Abstract
Cationic antimicrobial peptides are ubiquitous immune effectors of multicellular organisms. We previously reported, that in contrast to most of the classic antibiotics, cationic antimicrobial peptides (AMPs) do not increase mutation rates in E. coli. Here, we provide new evidence showing that AMPs do not stimulate or enhance bacterial DNA recombination in the surviving fractions. Recombination accelerates evolution of antibiotic resistance. Our findings have implications for our understanding of host–microbe interactions, the evolution of innate immune defences, and shed new light on the dynamic of antimicrobial-resistance evolution.
Keywords: cationic antimicrobial peptides, homologous recombination, antibiotic resistance
1. Introduction
Since the introduction of antibiotics to modern medicine, bacterial resistance has evolved and spread very rapidly [1]. In the last decades, it has become evident that antibiotics are not only selecting resistant variants, but they increase the probability of resistance evolution by elevating mutation rates, recombination frequency and horizontal gene transfer in bacteria [2].
When antibiotic treatment results in DNA damage, the SOS stress pathway is activated. DNA is repaired using error-prone alternative polymerases that introduce mutations [3]. The general stress response, controlled by RpoS in Gram-negative bacteria, also contributes to increasing mutation rates under antibiotic stress [4]. Independently of these two pathways some metabolic drugs, such as sulfonamides, can increase mutagenesis, for example, by causing an imbalance of nucleotide pool during replication [5].
Some antibiotics, such as the fluoroquinolone ciprofloxacin, stimulate recombination in bacteria [6] through mechanisms elicited by fluoroquinolones making the evolution of resistance a more dynamic process [7]. Recombination, in addition to gene duplication and amplification [8,9], is a very important mechanism resulting in antimicrobial-resistance evolution. Bacterial recombination can also promote the evolution of multi-drug-resistance in functionally diverse populations [10]. In pathogenic Escherichia coli, the recombination rate can be significantly higher than in commensals [11]. A recent study of the human pathogen Streptococcus pneumoniae found that recombination rates are higher in the most resistant lineages and less frequent in the least resistant variants [12].
Previously, we showed that a panel of natural cationic antimicrobial peptides (AMPs) covering diverse taxonomical origins, in contrast to antibiotics, does not increase mutation rates in bacteria [13]. As antibiotic resistance can result from point mutations but often is the result of recombination, we investigate if the effect of the envelope stress imposed by AMPs has an impact on recombination in bacteria. AMPs form not only a group of promising drug candidates, but they are also important immune effectors of multicellular organisms [14,15]. Therefore, the aim of this study was to determine whether cationic AMPs influence the recombination rate in the model bacterium E. coli K12 (MG1655).
To investigate our hypothesis, we selected a panel of AMPs and used ciprofloxacin, a recombination-stimulating antibiotic [6] as a positive control. We chose different cationic AMPs such as cecropin A, tenecin 1 and melittin (from insects) and magainin II and its derivative pexiganan, LL-37 from vertebrates to represent phylogenetic breadth. For mellitin, magainin, pexiganan and LL-37, the proposed killing mechanism is toroidal pore forming, while cecropin is considered to form a carpet on the bacterial cell envelopes [15]. We additionally included human serum, because most of its bactericidal effect comes from the activation of complement, which also is a pore-forming complex of cationic proteins and well conserved within vertebrates.
2. Material and methods
(a). Bacteria and growth conditions
Escherichia coli MG1655 was used as the bacterial model for all experiments and cultured in MHB devoid of cations. For genetic manipulation, Escherichia coli DH5α strain was used and routinely cultured in lysogeny broth (LB medium), supplemented with antibiotics when appropriate. All bacterial strains were cultured in lysogeny broth lenox (Carl Roth, Germany).
