Abstract
To determine whether overexpression of Fe-superoxide (SOD) dismutase would increase superoxide-scavenging capacity and thereby improve the winter survival of transgenic alfalfa (Medicago sativa L.) plants, two genotypes were transformed with the vector pEXSOD10, which contains a cDNA for Arabidopsis Fe-SOD with a chloroplast transit peptide and cauliflower mosaic virus 35S promoter. A novel Fe-SOD was detected by native PAGE in both greenhouse- and field-grown transgenic plants, but activity varied among independent transgenic plants. The increased Fe-SOD activity was associated with increased winter survival over 2 years in field trials, but not with oxidative stress tolerance as measured by resistance of leaves to methyl viologen, a superoxide generator. Total shoot dry matter production over 2 harvest years was not associated with Fe-SOD activity. There was no detectable difference in the pattern of primary freezing injury, as shown by vital staining, nor was there additional accumulation of carbohydrates in field-acclimated roots of the transgenic alfalfa plants. We did not detect any difference in growth of one transgenic plant with high Fe-SOD activity compared with a non-transgenic control. Therefore, the improvement in winter survival did not appear to be a consequence of improved oxidative stress tolerance associated with photosynthesis, nor was it a consequence of a change in primary freezing injury. We suggest that Fe-SOD overexpression reduced secondary injury symptoms and thereby enhanced recovery from stresses experienced during winter.
Oxidative stress is a common disorder in plants during or following exposure to adverse environmental conditions. Allen (1995) speculated that frequent, mild oxidative stresses occur even in a normal field situation, and that these stresses inhibit photosynthesis and therefore yield. Allen also predicted that yield enhancement may be achieved by the overexpression of antioxidant genes in transgenic annual crops. During the winter, perennial plants are exposed to even greater environmental extremes, including freezing, anoxia, and desiccation.
The enzyme superoxide dismutase (SOD; EC 1.15.1.1) is a metalloprotein that catalyzes the initial step in the Asada-Halliwell pathway in chloroplasts, the dismutation of superoxide to H2O2 and molecular oxygen (Scandalios, 1993; Bowler et al., 1994; Allen, 1995). The subsequent reduction of H2O2 to water through the ascorbate-glutathione cycle in the chloroplast uses reducing equivalents from NADPH (Foyer and Halliwell, 1976; Foyer et al., 1994b). SOD enzymes are classified according to their metal cofactor and their subcellular localization. The predominant forms are a mitochondrial Mn-SOD, a cytosolic Cu/Zn-SOD, and a chloroplastic Cu/Zn-SOD. In a number of plant species, chloroplasts also contain Fe-SOD. The four forms of SOD differ in their biochemical properties and inhibition by H2O2 and cyanide (Scandalios, 1993; Bowler et al., 1994; Allen, 1995). The native forms of SOD provide protection from activated oxygen during periods of environmental stress (Salin, 1991; Bowler et al., 1992; Scandalios, 1993; Smirnoff, 1993; Foyer et al., 1997). To test this relationship further, different SOD transgenes have been expressed in transgenic plants, but the results vary (Herouart et al., 1993; Scandalios, 1993; Foyer et al., 1994a; Allen, 1995). For example, Pitcher et al. (1991), Tepperman and Dunsmuir (1990), and Payton et al. (1997) found no improvements, whereas Sen Gupta et al. (1993a, 1993b), Bowler et al. (1991), Van Camp et al. (1994, 1996), Perl et al. (1993), and McKersie et al. (1993, 1996, 1999a) found significant improvements to oxidative or environmental stress tolerances. This disparity has usually been attributed to the complexity of the detoxification system, because changing one enzyme may not change the capacity of the pathway as a whole.
We have previously reported that transgenic alfalfa (Medicago sativa L.) plants expressing a Mn-SOD had increased vigor after freezing stress and increased winter survival under field conditions (McKersie et al., 1996, 1997, 1999a). However, tolerance of freezing stress measured at the cellular level by electrolyte leakage or by vital staining with tetrazolium was not affected by the presence of transfer DNA (T-DNA) in these alfalfa plants (McKersie et al., 1999a). Van Camp et al. (1996) demonstrated that Fe-SOD and Mn-SOD have different protective properties when targeted to the chloroplast in transgenic plants. This may occur because their biochemical properties differ or because their subcellular localization within the chloroplast is influenced by their different affinities for membranes. In this study, we investigated whether overexpression of Fe-SOD would mimic the effects of Mn-SOD and improve the winter hardiness of transgenic alfalfa plants. We observed that the level of increased winter survival was correlated with the level of increased Fe-SOD activity in the transgenic plants, but that this was apparently independent of any effect of the Fe-SOD transgene on photosynthesis, growth, or oxidative stress tolerance in leaves.
MATERIALS AND METHODS
Plant Transformation
Transformation of alfalfa (Medicago sativa L.) petiole explants by an overnight culture of Agrobacterium tumefaciens C58C1 pMP90 containing the pEXSOD10 binary vector (Van Camp et al., 1996) was conducted as previously described (McKersie et al., 1999a). Petiole explants and agrobacterium were co-cultivated for 3 d in the dark on SH induction medium (Shetty and McKersie, 1993) containing 288 mg L−1 Pro, 53 mg L−1 thioproline, 4.35 g L−1 K2SO4, and 100 μm acetosyringinone, then washed with one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962). The explants were plated on SH induction medium lacking acetosyringinone but with 500 mg L−1 claforan and 50 mg L−1 kanamycin. Somatic embryos were matured on BOi2Y development medium (Bingham et al., 1975) containing 50 g L−1 Suc but no growth regulators or antibiotics.
The putative transgenic plants were screened for the presence of T-DNA using PCR primers to amplify a region within nos-nptII (McKersie et al., 1999a). The sense primer was 5′-AGCTGTGCTCGACGTTGTCAC-3′ and the antisense primer was 5′-GGTGGGCGAAGAACTCCAGCA-3′. The amplification was carried out on a PCR system (GeneAmp 2400, Perkin-Elmer, Foster City, CA) as previously described (McKersie et al., 1999a). Only positive plants were transplanted to growth medium (Turface, Plant Products, Mississauga, Ontario, Canada) in a greenhouse at approximately 23°C/18°C (day/night) and a minimum 16-h photoperiod. Mercury-halogen lamps provided additional light. The plants were watered automatically two times daily with a dilute solution of 15-15-30 fertilizer (Plant Products, Brampton, Ontario, Canada).
DNA was isolated using the method described by Saghai-Maroof et al. (1984). Leaves (1 g) were ground in liquid nitrogen, and 27 mL of extraction buffer (100 mm Tris, 700 mm NaCl, 50 mm EDTA, 1% [w/v] cetyl-trimethyl-ammonium bromide [CTAB], and 140 mm β-mercaptoethanol) added and incubated at 65°C. After two separations with chloroform/octanol, the aqueous solution was treated with RNase A before precipitation with isopropanol. The DNA was washed with ethanol solutions, dried, and suspended in Tris-EDTA buffer. DNA was digested using the restriction enzyme HindIII, purified by alcohol precipitation, and separated on a 0.8% (w/v) agarose gel overnight. The gel was blotted onto a positively charged nylon membrane (Boehringer Mannheim, Basel) as described by Ausubel et al. (1991) and UV-crosslinked. Southern-blot analysis was carried out using the non-radioactive digoxygenin (DIG) chemiluminescent system (Boehringer Mannheim) as described by van Miltenburg (1995). The DIG-labeled hybridization probes were synthesized using DIG-dUTP (Boehringer Mannheim) in a PCR amplification using 5′-TCCCAGTGAAGAAGGTCAAC-3′, located within the Arabidopsis Fe-SOD sequence, and 5′-AATGGTAAGCAATGGGAAAG-3′, located within the chloroplast transit peptide sequence. The amplification used purified plasmid DNA as the template, 1 μm of each primer, and a 55°C annealing temperature, and the resulting PCR fragment was 536 bp in length.
