Abstract
Introgression is emerging as an important source of novel genetic variation, alongside standing variation and mutation. It is adaptive when such introgressed alleles are maintained by natural selection. Recently, there has been an explosion in the number of studies on adaptive introgression. In this review, we take a plant perspective centred on four lines of evidence: (i) introgression, (ii) selection, (iii) phenotype and (iv) fitness. While advances in genomics have contributed to our understanding of introgression and porous species boundaries (task 1), and the detection of signatures of selection in introgression (task 2), the investigation of adaptive introgression critically requires links to phenotypic variation and fitness (tasks 3 and 4). We also discuss the conservation implications of adaptive introgression in the face of climate change. Adaptive introgression is particularly important in rapidly changing environments, when standing genetic variation and mutation alone may only offer limited potential for adaptation. We conclude that clarifying the magnitude and fitness effects of introgression with improved statistical techniques, coupled with phenotypic evidence, has great potential for conservation and management efforts.
Keywords: adaptive introgression, natural selection, phenotypic variation, species boundaries, hybridization, climate change
1. Role of plants in the study of adaptive hybridization
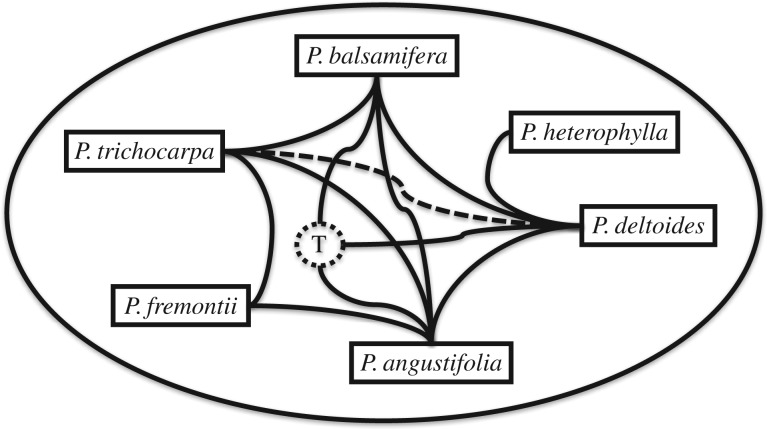
In the early twentieth century the botanist J.P. Lotsy vigorously championed the role of hybridization in evolution [1]. Although Lotsy's views now seem primitive in the light of modern genetics, he undoubtably primed a succeeding generation of botanists to look at hybridization seriously. Arguably his most enduring contribution was to coin the term ‘syngameon’ which he applied, for instance, to hybridizing European species of birch trees. This concept was later developed by Grant [2], who identified many plant syngameons, and this in turn influenced the development of the ecological species concept as exemplified by oaks [3]. A syngameon is a group of otherwise distinct species interconnected by limited gene exchange, i.e. the most inclusive interbreeding evolutionary unit (figure 1). The sharing of allelic diversity between a group of species in a syngameon has clear evolutionary consequences.
Figure 1.
Syngameon of North American poplars (Populus). Lines represent hybrid zones connecting the largely parapatric species. ‘T’ indicates the three-species hybrid zone in southern Alberta. P. trichocarpa × P. deltoides, although common in cultivation, is rare in the wild (indicated with dotted line). Adaptive introgression has been demonstrated between P. trichocarpa and P. balsamifera (see text) but potentially occurs more widely across the syngameon.
Another key concept to originate from plants was that of ‘introgression’, coined by Anderson and exemplified by his studies of hybridization in spiderworts [4]. Introgression refers to the transfer of a small amount of the genome from one parental taxon (usually species) to another by hybridization and repeated backcrossing. From the beginning, Anderson was clear about the potential adaptive effects of introgression: ‘the final result will depend upon the balance between the deleterious effects of the foreign germplasm and its advantageous effects in the areas where the hybridization has taken place or to which the hybrids may spread’ [4, p. 396]. Since Anderson, the evolutionary consequences of syngameons and introgression have been enormously developed and extended both in plant systems [5–8] and more widely [9].
This review focuses specifically on ‘adaptive introgression’, defined here as the transfer by introgression of relatively small genomic regions from a donor species that have positive fitness consequences in the recipient species. We discuss the key role that plants have had, and will have, in the development of the field, and outline the methodology required to demonstrate adaptive introgression, in both plant and animal systems, including requirements for the future.
2. Adaptive introgression
According to current views of divergence along the speciation continuum [10], species boundaries are characterized as being ‘porous’ or ‘semipermeable’, with the latter at least implying that differential introgression results from a selective process [11]. Although much of this recent work has been conducted in animals, there has also been considerable contribution from plant systems [12–14] and a plant perspective is particularly important owing to the frequency of plant hybridization. It is estimated that around 25% of plant species exchange genes with relatives, almost double the rate suggested for animals [15].
Although stochastic processes could predominate during introgressive hybridization in nature, adaptive introgression occurs when introgressed alleles are maintained by natural selection [16–18]. Adaptive introgression has now emerged as an important source of variation available for evolution, alongside de novo mutation and standing variation, widening the pool of genetic variation available for adaptation [19]. Furthermore, introgressed alleles may result in epistatically favourable gene combinations within the new genomic background and further accelerate adaptation [20].
