ABSTRACT
In Drosophila epithelial cells, apical exclusion of Bazooka (the Drosophila Par3 protein) defines the position of the zonula adherens (ZA), which demarcates the apical and lateral membrane and allows cells to assemble into sheets. Here, we show that the small GTPase Rap1, its effector Canoe (Cno) and the Cdc42 effector kinase Mushroom bodies tiny (Mbt), converge in regulating epithelial morphogenesis by coupling stabilization of the adherens junction (AJ) protein E-Cadherin and Bazooka retention at the ZA. Furthermore, our results show that the localization of Rap1, Cno and Mbt at the ZA is interdependent, indicating that their functions during ZA morphogenesis are interlinked. In this context, we find the Rap1-GEF Dizzy is enriched at the ZA and our results suggest that it promotes Rap1 activity during ZA morphogenesis. Altogether, we propose the Dizzy, Rap1 and Cno pathway and Mbt converge in regulating the interface between Bazooka and AJ material to promote ZA morphogenesis.
KEY WORDS: Epithelial polarity, Pak4, Par3, AFDN, Bazooka, Photoreceptor, Rap1, Zonula adherens
Summary: Converging pathways regulate adherens junction stability and retention of Bazooka to promote zonula adherens maturation during epithelial cell morphogenesis in Drosophila.
INTRODUCTION
The epithelial zonula adherens (ZA) enables cell–cell adhesion, allowing epithelial cells to assemble into sheets and form organs. Elucidating how ZA morphogenesis is regulated during epithelial cell morphogenesis remains an important goal in epithelial cell biology. The ZA includes the adhesion molecule E-Cadherin (E-Cad; known as Shotgun in flies) and its effector β-catenin [known as Armadillo (Arm) in flies], which are the main adherens junction (AJ) components that mediate cell–cell adhesion (Tepass, 2012). Several factors regulate the morphogenesis and accumulation of AJ material during ZA assembly. These include the small GTPase Rap1 and its effector actin-binding protein Canoe (Cno; the homolog of mammalian AF6, also known as AFDN) (Bos et al., 2001; Niessen and Gottardi, 2008; Pannekoek et al., 2009), the type 2 p21-activated kinase Mushroom bodies tiny (Mbt, the homolog of Pak4) (Menzel et al., 2007; Wallace et al., 2010; Walther et al., 2016) and the Par complex (Cdc42–Par6–aPKC–Bazooka) (McGill et al., 2009; Morais-de-Sá et al., 2010; Walther and Pichaud, 2010). However, we lack an integrated view of how these factors come together to regulate ZA morphogenesis and remodeling during epithelial cell morphogenesis.
The Drosophila pupal photoreceptor has long been used as a model system to study the genetic and molecular basis of the specification and morphogenesis of the epithelial apical, subapical and ZA membrane domains. In these cells, these domains are clearly separated along the apical basal (x–y) axis (Fig. 1A–C), and the apical organelle, called the rhabdomere, is analogous to the epithelial brush border and consists of ∼60,000 microvilli. The subapical membrane is called the stalk and can be up to 1.5 µm in length, and connects the rhabdomere to the more basal ZA. These three membrane domains are specified early during pupal development and undergo sustained morphogenesis as the cells elongate by ∼10-fold to generate the lens (proximal) to brain (distal) axis of the retina (Ready, 2002) (Fig. 1A,B). In pupal photoreceptors, the Par complex regulates the separation of the ZA from the stalk membrane (Hong et al., 2003; Nam and Choi, 2003; Walther et al., 2016; Walther and Pichaud, 2010). Concomitantly, the conserved transmembrane protein Crumbs (Crb) functions with the Par complex to drive stalk membrane and ZA morphogenesis as photoreceptors elongate along the proximal-distal axis of the retina (Izaddoost et al., 2002; Pellikka et al., 2002).
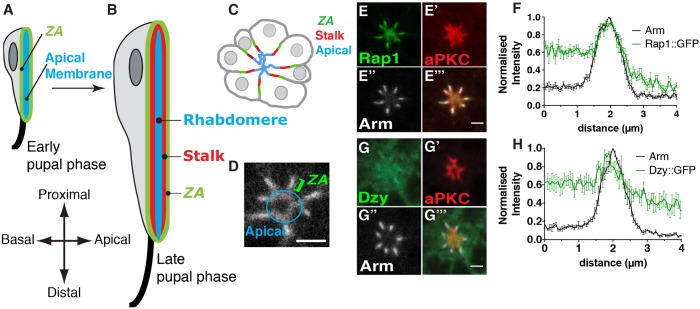
Fig. 1.
Dizzy and Rap1 are ZA-associated proteins. (A,B) Schematic representation of the developing pupal photoreceptor. (A) Early and (B) late stage pupal photoreceptors shown along the lens (top) to brain (bottom) axis of the retina. The apical membrane, which is clearly differentiated by mid pupation and by late pupation forms the rhabdomere, is depicted in blue. The stalk membranes are depicted in red and the ZA in green. The axon is depicted as a black line, at the bottom (brain or distal pole) of the cell. (C) Cross section of a cluster (ommatidium) of photoreceptors at mid pupation when the ZA (green), stalk membrane (red) and apical membrane (blue) have been specified. (D) Annotated magnification of the Rap1::GFP staining showing the apical membrane and the ZA. (E–E‴) Photoreceptors expressing Rap1::GFP (E), stained for aPKC (E′) and Arm (E″). Scale bar: 2 µm. (F) Intensity profiles of Rap1::GFP and Arm measured along the apical-basal axis. Results are mean±s.e.m. (n=8 cells from 3 retinas). (G–G‴) Photoreceptors expressing Dzy::GFP (G), stained for aPKC (G′) and Arm (G″). Scale bar: 1.5 µm. (H) Intensity profiles of Dzy::GFP and Arm measured along the apical-basal axis. Results are mean±s.e.m. (n=6 cells from 3 retinas).