SOD Activity and Oxidative Stress Tolerance
To measure SOD activity, leaf samples were collected from greenhouse-grown plants at the mid-vegetative stage of development (Kalu and Fick, 1983); the maturing stems were approximately 20 cm in height with no buds or flowers. Whole leaves were ground with liquid nitrogen in a mortar, and 0.4 mL of homogenizing buffer, 50 mm KH2PO4, pH 7.8, was added to extract the soluble proteins.
SOD activity was determined using the in situ staining technique (Beauchamp and Fridovich, 1971). The protein samples were separated by native PAGE on a separating gel of 13% (w/v) polyacrylamide in a tank buffer containing 25 mm Tris (pH 8.3) and 64 mm L-Ile. The gels were stained for 30 min in the dark using a 1:1 mixture of: (a) 0.06 mm riboflavin and 0.651% (w/v) TEMED, and (b) 2.5 mm nitroblue tetrazolium (NBT), both in 50 mm phosphate buffer at pH 7.8, and then developed for 20 min under moderate light conditions. The gels were digitally photographed using imaging software (Northern Exposure, ImagExperts, Mississauga, Ontario, Canada), and an analysis program (Quantity One, Bio-Rad, Hercules, CA) was used to measure the intensity of each band. The area of individual SOD isozymes was expressed relative to a standard of Escherichia coli Fe-SOD (Sigma Chemical, St. Louis) to calculate each enzyme's activity. One unit of enzyme activity is the amount that will inhibit the rate of reduction of cytochrome c by 50% in a coupled system with xanthine and xanthine oxidase at pH 7.8 at 25°C in a 3-mL reaction volume (McCord and Fridovich, 1969). The protein content was determined (Bradford, 1976), and SOD activity for each individual isozyme was expressed as units per milligram of protein.
The oxidative stress tolerance of the plants was assessed using the method described previously (Bowler et al., 1991). Leaf slices, 0.5 cm across the largest width of the leaflet, were incubated in aqueous solutions of methyl viologen (Paraquat, Sigma Chemical), with concentrations ranging from 0 to 16 μm, overnight in the dark at 21°C. The samples were exposed to light for 2 h (200 μmol m−2 s−1 photosynthetically active radiation) at 26°C, and were then allowed to develop in the dark for 20 h at 30°C. The methyl viologen-dependent oxygen radical damage was estimated by chlorophyll fluorescence determination of photochemical yield, Fv/Fm (Genty et al., 1989), using a portable chlorophyll fluorometer (Mini Pam photosynthesis yield analyzer, Heinz Walz BmbH, Effeltrich, Germany).
SOD and ascorbate peroxidase (APX) activity were determined on the same extracts from alfalfa leaves treated with methyl viologen. To quantify APX activity, the extract (25 μL) and 150 μL of 0.03% (w/v) H2O2 were added to 2 mL of assay buffer, containing 0.5 mm ascorbate and 0.1 mm EDTA in phosphate buffer at pH 7.0 (Nakano and Asada, 1981). APX was determined by measuring the rate of oxidation of ascorbate at 290 nm using an absorbance coefficient of 2.8 mm−1 cm−1. One unit of APX was defined as 1 μmol of ascorbate oxidized per minute at pH 7.0 and 25°C and is expressed per milligram of protein.
1996 Field Trial
The 1996 field evaluation of the transgenic plants was carried out at the Elora Research Station (Elora, Ontario, Canada) following protocols authorized by the Plant Products Division, Agriculture and Agri-Food Canada (test 96-UOG2-ALF05-ON0–1-01, approved April 9, 1996). Replicate plants were grown from rooted stem cuttings in the greenhouse and transplanted to the field in June, 1996. The soil at Elora is a clayed brunisolic gray-brown luvisol-London. Soil test analysis was done to determine the appropriate amount of fertilizer (P and K) to apply. Elora has on average 130 frost-free days, and the start of the critical fall harvest period for alfalfa is August 30. The test was arranged in a randomized complete block design with 15 cuttings of each control (non-transgenic) and five cuttings of each transgenic genotype as the experimental units and three replications (blocks). Five cuttings were planted sequentially in a row with 0.5-m spacing between plants and 1.5-m spacing between rows. Plants were harvested once in the year of transplanting. Survival (the no. of plants with green shoots) was recorded in the fall of the transplant year, and in the spring and summer of the 2 subsequent years. The herbage (shoot dry matter) was harvested from each surviving plant individually. Plants that did not survive the winter were not included in the analysis. Plants were defoliated to determine dry matter yields in three harvests per year on June 28, July 28, and August 28, 1997, and on May 27, July 1, and August 11, 1998. Yields per plant were the sum of the three harvests for each year.
SOD activity was measured in leaves harvested at the vegetative stage of development from field-grown plants in the 1996 field trial when they were approximately 20 cm in height. Three replications were taken from three blocks in the field experiment on three sequential days in July 1997. Leaves were randomly sampled from the five plants in each block.
1997 Field Trial
The 1997 field trial was also conducted at the Elora Research Station following protocols authorized by Plant Products Division, Agriculture and Agri-Food Canada (test no. 97–UOG1–075–ALF04–224–ONO1–01, approved May 12, 1997). Replicated plots were established on May 28, 1997 by transplanting rooted cuttings of each transgenic and control genotype at an adjacent site on the Elora Research Station. In the 1997 test, four replicated plots of 1- × 1.5-m rectangular plots were established by transplanting 100 rooted propagules of each transgenic per plot. Each plot consisted of a population of independent primary transgenic plants for each construct. Plants were harvested twice in the year of transplanting on July 1 and September 2, 1997. On November 19, 1997, samples were dug from the field, rinsed, separated into taproot, crown, and leaves, and immediately frozen in liquid nitrogen. The samples were ground and analyzed for carbohydrates and protein. Other samples were analyzed for SOD activity on native PAGE gels as described above. The plots were sampled again for soluble carbohydrates on April 14, 1998.
On November 19, 1997 and on December 2, 1997, whole plants were dug from the field, washed, shoots excised, and crowns with attached roots were placed in moist filter paper and subjected to freezing temperatures in a programmable freezer. The samples were frozen at −2°C overnight, and then cooled at 2°C per hour to −6°C, −8°C, −10°C, −12°C, or −14°C. The frozen samples were thawed for 1 d at 2°C and subsequently separated into crowns, taproots, and crown buds, bisected, and assessed by viability staining with tetrazolium as previously described (Tanino and McKersie, 1985).
1998 Field Trial
Permits to conduct the 1998 field trails were obtained from the Canadian Food Inspection Agency (nos. 98–UOG1–075–ALF–02–224–ON01–01 for Elora and 98–UOG1–075–ALF–02–224–ON30–01 for New Liskeard). The trials were established with two replications at Elora, Ontario, Canada, on May 20, 1998 and with two replications at New Liskeard, Ontario, Canada, on June 3, 1998. The site at Elora was adjacent to the previous 1996 and 1997 sites. The soil at New Liskeard is a clay loam (lacustrine light brown gray). New Liskeard is a short-season area, having only 99 frost-free days. The start of the critical fall harvest period for alfalfa is August 15.