Loci associated with broadly advantageous traits are expected to introgress more frequently [21–23], while alleles that contribute to reproductive isolation will introgress little or not at all [20,24,25]. Local adaptation can, therefore, potentially contribute to both reproductive isolation and its breakdown, depending on whether locally adapted alleles contribute to genetic incompatibilities, for instance as locally adaptive alleles may be specific to a genomic background and cause outbreeding depression in alternative backgrounds [26,27]. Where not, introgressed variation may readily facilitate local adaptation in the recipient species from the already adapted donor species [15,21], and this is the process we are primarily concerned with here.
3. Methods to detect adaptive introgression
The detection of introgression is generally straightforward, but showing that introgression is adaptive ideally requires multiple lines of evidence to demonstrate beyond reasonable doubt that the introgressed genomic regions from a donor species have a fitness effect in the receiving genomic background (tables 1 and 2). The evidence necessary to document adaptive introgression includes the following main steps:
Introgression—identifying true introgressed regions by distinguishing recent transfer from shared ancestral genetic variation [65,28]. A rapidly growing toolbox (table 1) facilitates tests for introgression of genomic blocks relative to alternative scenarios, such as incomplete lineage sorting [26,28,31,32,39]. Many of these approaches exploit the fact that introgressed blocks are generally longer than segments originated from incomplete lineage sorting and that the expected length of introgressed regions depends on the time since admixture [30,67].
Genomic signature of selection—uncovering the genomic signature of selection, for instance by demonstrating significantly greater persistence and spread of introgressed regions than expected by chance [33,38]. The environmental context in which hybridization occurs, together with dispersion distance and natural selection are key factors underlying the geographical patterns and spatial scale of introgression [66,68,69].
Adaptively relevant phenotypic variation—demonstrating that introgressed alleles have adaptively relevant phenotypic consequences [18,29]. A particularly promising approach that has been enabled by genome data is admixture mapping. Almost a decade ago, a review [39] discussed potential applications of admixture mapping in the plant genus Helianthus [64], in two European Populus species, Populus alba and Populus tremula [70], and the lake whitefish (Coregonus spp.) species complex [51]. This review noted other hybridizing taxa that showed promise for admixture mapping, including hybrid populations of Antirrhinum and of Silene, Peromyscus mice species, invasive and native sculpins, Heliconius butterflies and sticklebacks [39]. Recently, for instance, local ancestry and admixture mapping has been used in sticklebacks (Gasterosteus aculeatus) to identify genomic regions associated with divergence in male nuptial colour [52], in Mimulus to determine the genetic basis of trichome differentiation between Mimulus guttatus adapted to geothermally heated soils and nonthermal putative ancestors [63], and in two North American Populus species (Populus trichocarpa and Populus balsamifera) to explore if introgressed regions are driving variation in adaptive traits and contributing to the northern range expansion of P. trichocarpa [61,62] (table 2).
Fitness—direct measurement of a fitness effect of the introgressed region, and its resultant phenotype, in the receiving species [22,71]. This is the most critical step, yet often inadequately addressed. A strong demonstration of adaptive introgression ideally involves experimentally assaying the fitness effects of heterospecific allele combinations [72,73]. Plants are promising organisms for this as common garden trials can be readily established. These may easily be replicated in the case of clonally propagated plants. More experiments of this type would be highly desirable for validating putative cases of adaptive introgression, especially in study systems in which whole-genome sequencing of many individuals from admixed populations is feasible.
The dramatic increase in the amount of genomic data has allowed the rapid accumulation of evidence for introgression (step 1) as well as candidate genomic regions for adaptive introgression (step 2) but information on the genetic architecture of introgressed phenotypic traits (step 3) lags behind. Step 4 (direct measurement of fitness) has benefited least from genomics, and more work in this area is urgently required.
Table 1.
A summary of the different classes of investigation used in studies of adaptive introgression.