In Drosophila epithelia, Bazooka (Baz) phosphorylation at serine S980 (P-S980-Baz) by atypical protein kinase C (aPKC) is essential for specifying the ZA and subapical membrane. Baz phosphorylation occurs upon Par complex assembly and is thought to allow for Crb to capture the Cdc42–Par6–aPKC complex, thus leading to the apical exclusion of P-S980-Baz (Krahn et al., 2010; Morais-de-Sá et al., 2010; Walther and Pichaud, 2010). Confined to the apical-lateral border of the cell, P-S980-Baz is then thought to promote ZA assembly, at least in part through its ability to bind to Arm (Wei et al., 2005). In the pupal photoreceptor, Crb and Par6–aPKC accumulate at the stalk membrane and P-S980-Baz is found immediately basal to it, at the developing ZA (Fig. 1B,C). It is likely that Par3 phosphorylation and its concomitant apical exclusion play a similar role in vertebrate neuroepithelial cells. In vertebrates, Par3 is phosphorylated by aPKC (Nagai-Tamai et al., 2002), and in neuroepithelial cells, is found basal to aPKC and Par6, at the apical junctional complex (AJC), which contains cadherins (Aaku-Saraste et al., 1996; Afonso and Henrique, 2006).
In addition to Baz, the p21-activated kinase Mushroom bodies tiny (Mbt) and its vertebrate homolog Pak4 have also been shown to regulate ZA morphogenesis. In pupal photoreceptors, Mbt regulates ZA morphogenesis and overall apical membrane differentiation by promoting the accumulation of the E-Cad–Arm complex via phosphorylating Arm and regulating the F-actin cytoskeleton, which in turn is essential for the retention of Baz at the ZA (Jin et al., 2015; Law and Sargent, 2014; Menzel et al., 2008; Schneeberger and Raabe, 2003; Walther et al., 2016). In these cells, failure to retain AJ material, including Baz at the ZA, leads to a shortening of the ZA along the apical-basal axis of the cell. In addition, severe defects in polarized photoreceptor morphogenesis can occur (Walther et al., 2016). In vertebrate cells, Pak4 also regulates ZA maturation (Jin et al., 2015; Law and Sargent, 2014; Wallace et al., 2010), and its function during epithelial morphogenesis has been linked to that of the Par complex, as Pak4 phosphorylates Par6b (Jin et al., 2015; Wallace et al., 2010). While in flies Mbt does not phosphorylate Par6, Mbt and Baz are the main regulators of AJ material accumulation at the plasma membrane. In the absence of baz, AJ material can still be detected at the plasma membrane of pupal photoreceptors within the apical pole of the cell. Similarly, ZA domains are present in mbt-null mutant cells, although they are shorter and present less AJ material than in wild-type cells. However, no AJ domains are found in photoreceptors mutant for both baz and mbt, indicating that Baz and Mbt function in parallel pathways to promote AJ morphogenesis and/or stabilization at the plasma membrane (Walther et al., 2016).
Another conserved factor that regulates AJ material morphogenesis is Rap1, which in epithelia can be activated by the PDZ-containing guanine exchange factor (GEF) protein Dizzy (Dzy) (de Rooij et al., 1999; KawAJiri et al., 2000). Rap1 has been shown to localize at the AJ in various fly epithelia, and to be an essential AJ regulator (Boettner et al., 2003; Boettner and Van Aelst, 2007; Choi et al., 2013; Knox and Brown, 2002; O'Keefe et al., 2009; Spahn et al., 2012; Wang et al., 2013). In the fly embryo, Rap1 and its effector F-actin-binding protein Cno (Boettner et al., 2003; Mandai et al., 2013; Sawyer et al., 2009) regulate the apical localization of both Baz and Arm, with Baz reciprocally influencing Cno localization. Furthermore, Baz is required to capture preassembled AJ material, thus promoting the morphogenesis of spot AJs, which are precursors of the ZA in this tissue (McGill et al., 2009). In addition, work in human MCF7 cells has shown a role for Rap1 (which has two forms, Rap1a and Rap1b) during AJ maturation via promoting E-Cad recruitment at the sites of cell–cell contact, a function that has been shown to be mediated, at least in part, by Cdc42 (Hogan et al., 2004). However, how the functions of Rap1, Cno, Baz and Mbt relate to each other during ZA morphogenesis has not been examined. Here, we used the Drosophila photoreceptor system to investigate these relationships.
RESULTS
Rap1 regulates pupal photoreceptor ZA morphogenesis
In the fly retina, Rap1 has been previously shown to regulate AJ remodeling between newly specified photoreceptors, and between retinal accessory cells that surround the photoreceptors (cone and pigment cells) (O'Keefe et al., 2009). To examine the distribution of Rap1 and its GEF Dzy in the pupal photoreceptor (Fig. 1A–C), we made use of the rap1-Rap1::GFP and dzy-Dzy::GFP transgenes, which allow for expression of these proteins under the control of their endogenous promoter. We found that Rap1::GFP is present at the apical membrane and accumulates predominantly at the developing ZA (Fig. 1D–F). Dzy::GFP (Fig. 1G) shows a low level expression all over the apical membrane and presents a slight but reproducible enrichment at the developing ZA (Fig. 1G,H). These results suggest that Dzy and Rap1 might regulate apical membrane and ZA morphogenesis in the pupal photoreceptor.