The plots were established with six rooted propagules of the same transgenic plant per row, one row of each transgenic plant per plot and 24 plots per replication. Two replicate plots were dug on each sampling date to measure root, crown, and shoot dry weight. The plants were defoliated twice at Elora (July 6 and August 24, 1998) and twice at New Liskeard (July 20 and August 16, 1998) prior to flowering. The growth of the plants was followed through three growth cycles and in the subsequent spring at each location. At Elora, cycle 1 was from May 20 to July 20, with samples taken on June 8, June 29, July 6, July 13, and July 20; cycle 2 was from July 6 to August 17, with samples taken on July 27, August 4, August 10, and August 17; cycle 3 began August 24, with samples taken August 31, September 14, September 28, October 13, and October 26. A subsequent spring sample was taken with four replications on May 4. At New Liskeard, cycle 1 was June 3 to July 21, with samples taken on June 22, July 6, July 13, and July 21; cycle 2 was from July 20 to August 17, with samples taken on July 27, August 4, August 10, and August 17; cycle 3 began August 16, with samples taken on August 31, September 14, and September 28. A subsequent spring sample was taken with two replications each on May 14 and 17. Statistical analysis was conducted as a split plot in a time-factorial experiment with locations as main plots and entries as subplots (Steel et al., 1997).
Protein and Carbohydrate Analysis
Total storage proteins were extracted (Avice et al., 1997) and quantified colorimetrically (Bradford, 1976). Total soluble carbohydrates were extracted from leaf, stem, and root tissue of cold-acclimated and non-acclimated alfalfa plants according to the method of Chaplin (1986). Starch was extracted according to the method of Rose et al. (1991), digested with amyloglucosidase (Aspergillus niger; Sigma Chemical) and α-amylase (porcine; Sigma Chemical) prior to analysis, and analyzed as Glc using a Glc diagnostics kit (Sigma Chemical). Soluble sugars (Glc, Fru, Suc, and raffinose) were quantified by gas chromatography as previously described (Jones et al., 1999) using phenyl β-d glucoside (Sigma Chemical) as an internal standard.
Statistical Analysis
Analysis of variance was determined using SAS for Windows, Proc GLM (Version 6.11, SAS Institute, Cary, NC) and, due to missing values in some experiments, Type III sums of squares and least squares means were calculated. Significance was determined at the 5% level of probability.
RESULTS
Two clones of alfalfa, designated as N4-4-2 and V4-11-3, were transformed using A. tumefaciens with a binary vector containing cDNA for Fe-SOD from Arabidopsis with a chloroplast transit peptide under control of the cauliflower mosaic virus (CaMV) 35S promoter (Van Camp et al., 1996). All putative transgenic plants survived selection on medium containing kanamycin and were positive according to a PCR test for the presence of the nos-nptII transgene. Four of these plants from clone N4-4-2 were tested by Southern-blot analysis for the presence of the Fe-SOD in the T-DNA. DNA that was digested with HindIII, which has a single restriction site in the binary vector, had either one or two bands on the Southern blot, indicating that one or two copies of the T-DNA were integrated into the plant DNA (Table I).
Table I.
Fe-SOD activities of leaf extracts from four independent transgenic plants of alfalfa clone N4-4-2 containing T-DNA of pEXSOD10 grown in greenhouse and field conditions
| Plant | No. of T-DNA Inserts | Greenhouse | Field |
|---|---|---|---|
| units/mg protein | |||
| N4-4-2 | 0 | NDa | ND |
| N4-FeSOD-25 | 1 | 51 ± 7 | 97 ± 8 |
| N4-FeSOD-19 | 1 | 209 ± 35 | 259 ± 50 |
| N4-FeSOD-30 | 1 | 267 ± 37 | 194 ± 17 |
| N4-FeSOD-13 | 2 | 393 ± 122 | 410 ± 68 |
Plant DNA was digested with HindIII that cuts only once within the T-DNA borders of pEXSOD10. T-DNA insertions were determined by Southern-blot analysis using a dig-labeled probe to the Arabidopsis cDNA for Fe-SOD. Fe-SOD activities were determined on three separate extracts (replications) from primary transgenic plants and are reported relative to E. coli Fe-SOD on the same PAGE gel. Values for the transgenic plants are the mean ± se.
ND, Not detectable on native PAGE gels.
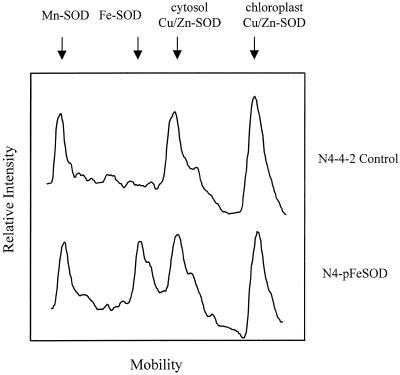
Native PAGE analysis of a leaf extract from these transgenic plants indicated the presence of a novel SOD enzyme (Fig. 1). Leaf extracts from non-transgenic alfalfa plants contained three forms of SOD: a slow-moving Mn-SOD, a cytosolic form of Cu/Zn-SOD, and a chloroplastic form of Cu/Zn-SOD (McKersie et al., 1993). The transgenic plants had an additional Fe-SOD intermediate in mobility between Mn-SOD and the cytosolic Cu/Zn-SOD. The additional SOD enzyme was also detected in extracts from leaf, crown, crown bud, and taproot of field-grown plants (data not shown).
Figure 1.
Linescan of a native PAGE gel for SOD activity in leaf extracts of transgenic alfalfa clone N4-4-2 expressing pExSOD10. Note the presence of the novel Fe-SOD band with a mobility between Mn-SOD and cytosol Cu/Zn-SOD in the N4-FeSOD plants, and its absence in the control N4-4-2 plants.
The activity of each of the SOD isoenzymes was quantified on native PAGE gels by comparing the intensity of SOD activity in the leaf extracts relative to a known standard. In leaf extracts from greenhouse-grown plants, the four transgenic plants tested had significantly different Fe-SOD activity, ranging from 51 to 393 units/mg protein (Table I). In contrast, leaf extracts of the control, non-transgenic N4-4-2 clone had no detectable Fe-SOD activity.
In leaf extracts from cuttings of the same plants grown in a field environment, Fe-SOD activity was similar quantitatively and in the relative ranking of the plants (Spearman's rank correlation r = 0.9, P = 0.037). The plant with the highest SOD activity in both greenhouse and field environments, designated as N4-FeSOD-13, had two insertions of the T-DNA according to Southern-blot analysis; the other three plants had one insertion (Table I).
The activities of the Mn-SOD, cytosolic, and chloroplastic forms of Cu/Zn-SOD did not differ among the four transgenic plants tested and, therefore, the values shown in Table II are averaged across the four transgenic plants listed in Table I. Although there was no significant difference between control and transgenic plants for Mn-SOD or either Cu/Zn-SOD in the greenhouse, the field-grown transgenic plants had lower cytosolic and higher chloroplastic Cu/Zn-SOD activity than the control plants. Presumably, this is a consequence of an environmental regulation of the native enzyme activities that has been modified by the introduced Fe-SOD transgene, but this mechanism is not fully understood.