| class of investigation | examples | notes |
|---|---|---|
| I. introgression (genome-wide-based methods to infer introgressed blocks) | Patterson's D statistic, fd | the Patterson's D statistic detects an excess of shared derived alleles between species but does not distinguish introgression from ancestral population structure or indicate which genomic regions show such an excess. fd, a modified version of a statistic developed to estimate the genome-wide fraction of admixture, provides an improved method of identifying candidate introgressed regions [28]. The joint distribution of fd and mean absolute genetic divergence (dXY) statistics may be useful for differentiating introgression from shared ancestral variation at individual loci |
| tract length and linkage disequilibrium (LD, S*statistics) | these statistics aim to capture events of introgression by incorporating information regarding tract length and linkage disequilibrium. These approaches exploit the fact that introgressed blocks are generally longer than segments originated from incomplete lineage sorting and that the expected length of introgressed regions depends on the time since introgression [30] | |
| probabilistic models such as hidden Markov models (HMM) | these methods have been used to reconstruct high-resolution recombination maps in admixed populations [31] and detect fragments introduced by introgression. Under this approach, initially proposed and implemented by [32], the ancestry of each genetic marker in the genome is a hidden random variable with two states (e.g. homospecific ancestry, mixed ancestry) which are estimated from the data | |
| II. genomic signature of selection | detection of locally varying selection: integrated haplotype score (iHS), extended haplotype homozygosity (EHH), Tajima's D, Fay and Wu's H, and variations on FST | these methods are based on the haplotype structure or the distribution of allele frequencies compared with that expected under neutrality [33–35]. However, the patterns in introgressed regions (selected or not), such as increased LD, are exactly those used by many statistical analyses to test for selection. Inferences of selection cannot, therefore, rely solely on the patterns generated by introgressed segments but rather should be accompanied by additional pieces of evidence [36] |
| III. adaptively relevant phenotypic variation | quantitative trait loci (QTL) studies, genome-wide association studies (GWAS) | adaptive introgression implies that the introgressed genomic regions produce phenotypic variation that can be selected on. It is, therefore, also necessary to find associations between introgressed DNA and a phenotype known or suggested to confer a fitness advantage. Formerly this was done by QTL analysis [37] but the advent of whole-genome data has made this even more powerful, for instance using genome-wide association scanning (GWAS) approaches [35,38] |
| molecular function | functional differences (such as differential expression or protein structural differences) between introgressed and non-introgressed alleles can be used to infer phenotypic effects, when the trait being controlled by an introgressed block is challenging to measure, or when there are complex interactions between genetic factors and the environment | |
| admixture mapping | admixture mapping is a powerful statistical approach to map genes associated with phenotypic traits in hybridizing or introgressed populations [39,40]. Correlating local ancestry with a particular phenotype requires genotype data from both the admixed and parental reference populations to quantify excess admixture as well as trait information from, at least, the admixed individuals. This method has been used extensively to map genes associated with diseases or traits with differential risk by ancestry in human populations [40]. However, the number of studies implementing this approach in non-human organisms, including plant species, is substantially lower | |
| IV. fitness | common garden and reciprocal transplant experiments | such experiments are best set up in a reciprocal manner [41], although experimental designs involving single environments (e.g. hybrid or parental habitats) can also be informative, as long as the tested biological material represents the whole breadth of genotypes relevant to the questions under study [42]. Common garden and transplant experiments involving hybrids (or crosses between highly divergent populations) have repeatedly uncovered important fitness effects of single traits or quantitative trait loci (QTL) in the wild [21,42–44] |
| polygenic modelling | a promising approach to link studies of phenotypes and fitness in an adaptive introgression context involves polygenic modelling via sparse whole-genome regression methods [45]. This family of methods potentially allows disentangling the effects of direct and indirect selection on individual loci from genome-wide marker data [46], analogous to very widely used methods of measuring selection on correlated characters [47] |
Table 2.
. Representative studies on adaptive introgression including examples in birds, fish, insects, mammals and plants. For each example, we note the type of evidence used, including tests for (1) introgression, (2) genomic signature of selection, (3) adaptively relevant phenotype and (4) direct fitness measurements.
| species group | organism | natural hybrids | (1) introgression | (2) selection | (3) phenotype | (4) fitness | references |
|---|---|---|---|---|---|---|---|
| bird | flycatcher (Zimmerius) | yes | yes | no | yes | no | [48] |
| fish | trout (Onchorhynchus) | yes | yes | no | no | no | [49] |
| fish | cichlid fishes (Astatotilapia) | yesa | yes | no | yes | no | [50] |
| fish | lake whitefish (Coregonus) | yes | yes | yes | yes | no | [51] |
| fish | stickleback (Gasterosteus) | yes | yes | yes | yes | no | [52] |
| insect | butterflies (Heliconius) | yes | yes | yes | yes | no | [23] |
| insect | malaria mosquitoes (Anopheles) | yes | yes | yes | yes | yes | [53] |
| mammal | Canis (wolf and coyote) | yes | yes | no | yes | no | [54] |
| mammal | pigs (Sus) | no | yes | yes | yes | no | [55] |
| mammal | house mice (Mus) | yes | yes | yes | yes | yes | [56] |
| mammal | humans (Homo) | yes | yes | yes | yes | yes | [57] |
| plant | Arabidopsis | yes | yes | yes | yes | no | [58] |
| plant | spruce (Picea) | yes | yes | yes | yes | no | [59] |
| plant | oak (Quercus) | yes | yes | yes | no | no | [60] |
| plant | poplar (Populus) | yes | yes | yes | yes | no | [61] |
| plant | poplar (Populus) | yes | yes | yes | yes | no | [62] |
| plant | monkeyflower (Mimulus) | yes | yes | yes | yes | no | [63] |
| plant | sunflower (Helianthus) | no | yes | no | yes | yes | [21,64] |
| plant | Iris | no | yes | no | yes | yes | [29] |
aHuman-mediated introgressive hybridization.
4. Adaptive introgression in plants: some recent examples
The phrase ‘adaptive introgression’ first appears in the Web of Science database in the year 2000, in a study of hybrid Rhododendron in the British Isles [74]. Since then (up to end of 2017) 163 papers have mentioned the phrase in key words, abstract or title, and 125 of these have appeared since the latest major review on adaptive introgression, which focused largely on animals [22]. The large increase of studies in this field has been strongly influenced by the increased availability of genomic information for an expanding range of species, which has very often revealed the importance of gene flow during or after speciation.