(b). Recombination frequency
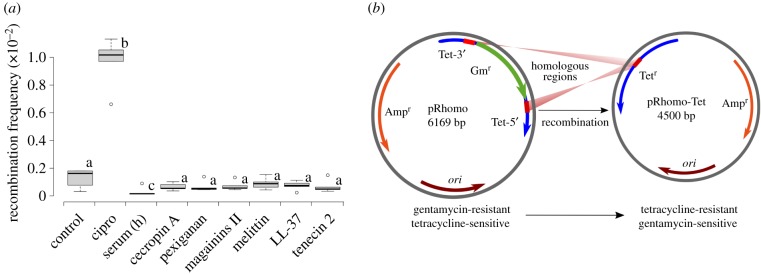
To estimate the impact of antimicrobial in recombination frequency, a genetic assay was carried out as previously described [11]. Briefly, the system consisted of a plasmid harbouring two truncated tetA alleles separated by an antibiotic-resistance cassette (aacC1, conferring gentamycin resistance). Recombination restores the functional tetA gene, thereby conferring tetracycline resistance, which can be selected for (figure 1b). Thus, this assay allows quantification of the frequency of recombinants. We exposed 1 ml of culture containing approximately 1–2 × 107 bacteria (dilution of 1 in 10 of mid exponential culture containing 1 × 108 cfu ml−1) to different concentrations of different antimicrobials over a period of 4 h as previously described [13]. These concentrations (see the electronic supplementary material) were taken to be half of the minimal inhibitory concentration (1/2 MIC). For selected peptides: LL-37, melittin and pexiganan, additional lower (1/4× and 1/8× MIC) and higher concentrations (1× MIC) were assayed. After incubation with gentle shaking, 9 ml of fresh MH were added to the cultures and the diluted cultures centrifuged at 3000g for 15 min to remove the antimicrobials. The pellets were resuspended in 2 ml of MH and cells were allowed to recover for 1 h before adding ampicillin (to maintain the plasmid) to a final concentration of 50 µg ml−1 and incubate with shaking at 37°C overnight. The next day, appropriate dilutions were plated in MH agar containing 50 µg ml−1 ampicillin, to estimate the viability, and MH agar containing 30 µg ml−1 of tetracycline to estimate the number of recombinants. Each experiment consisted of five replicas and was repeated twice. Recombinant frequencies were expressed as the ratio of medians of recombinant number by the number of viable bacteria.
Figure 1.
(a) Graphical representation of recombinant frequencies of E. coli MG1655 treated with different AMPs. Ciprofloxacin was used as a positive control. Boxplots represent the median values of recombination frequency of the bacteria exposed to the half-MIC during 4 h. Only ciprofloxacin b had a significantly higher recombination frequency (p = 0.006, Mann–Whitney test). The human serum c showed a significant lower recombination rate (p = 0.018, Mann–Whitney test) than the control group. The same letter represents no statistical differences while different ones indicate significant difference with the control (basal recombination rate value of the non-treated bacteria). Section (b) is a representation of the plasmid system to measure the recombination frequency (modified from reference [11]), explained in details in Material and methods.
(c). Overexpression of RpoE, recombination test and quality control by western blotting
Ten millilitres of samples were removed from non-induced and induced cultures (IPTG 0.1 mM) to 10 ml tubes of the E. coli MG1655 transformed with the pCA24N and pCA24N-RpoE from Aska collection [16] and harbouring the pRhomo plasmid [11]. One millilitre of each culture was used to measure recombination frequency as previously described in this section. Then, the tubes were centrifuged at 4000g for 10 min and the pellets were resuspended in 1 ml of distilled water. The cells were lysed and proteins extracted by adding 100 µl of cold 50% trichloroacetic acid to each tube. After a 5 min centrifugation step at 10 000g, the pellets were washed twice with 500 µl of ice-cold 80% acetone, air dried and resuspended in 1× SDS denaturing loading buffer (Bio-Rad, USA). Equal quantities of protein were separated on precast SDS-gradient acrylamide gels (7.5–15%) (Bio-Rad, USA) and transferred onto PVDF membrane filters (Novex, Life Technologies, Germany). Filters were incubated with anti-histidine tag monoclonal antibody at 1 : 1500 (CD Creative Diagnostic, USA). Immunoblots were developed by using horseradish peroxidase-conjugated goat anti-mouse IgG antibody, followed by on membrane developing using the metal enhanced DAB kit (Pierce, Thermo Scientific, Germany).
