Among the ten known D-type cyclins in Arabidopsis, CYCD7;1 was uniquely enriched in transcriptional profiles of fluorescence-activated cell sorting (FACS)-isolated cells of the late stomatal lineage (Adrian et al., 2015). We confirmed this predicted expression in GMCs with transcriptional and translational reporters (Fig. 1C-E) and observed that additional copies of CYCD7;1-YFP could force ectopic divisions in GCs, suggesting that the protein could play a role in regulating this division (Fig. 1C, white arrowhead). A translational reporter, pCYCD7;1:CYCD7;1-YFP, was characterized previously as peaking in GMCs (Adrian et al., 2015); however, the identity of CYCD7;1 expressing cells was only assessed by morphology. To refine the expression pattern, we co-expressed pCYCD7;1:CYCD7;1-YFP with CFP reporters for SPCH, MUTE and FAMA (Fig. 1F-N). SPCH-CFP and CYCD7;1-YFP expression appear to be mutually exclusive, suggesting that CYCD7;1 is not expressed in young meristemoids (Fig. 1F-H). MUTE-CFP, a marker of late meristemoids and GMCs, partially overlaps with CYCD7;1-YFP. Closer analysis of cell morphologies indicated that cells expressing MUTE, but not CYCD7;1, were meristemoids, never GMCs, suggesting that MUTE is expressed before CYCD7;1 (Fig. 1I-K). CYCD7;1-YFP is visible before FAMA-CFP in GMCs, and is expressed together with FAMA in newly divided GCs. FAMA, but not CYCD7;1, persists into maturing GCs (Fig. 1L-N) (Ohashi-Ito and Bergmann, 2006). Thus, the expression of CYCD7;1 in the stomatal lineage is temporally and spatially controlled. It commences after MUTE expression and is extinguished before the end of FAMA expression (Fig. 1A).
We did not observe expression of CYCD7;1-YFP in any vegetative tissue from the seedling stage through flowering (data not shown). In adult plants, CYCD7;1-YFP was expressed in pollen sperm cells at anthesis, but not in the vegetative nucleus (Fig. S1A,B). The expression of a D-type cyclin (typically expressed at G1/S) is consistent with the observations that sperm cells undergo an extended S phase in mature pollen grains (Friedman, 1999; Zhao et al., 2012).
Why does CYCD7;1 have such a restricted expression pattern in the stomatal lineage? One possible explanation is that CYCD7;1 has a unique function in GMC divisions. A second possibility is that CYCD7;1 has a canonical role, i.e. it acts like other cyclins in promoting cell divisions, but it is important to be able to tightly control deployment of that role in the stomatal lineage. To distinguish between these models, we characterized plants missing or misexpressing CYCD7;1, tested relationships between CYCD7;1 and other cell cycle regulators, and defined how CYCD7;1 expression was constrained by stomatal lineage transcription factors.
Ectopic expression of CYCD7;1 triggers divisions; cycd7;1 mutants decelerate GMC divisions
If CYCD7;1 has canonical CYCD activity, it should be able to promote cell divisions outside its normal expression window. To test this, we expressed CYCD7;1 and CYCD7;1-YFP with the pan-epidermal promoter ML1 (Roeder et al., 2010). Ectopic expression of CYCD7;1 (YFP-tagged or untagged) induced cell divisions of pavement cells in the leaf (Fig. 2A-D), indicating that CYCD7;1 can function as a canonical D-type cyclin.
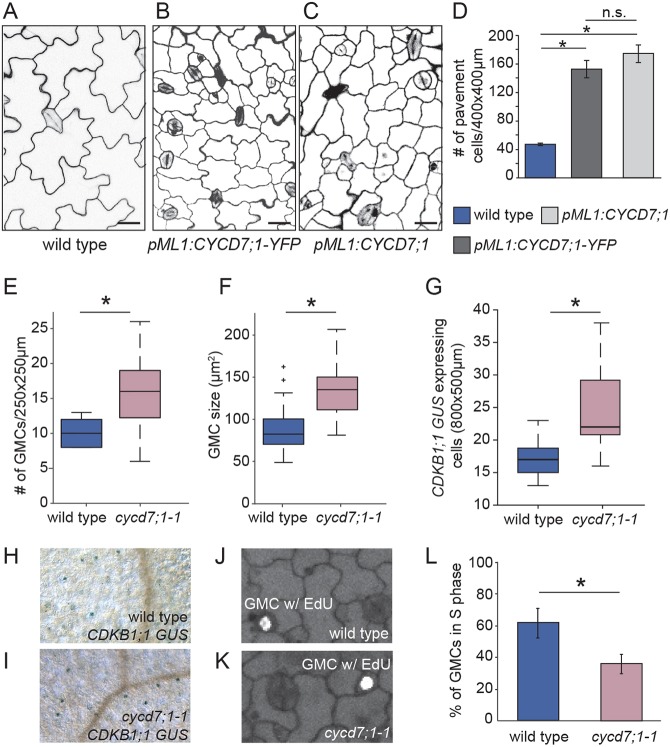
Fig. 2.
CYCD7;1 promotes cell divisions. (A-C) Confocal images of adaxial cotyledon epidermes of wild type, and plants expressing pML1:CYCD7;1-YFP and pML1:CYCD7;1 at 6 dag. Cell outlines were visualized with propidium iodide (magenta). Scale bars: 20 μm. (D) Quantification of extra divisions in pML1:CYCD7;1 and pML1:CYCD7;1-YFP pavement cells compared with those of wild-type cotyledons at 5 dag. Error bars represent standard error; *P<0.005, Student’s t-test; n.s., nonsignificant. (E) Quantification of the number of GMCs in wild-type and cycd7;1-1 cotyledons at 4 dag. *P=0.0032, Mann–Whitney U-test. (F) Quantification of GMC area in wild-type (n=55) and cycd7;1-1 (n=51) cotyledons at 4 dag. *P=6.76×10–13, Mann–Whitney U-test. (G) Quantification of cells expressing the CDKB1;1-GUS marker in wild-type and cycd7;1-1 cotyledons at 5 dag. *P=0.0023, Mann–Whitney U-test. (H) Image of a wild-type cotyledon expressing CDKB1;1-GUS marker at 5 dag. (I) Image of a cycd7;1-1 cotyledon expressing CDKB1;1-GUS marker at 5 dag. (J) Image of a wild-type GMC with EdU labeling in 4-day-old cotyledons. (K) Image of a cycd7;1-1 GMC with EdU labeling in 4-day-old cotyledons. (L) Quantification of EdU labeling in wild-type and cycd7;1-1 mutants. Graph shows the percentage of GMCs in S phase during a 90-min incubation with EdU. Error bars indicate the 95% confidence interval. *P=7×10–6, Fisher's exact test. Center lines in box plots show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles.