Introgression of adaptive genetic variation has now been well documented in a number of plant species, such as Senecio [75] and Helianthus [21,64,76]. For example, Helianthus annuus ssp. texanus (a hybrid of Helianthus debilis and H. annuus) gained increased herbivore resistance from its H. debilis parent, suggesting that introgression of biotic resistance traits was important in the adaptation of this hybrid subspecies [21].
Adaptive trait transfer has also been reported in the flood-tolerant Iris fulva and the dry-adapted Iris brevicaulis [29]. In artificial backcrosses of these two species, the ability to survive extreme flooding conditions was strongly influenced by the presence of introgressed Iris fulva alleles located throughout the genome [29]. In a serpentine autotetraploid, Arabidopsis arenosa, adaptation to drought, as well as mineral nutrient deficiency and phytotoxic levels of metals, appears to have been driven by genetic variants arising locally but also by capturing alleles from Arabidopsis lyrata, a diploid that independently colonized serpentine barrens [58].
In long-lived tree species, a number of studies have reported the contribution of interspecific hybridization to local adaptation. Examples include spruce [59,77], Eucalyptus [78], oak [79] and poplars. Poplar trees (Populus spp.) have emerged as models for population genomic studies of adaptation owing to porous species barriers and a wealth of genomic resources available, including sequenced genomes and annotated gene models [80,81]. Large ‘mosaic’ hybrid zones of P. alba and P. tremula along European river systems have been used in admixture mapping of the sort described above (in §3). Hybrids of these poplar species revealed conspicuous additive and overdominant genetic effects [82] as well as heterozygote excess for markers tagging phytochemical defence trait QTL [83]. Barriers to introgression appear strong but nevertheless permeable between these species [84–86].
The first study documenting fine-scale genomic introgression patterns and identifying adaptive introgression at the gene level in forest trees was carried out in two sibling poplar species, ecologically divergent and adapted to strongly contrasting environments, P. trichocarpa and P. balsamifera [61,62]. This work was carried out within the context of a large-scale genomics project that included landscape genomics [34] and phenotypic [87] components as relevant background to studies of adaptive introgression.
5. The conservation implications of adaptive introgression
Should conservationists try to prevent or encourage gene exchange between species? This important question has been debated widely in terms of the risks of genetic swamping and outbreeding depression on the one hand, and the expression of transgressive advantageous characters in hybrids and their potential transfer between species on the other [88]. Forests in particular are increasingly threatened by a changing climate and maintaining the health of tree populations is particularly challenging, as rapid climate change threatens to disrupt the match between local populations and climate [89]. Improving our understanding of the importance of adaptive introgression as a source of variation for genetic adaptation will allow better prediction of responses to conservation threats [90]. This is likely to become an increasingly urgent research topic in the future.
6. Future
While advances in genomics have contributed to our understanding of introgression and porous species boundaries (task 1) and the detection of signatures of selection in introgression (task 2), the investigation of adaptive introgression requires more than this: specifically, links to phenotype and fitness (tasks 3 and 4, as discussed above). There is currently a relative dearth of studies that directly determine the fitness consequences of genomic regions exchanged between divergent populations or species (table 2), although useful analytical tools are starting to emerge [45,46].
To better determine adaptive introgression, improved statistical techniques, coupled with more phenotypic data, could help. This type of method requires a realistic demographic scenario to distinguish introgression from other processes [91]. Once these complexities have been dealt with, phenotypic and fitness effects can readily be mapped by extending available genome-wide association study (GWAS) approaches [45,46]. The gold standard will be to determine experimentally whether introgression confers increased fitness in a relevant ecological context. Here, plants may play an important role as they are relatively amenable to experiments through common garden and reciprocal transplant approaches and ease of clonal propagation.
The widespread occurrence of natural hybridization in plants and the frequent high fertility of interspecific plant hybrids will provide a multiplicity of experimental possibilities. A ‘breeding’ approach, was used in Mimulus to transfer a flower colour allele between species and demonstrate a strong impact on pollinator visits under natural conditions [92]. Similarly, applied plant breeding provides a large number of analogous experiments: it is commonplace to transfer fitness traits from wild species into domesticated crop species by ‘introgression breeding’ under strong artificial selection. Resistance traits to more than seven pests have been transferred to rice from wild species [93]. By contrast, interspecific crosses are rather rarely used in animal breeding.
Lastly, it can be noted that closely related plant species vary in barriers to gene exchange, from complete cross-fertility to the major sterility barrier provided by differences in ploidy level (an easily studied sterility barrier that is extremely common in plants but rarer in animals). Remarkably, in plants at least, natural selection can introgress genes across the almost complete sterility barrier provided by ploidy [94]. This shows that even undetectable levels of gene flow are little impediment to adaptive introgression at an evolutionary time-scale and consequently adaptive introgression may be far more pervasive than we currently imagine.
Acknowledgements
We thank the many people who have kindly shared ideas on adaptive introgression with us, especially Armando Geraldes, Sally Aitken, Loren Rieseberg, Rob Guy and Alex Buerkle. We also apologize to those authors of notable papers on adaptive introgression for which space constraints have prevented inclusion here.
Data accessibility
This article has no additional data.