(d). Statistical analysis
To compare experimental several experimental groups, a Kruskal–Wallis test was performed. In the case of significance, Bonferroni-corrected one-tailed Mann–Whitney U-test was used to compare each treatment with the control group. P-values less than or equal to 0.05, after correction if needed, were considered statistically significant. All tests were performed with the statistic software R [17].
3. Results and discussion
None of the tested AMPs increased the recombination frequency (figure 1a) using a plasmid system that reconstitutes a disrupted resistance cassette by recombination (figure 1b) [11]. By contrast, ciprofloxacin induced an almost 10-fold increase in recombination frequency when compared to the control (p = 0.006, Mann–Whitney test, figure 1a). Interestingly, we found that human serum caused a slight but significant decrease in the basal recombination frequency (p = 0.018, Mann–Whitney test). We tested several concentration ranges, by repeating the same experiment for selected peptides at different sub-lethal concentrations (MIC, 1/2MIC, 1/4MIC and 1/8MIC). We found no significant changes in recombination rates (p-value = 0.7818 for LL-37, p-value = 0.7814 for melittin, p-value = 0.6217 for pexiganan, Kruskal–Wallis test, electronic supplementary material, figure S1). MIC values can be found in electronic supplementary material, table S1.
The sigma E regulon is a major factor in E. coli responses to LPS and external membrane disruption, including other chemical stresses. As cationic AMPs mostly attack the envelope [15], we reasoned that sigma E could influence recombination because is a required factor for mutagenesis and gene amplification during SOS response in E. coli [18]. We overexpressed the sigma E factor and checked the sensitivity to the peptides and measured the recombination rate of the overexpressing strain. While, the overexpression of this factor (determined by western blot; electronic supplementary material, figure S2), decreased sensitivity to human serum, LL-37, magainin II and pexiganan (electronic supplementary material, table S2), the overexpressing strain did not show any significant difference in the recombination frequency with control cultures (p-value = 0.1264, Kruskal–Wallis test; electronic supplementary material, figure S3), indicating a missing link between envelope stress response and recombination machinery.
Recombination plays a crucial role in the evolution of antibiotic resistance, and previous research has shown that many antibiotics also increase the recombination rate of bacteria [6,12]. Taken together with the finding that many antibiotics but not the panel of AMPs tested here elevate bacterial mutation rates, supports the view that resistance evolution against AMPs has a lower probability than against antibiotics [14]. AMPs have a number of other properties including their pharmacodynamics [19,20], that should result in lower probabilities of drug-resistance evolution than against antibiotics. Whether this makes AMPs potentially more sustainable drugs remains to be shown. It is intriguing though that AMPs are important components of innate immune systems and are also important players to police microbial symbionts of multicellular hosts [21].
Supplementary Material
Acknowledgements
We are indebted to Jesús Blázquez and Jerónimo Rodríguez-Beltran from the Spanish National Centre for Biotechnology (Spanish National Research Council, CSIC), for providing us with the construct tool to determine recombination frequency in E. coli.
Data accessibility
All data underlying the manuscript are either herein presented or available as the electronic supplementary material.
Authors' contributions
A.R.R. and J.R. conceived the study; J.M.M. and A.R.R. performed the experiments and collected the data; A.R.R., J.M.M., J.R. and J.M. analysed the data; A.R.R., J.M.M., J.R. and J.M. drafted the manuscript and revised the final document. All authors agree to be held accountable for the content therein and approved the final version.
Competing interests
The authors declare no competing interest.
Funding
A.R.R. and J.R. were supported SFB 973 (Deutsche Forschungsgemeinschaft), project C5).