Next, we asked whether mutations of CYCD7;1 result in abnormal phenotypes. We obtained five insertional mutant alleles of CYCD7;1: FLAG_369E02 (cycd7;1-1) (Collins et al., 2012), FLAG_498H08 (cycd7;1-2), GK_496G06-019628, SALK_068423 and SALK_068526. Two of these alleles, cycd7;1-1 and cycd7;1-2, were outcrossed twice to Columbia plants (Col-0) and we determined that neither produced transcript by qRT-PCR (Fig. S2B,C) in 6-day-old seedlings. On a whole-plant level, we could not detect any abnormalities in cycd7;1-1 compared with wild type (Fig. S2D). Because CYCDs promote G1/S transitions and CYCD7;1 is specifically expressed during the GMC divisions, we asked whether cycd7;1-1 mutants halt the GMC to GC transition by counting the number of GCs in cotyledons. In maturing cotyledons 7 days after germination (dag), we count no difference in GC numbers between cycd7;1-1 and wild type (Fig. S2E-G). However, when monitored at 4 dag, when the stomatal lineage is still proliferating and GC precursors (meristemoids and GMCs) are abundant, cycd7;1-1 cotyledons have more GMCs than wild-type cotyledons (Fig. 2E). This suggests that cycd7;1-1 does not block the development of GCs from GMCs, but might be required to promote timely transition through the GMC stage. Interestingly, the average size of cycd7;1-1 GMCs is larger than that of wild-type GMCs (Fig. 2F). Plant cells are known to increase in size during G1, so this phenotype also suggests that CYCD7;1 hastens cell cycle progression in the GMC to GC transition. We confirmed that these GMC abundance and size phenotypes were present in plants bearing the cycd7;1-2 allele (Fig. S2C,H,I). Because cycd7;1-1 and cycd7;1-2 were originally in Wassilewskija (Ws) background and outcrossed to Col-0 twice, we tested whether any of the stomatal phenotypes were caused by ecotype background effects. Quantification of GMC number and size revealed no significant differences between Ws and Col-0, indicating that the phenotype could be attributed to the cycd7;1 mutations and not to ecotype differences (Fig. S2J,K).
We characterized the cycd7;1-1 mutant in more detail. We introgressed pCDKB1;1:GUS, which labels the transition from GMC to GCs (Boudolf et al., 2004), into cycd7;1-1 mutants. Compared with wild type, cycd7;1-1 mutants show increased numbers of GUS-positive cells, suggesting that these cells remain longer in GMC fate before they divide into GCs (Fig. 2G-I). To directly test this hypothesis, we labeled S phases with 5-ethynyl-2′-deoxyuridine (EdU), a thymidine analogue readily incorporated during DNA replication (Fig. 2J-L). Strikingly, significantly fewer GMCs in cycd7;1-1 showed EdU labeling (indicating that they were in S phase during the EdU pulse) compared with wild-type GMCs (Fig. 2L). Together, these data suggest that CYCD7;1 is required for GMCs to make a timely entry into S phase before their transition into GCs.
CYCD7;1 interacts with RBR1
Typically, CYCDs drive the G1/S transition through inactivation of RBR1, and RBR1 activity was previously shown to be essential for repressing divisions in the stomatal lineage (Borghi et al., 2010; Matos et al., 2014). If CYCD7;1 and RBR1 function together, we would expect them to be co-expressed, to physically interact, and for there to be a phenotypic consequence of disrupting the interaction. Indeed, CYCD7;1 and RBR1 were shown to physically interact in bimolecular fluorescence complementation (BIFC) and yeast two-hybrid (Y2H) assays, dependent on the presence of the RBR1 binding motif LxCxE in CYCD7;1 (Matos et al., 2014). In addition, CYCD7;1 and RBR1 are co-expressed in GMCs (Fig. 3A-C). To test whether this interaction is functionally important, we took advantage of the fact that our translational reporter of CYCD7;1 triggers extra cell divisions in GCs (Figs 1C and 3D,E). Approximately 24% of GCs have one, and 18% have two, ectopic divisions in pCYCD7;1:CYCD7;1-YFP plants at 5 dag (Fig. 3G). If the RBR1 interaction is important for CYCD7;1 function, then mutation of the RBR1 binding motif LxCxE into LxGxK in CYCD7;1 should abrogate this division-promoting activity. Strikingly, we found that pCYCD7;1:CYCD7;1LGK-YFP no longer triggers ectopic cell divisions in GCs (Fig. 3F,G). This effect was not caused by differences in expression levels between CYCD7;1-YFP and CYCD7;1LGK-YFP (Fig. S1C). Production of ectopic cell divisions in GCs, therefore, depends on the RBR1 binding residues in CYCD7;1.
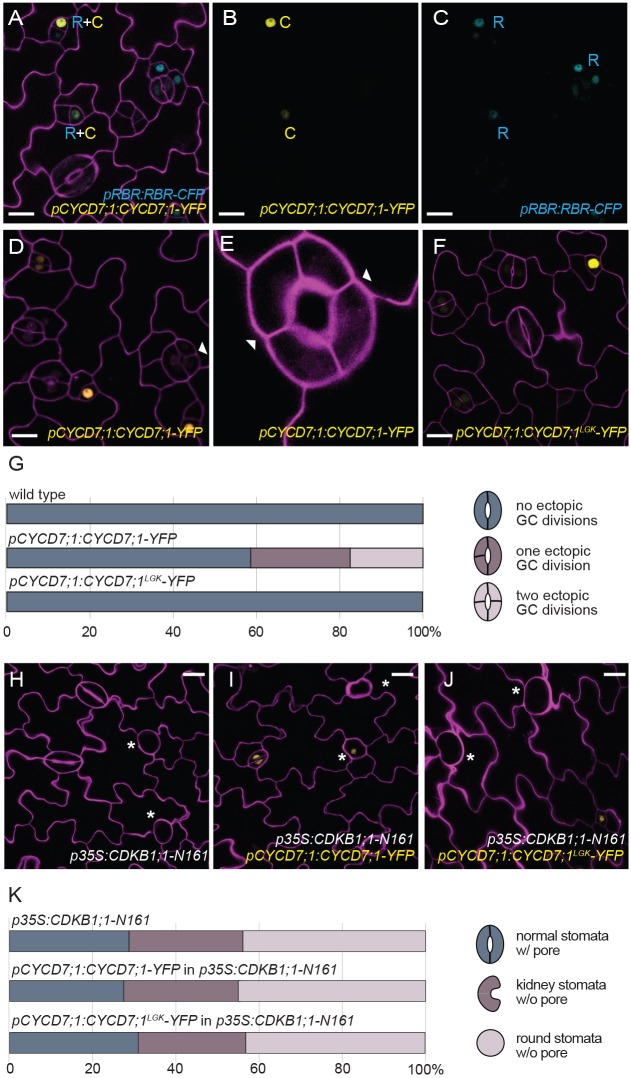
Fig. 3.