Authors' contributions
A.S.-G. performed a literature review and wrote the initial manuscript with substantial contributions from Q.C.B.C. and CL. All the authors approved the final version.
Competing interests
We declare we have no competing interests.
Funding
The laboratory of Q.C.B.C. is funded by the Natural Science and Engineering Research Council (NSERC) of Canada (Discovery Grant no. RGPIN-2014-05820) and that of C.L. by grants from the Swiss National Science Foundation (SNSF) and the University of Vienna.
References
- 1.Lotsy JP. 1916. Evolution by means of hybridization. The Hague, The Netherlands: M. Nijhoff. [Google Scholar]
- 2.Grant V. 1971. Plant speciation. New York, NY: Columbia University Press. [Google Scholar]
- 3.Van Valen L. 1976. Ecological species, multispecies, and oaks. Taxon 25, 233–239. ( 10.2307/1219444) [DOI] [Google Scholar]
- 4.Anderson E, Hubricht L. 1938. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am. J. Bot. 25, 396–402. ( 10.2307/2436413) [DOI] [Google Scholar]
- 5.Abbott R, et al. 2013. Hybridization and speciation. J. Evol. Biol. 26, 229–246. ( 10.1111/j.1420-9101.2012.02599.x) [DOI] [PubMed] [Google Scholar]
- 6.Arnold ML. 2004. Transfer and origin of adaptations through natural hybridization: were Anderson and Stebbins right? Plant Cell 16, 562–570. ( 10.1105/tpc.160370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieseberg LH, Wendel JF. 1993. Introgression and its consequences in plants. In Hybrid zones and the evolutionary process (ed. Harrison RG.), pp. 70–109. New York, NY: Oxford University Press. [Google Scholar]
- 8.Arnold ML, Kunte K. 2017. Adaptive genetic exchange: a tangled history of admixture and evolutionary innovation. Trends Ecol. Evol. 32, 601–611. ( 10.1016/j.tree.2017.05.007) [DOI] [PubMed] [Google Scholar]
- 9.Seehausen O. 2004. Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207. ( 10.1016/j.tree.2004.01.003) [DOI] [PubMed] [Google Scholar]
- 10.Feder JL, Egan SP, Nosil P. 2017. The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350. ( 10.1016/j.tig.2012.03.009) [DOI] [PubMed] [Google Scholar]
- 11.Harrison RG, Larson EL. 2014. Hybridization, introgression, and the nature of species boundaries. J. Hered. 105, 795–809. ( 10.1093/jhered/esu033) [DOI] [PubMed] [Google Scholar]
- 12.Rieseberg L, Whitton J, Gardner K. 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152, 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, Bodénès C, Burg K, Kremer A. 2004. Genome scanning for interspecific differentiation between two closely related oak species [Quercus robur L. and Q. petraea (Matt.) Liebl.]. Genetics 166, 1615–1626. ( 10.1534/genetics.104.026849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lexer C, Widmer A. 2008. The genic view of plant speciation: recent progress and emerging questions. Phil. Trans. R. Soc. B 363, 3023–3036. ( 10.1098/rstb.2008.0078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallet J, Besansky N, Hahn MW. 2016. How reticulated are species? Bioessays 38, 140–149. ( 10.1002/bies.201500149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twyford AD, Ennos RA. 2012. Next-generation hybridization and introgression. Heredity 108, 179–189. ( 10.1038/hdy.2011.68) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slatkin M. 2016. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16, 393–430. ( 10.1146/annurev.es.16.110185.002141) [DOI] [Google Scholar]
- 18.Rieseberg LH, et al. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216. ( 10.1126/science.1086949) [DOI] [PubMed] [Google Scholar]
- 19.Tigano A, Friesen VL. 2016. Genomics of local adaptation with gene flow. Mol. Ecol. 25, 2144–2164. ( 10.1111/mec.13606) [DOI] [PubMed] [Google Scholar]
- 20.Barton NH. 2001. The role of hybridization in evolution. Mol. Ecol. 10, 551–568. ( 10.1046/j.1365-294x.2001.01216.x) [DOI] [PubMed] [Google Scholar]
- 21.Whitney K, Randell RA, Rieseberg L. 2006. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am. Nat. 167, 794–807. ( 10.1086/504606) [DOI] [PubMed] [Google Scholar]
- 22.Hedrick PW. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22, 4606–4618. ( 10.1111/mec.12415) [DOI] [PubMed] [Google Scholar]
- 23.Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, McMillan WO, Jiggins CD. 2012. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet. 8, e1002752 ( 10.1371/journal.pgen.1002752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton NH. 1979. The dynamics of hybrid zones. Heredity 43, 341–359. ( 10.1038/hdy.1979.87) [DOI] [Google Scholar]
- 25.Shaw KL, Mullen SP. 2011. Genes versus phenotypes in the study of speciation. Genetica 139, 649–661. ( 10.1007/s10709-011-9562-4) [DOI] [PubMed] [Google Scholar]
- 26.Nolte AW, Tautz D. 2017. Understanding the onset of hybrid speciation. Trends Genet. 26, 54–58. ( 10.1016/j.tig.2009.12.001) [DOI] [PubMed] [Google Scholar]