Table II.
Mn- and Cu/Zn-SOD activities of leaf extracts from transgenic plants of alfalfa clone N4-4-2 containing T-DNA of pEXSOD10 grown in greenhouse and field conditions
| Isozyme | Greenhouse
|
Field
|
||
|---|---|---|---|---|
| N4-4-2 | Transgenic | N4-4-2 | Transgenic | |
| units/mg protein | ||||
| No. of observations | 3 | 12 | 3 | 15 |
| Mn-SOD | 130 ± 80 | 150 ± 40 | 370 ± 78 | 200 ± 34 |
| Cytosol Cu/Zn-SOD | 180 ± 24 | 210 ± 12 | 560 ± 52 | 350 ± 25 |
| Chloroplast Cu/Zn-SOD | 330 ± 62 | 363 ± 31 | 200 ± 64 | 690 ± 36 |
Each plant was sampled as three separate extracts from greenhouse or field plants (blocks). Values for the transgenic plants are the mean ± se of four (greenhouse) or five (field) independent transgenic plants that were not significantly different at the 5% level of probability.
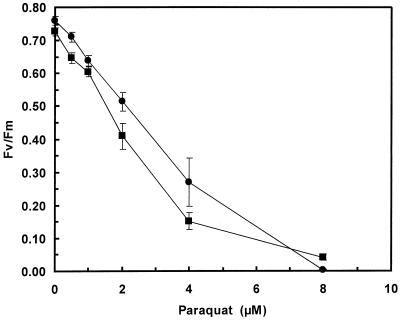
The leaves of greenhouse-grown transgenic plants expressing the Fe-SOD transgene did not have greater tolerance of the free-radical-generating herbicide methyl viologen (Fig. 2). The photosynthetic efficiency (Fv/Fm) of treated leaves of both control and transgenic plants declined in parallel with the concentration of methyl viologen in the treatment solution increased. In a similar test, the leakage of electrolytes was measured but did not detect any difference between transgenic and control plants when treated with a similar range of methyl viologen concentrations (data not shown). Based on these tests, expression of the Fe-SOD transgene apparently did not increase tolerance to oxidative stress in photosynthetic tissues.
Figure 2.
Photosynthetic efficiency of leaves from control and transgenic alfalfa clone N4-4-2 expressing pExSOD10 following exposure to oxidative stress produced by methyl viologen. Photosynthetic efficiency was measured by chlorophyll fluorescence as the Fv/Fm ratio following treatment of leaves from control (●) and transgenic (▪) plants with various concentrations of methyl viologen. The experiment was repeated twice, with five replications per experiment. Values of the transgenic plants are the least squares means of three independent transgenic plants (designated as Fe-SOD-13, -25, and -30 from Table I) that were not significantly different according to analysis of variance. se = 0.037 (n = 10 control) and 0.021 (n = 30 transgenic).
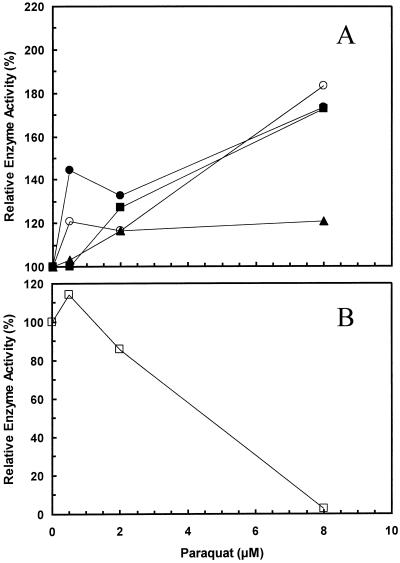
Activities of the three native forms of SOD and APX in the N4-4-2 control plant were compared with the enzyme activities in three transgenic N4-pFeSOD plants after oxidative stress treatment with methyl viologen. Methyl viologen treatment increased the activity of these enzymes in both control and transgenic plants, but there was no statistically significant difference in the response among plants, nor was there a significant statistical interaction (Fig. 3A). Compared with the activity in non-treated leaves, Mn-SOD, cytosolic Cu/Zn-SOD, and APX increased almost 180% following treatment with 8 μm methyl viologen, whereas chloroplastic Cu/Zn-SOD increased to 120%.
Figure 3.
Relative activity of superoxide dismutase isozymes and ascorbate peroxidase in leaves from control and transgenic alfalfa clone N4-4-2 expressing pExSOD10 following exposure to oxidative stress produced by methyl viologen. A, Enzyme activities are the mean of control and three transgenic plants expressing pEXSOD10 (13, 25, and 30) expressed relative to the non-stress treatment (100%). With no methyl viologen treatment the enzyme activities (in units mg−1 protein ± se) were: Mn-SOD (●), 589 ± 156; cytosolic Cu/Zn-SOD (▪), 1,100 ± 170; chloroplastic Cu/Zn-SOD (▴), 1,630 ± 62; and APX (○), 4.8 ± 0.5. B, Fe-SOD activity of three transgenic plants expressed relative to the non-stress treatment (100%). No Fe-SOD activity was detected in the control plants. With no methyl viologen treatment the enzyme activity (in units mg−1 protein ± se) was 700 ± 150.
In contrast, the Fe-SOD activity in the three transgenic plants declined to almost no activity with the same methyl viologen treatment (Fig. 3B). Fe-SOD was not detectable in the control plants. This presumably reflects an increased degradation of the Fe-SOD enzyme by this oxidative stress. Since the response of the CaMV35S promoter to this stress treatment is unknown, it is also possible that the loss of activity was due to inhibition of transcription, or translation of the transgene.
All putative primary transgenic plants that were PCR positive for the nos-nptII transgene were propagated by cuttings to create replicated samples and transplanted into the field in the spring of 1996. The plants were defoliated during the growing season prior to flowering and entered the fall acclimation period in the vegetative stage, as typically occurs in production fields. In the fall of 1996 and again in the spring of 1997, counts were made of all green plants. These counts were repeated at first harvest in the summer of 1997 and again in the spring and summer of 1998. The winter of 1996/97 was particularly harsh at this location, and survival of the control plants, N4-4-2 and V4-11-3, was less than typically seen. In contrast, the winter of 1997/1998 was quite mild, and there was no further reduction in the stand counts of clone N4-4-2; however, there was further reduction in both control and transgenic V4-11-3. The average winter survival of the alfalfa plants containing T-DNA of pEXSOD10 was significantly greater than the non-transgenic control plants for both clones (Table III).
Table III.
Winter survival (percentage) in spaced plantings in a field trial at Elora, Ontario, of transgenic plants of alfalfa clones N4-4-2 and V4-11-3 containing T-DNA from the vector pEXSOD10
| Parent | Vector | n | 1997
|
1998
|
||
|---|---|---|---|---|---|---|
| Spring | Summer | Spring | Summer | |||
| N4-4-2 | Control | 8 | 52 ± 7 | 52 ± 7 | 52 ± 8 | 51 ± 8 |
| pEXSOD10 | 94 | 83 ± 2 | 83 ± 2 | 75 ± 2 | 74 ± 2 | |
| V4-11-3 | Control | 8 | 46 ± 7 | 58 ± 7 | 26 ± 8 | 27 ± 8 |
| pEXSOD10 | 21 | 60 ± 5 | 60 ± 5 | 47 ± 5 | 47 ± 5 | |
The experiment was established in spring 1996, with three blocks each containing three controls and 32 (N4-4-2) and seven (V4-11-3) independent transgenic plants randomly arranged within each block. Values are the least squares means of all transgenic plants ± se. Survival was scored visually as the appearance of green shoots in spring and summer. n is the number of observations (replications × no. of independent transgenic plants minus missing plots) in each mean.