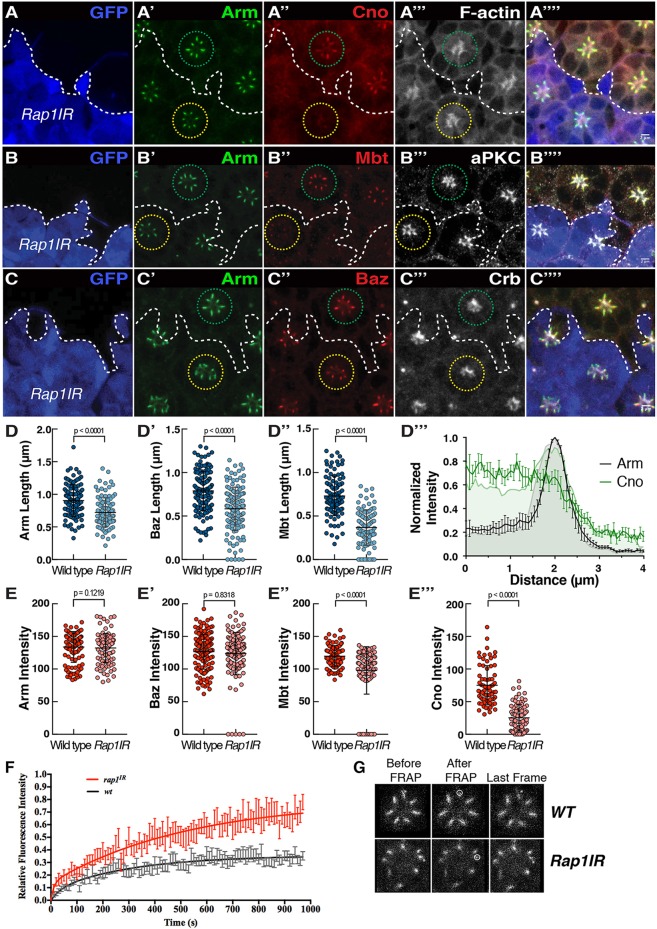
To assess the function of Rap1 during photoreceptor morphogenesis, we made use of available Rap1 loss-of-function alleles. We found that generating mutant clones using the strong allele Rap1CD3, or expressing high levels of a previously validated Rap1IR (Rap1 RNAi) construct (O'Keefe et al., 2009), leads to severe defects in recruiting the full complement of retinal accessory cells including the cone cells (Fig. S1A). Missing cone and pigment cells lead to retinal cell delamination, with many photoreceptors found below the floor of the retina (Fig. S1B–D), preventing us from assessing polarity and ZA morphogenesis. In order to bypass this strong phenotype, we limited the expression of Rap1IR. Decreasing the expression of Rap1 at pupal stages did not affect photoreceptor apical-basal polarity in the majority of ommatidia examined (Fig. 2A–C). However, quantification revealed that the length of the Arm, Mbt and Baz domains, measured along the apical-basal axis, were significantly reduced when compared to those in wild type (Fig. 2D–D″). In addition, while the levels of Arm and Baz were comparable to those measured in wild-type cells (Fig. 2E,E″), we found that Cno accumulation at the ZA was nearly abolished (Fig. 2A″,D‴,E‴) and Mbt levels were significantly decreased when compared to wild type (Fig. 2B″,E′). We also note that apical levels of F-actin (Fig. 2A‴), aPKC (Fig. 1B‴) and Crb (Fig. 2C‴), were not affected in Rap1IR photoreceptors when compared to those in wild type. These data indicate that Rap1 is required for the accumulation or retention of Cno and Mbt at the developing ZA and for regulating the length of the ZA along the apical-basal axis.
Fig. 2.
Rap1 regulates the accumulation of AJ material during ZA morphogenesis. (A–C) Rap1IR cells positively labeled for GFP (blue, the edge of which is denoted by the dashed line) and stained for Arm (A′,B′,C′), Cno (A″), Mbt (B″), Baz (C″), F-actin (A‴), aPKC (B‴) and Crb (C‴). Green circle, outline of wild-type ommatidia; yellow circle, outline of Rap1IR ommatidia. Scale bars: 2 μm. (D–D″) Quantification of Arm (D), Mbt (D′), Baz (D″) domain length at the ZA. Results are mean±s.d. (n≥105). (D‴) Normalized intensity profiles of Cno (green) and Arm (gray) in WT photoreceptors (shaded profiles) and Rap1IR photoreceptors. Results are mean±s.e.m. (n=7 cells from 3 retinas). (E–E‴) Quantification of Arm (E), Mbt (E′), Baz (E″) and Cno (E‴) mean pixel intensity at the ZA. Results are mean±s.d. (n≥105). (F) FRAP curve fit for E-Cad::GFP in wild-type (black) and Rap1IR (red) photoreceptors. For both genotypes, the basal end of the developing ZA (dashed circle) was photo-bleached (G). For wild-type ZA FRAP, n=14 and for Rap1IR, n=12. Error bars are s.e.m.
Rap1 promotes E-Cadherin stabilization at the ZA
We have previously shown that, in pupal photoreceptors, loss of mbt function leads to an increase in the mobile fraction of E-Cad at the ZA when compared to wild type as measured over 250 s (Walther et al., 2016). Our analysis of Rap1IR indicates that Mbt accumulation is strongly reduced at the ZA (Fig. 2B″,E′), which should therefore be accompanied by an increase in E-Cad mobility. To assess whether this is the case, we made use of fluorescence recovery after photobleaching (FRAP) and compared the recovery of an ubi-E-Cad::GFP transgene in wild-type and Rap1IR photoreceptors. In wild-type cells, over ∼250 s, we estimated that 25% of E-Cad::GFP was mobile (data not shown), which is consistent with previous estimations from our laboratory (Walther et al., 2016). However, while E-Cad::GFP shows a stronger recovery over this relatively short time scale in Rap1IR cells than in wild-type cells, the GFP signal failed to plateau (data not shown), preventing us from extrapolating the mobile fraction. We therefore performed FRAP over a longer time scale (1000 s). Over this long time scale, we found that ∼35% of E-Cad::GFP was mobile in the wild-type ZA, while ∼70% was mobile in Rap1IR photoreceptors (Fig. 2F,G). These data indicate that Rap1 promotes E-Cad stabilization at the ZA, and are compatible with Mbt mediating part of the Rap1 function during this process.