- 27.Gompert Z, Parchman TL, Buerkle CA. 2011. Genomics of isolation in hybrids. Phil. Trans. R. Soc. B 367, 439–450. ( 10.1098/rstb.2011.0196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin SH, Davey JW, Jiggins CD. 2015. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257. ( 10.1093/molbev/msu269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin N, Bouck A, Arnold M. 2006. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics 172, 2481–2489. ( 10.1534/genetics.105.053538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman NH, Thompson EA. 2002. The effect of population history on the lengths of ancestral chromosome segments. Genetics 162, 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wegmann D, et al. 2011. Recombination rates in admixed individuals identified by ancestry-based inference. Nat. Genet. 43, 847–853. ( 10.1038/ng.894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. 2007. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 8, 857–868. ( 10.1038/nrg2187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geraldes A, Farzaneh N, Grassa CJ, McKown AD, Guy RD, Mansfield SD, Douglas CJ, Cronk QCB. 2014. Landscape genomics of Populus trichocarpa: the role of hybridization, limited gene flow, and natural selection in shaping patterns of population structure. Evolution 68, 3260–3280. ( 10.1111/evo.12497) [DOI] [PubMed] [Google Scholar]
- 35.Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. 2011. A map of local adaptation in Arabidopsis thaliana. Science 334, 86–89. ( 10.1126/science.1209271) [DOI] [PubMed] [Google Scholar]
- 36.Hoban S, et al. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am. Nat. 188, 379–397. ( 10.1086/688018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradshaw HD, Otto KG, Frewen BE, McKay JK, Schemske DW. 1998. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149, 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigel D, Nordborg M. 2015. Population genomics for understanding adaptation in wild plant species. Annu. Rev. Genet. 49, 315–338. ( 10.1146/annurev-genet-120213-092110) [DOI] [PubMed] [Google Scholar]
- 39.Buerkle CA, Lexer C. 2008. Admixture as the basis for genetic mapping. Trends Ecol. Evol. 23, 686–694. ( 10.1016/j.tree.2008.07.008) [DOI] [PubMed] [Google Scholar]
- 40.Shriner D. 2013. Overview of admixture mapping. Curr. Protocols Hum. Genet.94, 1.23.1–1.23.8. ( 10.1002/cphg.44) [DOI]
- 41.Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241. ( 10.1111/j.1461-0248.2004.00684.x) [DOI] [Google Scholar]
- 42.Lexer C, Welch ME, Durphy JL, Rieseberg LH. 2003. Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol. Ecol. 12, 1225–1235. ( 10.1046/j.1365-294X.2003.01803.x) [DOI] [PubMed] [Google Scholar]
- 43.Agren J, Oakley CG, McKay JK, Lovell JT, Schemske DW. 2013. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 110, 21 077–21 082. ( 10.1073/pnas.1316773110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gompert Z, Comeault AA, Farkas TE, Feder JL, Parchman TL, Buerkle CA. 2014. Experimental evidence for ecological selection on genome variation in the wild. Ecol. Lett. 17, 369–379. ( 10.1111/ele.12238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Carbonetto P, Stephens M. 2013. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 9, e1003264 ( 10.1371/journal.pgen.1003264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gompert Z, Egan SP, Barrett RDH, Feder JL, Nosil P. 2017. Multilocus approaches for the measurement of selection on correlated genetic loci. Mol. Ecol. 26, 365–382. ( 10.1111/mec.13867) [DOI] [PubMed] [Google Scholar]
- 47.Lande R, Arnold SJ. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226. ( 10.1111/j.1558-5646.1983.tb00236.x) [DOI] [PubMed] [Google Scholar]
- 48.Rheindt FE, Fujita MK, Wilton PR, Edwards SV. 2014. Introgression and phenotypic assimilation in Zimmerius flycatchers (Tyrannidae): population genetic and phylogenetic inferences from genome-wide SNPs. Syst. Biol. 63, 134–152. ( 10.1093/sysbio/syt070) [DOI] [PubMed] [Google Scholar]
- 49.Hohenlohe PA, et al. 2013. Genomic patterns of introgression in rainbow and westslope cutthroat trout illuminated by overlapping paired-end RAD sequencing. Mol. Ecol. 22, 3002–3013. ( 10.1111/mec.12239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichols P, Genner MJ, van Oosterhout C, Smith A, Parsons P, Sungani H, Swanstrom J, Joyce DA. 2014. Secondary contact seeds phenotypic novelty in cichlid fishes. Proc. R. Soc. B 282, 20142272 ( 10.1098/rspb.2014.2272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers SM, Campbell D, Baird SJ, Danzmann RG, Bernatchez L. 2001. Combining the analyses of introgressive hybridisation and linkage mapping to investigate the genetic architecture of population divergence in the lake whitefish (Coregonus clupeaformis, Mitchill). Genetica 111, 25–41. ( 10.1023/A:1013773600304) [DOI] [PubMed] [Google Scholar]