CYCD7;1 requires RBR1 binding and CDKB1;1 activity for ectopic cell divisions. (A-C) Co-expression of pCYCD7;1:CYCD7;1-YFP (yellow, C) and pRBR1:RBR1-CFP (cyan, R) in GMCs at 5 dag. (D,E) Expression of pCYCD7;1:CYCD7;1-YFP drives ectopic cell divisions (white arrowheads). (F) Expression of pCYCD7;1:CYCD7;1LGK-YFP (yellow) does not drive ectopic cell divisions. (G) Quantification of ectopic cell divisions in GCs at 5 dag in cotyledons in wild type (n=173), pCYCD7;1:CYCD7;1-YFP (n=306) and pCYCD7;1:CYCD7;1LGK-YFP (n=288); P=2.7343×10–22, Mann–Whitney U-test. (H) Phenotype of dominant-negative p35S:CDKB1;1-N161 at 6 dag. Asterisks label arrested GMCs. (I,J) Failure of pCYCD7;1:CYCD7;1-YFP (I) and pCYCD7;1:CYCD7;1LGK-YFP (J) to suppress CDKB1;1-N161 phenotype at 6 dag. Asterisks label arrested GMCs. (K) Quantification of stomata phenotypes in cotyledons in p35S:CDKB1;1-N161 (n=238), pCYCD7;1:CYCD7;1-YFP in p35S:CDKB1;1-N161 (n=296) and pCYCD7;1:CYCD7;1LGK-YFP in p35S:CDKB1;1-N161 (n=217) at 6 dag. Confocal images show cell outlines (magenta) stained with propidium iodide. Scale bars: 10 μm (A-D,F) and 20 μm (H-J).
CYCD7;1 needs CDKB1 activity to drive ectopic divisions
Cyclins bind to CDKs to ensure kinase activity and completion of cell division. It was previously shown that CDKB1;1 is enriched in late stomatal lineage cells (Boudolf et al., 2004) and that CDKA;1 is expressed in all dividing cells (Adachi et al., 2009), making these potential CYCD7;1 partners. In addition, reduction or loss of CDK activity (e.g. dominant-negative CDKB1;1-N161 (Boudolf et al., 2004), cdkb1;1 cdkb1;2 double mutants (Xie et al., 2010) or hypomorphic cdka;1 mutants (Weimer et al., 2012) results in undivided GCs, although these cells might still express GC fate markers. To test whether CYCD7;1 required CDK activity to drive divisions, we expressed CYCD7;1-YFP and CYCD7;1LGK-YFP under the CYCD7;1 promoter in plants bearing a dominant-negative version of CDKB1;1 (CDKB1;1-N161, Fig. 3H-J). Although we could see expression of both CYCD7;1 markers in arrested GMCs, they could neither rescue the phenotype nor trigger ectopic cell divisions (Fig. 3I-K). Thus, CYCD7;1 requires CDKB1 activity either as a partner, or downstream at the G2/M transition, for completion of the division.
CYCD7;1 expression domain is constrained by stomatal lineage transcription factors
Our evidence points to CYCD7;1 acting like a canonical CYCD; therefore, we turned our attention to regulation of its highly restricted expression pattern. Transcription factors MUTE and FAMA partially overlap CYCD7;1 expression (Fig. 1I-N), but MUTE precedes CYCD7;1, while FAMA persists longer. In addition, the R2R3 MYB transcription factor, FOUR LIPS (FLP), is expressed in GMCs and young GCs (Lai et al., 2005; Lee et al., 2014; Xie et al., 2010), and has been associated with cell cycle control in GCs through its repression of CDKB1;1 (Lee et al., 2013; Vanneste et al., 2011; Xie et al., 2010). Given these patterns, we tested whether MUTE was necessary to induce CYCD7;1 expression and whether FLP and FAMA repressed it. When pCYCD7;1:CYCD7;1-YFP was crossed into the mute mutant, we could observe the typical mute phenotype of many small meristemoid-like cells that fail to differentiate into GMCs (Pillitteri et al., 2007). In a few of these meristemoid-like cells, we detected weak CYCD7;1-YFP signal (Fig. 4A,B). Fluorescence intensity measurements showed that CYCD7;1-YFP signals in mute are ∼50% reduced (Fig. 4C-E), indicating that MUTE promotes CYCD7;1 expression, although it is not absolutely essential for it. In none of these images did we observe any ectopic divisions of the meristemoid-like cells. Mutations in FLP and its redundantly acting homolog MYB88 result in GMCs dividing multiple times before transitioning into GCs (Fig. 4F,I). CYCD7;1-YFP (and CYCD7;1LGK-YFP) translational reporters are highly expressed in flp/myb88, a result consistent with FLP/MYB88 repressing CYCD7;1 (Fig. 4F-L). Interestingly CYCD7;1-YFP, but not CYCD7;1LGK_YFP, induces ectopic divisions in the flp/myb88 GMCs and GCs (Fig. 4F-L).
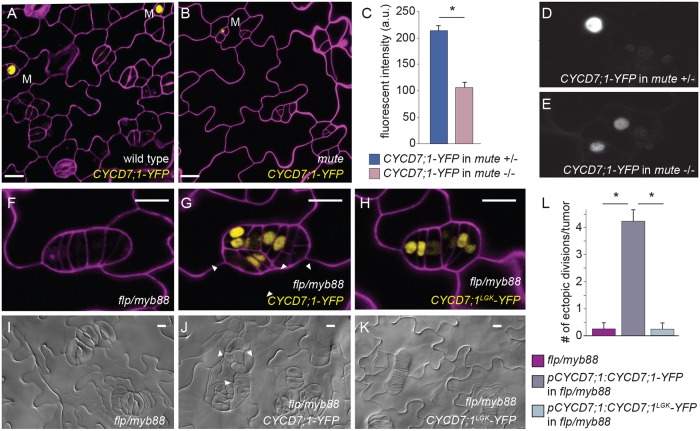
Fig. 4.
CYCD7;1-YFP is expressed at low levels in mute mutants and persists and drives ectopic divisions in flp/myb88 mutants. (A,B) Wild type and mute mutants expressing pCYCD7;1:CYCD7;1-YFP in 6-day-old cotyledons. M, meristemoid. (C-E) Quantification of fluorescence intensity of CYCD7;1-YFP in homozygous mute mutants (n=27) and their heterozygous or wild-type sister plants (n=21) (a.u., arbitrary units). Images of cotyledons were taken at 4 dag. Error bars show standard error; *P<0.0001, Student's t-test. (F) Phenotype of the double mutant flp/myb88 at 6 dag. (G) Expression of pCYCD7;1:CYCD7;1-YFP in flp/myb88 drives ectopic divisions in tumors at 6 dag. (H) Expression of pCYCD7;1:CYCD7;1LGK-YFP in flp/myb88 is less able to drive ectopic divisions at 6 dag. (I) DIC images of the phenotype of the double mutant flp/myb88 at 12 dag. (J) Expression of pCYCD7;1:CYCD7;1-YFP in flp/myb88 drives ectopic divisions in tumors at 12 dag. (K) Expression of pCYCD7;1:CYCD7;1LGK-YFP in flp/myb88 is less able to drive ectopic divisions at 12 dag. (L) Quantification of ectopic divisions in tumors of flp/myb88 mutants, pCYCD7;1:CYCD7;1-YFP in flp/myb88 mutant background and pCYCD7;1:CYCD7;1LGK-YFP in flp/myb88 mutant background. *P<0.0001, Student's t-test. White arrowheads label ectopic divisions. Confocal images show cell outlines (magenta) stained with propidium iodide. Scale bars: 10 μm.