Herbage (total shoot dry matter) production was measured individually for all surviving plants. Plants that did not survive the winter were not included in this analysis; consequently, fewer observations (n) were made in Table IV than in Table III. The data in Table IV measure an individual plant's vigor independent of winter survival. The average yields in the 2 years were very similar. The two clones N4-4-2 and V4-11-3 differed in yield but, on average, the presence of the T-DNA in either clone did not increase total shoot dry matter production.
Table IV.
Total shoot dry matter yields in a field trial at Elora, Ontario, of spaced transgenic plants of alfalfa clones N4-4-2 and V4-11-3 containing T-DNA from the vector pEXSOD10
| Parent | Vector | 1997
|
1998
|
||
|---|---|---|---|---|---|
| n | g/plant | n | g/plant | ||
| N4-4-2 | Control | 6 | 47 ± 10.8 | 6 | 46 ± 11.7 |
| pEXSOD10 | 91 | 46 ± 2.8 | 89 | 46 ± 3.0 | |
| V4-11-3 | Control | 6 | 24 ± 10.8 | 4 | 26 ± 14.5 |
| pEXSOD10 | 17 | 28 ± 6.4 | 16 | 20 ± 7.1 | |
The experiment was established in spring 1996, with three blocks each containing three controls and 32 (N4-4-2) and seven (V4-11-3) independent transgenic plants randomly arranged within each block. Values are the least squares means ± se for all independent transgenic plants surviving the winter. Total shoot dry matter yields are the sum of three harvests in each year. n is the number of observations in each mean (replications × no. of independent transgenic plants minus missing plots and minus plots not surviving the winter).
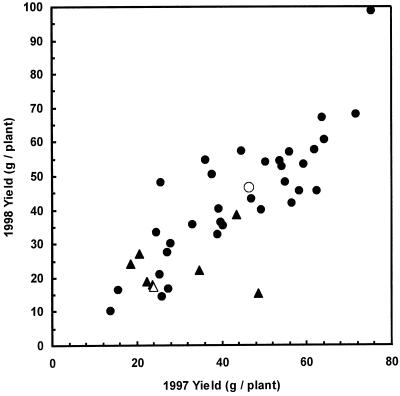
There was considerable variability observed among individual transgenic plants for both yield and winter survival. To illustrate that variability, Figure 4 compares the 1997 and 1998 herbage yields of all plants in the field trial. The ranking of an individual transgenic plant in 1998 was very similar to its ranking in 1997, indicating reproducible relative yields in the 2 years. Individual transgenic plants had both higher and lower yields than the control clones, N4-4-2 and V4-11-3, and in the extreme cases, these differences were statistically significant.
Figure 4.
Relationship between herbage dry matter production in 1997 and 1998 of control and independent transgenic alfalfa clones N4-4-2 and V4-11-3 containing pExSOD10. Control plants are shown as white symbols and transgenic plants as black symbols for clones N4-4-2 (circles) and V4-11-3 (triangles). The total herbage dry matter from three harvests was determined for each independent transgenic plant in the field trial at Elora Ontario. The positive relationship indicates that herbage production was consistent in the 2 years for individual transgenic plants. Plants that did not survive the winter were not included in the analysis and therefore these data are a measure of individual plant vigor. se = 10.3 (1997) and 10.9 (1998).
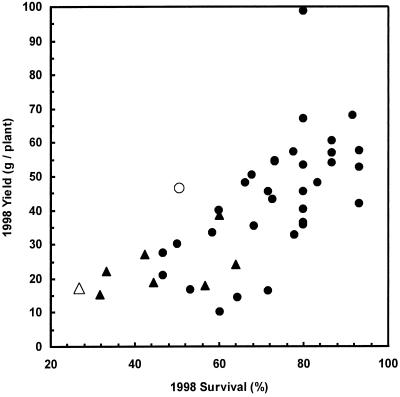
A slightly different relationship was observed with winter survival (Fig. 5). Higher yields tended to be associated with higher survival, even though yield is reported as dry matter production from surviving plants only. In other words, to calculate yield per plant, the yield per plot was divided by the number of surviving plants, not by the number originally planted in the plot. This observation was not unexpected, because winter injury is often sublethal. In alfalfa, sublethal injury may reduce the number of crown buds that establish new shoots or the vigor of the new shoot growth. Most transgenic plants from both clones had higher survival than the corresponding non-transgenic controls. Therefore, the relationship between yield compared with survival was skewed toward increased survival. Yield was normally distributed around the non-transgenic control, whereas survival was shifted higher (compare the white and black symbols in Fig. 5).
Figure 5.
Relationship between winter survival after two winters and herbage dry matter production in 1998 of control and independent transgenic alfalfa clones N4-4-2 and V4-11-3 expressing pExSOD10. Control plants are shown as white symbols and transgenic plants as black symbols for clones N4-4-2 (circles) and V4-11-3 (triangles). Winter survival was determined by stand counts in spring 1998, 2 years after transplanting. The total herbage dry matter from three harvests was determined for each independent transgenic plant in the field trial at Elora Ontario. Plants that did not survive the winter were not included in the herbage yield analysis. The relationship indicates that the Fe-SOD transgene had a greater impact on winter survival than on herbage yield. se = 19.1 (survival) and 10.9 (yield).
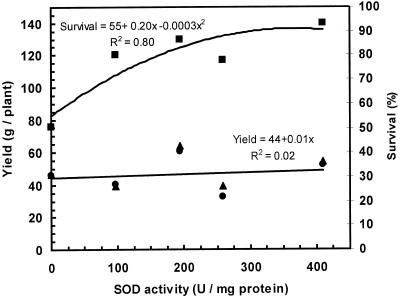
The higher relative winter survival of the transgenic plants was associated with higher activity of the Fe-SOD transgene in the four transgenic plants shown in Table I (Fig. 6). If the non-transgenic control was included in the regression analysis, the relationship was exponential (r2 = 0.80), but if only the transgenic plants were included the relationship was linear (r2 = 0.48). In contrast, the variation in herbage yield was not associated with differences in Fe-SOD activity.
Figure 6.
Relationship between winter survival, herbage dry matter production, and Fe-SOD activity among control and transgenic alfalfa expressing pExSOD1. SOD activity was determined from Table I. Winter survival (▪) and total herbage dry matter in 1997 (▴) and in 1998 (●) were determined from Figures 4 and 5.
In a second field trial, a random selection of primary transgenic and control alfalfa plants was propagated by cuttings, transplanted to the field in the spring, and sampled in November 1997, before the plants had experienced a severe freezing or winter stress. The root systems of these transgenic plants were larger than in the control, non-transgenic N4-4-2 (1.93 g compared to 1.31 g, respectively), but on a dry weight basis, the roots and crowns of transgenic plants contained the same amount of Glc, Fru, raffinose, starch, and protein as the control (data not shown). Samples were taken from the same field plots in spring 1998, and, again, the soluble carbohydrate analysis did not detect any difference between control and transgenic plants (data not shown).