- 52.Malek TB, Boughman JW, Dworkin I, Peichel CL. 2012. Admixture mapping of male nuptial colour and body shape in a recently formed hybrid population of threespine stickleback. Mol. Ecol. 21, 5265–5279. ( 10.1111/j.1365-294X.2012.05660.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norris LC, Main BJ, Lee Y, Collier TC, Fofana A, Cornel AJ, Lanzaro GC. 2015. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc. Natl Acad. Sci. USA 112, 815–820. ( 10.1073/pnas.1418892112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.vonHoldt BM, Kays R, Pollinger JP, Wayne RK. 2016. Admixture mapping identifies introgressed genomic regions in North American canids. Mol. Ecol. 25, 2443–2453. ( 10.1111/mec.13667) [DOI] [PubMed] [Google Scholar]
- 55.Ai H, et al. 2017. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 47, 217–225. ( 10.1038/ng.3199) [DOI] [PubMed] [Google Scholar]
- 56.Song Y, Endepols S, Klemann N, Richter D, Matuschka F, Shih C, Nachman MW, Kohn MH. 2016. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between Old World mice. Curr. Biol. 21, 1296–1301. ( 10.1016/j.cub.2011.06.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huerta-Sanchez E, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197. ( 10.1038/nature13408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arnold BJ, Lahner B, DaCosta JM, Weisman CM, Hollister JD, Salt DE, Bomblies K, Yant L. 2016. Borrowed alleles and convergence in serpentine adaptation. Proc. Natl Acad. Sci. USA 113, 8320–8325. ( 10.1073/pnas.1600405113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamilton JA, De la Torre AR, Aitken SN. 2014. Fine-scale environmental variation contributes to introgression in a three-species spruce hybrid complex. Tree Genet. Genomes 11, 1–14. [Google Scholar]
- 60.Lind-Riehl F, Gailing O. 2017. Adaptive variation and introgression of a CONSTANS-like gene in North American red oaks. Forests 8, 3 ( 10.3390/f8010003) [DOI] [Google Scholar]
- 61.Suarez-Gonzalez A, Hefer CA, Christe C, Corea O, Lexer C, Cronk QCB, Douglas CJ. 2016. Genomic and functional approaches reveal a case of adaptive introgression from Populus balsamifera (balsam poplar) in P. trichocarpa (black cottonwood). Mol. Ecol. 25, 2427–2442. ( 10.1111/mec.13539) [DOI] [PubMed] [Google Scholar]
- 62.Suarez-Gonzalez A, Hefer CA, Lexer C, Douglas CJ, Cronk QCB. 2018. Introgression from Populus balsamifera underlies adaptation and range boundaries in P. trichocarpa. New Phytol. 217, 416–427. ( 10.1111/nph.14779) [DOI] [PubMed] [Google Scholar]
- 63.Hendrick MF, Finseth FR, Mathiasson ME, Palmer KA, Broder EM, Breigenzer P, Fishman L. 2016. The genetics of extreme microgeographic adaptation: an integrated approach identifies a major gene underlying leaf trichome divergence in Yellowstone Mimulus guttatus. Mol. Ecol. 25, 5647–5662. ( 10.1111/mec.13753) [DOI] [PubMed] [Google Scholar]
- 64.Kim S, Rieseberg LH. 1999. Genetic architecture of species differences in annual sunflowers: implications for adaptive trait introgression. Genetics 153, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lexer C, Kremer A, Petit RJ. 2006. Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Mol. Ecol. 15, 2007–2012. ( 10.1111/j.1365-294X.2006.02896.x) [DOI] [PubMed] [Google Scholar]
- 66.Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto FJ, Kremer A, Gerber S. 2009. Species relative abundance and direction of introgression in oaks. Mol. Ecol. 18, 2228–2242. ( 10.1111/j.1365-294X.2009.04137.x) [DOI] [PubMed] [Google Scholar]
- 67.Zhang W, Dasmahapatra KK, Mallet J, Moreira GRP, Kronforst MR. 2016. Genome-wide introgression among distantly related Heliconius butterfly species. Genome Biol. 17, 25 ( 10.1186/s13059-016-0889-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buerkle CA. 2009. Ecological context shapes hybridization dynamics. Mol. Ecol. 18, 2077–2079. ( 10.1111/j.1365-294X.2009.04138.x) [DOI] [PubMed] [Google Scholar]
- 69.The Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98. ( 10.1038/nature11041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lexer C, Buerkle CA, Joseph JA, Heinze B, Fay MF. 2006. Admixture in European Populus hybrid zones makes feasible the mapping of loci that contribute to reproductive isolation and trait differences. Heredity 98, 74–84. ( 10.1038/sj.hdy.6800898) [DOI] [PubMed] [Google Scholar]
- 71.Rieseberg L. 2011. Adaptive introgression: the seeds of resistance. Curr. Biol. 21, R581–R583. ( 10.1016/j.cub.2011.06.038) [DOI] [PubMed] [Google Scholar]
- 72.Arnold ML, Martin NH. 2010. Hybrid fitness across time and habitats. Trends Ecol. Evol. 25, 530–536. ( 10.1016/j.tree.2010.06.005) [DOI] [PubMed] [Google Scholar]