The elevation of CYCD7;1 levels in flp/myb88 suggests that CYCD7;1 is repressed to limit its expression domain. We next tested whether this was true for FAMA. Additionally, because FAMA is a master transcriptional regulator of stomatal division and differentiation, we tested whether its regulation of CYCD7;1 was direct. In fama mutants, GMCs divide repeatedly without attaining GC fate (Fig. 5A-E). We found that these ‘tumors’ express CYCD7;1-YFP (Fig. 5B,C), as would be expected if FAMA acted to repress CYCD7;1. It is important to note, however, that after its initial upregulation, CYCD7;1-YFP expression fades in the fama ‘tumors’ in older leaves, suggesting that CYCD7;1 is also subject to post-translational regulation (Fig. 5D,E). In the fama ‘tumors’, pCYCD7;1:CYCD7;1-YFP also drives ectopic divisions (Fig. 5B,D,F, white arrowheads), but the CYCD7;1LGK version that cannot bind RBR1 does not (Fig. 5C,E,F). To test whether FAMA might directly regulate CYCD7;1, we extracted reads from a FAMA chromatin immunoprecipitation with sequencing (ChIP-seq) experiment, performed under similar conditions as in Lau and Bergmann (2015) and Lau et al. (2014), and found FAMA associated with the promoter region and gene body of CYCD7;1 (Fig. 5G, Fig. S3).
Fig. 5.
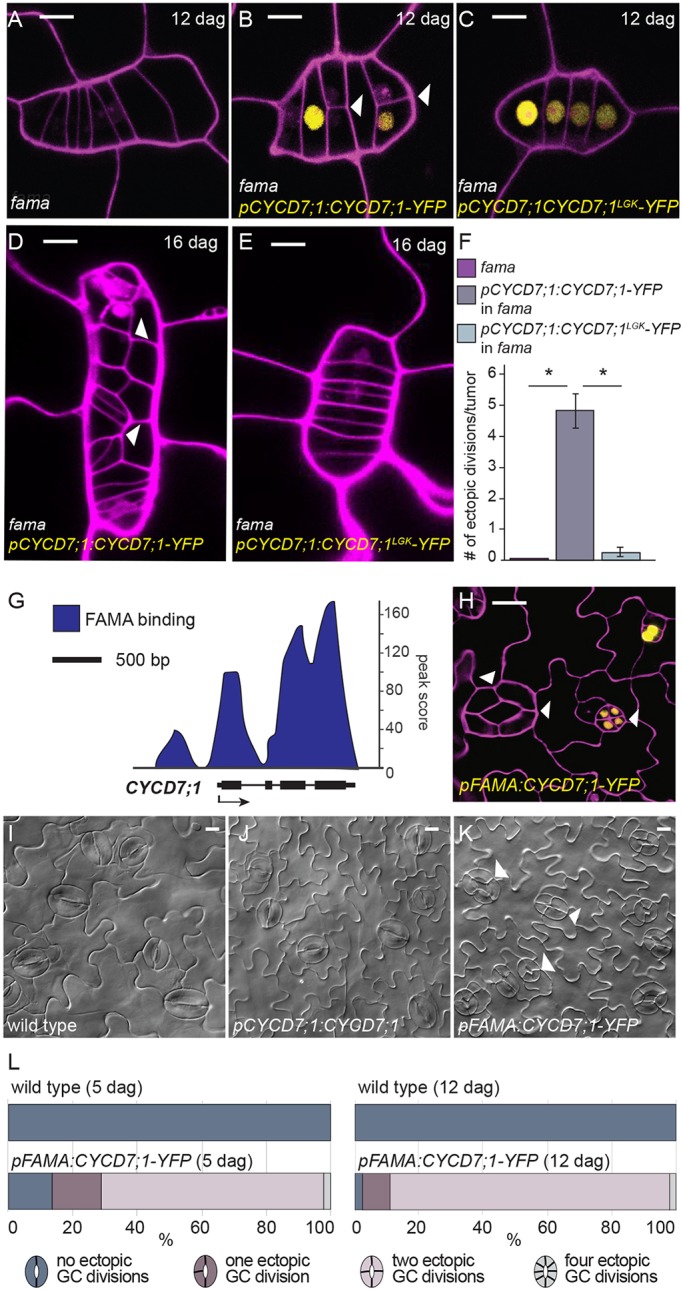
CYCD7;1 expression is regulated by FAMA, which serves to constrain CYCD7;1 activity. (A-E) Confocal images of fama at 12 dag, and pCYCD7;1:CYCD7;1-YFP in fama mutant background and pCYCD7;1:CYCD7;1LGK-YFP in fama mutant background at 12 dag and 16 dag. (F) Quantification of ectopic divisions in tumors of fama mutants, pCYCD7;1:CYCD7;1-YFP in fama mutant background and pCYCD7;1:CYCD7;1LGK-YFP in fama mutant background. *P<0.0001, Student's t-test. (G) ChIP-seq profile of FAMA binding to the promoter and gene body of CYCD7;1. Black arrow indicates gene orientation and transcriptional start sites. (H) Confocal image of pFAMA:CYCD7;1-YFP at 5 dag. White arrowheads show ectopic division and prolonged CYCD7;1-YFP presence. (I-K) DIC images of abaxial cotyledon epidermis of wild type, pCYCD7;1:CYCD7;1 and pFAMA:CYCD7;1-YFP at 12 dag. White arrowheads point at ectopic cell divisions. (L) Quantification of ectopic cell divisions in wild type (n=142) and pFAMA:CYCD7;1-YFP (n=237) at 5 dag (P=1.4371×10–54, Mann–Whitney U-test), and in wild type (n=125) and pFAMA:CYCD7;1-YFP (n=153) at 12 dag (P=1.3625×10–55, Mann–Whitney U-test). Differences between pFAMA:CYCD7;1-YFP at 5 dag and 12 dag are significant (P=6.4773×10–05, Mann–Whitney U-test). Differences between pFAMA:CYCD7;1-YFP at 5 dag and pCYCD7;1:CYCD7;1-YFP at 5 dag are also significant (Fig. 3G; P=1.0473×10–37, Mann–Whitney U-test). Confocal images show cell outlines (magenta) stained with propidium iodide. Scale bars: 10 µm.