The crowns and roots of field-acclimated plants were sampled from the 1997 field trial in November and again in December, and then subjected to freezing temperatures. Viability was determined by vital staining with tetrazolium. At −14°C, the bud axis, inner scales, and vascular cylinder of the taproots were unable to reduce tetrazolium, but the endodermis of the root, the vascular system of the crown, and the cortex of the crown were viable and stained red. No differences in this pattern of freezing injury were found between the non-transgenic and any of transgenic plants (data not shown).
To determine if the increased Fe-SOD activity in the transgenic plants improved photosynthesis, as measured by accumulation of dry matter in a field environment, a third field trial was established in 1998 in which one primary transgenic alfalfa plant with T-DNA from pEXSOD10, N4-FeSOD-13 (Table I), and the control N4-4-2 were propagated by cuttings and transplanted to establish the 1998 field trial at two locations in Ontario, Canada. The plots were sampled for shoot and for root and crown biomass at regular intervals during the growing season of 1998. In the year of transplanting (i.e. before exposure to any winter stress) the shoot dry weight of the control and transgenic plants were not significantly different at any growth stage at either location (Table V). Similarly, in the spring of 1999, after one winter, there was no difference in the growth of new shoots from the overwintering crown buds at either location. The crown and root dry weights of the alfalfa plants were greater at New Liskeard than at Elora (data not shown), but, again, there was no significant difference between control and transgenic plants at any of the sampling times (Table V).
Table V.
Dry weight (g/plant) of transgenic alfalfa N4-FeSOD-13 and non-transgenic N4-4-2 grown in field plots at two locations
| Growth Cycle | n | N4-4-2 Control
|
Transgenic N4-FeSOD-13
|
||
|---|---|---|---|---|---|
| Shoot | Root | Shoot | Root | ||
| 1 | 16–20 | 1.38 ± 0.24 | 0.47 ± 0.21 | 1.18 ± 0.30 | 0.36 ± 0.28 |
| 2 | 16 | 2.39 ± 0.25 | 1.24 ± 0.22 | 2.58 ± 0.25 | 1.40 ± 0.22 |
| 3 | 15–16 | 3.46 ± 0.29 | 3.16 ± 0.26 | 3.35 ± 0.27 | 3.42 ± 0.24 |
| Spring | 7–8 | 4.85 ± 0.35 | 4.08 ± 0.32 | 4.45 ± 0.38 | 4.07 ± 0.35 |
Plants were defoliated at 1- or 2-week intervals during each growth cycle, and values are the least squares means of shoot dry weight during that growth cycle at both locations ± se of the mean; n is the number of observations. The values between control and transgenic plants were not significantly different at the 5% level of probability at any growth cycle at either location.
DISCUSSION
Two alfalfa clones were transformed using A. tumefaciens with T-DNA from the binary vector pEXSOD10 (Van Camp et al., 1996) containing a Fe-SOD transgene with a chloroplast transit peptide and the constitutive CaMV35S promoter. We observed a novel form of SOD by native PAGE in both greenhouse- and field-grown transgenic plants, but expression of the SOD transgene varied among independent transgenic plants. The increased Fe-SOD activity was associated with variability in the winter survival of the transgenic plants but, surprisingly, not in oxidative stress tolerance as measured by response of leaves to methyl viologen, a superoxide generator. In previous studies, incorporation of the same T-DNA protected both the plasmalemma and thylakoids of tobacco against superoxide generated by illumination in the presence of methyl viologen (Van Camp et al., 1996). We attempted to reproduce the treatment and measurement conditions described by Van Camp et al. (1996) as closely as possible, so these conflicting results may be due to differences between species or to the relatively small number of transgenic plants tested in both studies.
The observation that increased Fe-SOD activity in the transgenic plants in both alfalfa clones was positively related to the improvement in winter survival supports our original hypothesis that perennial and winter annual plants experience oxidative stress during winter, and that antioxidant defense systems contribute to winter hardiness. The mechanism of this protection is unknown. Winter hardiness is a composite of tolerances to freezing, anoxia, desiccation, and disease stresses, although freezing is usually considered to be the predominant stress in most environments. Freezing injury occurs in two phases. Primary freezing injury is caused by cellular dehydration that promotes lipid phase transitions and membrane damage (Thomashow, 1999); subsequently, secondary injuries occur to other physiological processes that culminate in cell and tissue death. Also, primary freezing injury measured by vital staining is not uniform within a heterogeneous tissue such as an acclimated crown, but instead occurs only in specific cell types (Tanino and McKersie, 1985; McKersie et al., 1999). Our tetrazolium tests did not detect any difference in freezing injury between control and transgenic plants immediately after a freezing stress imposed by a programmable freezer, and this suggests that there is not a difference in the primary freezing injury between control and Fe-SOD transgenic plants. Instead, the present data suggest that in the transgenic plants expressing a novel SOD, secondary injury is reduced—in other words, the recovery from the primary freezing injury is enhanced.
An alternative explanation for our field observations is that increased Fe-SOD activity in the alfalfa leaves enhanced photosynthesis under mild stress conditions, as was originally proposed by Allen (1995). This may have enabled the transgenic plants to allocate more sugars to the root compared with the control plants, and therefore further increase root carbohydrate levels during cold acclimation. Since soluble carbohydrate levels in the roots of alfalfa have been correlated with the level of freezing tolerance across different cultivars and across stages of acclimation (Castonguay et al., 1995, 1997), this would predict increased winter survival in the transgenic plants. Our results do not support this mechanism of action for SOD in these transgenic plants.
First, the carbohydrate analysis did not detect any additional accumulation of carbohydrates in the field-acclimated roots of the transgenic alfalfa plants. Second, although statistically significant and consistent differences in herbage yield were found among the transgenic plants in the 2 production years of the 1996 field trial, the differences were not directly related to increased Fe-SOD activity. The growth analysis experiment showed very convincingly that one transgenic plant that had high Fe-SOD expression, N4-FeSOD-13 (Table I), did not have increased dry matter production (Table V). Third, our transgenic plants were not more tolerant of oxidative stress to photosynthesis, as determined using the standard test of methyl viologen tolerance measured by chlorophyll fluorescence. Fourth, the activities of the free-radical-scavenging enzymes were induced to a similar extent following treatment with methyl viologen in both control and transgenic plants, suggesting that plants with and without the Fe-SOD transgene received similar oxidative stress from methyl viologen. Finally, Fe-SOD activity was inactivated by the oxidative stress imposed by methyl viologen, and therefore this transgene may provide only very limited protection to oxidative stress in leaves.
The SOD enzyme not only consumes superoxide and thereby provides tolerance to oxidative stress, but also produces H2O2. It is tempting to speculate that an increased steady-state level of H2O2 or an increased flux through the H2O2 pool enhanced an acclimation process that enabled the plants to tolerate or repair freezing injury more effectively and as a result improved winter survival. H2O2 has potential toxicity in plants, but it may also have a number of regulatory roles. Recent reports suggest that H2O2 mediates some responses to pathogens (Chen et al., 1993), produces a transient Ca2+ surge, which is a known signaling component (Price et al., 1994), and initiates the production of other antioxidant enzymes during acclimation (Prasad, 1997). H2O2 is metabolized by a number of peroxidases using reducing equivalents to form water. When plant cells are treated with elicitors, H2O2 rises, ATP is depleted, and the NADH/NAD ratio transiently drops (Robertson et al., 1995). A similar experiment noted that in the long term (hours rather than minutes), the redox potential (NADPH/NADP) did not change but genes in the pentose-phosphate pathway were activated, which suggests a greater supply of NADPH (Fahrendorf et al., 1995). Cellular redox is an important contributing factor in the acclimation process. For example, plants grown at low temperatures have a prostrate growth habit, which can also be induced at high light intensities (Huner et al., 1996). In both environments, the plants have excess amounts of NAD(P)H. Therefore, it has been proposed that plants respond to changes in the redox state of photosynthetic electron transport, and this redox sensing mechanism in the chloroplast may be an important component for sensing the plant's environment.