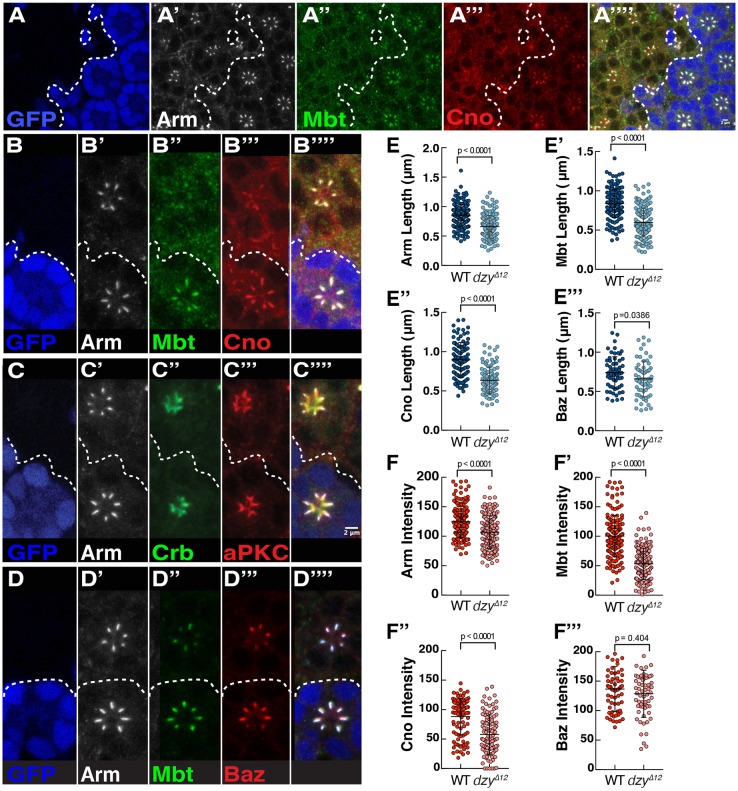
Dzy regulates ZA morphogenesis through Rap1
To examine the function of the Rap1-GEF dzy during photoreceptor morphogenesis, we made use of the strong dzyΔ12 allele. We found that reducing dzy expression leads to a phenotype similar to that seen in the hypomorphic Rap1IR photoreceptors (Fig. 3A), including a notable decrease the length of the Arm, (Fig. 3A′,B′,C′,D′,E), Mbt (Fig. 3A″,B″,D″,E′), Cno (Fig. 3A‴,B‴,E″) and Baz (Fig. 3D‴,E‴) domains along the apical basal axis of the cell. In addition, the levels of Arm, Mbt and Cno are significantly reduced at the ZA when compared to those in wild-type cells (Fig. 3F–F″), but those of Baz were similar to that measured in wild-type cells (Fig. 3F‴). Consistent with Dzy acting as a Rap1-GEF in photoreceptors, removing a copy of the dzy locus enhances the mild rough-eye phenotype obtained when reducing the expression of Rap1IR (Fig. S2). However, we note that the dzy loss-of-function phenotype is much milder than that of Rap1CD3 and that seen upon strong Rap1IR, in that no cells delaminate below the floor of the retina in dzy mutant clones. Other Rap1-GEFs must therefore be at play in the developing retina.
Fig. 3.
Dzy regulates Cno and Mbt accumulation at the ZA. (A–A‴) dzyΔ12 mutant clone labeled by the lack of nuclear GFP (blue, the contour of which is denoted by the dashed line), stained for Arm (A′), Mbt (A″) and Cno (A″″). (B–B‴) An ommatidium mutant for dzy (lacking GFP, blue, B), stained for Arm (B′), Mbt, (B″) and Cno (B‴). (C–C‴′) Ommatidium mutant for dzy (lacking GFP, blue, C), stained for Arm (C′), Crb (C″) and aPKC (C‴). (D–D‴′) Ommatidium mutant for dzy (lacking GFP, blue, D), stained for Arm (D′), Mbt (D″) and Baz (D‴). Scale bars: 2 µm. (E–E‴) Quantification of Arm (E), Mbt (E′), Cno (E″) and Baz (E‴) domain length at the ZA. (F–F‴) Quantification of Arm (F), Mbt (F′), Cno (F″) and Baz (F‴) mean pixel intensity at the ZA. All error bars are s.d. (n≥70 from 4 retinas).
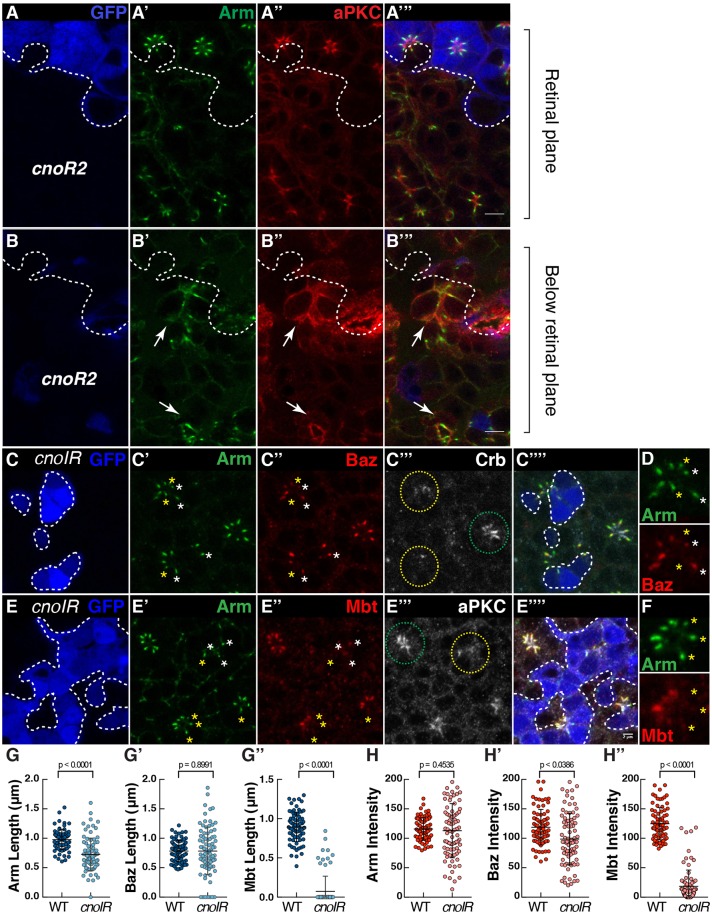
Cno couples Arm and Baz at the ZA and is required for the apical accumulation of aPKC and Crb
As well as regulating Mbt accumulation at the ZA, one likely mechanism whereby Rap1 might promote E-Cad stabilization is through the F-actin linker Cno. In the pupal photoreceptor, Cno is enriched at the ZA and found at low levels at the apical membrane, which is similar to the Arm expression pattern (Fig. 2A″,D‴). To assess Cno function, we made use of the strong cnoR2 allele. We found that cnoR2 mutant photoreceptors delaminate through the floor of the retina (Fig. 4A,B), a phenotype resembling that obtained when strongly reducing Rap1 expression. As with Rap1CD3, the polarity of the delaminated photoreceptors is strongly compromised in cnoR2 mutant cells, and the delamination phenotype is likely due to defects in assembling the full complement of interommatidial accessory cells. In order to circumvent the delamination phenotype, we made use of cnoIR (cno RNAi). Examining retinas mosaic for cnoIR revealed that Cno regulates the length of the ZA and is required for the accumulation of Arm (Fig. 4C′,4E′,G,H), Baz (Fig. 4C″,H′) and Mbt (Fig. 4E″,G″,H″) at the developing ZA. We also noted instances where Arm was present at the ZA but Mbt was absent (Fig. 4D,F). The similarity between the Rap1IR and cnoIR