Because CYCD7;1-YFP expression is extinguished before FAMA-YFP (Fig. 1L-N) and prolonged in fama, FAMA is expected to repress CYCD7;1 expression in GMCs. To test this, we transformed an estradiol-inducible version of FAMA (Hachez et al., 2011; Ohashi-Ito and Bergmann, 2006) into plants harboring the pCYCD7;1:YFP reporter line. This enabled us to provide a pulse of FAMA overexpression and follow CYCD7;1 response in the appropriate cells over time in intact leaves. We observed a significant reduction of YFP fluorescence in GMCs during an 8 h time course following estradiol (but not mock) induction of FAMA (Fig. S4 and supplementary Materials and Methods).
The phenotypes of loss and gain of CYCD7;1 activity suggest that its narrow window of expression is essential to guarantee a two-celled stomatal complex. Using the FAMA promoter in wild type, thus driving CYCD7;1 slightly later than under its endogenous cis-regulatory control, we find a dramatic enhancement of ectopic divisions (Fig. 5H-L). Compared with pCYCD7;1:CYCD7;1-YFP, in which ∼18% of stomata were four-celled at 5 dag (Fig. 3G), in pFAMA:CYCD7;1-YFP, that number was ∼70%, with 2% of stomata being eight-celled (n=237). The amount of four-celled stomata increases to 87% at 12 dag, with another 2% being eight-celled (n=153) (Fig. 5L). Quantification of fluorescence intensity indicates that expression with FAMA and CYCD7;1 promoters yields equivalent levels of CYCD7;1-YFP in GMCs (Fig. S1C); however, this fusion protein persists in ectopically divided GCs when expressed under the FAMA promoter (Fig. 5H). This directly links the activity of FAMA as a lineage specific transcription factor with the cell cycle regulator CYCD7;1 to ensure ‘one and only one division’ to create a pair of GCs.
DISCUSSION
We have shown that CYCD7;1 is specifically expressed in GMCs prior to the last symmetric cell division that forms the two-celled stomatal complex. Depletion of CYCD7;1 slows down this cell division, whereas ectopic expression of CYCD7;1 can trigger cell divisions in GCs. Mutation of the RBR1 binding motif in CYCD7;1 disrupts its interaction with RBR1 and renders CYCD7;1LGK incapable of driving ectopic division. The connection to RBR1 fits with previous work showing that CYCD7;1 interacts with CDKA;1 (Van Leene et al., 2010), together supporting a role for CYCD7;1 in the canonical regulatory complex for G1/S transitions and the commitment to divide. CYCD7;1 activity in cell cycles, however, is directly repressed by the lineage specific transcription factor FAMA to ensure a coupling between the cell division, which terminates the stomatal lineage, and the formation of terminally fated GCs. This interconnection represents a direct link between cell cycle regulators and developmental decisions (Fig. 6).
Fig. 6.
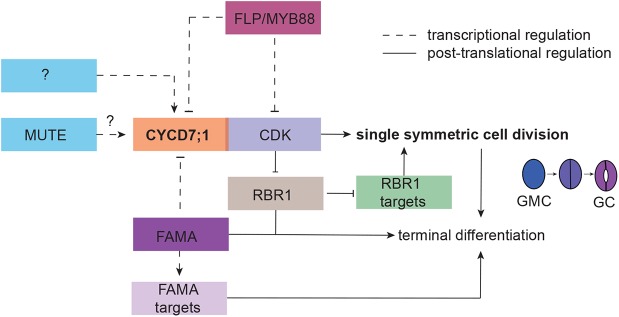
Model of the developmental integration of CYCD7;1 to ensure lineage-specific cell cycle regulation. Cell cycle regulators are integrated with stomatal specific transcription factors to ensure that the last formative division of the lineage creates a pair of symmetric GCs. Initiation of CYCD7;1 expression in GMCs requires MUTE and additional unknown factors (question mark). CYCD7;1 together with its CDK partner executes the formative division of the GMC. Owing to the observation that this last division is not completely abolished in cycd7;1 mutants, other D-type cyclins likely promote the G1-S phase transition. CDK/CYCD complexes phosphorylate RBR1 in order to release its negative function on S phase-promoting factors. To ensure termination of the lineage, the transcription factor FAMA, itself slightly later expressed than CYCD7;1, binds to the CYCD7;1 promoter to temporally restrict CYCD7;1 expression to GMCs and to restrict the cell cycle right after the last division. Transcriptional regulation is marked by dashed lines. This regulatory network ensures high cell cycle activity for the last formative division in the stomatal lineage and terminates cell division activity to ‘one and only one’ division.
CYCDs are crucial for the G1/S transition and commitment to divide, and are therefore interesting candidate hubs for the integration of developmental control with the cell cycle machinery. In Arabidopsis, there are 10 D-type cyclins, some active in multiple tissues (CYCD3s, CYCD4s, CYCD2;1), and others for which activity is linked to specific cell types (CYCD6;1 and CYCD7;1) or cell cycle behaviors (CYCD5;1 in endoreplication) (Dewitte et al., 2007; Kono et al., 2007; Sanz et al., 2011; Sterken et al., 2012; Adrian et al., 2015; Sozzani et al., 2010). Phylogenetic analyses showed that CYCD6;1 and CYCD7;1 proteins diverge from other D-type cyclins in Arabidopsis (Wang et al., 2004), but also that CYCD7;1 most closely resembles the single D-type cyclin in Physcomitrella (Menges et al., 2007), consistent with our observation that it could promote G1/S transitions (a core D-type activity) in multiple cell types.
Interestingly, both CYCD6;1 and CYCD7;1 are limiting for essential formative divisions during development. In the root, CYCD6;1 is important for the cortex endodermis initial daughter (CEID) cell divisions (Sozzani et al., 2010; Weimer et al., 2012). Here, SHORTROOT (SHR) directly activates expression of CYCD6;1, which works in concert with CDKA;1 to trigger the formative division of the CEID (Cruz-Ramírez et al., 2012; Sozzani et al., 2010; Weimer et al., 2012). This interaction promotes the initiation of an asymmetric stem-cell division program. By contrast, CYCD7;1 expression marks the boundary between two types of divisions: the continual asymmetric divisions of meristemoids versus the single symmetric division of a GMC. Here, we find a quantitative requirement for MUTE to promote full CYCD7;1 expression, but a clear requirement for FAMA and FLP/MYB88 to repress CYCD7;1 after GMC division. The low expression level of CYCD7;1 in the absence of MUTE might point to a direct role for MUTE in activating CYCD7;1 expression. MUTE is structurally similar to FAMA, and therefore might be able to interact with CYCD7;1 regulatory sequences. Alternatively, as meristemoid cells in mute never transition into GMCs, low CYCD7;1 levels might be an indirect consequence of altered cell fate. In either case, it is notable that the introduction of CYCD7;1-YFP in mute did not drive additional meristemoid cell divisions, suggesting that CYCD7;1 division-promoting behavior requires a threshold level not reached in this genetic background.