Several other genes have recently been reported to enhance tolerance of various environmental stresses when overexpressed in transgenic plants. One of these is the codA gene encoding choline oxidase, which produces Gly betaine and provides protection to high light and other stresses (Alia et al., 1999). However, like Fe-SOD, choline oxidase produces H2O2. Although H2O2 did not accumulate to high levels, an increased flux through the H2O2 pool may have altered redox. Although speculative, perhaps transgenes such as SOD and codA act by a common indirect mechanism through changes in redox. The observation that transgenic alfalfa plants expressing either Mn-SOD or Fe-SOD have less tolerance to pathogens causing verticillium wilt and bacterial wilt (McKersie et al., 1999b) supports our assumption that the expression of the SOD transgenes may alter H2O2 flux and therefore cell signaling processes.
The alfalfa system that we have used provides several advantages as a model to study the effects of transgenes on winter hardiness. It is easily transformed by A. tumefaciens; it is perennial and acclimates in the autumn as part of its life cycle; it can be vegetatively propagated to maintain and to replicate individual transgenic plants or cross-pollinated to produce seed; a single plant can be maintained in a vegetative state by defoliation, and therefore repeated measurements can be made on the same plant; and, finally, it can be grown in a field environment and subjected to complex winter stress conditions. Nonetheless, it is not a perfect model because cultivated alfalfa is a cross-pollinating autotetraploid species. It therefore has limitations for genetic studies, most notably that it cannot be self-pollinated to establish pure lines. Recognizing this limitation, we chose to study the effects of the Fe-SOD transgene in primary transgenic plants because a large number of independent transgenic plants could be compared.
We used two types of controls: a non-transgenic control that did not pass through tissue culture, and transgenic plants that exhibited variation in transgene expression. Although somaclonal variation may have occurred in some transgenic plants due to the tissue culture process, we do not believe that this contributed to our observations. Somaclonal variation is caused by random mutations and is not normally observed in a tetraploid species in the first generation after tissue culture, because most of the induced mutations are recessive. We did not consider a control plant that was regenerated from cell culture to be an effective control, even if it displayed somaclonal variation, because the comparison would simply be one randomly generated mutation to another. Therefore, the best comparison for the determination of transgene effects are between F1 progeny with and without the transgene, but because of alfalfa's autotetraploid genetics, this comparison requires the generation of populations of plants, not individual pure lines. These comparisons are now in progress for the N4-FeSOD-13 plant, but comparison of populations from all 39 independent transformation events was not feasible.
We examined a large number (39) of independent primary transgenic plant created in two genetic backgrounds. Although somaclonal variation or similar random mutation may explain a single novel event, this explanation is inconsistent with the range of variability observed among these transgenic plants, particularly because all of the transgenic plants shown in Figure 5 had similar or improved winter survival compared with the control. Alternatively, tissue culture may have caused epigenetic effects, virus elimination, or imbalances in growth regulators, but if these factors contributed to improved winter survival, they were maintained for 3 years after the transformation event in plants that were repeatedly defoliated.
We propose that increased Fe-SOD activity in transgenic plants increased winter survival in alfalfa. We also propose that this may have occurred by two non-mutually exclusive mechanisms. Fe-SOD may have acted directly by increasing the scavenging capacity for superoxide produced following primary freezing injury in the root and/or indirectly by increasing the flux through the H2O2 pool, thereby modifying redox and cell signaling processes. This model predicts that SOD has its effect directly in the cells of the root, not in photosynthesis in the shoot, and this is consistent with our experimental observations on methyl viologen tolerance. Transformations with T-DNA containing root- and shoot-specific promoters controlling SOD expression are now in progress to test this proposal further.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Dr. Dirk Inzé, Universiteit Gent, Belgium, for providing the binary vector pEXSOD10, Cecilio Gregorio for conducting the alfalfa transformations, Lori Wright for maintaining the transgenic plants in the greenhouse, and Donna Hancock, Matt Bowman, and Ning Chen for conducting the transgenic field trials.
Footnotes
Financial support for this research was provided by a research grant from the Natural Sciences and Engineering Research Council of Canada and by the Ontario Ministry of Agriculture Food and Rural Affairs.
LITERATURE CITED
- Alia, Kondo Y, Sakamoto A, Nonaka H, Hayashi H, Saradhi PP, Chen THH, Murata N. Enhanced tolerance to light stress of transgenic arabidopsis plants that express the codA gene for a bacterial choline oxidase. Plant Mol Biol. 1999;40:279–288. doi: 10.1023/a:1006121821883. [DOI] [PubMed] [Google Scholar]
- Allen RD. Dissection of oxidative stress tolerance using transgenic plants. Plant Physiol. 1995;107:1049–1054. doi: 10.1104/pp.107.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Roger B, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Toronto, Ontario, Canada: Green Publishing Associates and Wiley-Interscience; 1991. [Google Scholar]
- Avice JC, Ourry A, Lemaire G, Volenec JJ, Boucaud J. Root protein and vegetative storage protein are key organic nutrients for alfalfa shoot regrowth. Crop Sci. 1997;37:1187–1193. [Google Scholar]
- Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- Bingham ET, Hurley LV, Kaatz DM, Saunders JW. Breeding alfalfa which regenerates from callus tissue in culture. Crop Sci. 1975;15:719–721. [Google Scholar]
- Bowler C, Montagu Mv, Inzé D, Van Montagu M. Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:83–116. [Google Scholar]
- Bowler C, Slooten L, Vandenbranden S, de Rycke R, Botterman J, Sybesma C, van Montagu M, Inzé D. Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J. 1991;10:1723–1732. doi: 10.1002/j.1460-2075.1991.tb07696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Van Camp W, Van Montagu M, Inzé D. Superoxide dismutase in plants. Crit Rev Plant Sci. 1994;13:199–218. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Castonguay Y, Nadeau P, Laberge S, Vezina LP. Changes in gene expression in six alfalfa cultivars acclimated under winter hardening conditions. Crop Sci. 1997;37:332–342. [Google Scholar]
- Castonguay Y, Nadeau P, Lechasseur P, Chouinard L. Differential accumulation of carbohydrates in alfalfa cultivars of contrasting winterhardiness. Crop Sci. 1995;35:509–516. [Google Scholar]
- Chen Z, Silva H, Klessig DF. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science. 1993;262:1883–1886. doi: 10.1126/science.8266079. [DOI] [PubMed] [Google Scholar]