ZA phenotypes suggests that the function of Rap1 and Cno during ZA morphogenesis are linked. However, in the case of cnoIR, we also detect ZAs without Baz, a phenotype not detected in Rap1IR and indicative of a failure in retaining Baz at the developing ZA. Lack of Baz at the ZA is seen when overexpressing a version of Arm that cannot be phosphorylated by Mbt (ArmSAmbt) raising the possibility Mbt mediates Cno function (Walther et al., 2016). To test this possibility we expressed a phospho-mimetic version of Arm (ArmSEmbt) in cnoIR retinas. However, this did not ameliorate the cnoIR phenotype when considering ZA length along the apical-basal axis or Baz retention at the ZA (Fig. S3).
Fig. 4.
Cno regulates the coupling of Arm, Baz and Mbt at the developing ZA. (A–B) cnoR2 mutant cells (lacking GFP, blue, the contour of which is denoted by the dashed line, A,B) stained for Arm (A′,B′) and aPKC (A″,B″). White arrows indicate cnoR2 mutant photoreceptors that have delaminated from the retinal neuroepithelium. (C–F) cnoIR clones positively labeled by GFP (blue, C,E) and stained for Arm (C′,E′), Baz (C″), Crb (C‴), Mbt (E″) and aPKC (E‴). Green and yellow circles outline wild-type and cnoIR ommatidia, respectively. (D,F) A magnification of one mosaic ommatidium to highlight the absence of Baz (D) or Mbt (F) in some of the Arm domains. White stars label ZAs containing both Arm and Baz, while yellow stars indicate ZAs containing Arm but depleted for Baz (D) or containing Arm but depleted for Mbt (F). Scale bars: 2 μm. (G–G″) Quantification of Arm (G), Baz (G′) and Mbt (G″) domain length at the ZA. (H–H″) Quantification of Arm (H), Baz (H′) and Mbt (H″) mean pixel intensity at the ZA. All error bars are s.d. (n≥77 from 5 retinas).
In addition, we observed that unlike for Rap1IR, levels of Crb and aPKC were decreased in cnoIR mutant cells (Fig. 4C‴,E‴), indicating that Cno might regulate the accumulation of these factors during apical membrane morphogenesis. However, we note that our manipulation of Rap1 levels using Rap1IR does not lead to a complete loss of Cno at the ZA (Fig. 2A″,E‴), while Cno is virtually undetectable in our cnoIR experiments (Fig. S4). We therefore envisage that residual Cno in Rap1IR is sufficient to support the retention of Baz at the ZA, and the apical accumulation of Crb and aPKC.
Mbt is required for the accumulation of Cno and enrichment of Rap1 at the ZA
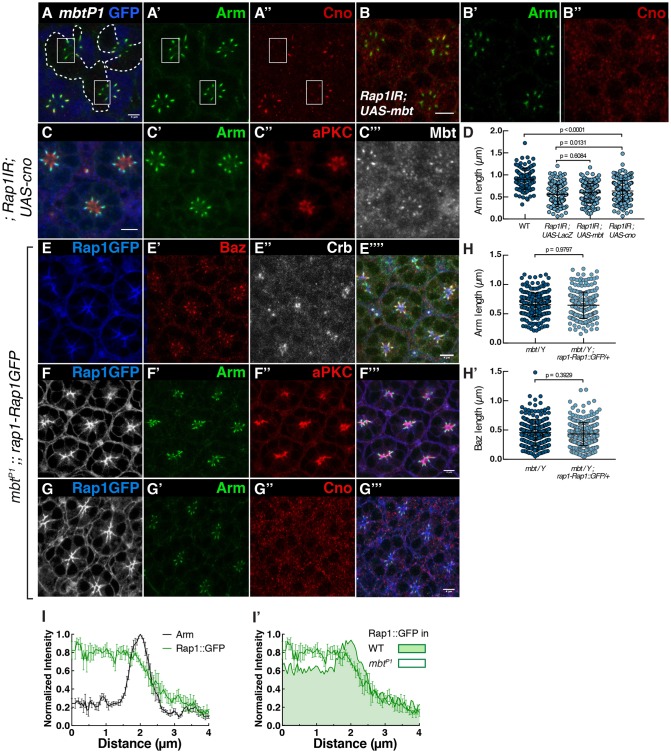
Our results indicate that Rap1 is required for the recruitment of Cno and Mbt at the photoreceptor ZA. Mbt is strongly decreased in cnoIR photoreceptors (Fig. 4E″,H″), which is compatible with Cno mediating Rap1 function in promoting Mbt accumulation at the ZA. Conversely, we find that Cno accumulation at the ZA depends on mbt (Fig. 5A–A″). Therefore, the localization of Cno and Mbt at the ZA are interlinked. To examine the functional relationship between Rap1, Cno and Mbt, and to test whether Cno and Mbt mediate Rap1 function during ZA morphogenesis, we asked whether expressing Mbt or Cno could ameliorate the Rap1IR ZA phenotype. We found even when expressing high levels of mbt (Fig. S5), the Rap1IR phenotype was not ameliorated (Fig. 5B,D). Similarly, expressing cno in Rap1IR cells did not restore Mbt accumulation to wild-type levels and did not ameliorate the length of the ZA (Fig. 5C,D).