It is tempting to speculate that spatiotemporal restriction of CYCDs could be a mechanism to control the cell cycle machinery more efficiently, and to cope with different developmental programs. For example, leaves lose overall division competency and general cell cycle gene expression as they mature, leading to a situation in which GMCs are caught in a largely postmitotic zone. Formation of functional stomata, however, requires that the GMC divides again, though only once, as there have yet to be found plants in which stomatal pores are flanked by more than two GCs (McElwain et al., 2016). The importance of specialized CYCDs, however, must be squared with the relatively minor phenotypes associated with their loss; neither CYCD7;1 nor CYCD6;1 mutants abolish the production of specialized cells or tissue layers (Fig. 2) (Sozzani et al., 2010). Most likely, CYCD6;1 and CYCD7;1 assist other, more general, cyclins in executing cell division programs, or they might ensure particularly high cell cycle kinase activity. For example, in leaves, CYCD3;1 and CYCD3;2, despite being considered general G1/S cyclins (Dewitte et al., 2007, 2003; Menges et al., 2006), show high expression in the stomatal lineage (Adrian et al., 2015) and could be partially redundant with CYCD7;1. It is also important to recognize that CYCD/CDKA complexes likely have various downstream targets, and that increased kinase activity could induce different downstream processes, either in a feedback loop or for differentiation processes. In plants, specific CDK/cyclin complexes can have differential activity toward individual substrates. Both CDK and cyclin proteins contribute to substrate recognition (Harashima and Schnittger, 2012), but there is evidence that the cyclin plays the more prominent role (Weimer et al., 2016). Specific expression of individual cyclins, such as CYCD7;1, in the stomatal lineage could, therefore, contribute to fine-tuning of cell division control and downstream substrate recognition.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana Columbia (Col-0) was used as wild type in all experiments except as noted in Fig. S2. Seedlings were grown on half-strength Murashige and Skoog (MS) medium (Caisson Labs) at 22°C under 16 h-light/8 h-dark cycles and were examined at the indicated times. The following previously described mutants and reporter lines were used in this study: mute (Pillitteri et al., 2007); fama-1 (Ohashi-Ito and Bergmann, 2006); flp;myb88 (Lai et al., 2005); proSPCH:SPCH:CFP and proMUTE:MUTE-YFP (Davies and Bergmann, 2014); proRBR1:RBR1-CFP (Cruz-Ramírez et al., 2012); pro35S:CDKB1;1-N161 (Boudolf et al., 2004); and proCDKB1;1:GUS (Boudolf et al., 2004).
CYCD7;1 mutants
CYCD7;1 mutants FLAG_369E02 (cycd7;1-1) and FLAG_498H08 (cycd7;1-2) were derived from the INRA/Versailles collection (Versailles, France) and both lines were outcrossed to Col-0 twice. GK_496G06-019628 was derived from the GABI-Kat collection (Cologne, Germany). SALK_068423 and SALK_068526 were obtained from the Arabidopsis Biological Resource Center (Columbus, USA).
Vector construction and plant transformation
Constructs were generated using the Gateway system (Invitrogen). Appropriate genome sequences (PCR amplified from Col-0 or from entry clones) were cloned into Gateway-compatible entry vectors, typically pENTR/D-TOPO (Life Technologies), to facilitate subsequent cloning into plant binary vectors pHGY (Kubo et al., 2005) or R4pGWB destination vector system (Nakagawa et al., 2008; Tanaka et al., 2011). The translational reporter for CYCD7;1 was generated by cloning the genomic fragment (promoter+CDS) into the entry vector pENTR to generate the entry vector CYCD7;1-genomic-pENTR, followed by LR recombination into the destination vector pHGY to generate the final construct. For the translational reporter for CYCD7;1LGK, the LxCxE motif of CYCD7;1-genomic-pENTR was mutated to LxGxK by site-directed mutagenesis using the QuikChange II Kit (Agilent) to generate the entry clone CYCD7;1-genomic-pENTR and then recombined into pHGY. The transcriptional reporters for CYCD7;1 were generated by cloning the CYCD7;1 promoter region into pENTR, then recombined into the destination vectors pHGY (cytosolic YFP). The other constructs generated in this study, proCYCD7;1:YFP-YFPnls (a transcriptional reporter fused to YFP and a second nlsYFP), proFAMA:FAMA-CFP, proML1:CYCD7;1-YFP, proML1:CYCD7;1, proCYCD7;1:CYCD7;1 and proFAMA:CYCD7;1-YFP, were generated with the tripartite recombination of the plant binary vector series R4pGWB (Nakagawa et al., 2008; Tanaka et al., 2011), with the Gateway entry clones of the promoters and coding sequences compatible with the binary R4pGWB destination vector system. Primer sequences used for entry clones are provided in Table S1. Transgenic plants were generated by Agrobacterium-mediated transformation (Clough, 2005), and transgenic seedlings were selected by growth on half-strength MS plates supplemented with 50 mg/l Hygromycin (pHGY-, p35HGY-, pGWB1-, pGWB540-based constructs), 100 mg/l Kanamycin (pGWB440- and pGWV401-based constructs) or 12 mg/l Basta (pGWB640-based constructs).
Confocal and differential interference contrast microscopy
For confocal microscopy, images were taken with a Leica SP5 microscope and processed in ImageJ. Cell outlines were visualized by 0.1 mg/ml propidium iodide in water (Molecular Probes). Seedlings were incubated for 10 min in the staining solution and then rinsed once in H2O. For differential interference contrast (DIC) microscopy, samples were cleared in 7:1 ethanol:acetic acid, treated for 30 min with 1N potassium hydroxide, rinsed in water and mounted in Hoyer's medium. DIC images were obtained from the middle region of adaxial epidermis of cotyledons on a Leica DM2500 microscope or a Leica DM6 B microscope.
Quantification of fluorescence intensity
Images of GMCs in cotyledons were taken at 4 dag with identical settings between mute mutants and their sister plants from a segregating population and processed in ImageJ. Fluorescence intensity was measured as mean gray value in the nucleus, subtracted by the background. Measurements were averaged for mutant and control experiments, and Student's-t-test was used to determine the statistical significance.
GUS staining
The cycd7;1-1 mutant (which had been outcrossed two times to Col-0), was introgressed into CDKB1;1-GUS marker lines. Five-day-old seedlings were incubated in staining solution for 12 h and destained in 70% ethanol at 60-70°C for 4 h. The staining solution (5 ml) contained 100 µl 10% Triton X-100, 250 µl 1 M NaPO4 (pH 7.2), 100 µl 100 mM potassium ferrocyanide, 100 µl potassium ferricyanide, 400 µl 25 mM X-Gluc and 4050 µl dH2O. Images were taken with a Leica DM6 B microscope.