- 73.Arnold ML, Ballerini ES, Brothers AN. 2012. Hybrid fitness, adaptation and evolutionary diversification: lessons learned from Louisiana irises. Heredity 108, 159–166. ( 10.1038/hdy.2011.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milne RI, Abbott RJ. 2000. Origin and evolution of invasive naturalized material of Rhododendron ponticum L. in the British Isles. Mol. Ecol. 9, 541–556. ( 10.1046/j.1365-294x.2000.00906.x) [DOI] [PubMed] [Google Scholar]
- 75.Kim M, Cui ML, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E. 2008. Regulatory genes control a key morphological and ecological trait transferred between species. Science 322, 1116–1119. ( 10.1126/science.1164371) [DOI] [PubMed] [Google Scholar]
- 76.Whitney KD, Randell RA, Rieseberg LH. 2010. Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. New Phytol. 187, 230–239. ( 10.1111/j.1469-8137.2010.03234.x) [DOI] [PubMed] [Google Scholar]
- 77.Lafontaine G, Prunier J, Gérardi S, Bousquet J. 2015. Tracking the progression of speciation: variable patterns of introgression across the genome provide insights on the species delimitation between progenitor–derivative spruces (Picea mariana × P. rubens). Mol. Ecol. 24, 5229–5247. ( 10.1111/mec.13377) [DOI] [PubMed] [Google Scholar]
- 78.Larcombe MJ, Holland B, Steane DA, Jones RC, Nicolle D, Vaillancourt RE, Potts BM. 2015. Patterns of reproductive isolation in Eucalyptus–a phylogenetic perspective. Mol. Biol. Evol. 32, 1833–1846. ( 10.1093/molbev/msv063) [DOI] [PubMed] [Google Scholar]
- 79.Abadie P, Roussel G, Dencausse B, Bonnet C, Bertocchi E, Louvet J, Kremer A, Garnier-Géré P. 2012. Strength, diversity and plasticity of postmating reproductive barriers between two hybridizing oak species (Quercus robur L. and Quercus petraea (Matt.) Liebl.). J. Evol. Biol. 25, 157–173. ( 10.1111/j.1420-9101.2011.02414.x) [DOI] [PubMed] [Google Scholar]
- 80.Cronk QCB. 2005. Plant eco-devo: the potential of poplar as a model organism. New Phytol. 166, 39–48. ( 10.1111/j.1469-8137.2005.01369.x) [DOI] [PubMed] [Google Scholar]
- 81.Jansson S, Douglas CJ. 2013. Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458. ( 10.1146/annurev.arplant.58.032806.103956) [DOI] [PubMed] [Google Scholar]
- 82.Lindtke D, Gonzalez-Martinez S, Macaya-Sanz D, Lexer C. 2013. Admixture mapping of quantitative traits in Populus hybrid zones: power and limitations. Heredity 111, 474–485. ( 10.1038/hdy.2013.69) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caseys C, Stritt C, Glauser G, Blanchard T, Lexer C. 2015. Effects of hybridization and evolutionary constraints on secondary metabolites: the genetic architecture of phenylpropanoids in European Populus species. PLoS ONE 10, e0128200 ( 10.1371/journal.pone.0128200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Christe C, Stölting KN, Bresadola L, Fussi B, Heinze B, Wegmann D, Lexer C. 2016. Selection against recombinant hybrids maintains reproductive isolation in hybridizing Populus species despite F1 fertility and recurrent gene flow. Mol. Ecol. 25, 2482–2498. ( 10.1111/mec.13587) [DOI] [PubMed] [Google Scholar]
- 85.Christe C, Stölting KN, Paris M, Fraїsse C, Bierne N, Lexer C. 2017. Adaptive evolution and segregating load contribute to the genomic landscape of divergence in two tree species connected by episodic gene flow. Mol. Ecol. 26, 59–76. ( 10.1111/mec.13765) [DOI] [PubMed] [Google Scholar]
- 86.Macaya-Sanz D, Heuertz M, Lindtke D, Vendramin GG, Lexer C, Gonzalez-Martinez SC. 2016. Causes and consequences of large clonal assemblies in a poplar hybrid zone. Mol. Ecol. 25, 5330–5344. ( 10.1111/mec.13850) [DOI] [PubMed] [Google Scholar]
- 87.McKown AD, et al. 2014. Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytol. 203, 535–553. ( 10.1111/nph.12815) [DOI] [PubMed] [Google Scholar]
- 88.Hamilton JA, Miller JM. 2015. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 30, 33–41. ( 10.1111/cobi.12574) [DOI] [PubMed] [Google Scholar]
- 89.Aitken SN, Yeaman S, Holliday JA, Wang TL, Curtis-McLane S. 2008. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1, 95–111. ( 10.1111/j.1752-4571.2007.00013.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alberto FJ, et al. 2013. Potential for evolutionary responses to climate change–evidence from tree populations. Glob. Change Biol. 19, 1645–1661. ( 10.1111/gcb.12181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Racimo F, Sankararaman S, Nielsen R, Huerta-Sanchez E. 2015. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359–371. ( 10.1038/nrg3936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bradshaw HD, Schemske DW. 2003. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426, 176–178. ( 10.1038/nature02106) [DOI] [PubMed] [Google Scholar]
- 93.Zamir D. 2001. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2, 983–989. ( 10.1038/35103590) [DOI] [PubMed] [Google Scholar]
- 94.Chapman MA, Abbott RJ. 2010. Introgression of fitness genes across a ploidy barrier. New Phytol. 186, 63–71. ( 10.1111/j.1469-8137.2009.03091.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.