Fig. 5.
Mbt is required for the accumulation of Cno and enrichment of Rap1 at the ZA. (A) mbtP1 mutant photoreceptors (lacking GFP, blue, the contour of which is denoted by the dashed line, A) and stained for Arm (A′) and Cno (A″). White boxes highlight ZAs within the mbtP1 mutant tissue. (B) Rap1IR photoreceptors expressing Mbt and stained for Arm (B′) and Cno (B″). (C) Rap1IR photoreceptors expressing Cno and labeled for Arm (C′), aPKC (C″) and Mbt (C‴). (D) Quantification of the Arm domain length at the ZA in wild-type photoreceptors, and for Rap1IR photoreceptors co-expressing UAS-LacZ, UAS-mbt or UAS-cno driven by NP-Gal42631. Results are mean±s.d. (n≥105 from 4 retinas). (E–G) mbtP1 mutant photoreceptors expressing rap1-Rap1::GFP (E,F,G) stained for Baz (E′), Arm (F′,G′), Crb (E″), aPKC (F″) and Cno (G″). (H) Quantification of the length of the Arm (H) and Baz (H′) domains at the ZA in the mbtP1 mutant and mbtP1 mutants expressing rap1-Rap1::GFP. Results are mean±s.d. (n≥187 from 4 retinas). (I) Intensity profiles measured for Rap1::GFP and Arm along the apical-basal axis in mbtP1 photoreceptors. (I′) Comparison of intensity profiles of Rap1::GFP measured in mbtP1 photoreceptors compared to that of wild-type photoreceptors (shaded). Results are mean±s.e.m. (n≥6 cells from 3 retinas). Scale bars: 2 μm.
Next, we assessed whether Rap1 could mediate some of the mbt function by expressing the rap1-Rap1::GFP transgene in mbtP1-null mutant cells. mbtP1 mutant cells are characterized by a decreased accumulation of Arm, Baz (Walther et al., 2016) (Fig. S6A,B) and Cno (Fig. 5A″) at their ZA. When expressing rap1-Rap1::GFP in mbtP1 mutant cells (Fig. 5E–G), we did not measure any significant recovery in the length of the Arm (Fig. 5F′,G′,H) or Baz domains (Fig. 5E′,H′), when compared to that in mbtP1 mutant cells, and Cno levels were not restored (Fig. 5G″). However, we noted that Rap1::GFP lacked the relative enrichment at the ZA normally detected in wild-type cells at this developmental stage (Figs 1F and 5I,I′).
One possibility is that Mbt might regulate the localization of Dzy, which in turn could shape that of Rap1. To test this possibility, we examined the localization of Dzy::GFP in mbtP1 mutant photoreceptors and found it was much reduced when compared to wild type (Fig. S6C,D). It is therefore possible that Mbt influences Rap1 distribution along the apical-basal axis through Dzy.
Dzy, Rap1, Cno synergize with Baz to promote AJ accumulation at the plasma membrane
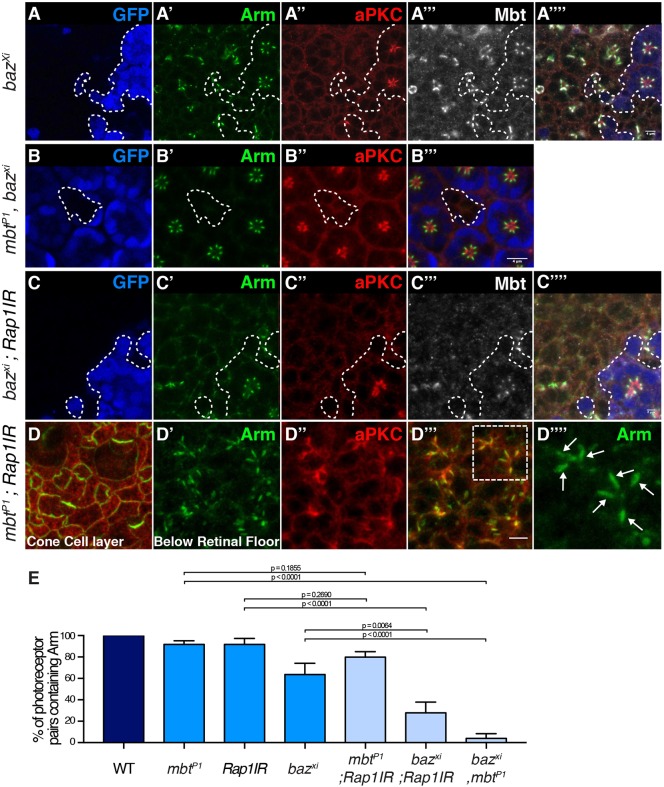
In order to test whether the Rap1–Cno pathway mediates some of the Baz function in promoting AJ material accumulation at the plasma membrane, we made use of genetics to probe the relationship between Rap1 and baz. First, we found that Rap1 and baz genetically interact during eye development, as decreasing the expression of baz by using RNAi (bazIR), enhances the Rap1IR rough eye phenotype (Fig. S2A,B,E,F). Second, to assay whether Rap1 function during AJ morphogenesis relates to that of Baz we generated photoreceptors deficient for both baz (using the bazxi106 allele) and Rap1 (using the NP-Gal42631-Rap1IR strain) (O'Keefe et al., 2009). As we have shown previously (Walther et al., 2016), AJ material, such as Arm, is detected at the plasma membrane in bazxi106 and mbtP1 single mutant cells (Fig. 6A′; Fig. S6B′). However, no AJ material is detected in bazxi106 mbtP1 double-mutant cells (Fig. 6B) indicating that baz and mbt function through parallel pathways to promote AJ material accumulation at the plasma membrane. We found that expressing Rap1IR in bazxi106 photoreceptors led to fewer cortical domains positive for Arm that are shared by flanking photoreceptors when compared to bazxi106 and Rap1IR single mutant cells (Figs 6C″, 5E). This was accompanied by a loss of Mbt (Fig. 6C‴), which is consistent with our observation that Rap1 is required for the accumulation of Mbt at the ZA (Fig. 2B″,E′). In contrast, AJ domains containing Arm are still present in double mbtP1 Rap1IR cells (Fig. 6D,E). Taken together, these data argue that while the respective functions of Rap1, Cno, Mbt and Baz converge during ZA morphogenesis, Rap1, Cno and Mbt function in parallel to Baz to promote AJ accumulation at the plasma membrane.