EdU labeling
EdU labeling was performed using the Click-iT EdU Alexa Fluor 488 Imaging Kit (ThermoFisher Scientific). Four-day-old seedlings were incubated in 20 µM EdU solution in half-strength MS for 90 min at room temperature. Seedlings were transferred to new tubes and washed three times with wash buffer (1% BSA in PBS). Wash buffer was removed and fixation buffer was added (3.7% formaldehyde in PBS) for 30 min at room temperature. Seedlings were transferred to new tubes and washed two times with permeabilization buffer (0.5% Triton X-100 in PBS) for 10 min each, protected from light on a slow rocking platform. Plants were transferred to new tubes and incubated in reaction cocktail (455 µl Click-IT reaction buffer, 20 µl CuSO4, 2 µl Alexa Fluor Azide 488, 25 µl 1× Click-IT EdU additive) for 1 h at room temperature, protected from light, without agitation. Seedlings were transferred to new tubes and washed twice for 10 min at room temperature with wash buffer on slow rocking platforms, protected from light. Cotyledons were imaged using a Leica SP5 microscope not more than 2 h after the completion of washes, and images were processed in ImageJ.
qPCR
For qPCR, 100 mg ground frozen material from 6-day-old plants was used for RNA extraction according to the manufacturer's manual (RNeasy Mini Kit, Qiagen), with 1 µg total RNA used as a template for cDNA synthesis (iScript cDNA synthesis kit, Bio-Rad). qPCR setup was according to the manual of the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). qPCR was performed by a CFX96 Real Time C1000 Thermal Cycler (Bio-Rad) according to the following reaction conditions: 95°C for 30 s, followed by 39 cycles at 95°C for 10 s and at 60°C for 30 s. ACTIN was used as a reference gene for all qPCRs performed. Primers are listed in Table S1.
Supplementary Material
Acknowledgements
We thank members of the Bergmann laboratory for helpful comments on the manuscript and Charles Hachez (UCLouvain) for his contributions to the initiation of the CYCD7;1 project.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.K.W., J.L.M., D.C.B.; Methodology: A.K.W., J.L.M., F.P., D.C.B.; Validation: A.K.W., J.L.M., D.C.B.; Formal analysis: A.K.W., J.L.M., N.S.; Investigation: A.K.W., J.L.M., N.S., F.P., D.C.B.; Resources: J.A.H.M., W.D., D.C.B.; Writing - original draft: A.K.W., D.C.B.; Writing - review & editing: A.K.W., J.L.M., J.A.H.M., W.D., D.C.B.; Visualization: A.K.W., J.L.M.; Supervision: D.C.B.; Project administration: A.K.W., J.L.M., D.C.B.; Funding acquisition: A.K.W., D.C.B.
Funding
A.K.W. is supported by a postdoctoral fellowship from Deutsche Forschungsgemeinschaft. D.C.B. is an investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.160671.supplemental
References
- Adachi S., Nobusawa T. and Umeda M. (2009). Quantitative and cell type-specific transcriptional regulation of A-type cyclin-dependent kinase in Arabidopsis thaliana. Dev. Biol. 329, 306-314. 10.1016/j.ydbio.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Adrian J., Chang J., Ballenger C. E., Bargmann B. O. R., Alassimone J., Davies K. A., Lau O. S., Matos J. L., Hachez C., Lanctot A. et al. (2015). Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev. Cell 33, 107-118. 10.1016/j.devcel.2015.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L., Gutzat R., Fütterer J., Laizet Y., Hennig L. and Gruissem W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22, 1792-1811. 10.1105/tpc.110.074591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V., Barrôco R., Engler Jde A., Verkest A., Beeckman T., Naudts M., Inzé D. and De Veylder L. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945-955. 10.1105/tpc.021774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J., Wester K., Weinl C., Roodbarkelari F., Kasili R., Larkin J. C., Hülskamp M. and Schnittger A. (2010). Endoreplication controls cell fate maintenance. PLoS Genet. 6, e1000996 10.1371/journal.pgen.1000996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J. (2005). Floral dip: agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286, 91-102. [DOI] [PubMed] [Google Scholar]
- Collins C., Dewitte W. and Murray J. A. H. (2012). D-type cyclins control cell division and developmental rate during Arabidopsis seed development. J. Exp. Bot. 63, 3571-3586. 10.1093/jxb/ers015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A., Díaz-Triviño S., Blilou I., Grieneisen V. A., Sozzani R., Zamioudis C., Miskolczi P., Nieuwland J., Benjamins R., Dhonukshe P. et al. (2012). A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150, 1002-1015. 10.1016/j.cell.2012.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K. A. and Bergmann D. C. (2014). Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc. Natl. Acad. Sci. USA 111, 15585-15590. 10.1073/pnas.1411766111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B., Ramirez-Parra E., Xie Q., Chua N.-H. and Gutierrez C. (2006). Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140, 67-80. 10.1104/pp.105.071027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T. and Inzé D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8, 655-665. 10.1038/nrm2227 [DOI] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J. M. S., Jacqmard A., Kilby N. J. and Murray J. A. H. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15, 79-92. 10.1105/tpc.004838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A. A., Maughan S. C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V. et al. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104, 14537-14542. 10.1073/pnas.0704166104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman W. E. (1999). Expression of the cell cycle in sperm of Arabidopsis: implications for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development 126, 1065-1075. [DOI] [PubMed] [Google Scholar]
- Gutzat R., Borghi L. and Gruissem W. (2012). Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci. 17, 139-148. 10.1016/j.tplants.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Hachez C., Ohashi-Ito K., Dong J. and Bergmann D. C. (2011). Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol. 155, 1458-1472. 10.1104/pp.110.167718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H. and Schnittger A. (2012). Robust reconstitution of active cell-cycle control complexes from co-expressed proteins in bacteria. Plant Methods 8, 23 10.1186/1746-4811-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H., Dissmeyer N. and Schnittger A. (2013). Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 23, 345-356. 10.1016/j.tcb.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Harbour J. W. and Dean D. C. (2000). The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14, 2393-2409. 10.1101/gad.813200 [DOI] [PubMed] [Google Scholar]
- Inzé D. and De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77-105. 10.1146/annurev.genet.40.110405.090431 [DOI] [PubMed] [Google Scholar]
- Kanaoka M. M., Pillitteri L. J., Fujii H., Yoshida Y., Bogenschutz N. L., Takabayashi J., Zhu J.-K. and Torii K. U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20, 1775-1785. 10.1105/tpc.108.060848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri Y., Hasegawa J., Fujikura U., Hoshino R., Matsunaga S. and Tsukaya H. (2016). The coordination of ploidy and cell size differs between cell layers in leaves. Development 143, 1120-1125. 10.1242/dev.130021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A., Umeda-Hara C., Adachi S., Nagata N., Konomi M., Nakagawa T., Uchimiya H. and Umeda M. (2007). The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell 19, 1265-1277. 10.1105/tpc.106.046763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H. and Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855-1860. 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L. B., Nadeau J. A., Lucas J., Lee E.-K., Nakagawa T., Zhao L., Geisler M. and Sack F. D. (2005). The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17, 2754-2767. 10.1105/tpc.105.034116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S. and Bergmann D. C. (2015). MOBE-ChIP: a large-scale chromatin immunoprecipitation assay for cell type-specific studies. Plant J. 84, 443-450. 10.1111/tpj.13010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S., Davies K. A., Chang J., Adrian J., Rowe M. H., Ballenger C. E. and Bergmann D. C. (2014). Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345, 1605-1609. 10.1126/science.1256888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. K., Liu X., Eglit Y. and Sack F. (2013). FOUR LIPS and MYB88 conditionally restrict the G1/S transition during stomatal formation. J. Exp. Bot. 64, 5207-5219. 10.1093/jxb/ert313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. K., Lucas J. R. and Sack F. D. (2014). Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. Plant J. 78, 555-565. 10.1111/tpj.12489 [DOI] [PubMed] [Google Scholar]