References
- 1.Baker S. 2015. A return to the pre-antimicrobial era? Science 347, 1064–1066. ( 10.1126/science.aaa2868) [DOI] [PubMed] [Google Scholar]
- 2.Rodríguez-Rojas A, Rodríguez-Beltrán J, Couce A, Blázquez J. 2013. Antibiotics and antibiotic resistance: a bitter fight against evolution. Int. J. Med. Microbiol. 303, 293–297. ( 10.1016/j.ijmm.2013.02.004) [DOI] [PubMed] [Google Scholar]
- 3.Blázquez J, Couce A, Rodríguez-Beltrán J, Rodríguez-Rojas A, Blazquez J, Couce A, Rodriguez-Beltran J, Rodriguez-Rojas A. 2012. Antimicrobials as promoters of genetic variation. Curr. Opin. Microbiol. 15, 561–569. ( 10.1016/j.mib.2012.07.007) [DOI] [PubMed] [Google Scholar]
- 4.Gutierrez A, et al. 2013. β-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 4, 1610 ( 10.1038/ncomms2607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordman J, Wright A. 2008. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc. Natl Acad. Sci. USA 105, 10 197–10 202. ( 10.1073/pnas.0802816105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez E, Blazquez J. 2009. Effect of subinhibitory concentrations of antibiotics on intrachromosomal homologous recombination in Escherichia coli. Antimicrob. Agents Chemother. 53, 3411–3415. ( 10.1128/AAC.00358-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478. ( 10.1038/nrmicro3270) [DOI] [PubMed] [Google Scholar]
- 8.Sandegren L, Andersson DI. 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 578–588. ( 10.1038/nrmicro2174) [DOI] [PubMed] [Google Scholar]
- 9.Elliott KT, Cuff LE, Neidle EL. 2013. Copy number change: evolving views on gene amplification. Future Microbiol. 8, 887–899. ( 10.2217/fmb.13.53) [DOI] [PubMed] [Google Scholar]
- 10.Perron GG, Lee AEG, Wang Y, Huang WE, Barraclough TG. 2012. Bacterial recombination promotes the evolution of multi-drug-resistance in functionally diverse populations. Proc. R. Soc. B 279, 1477–1484. ( 10.1098/rspb.2011.1933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Beltrán J, Tourret J, Tenaillon O, López E, Bourdelier E, Costas C, Matic I, Denamur E, Blázquez J. 2015. High recombinant frequency in extraintestinal pathogenic Escherichia coli strains. Mol. Biol. Evol. 32, 1708–1716. ( 10.1093/molbev/msv072) [DOI] [PubMed] [Google Scholar]
- 12.Mostowy R, Croucher NJ, Hanage WP, Harris SR, Bentley S, Fraser C. 2014. Heterogeneity in the frequency and characteristics of homologous recombination in pneumococcal evolution. PLoS Genet. 10, e1004300 ( 10.1371/journal.pgen.1004300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Rojas A, Makarova O, Rolff J. 2014. Antimicrobials, stress and mutagenesis. PLoS Pathog. 10, e1004445 ( 10.1371/journal.ppat.1004445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. ( 10.1038/415389a) [DOI] [PubMed] [Google Scholar]
- 15.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. ( 10.1038/nrmicro1098) [DOI] [PubMed] [Google Scholar]
- 16.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299. ( 10.1093/dnares/dsi012) [DOI] [PubMed] [Google Scholar]
- 17.R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 18.Gibson JL, et al. 2010. The sigma(E) stress response is required for stress-induced mutation and amplification in Escherichia coli. Mol. Microbiol. 77, 415–430. ( 10.1111/j.1365-2958.2010.07213.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Baeder DY, Regoes RR, Rolff J. 2016. Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 60, 1717–1724. ( 10.1128/AAC.02434-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Baeder DY, Regoes RR, Rolff J. 2018. Predicting drug resistance evolution: insights from antimicrobial peptides and antibiotics. Proc. R. Soc. B 283, 20172687 ( 10.1098/rspb.2017.2687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Login FH, Balmand S, Vallier A, Vincent-Monégat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. ( 10.1126/science.1209728) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underlying the manuscript are either herein presented or available as the electronic supplementary material.