Fig. 6.
Rap1, Cno and Mbt synergize with Baz to promote AJ accumulation at the plasma membrane. (A–A‴′) bazxi106 mutant cells (lacking GFP, blue, the contour of which is denoted by the dashed line, A) and stained for Arm (A′), aPKC (A″) and Mbt (A‴). (B–B‴) mbtP1, bazxi106 double mutant cells (lacking GFP, blue, B) and stained for Arm (B′) and aPKC (B″). (C–C‴′) bazxi106, Rap1IR double mutant cells (lacking GFP, blue, C) and stained for Arm (C′), aPKC (C″) and Mbt (C‴). (D) Confocal section of the cone and pigment cells in an mbtP1; Rap1IR retina stained for Arm (green) and aPKC (red). (D′–D‴′) View of the delaminated photoreceptor proximal to D. (D′) Arm, (D″) aPKC, (D‴), merge (D″,D‴); a white-dashed rectangle highlights two ommatidia that are magnified in D‴′. The white arrows point to ZA domains between flanking photoreceptors. (E) Quantification of the percentage of pairs of photoreceptors sharing a lateral Arm domain in wild-type, mbtP1, Rap1IR, baz xi106, double mbtP1; Rap1IR, double baz xi106; Rap1IR and double baz xi106, mbtP1 cells. Results are mean±s.e.m. (n≥180 from 5 retinas). Scale bars: 4 μm.
DISCUSSION
In the pupal photoreceptor, ZA morphogenesis is orchestrated by a conserved protein network that includes Cdc42, Par6, aPKC, Baz, Crb and its binding partner Sdt, and Par1 (Berger et al., 2007; Hong et al., 2003; Izaddoost et al., 2002; Nam and Choi, 2003; Pellikka et al., 2002; Richard et al., 2006; Walther et al., 2016; Walther and Pichaud, 2010). In turn, AJ material is an essential part of the regulatory network that orchestrates polarity (Walther et al., 2016). We and others have previously shown that Mbt regulates pupal photoreceptor development by promoting ZA morphogenesis (Menzel et al., 2007; Walther et al., 2016). During this process Mbt contributes in preventing Baz from spreading to the lateral membrane, a regulation that we have found depends in part on the phosphorylation of Arm by Mbt at S561 and S688. We proposed that Mbt regulates photoreceptor polarity by promoting the retention of Baz at the developing ZA. Failure in ZA retention leads to Baz spreading to the lateral membrane where it is eliminated through Par1-mediated displacement. In these cells, failure to retain AJ material, including Baz, at the ZA leads to its shortening along the apical basal axis and can impact on the polarization program of the photoreceptor (Walther et al., 2016).
In the present study, we show that Mbt function is linked to that of Dzy, Rap1 and Cno. First, Cno and Mbt accumulation at the ZA is interdependent, reflecting a tight coupling between the Rap1 and Cno pathway and Mbt. Second, we find that Cno promotes Baz retention at the ZA, as cnoIR leads to shorter ZAs that can be depleted of Arm and Baz. This phenotype resembles that of mbt mutant cells and is also seen when overexpressing a version of Arm that cannot be phosphorylated by Mbt (Walther et al., 2016). These observations prompted us to test the hypothesis that Rap1, Cno and Mbt might function as part of a linear pathway promoting Baz retention at the ZA. In this pathway, we reasoned that Mbt could mediate Rap1 function through Arm phosphorylation. In testing this hypothesis, we found that this is not the case. Instead, the observation that expressing a version of Arm that mimics its constitutive phosphorylation by Mbt does not ameliorate the cnoIR phenotype suggests that Rap1, Cno, and Mbt converge in promoting Baz retention at the ZA, and cannot compensate for each other during this process. This conclusion is well supported by the finding that overexpressing cno in mbt mutant cells does not lead to an amelioration of the mbt phenotype. Third, we found that Mbt influences the distribution of Rap1 along the apical-basal axis of the cell in that Rap1::GFP no longer accumulates preferentially at the ZA. This correlates with a loss of Dzy::GFP at the plasma membrane, raising the possibility that Mbt might regulate Rap1 through Dzy. However, the dzy phenotype is milder than that seen with Rap1 or cno, in that loss of dzy does not lead to cell delamination from the retina. This suggests that, as recently reported in the cellularizing embryo (Bonello et al., 2018; Schmidt et al., 2018), other GEFs regulate Rap1 during epithelial morphogenesis.
An interesting aspect of the cnoIR phenotype is the defects in apical accumulation of aPKC and Crb. These defects are not observed in the dzy mutant or Rap1IR cells, indicating that Cno might function independently of Rap1 during this process. However, we note that while we cannot detect Cno at the ZA of cnoIR cells, we still detect it in Rap1IR cells. We therefore hypothesize that residual Cno in Rap1IR cells supports optimum aPKC and Crb accumulation at the apical membrane. In our model, Dzy, Rap1 and Cno function as part of the same pathway, which includes a function in promoting optimum apical accumulation of Crb and aPKC. Baz is required for Par complex assembly and associated aPKC and Crb recruitment at photoreceptor apical membrane (Walther et al., 2016; Walther and Pichaud, 2010). We hypothesize that the defects in Crb and aPKC that we detect in cnoIR cells are linked to the failure in retaining Baz at the ZA, which leads to its elimination from the lateral membrane by Par1. More work will be required to understand how exactly AJ material and ZA retention of Baz influences apical membrane specification.