- MacAlister C. A., Ohashi-Ito K. and Bergmann D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537-540. 10.1038/nature05491 [DOI] [PubMed] [Google Scholar]
- Matos J. L. and Bergmann D. C. (2014). Convergence of stem cell behaviors and genetic regulation between animals and plants: insights from the Arabidopsis thaliana stomatal lineage. F1000Prime Rep. 6, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J. L., Lau O. S., Hachez C., Cruz-Ramírez A., Scheres B. and Bergmann D. C. (2014). Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. Elife 3, 1792 10.7554/eLife.03271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwain J. C., Yiotis C. and Lawson T. (2016). Using modern plant trait relationships between observed and theoretical maximum stomatal conductance and vein density to examine patterns of plant macroevolution. New Phytol. 209, 94-103. 10.1111/nph.13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., Hennig L., Gruissem W. and Murray J. A. H. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53, 423-442. 10.1023/B:PLAN.0000019059.56489.ca [DOI] [PubMed] [Google Scholar]
- Menges M., Samland A. K., Planchais S. and Murray J. A. H. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18, 893-906. 10.1105/tpc.105.039636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., Pavesi G., Morandini P., Bögre L. and Murray J. A. H. (2007). Genomic organization and evolutionary conservation of plant D-type cyclins. Plant Physiol. 145, 1558-1576. 10.1104/pp.107.104901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Kawamura K., Sugisaka K., Sekine M. and Shinmyo A. (2002). Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14, 1847-1857. 10.1105/tpc.002550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Nakamura S., Tanaka K., Kawamukai M., Suzuki T., Nakamura K., Kimura T. and Ishiguro S. (2008). Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci. Biotechnol. Biochem. 72, 624-629. 10.1271/bbb.70678 [DOI] [PubMed] [Google Scholar]
- Nowack M. K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A. K., De Winter F., Yang F. and Schnittger A. (2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22, 1030-1040. 10.1016/j.devcel.2012.02.015 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K. and Bergmann D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493-2505. 10.1105/tpc.106.046136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J. and Torii K. U. (2007). Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. BioEssays 29, 861-870. 10.1002/bies.20625 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Sloan D. B., Bogenschutz N. L. and Torii K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501-505. 10.1038/nature05467 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Menges M., Healy J. M. S. and Murray J. A. H. (2000). Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20, 4513-4521. 10.1128/MCB.20.13.4513-4521.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A. H. K., Chickarmane V., Cunha A., Obara B., Manjunath B. S. and Meyerowitz E. M. (2010). Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 8, e1000367 10.1371/journal.pbio.1000367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Dewitte W., Forzani C., Patell F., Nieuwland J., Wen B., Quelhas P., De Jager S., Titmus C., Campilho A. et al. (2011). The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23, 641-660. 10.1105/tpc.110.080002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G. and Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036-5046. 10.1093/emboj/cdf524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M. A., Busch W., Van Norman J. M., Vernoux T., Brady S. M., Dewitte W., Murray J. A. H. and Benfey P. N. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466, 128-132. 10.1038/nature09143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken R., Kiekens R., Boruc J., Zhang F., Vercauteren A., Vercauteren I., De Smet L., Dhondt S., Inzé D., De Veylder L. et al. (2012). Combined linkage and association mapping reveals CYCD5;1 as a quantitative trait gene for endoreduplication in Arabidopsis. Proc. Natl. Acad. Sci. USA 109, 4678-4683. 10.1073/pnas.1120811109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Nakamura S., Kawamukai M., Koizumi N. and Nakagawa T. (2011). Development of a series of gateway binary vectors possessing a tunicamycin resistance gene as a marker for the transformation of Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 75, 804-807. 10.1271/bbb.110063 [DOI] [PubMed] [Google Scholar]
- Uemukai K., Iwakawa H., Kosugi S., de Uemukai S., Kato K., Kondorosi E., Murray J. A. H., Ito M., Shinmyo A. and Sekine M. (2005). Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: cyclin D expression overcomes this repressor activity. Plant Mol. Biol. 57, 83-100. 10.1007/s11103-004-6601-x [DOI] [PubMed] [Google Scholar]
- Umen J. G. and Goodenough U. W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15, 1652-1661. 10.1101/gad.892101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S. and Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903-916. 10.1105/tpc.010445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Hollunder J., Eeckhout D., Persiau G., Van De Slijke E., Stals H., Van Isterdael G., Verkest A., Neirynck S., Buffel Y. et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6, 397 10.1038/msb.2010.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Coppens F., Lee E., Donner T. J., Xie Z., Van Isterdael G., Dhondt S., De Winter F., De Rybel B., Vuylsteke M. et al. (2011). Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30, 3430-3441. 10.1038/emboj.2011.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Kong H., Sun Y., Zhang X., Zhang W., Altman N., DePamphilis C. W. and Ma H. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135, 1084-1099. 10.1104/pp.104.040436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer A. K., Nowack M. K., Bouyer D., Zhao X., Harashima H., Naseer S., De Winter F., Dissmeyer N., Geldner N. and Schnittger A. (2012). Retinoblastoma related1 regulates asymmetric cell divisions in Arabidopsis. Plant Cell 24, 4083-4095. 10.1105/tpc.112.104620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer A. K., Biedermann S., Harashima H., Roodbarkelari F., Takahashi N., Foreman J., Guan Y., Pochon G., Heese M., Van Damme D. et al. (2016). The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 35, 2068-2086. 10.15252/embj.201593083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Lee E., Lucas J. R., Morohashi K., Li D., Murray J. A. H., Sack F. D. and Grotewold E. (2010). Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22, 2306-2321. 10.1105/tpc.110.074609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Harashima H., Dissmeyer N., Pusch S., Weimer A. K., Bramsiepe J., Bouyer D., Rademacher S., Nowack M. K., Novak B. et al. (2012). A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet. 8, e1002847 10.1371/journal.pgen.1002847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.