- Fahrendorf T, Ni WT, Shorrosh BS, Dixon RA. Stress responses in alfalfa (Medicago sativa L.)19: transcriptional activation of oxidative pentose phosphate pathway genes at the onset of the isoflavonoid phytoalexin response. Plant Mol Biol. 1995;28:885–900. doi: 10.1007/BF00042073. [DOI] [PubMed] [Google Scholar]
- Foyer C, Noctor G, Morotgaudry JF. Oxygen: friend or foe for plants. Biofutur. 1997;169:27–29. [Google Scholar]
- Foyer CH, Descourvieres P, Kunert KJ. Protection against oxygen radicals: an important defence mechanism studied in transgenic plants. Plant Cell Environ. 1994b;17:507–523. [Google Scholar]
- Foyer CH, Halliwell B. The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta. 1976;133:21–25. doi: 10.1007/BF00386001. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Lelandais M, Kunert KJ. Photoxidative stress in plants. Physiol Plant. 1994a;92:696–717. [Google Scholar]
- Genty B, Briantais JM, Baker NR. The relationship between quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochem Biophys Acta. 1989;990:87–92. [Google Scholar]
- Herouart D, Bowler C, Willekens H, Van Camp W, Slooten L, Van Montagu M, Inzé D. Genetic engineering of oxidative stress resistance in higher plants. Phil Trans R Soc London Series B Biol Sci. 1993;342:235–240. [Google Scholar]
- Huner NPA, Maxwell DP, Gray GR, Savitch LV, Krol M, Ivanov AG, Falk S. Sensing environmental temperature change through imbalances between energy supply and energy consumption: redox state of photosystem II. Physiol Plant. 1996;98:358–364. [Google Scholar]
- Jones KS, Paroschy J, McKersie BD, Bowley SR. Carbohydrate composition and freezing tolerance of canes and buds in Vitis vinifera. J Plant Physiol. 1999;155:101–106. [Google Scholar]
- Kalu BA, Fick GW. Morphological stage of development as a predictor of alfalfa herbage quality. Crop Sci. 1983;23:1167–1172. [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase: an enzymic function for ethrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- McKersie BD, Bowley SR, Harjanto E, Leprince O. Water-deficit tolerance and field performance of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiol. 1996;111:1177–1181. doi: 10.1104/pp.111.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie BD, Bowley SR, Jones KS. Winter survival of transgenic alfalfa overexpressing superoxide dismustase. Plant Physiol. 1999a;119:839–847. doi: 10.1104/pp.119.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie BD, Bowley SR, Jones KS, Gossen B. Winter survival of transgenic alfalfa over-expressing superoxide dismustase. In: Smallwood MF, Calvert CM, Bowles DJ, editors. Plant Responses to Environmental Stress. Oxford: BIOS Scientific Publishers; 1999b. pp. 117–126. [Google Scholar]
- McKersie BD, Chen YR, deBeus M, Bowley SR, Bowler C, Inzé D, D'Halluin K, Botterman J. Superoxide dismutase enhances tolerance of freezing stress in transgenic alfalfa (Medicago sativa L.) Plant Physiol. 1993;103:1155–1163. doi: 10.1104/pp.103.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie BD, Murnaghan J, Bowley SR. Manipulating freezing tolerance in transgenic plants. Acta Physiol Plant. 1997;19:485–495. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco cultures. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascorbate specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981;22:867–880. [Google Scholar]
- Payton P, Allen RD, Trolinder N, Holaday AS. Over-expression of chloroplast-targeted Mn superoxide dismutase in cotton (Gossypium hirsutum L., cv. Coker 312) does not alter the reduction of photosynthesis after short exposures to low temperature and high light intensity. Photosynth Res. 1997;52:233–244. [Google Scholar]
- Perl A, Perl Treves R, Galili S, Aviv D, Shalgi E, Malkin S, Galun E. Enhanced oxidative-stress defense in transgenic potato expressing tomato Cu, Zn superoxide dismutases. Theor Appl Genet. 1993;85:568–576. doi: 10.1007/BF00220915. [DOI] [PubMed] [Google Scholar]
- Pitcher LH, Brennan E, Hurley A, Dunsmuir P, Tepperman JM, Zilinskas BA. Overproduction of petunia chloroplastic copper/zinc superoxide dismutase does not confer ozone tolerance in transgenic tobacco. Plant Physiol. 1991;97:452–455. doi: 10.1104/pp.97.1.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TK. Role of catalase in inducing chilling tolerance in pre- emergent maize seedlings. Plant Physiol. 1997;114:1369–1376. doi: 10.1104/pp.114.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AH, Taylor A, Ripley SJ, Griffiths A, Trewavas AJ, Knight MR. Oxidative signals in tobacco increase cytosolic calcium. Plant Cell. 1994;6:1301–1310. doi: 10.1105/tpc.6.9.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D, Davies DR, Gerrish C, Jupe SC, Bolwell GP. Rapid changes in oxidative metabolism as a consequence of elicitor treatment of suspension-cultured cells of French bean (Phaseolus vulgaris L) Plant Mol Biol. 1995;27:59–67. doi: 10.1007/BF00019178. [DOI] [PubMed] [Google Scholar]
- Rose R, Rose C, Omi SK, Forry KR, Durall DM, Gigg WL. Starch determination by perchloric acid versus enzymes: evaluating the accuracy and precision of six colorimetric methods. J Agric Food Chem. 1991;39:2–11. [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salin ML. Chloroplast and mitochondrial mechanisms for protection against oxygen toxicity. Free Rad Res Comm. 1991;12:851–858. doi: 10.3109/10715769109145867. [DOI] [PubMed] [Google Scholar]
- Scandalios JG. Oxygen stress and superoxide dismutases. Plant Physiol. 1993;101:7–12. doi: 10.1104/pp.101.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Gupta A, Heinen JL, Holaday AS, Burke JJ, Allen RD. Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA. 1993a;90:1629–1633. doi: 10.1073/pnas.90.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Gupta A, Webb RP, Holaday AS, Allen RD. Over-expression of superoxide dismutase protects plants from oxidative stress: induction of ascorbate peroxidase in superoxide dismutase-overexpressing plants. Plant Physiol. 1993b;103:1067–1073. doi: 10.1104/pp.103.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty K, McKersie BD. Proline, thioproline and potassium mediated stimulation of somatic embryogenesis in alfalfa (Medicago sativa L.) Plant Sci. 1993;88:185–193. [Google Scholar]
- Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Steel RGD, Torrie JH, Dickey DA. Principles and Procedures of Statistics. Ed 3. New York: McGraw-Hill; 1997. [Google Scholar]
- Tanino KK, McKersie BD. Injury within the crown of winter wheat seedlings after freezing and icing stress. Can J Bot. 1985;63:432–436. [Google Scholar]
- Tepperman JM, Dunsmuir P. Transformed plants with elevated levels of chloroplastic SOD are not more resistant to superoxide toxicity. Plant Mol Biol. 1990;14:501–511. doi: 10.1007/BF00027496. [DOI] [PubMed] [Google Scholar]
- Thomashow M. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Van Camp W, Capiau K, Van Montagu M, Inzé D, Slooten L. Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiol. 1996;112:1703–1714. doi: 10.1104/pp.112.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp W, Willekens H, Bowler C, Montagu Mv, Inzé D, Reupold Popp P, Sandermann H, Jr, Langebartels C. Elevated levels of superoxide dismutase protect transgenic plants against ozone damage. Bio/Technology. 1994;12:165–168. [Google Scholar]
- van Miltenburg R, Ruger B, Grunewald-Janho S, Leons M, Schroder C. The DIG System User's Guide for Filter Hybridization. Indianapolis: Boehringer Mannheim; 1995. [Google Scholar]