Rap1 and cno have been shown to regulate apical-basal polarity in the cellularizing embryo. In this model system, Rap1 and Cno regulate the apical localization of Baz and Arm, which precedes the apical recruitment of Crb. In turn, Baz influences the localization of Cno (Choi et al., 2013). Our work indicates that similar complex regulations are at play in the pupal photoreceptor. However, unlike in the early embryo, AJ material (Arm) is absolutely required for Baz (and Par6–aPKC) accumulation or retention at the cell cortex in the developing pupal photoreceptor (Walther et al., 2016). We therefore favor a model whereby Mbt, Rap1 and Cno influence ZA morphogenesis primarily through regulating the interface between E-Cad or Arm, Baz and the F-actin cytoskeleton. In this model, Mbt regulates this interface both through Arm phosphorylation and cofilin-dependent regulation of F-actin (Walther et al., 2016), and Cno contributes to this process, at least in part, through its ability to bind to F-actin.
To probe Rap1 and Cno function during photoreceptor ZA morphogenesis, we assessed the effect of decreasing Rap1 expression on E-Cad stability. Consistent with the notion that the function of mbt and Rap1 are linked during ZA morphogenesis, we find that, as it is the case for Mbt (Walther et al., 2016), Rap1 is required to stabilize E-Cad::GFP at the photoreceptor ZA. However, the mobile fraction estimated for E-Cad is much higher in Rap1IR cells than in mbtP1 null cells – evaluated at ∼70% for Rap1IR and 45% for mbtP1 (Walther et al., 2016). Together with our finding that Mbt accumulation at the ZA is decreased in Rap1IR cells, our FRAP data are therefore compatible with Mbt mediating part of the function of Rap1 in promoting E-Cad stability. However, the much larger mobile fraction we estimate in the Rap1IR genotype when compared to mbtP1 photoreceptors indicates that Rap1 must also regulate E-Cad stability independently of Mbt. The longer time scale for E-Cad::GFP to recover in Rap1IR cells when compared to mbtP1 mutant cells is compatible with Rap1 functioning, in part, through promoting E-Cad delivery.
MATERIALS AND METHODS
Fly strains
The following fly strains were used: rap1-Rap1::GFP and NP-Gal42631, Rap1IR (O'Keefe et al., 2009; BL #29434); bazIR (BL #39072), cnoIR (BL #33367) and UAS-lacZ (Bloomington Stock Center BL #3956); dzyΔ12, FRT40A (Huelsmann et al., 2006); dzy-Dzy::GFP (Boettner and Van Aelst, 2007); ubi-Cad::GFP (Oda and Tsukita, 2001); mbtP1 and UAS-Mbt (Schneeberger and Raabe, 2003); mbtP1, FRT19A;, mbtP1, bazxi106, FRT9.2;, ;;UAS-Arm, ;;UAS-ArmSAmbt and ;;UAS-ArmSEmbt (Walther et al., 2016); w,bazxi106, FRT9.2 (Nusslein-Volhard et al., 1987); FRT82B, cnoR2 (Sawyer et al., 2009); UAS-Cno (Matsuo et al., 1997); GMR-Gal4 (Freeman, 1996); and NP-Gal42631 (Drosophila Genomics Resource Center #104266) (Hayashi et al., 2002).
Analysis of gene function
Clonal analysis of mutant alleles in the retina was performed using the standard FLP-FRT technique (Xu and Rubin, 1993) with appropriate FRT, ubi-GFP chromosomes used to generate negatively marked mutant tissue in combination with eyFLP (Newsome et al., 2000). Retina expressing RNAi in clones were generated by using the coinFLP system (Bosch et al., 2015). Clones of retinal tissue expressing RNAi against Rap1 were generated both with and without UAS-dicer, while clones of retinal tissue expressing RNAi against cno were generated without UAS-dicer only. In order to mitigate the strong Rap1 loss-of-function phenotype, Rap1IR animal were raised at 20°C and shifted to the appropriate temperature (25 or 29°C) at puparium formation. UAS transgenes were co-expressed with UAS-Rap1IR or UAS-cnoIR under the control of the NP-Gal42631 or GMR-Gal4 drivers, respectively.
Antibodies and immunological methods
Whole mount retinas at 40% after puparium formation (APF) were prepared as previously described (Walther and Pichaud, 2006). The following antibodies were used: rabbit anti-PKCζ (1:600, SAB4502380, Sigma), mouse anti-Arm (1:200, N27-A1, Developmental Studies Hybridoma Bank), rat anti-Baz, (1:1000, a gift from Andreas Wodarz, University of Cologne, Germany), rabbit anti-Cno, (1:200, a gift from Linda Van Aelst, Cold Spring Harbor Laboratory, New York, USA Boettner et al., 2003), rabbit anti-Baz (1:2000), rat anti-Crb (1:200), guinea pig anti-Mbt (1:200) (Walther et al., 2016), with the appropriate combination of mouse, guinea pig, rabbit and rat secondary antibodies conjugated to Dy405, Alexa Fluor 488, Cy3 or Cy5 as appropriate at 1:200 each (Jackson ImmunoResearch) or TRITC-conjugated Phalloidin (P1951, Sigma) at 2 μg/ml. Retinas were mounted in VectaShield™ with or without DAPI as appropriate, and imaging was performed by using a Leica SP5 confocal microscope. Images were edited with ImageJ and Adobe Photoshop 7.0.
Western blot analysis
Pupal retina were dissected at 40% APF. For each genotype, ten retina were snap-frozen in PBS and SDS sample buffer was added to a final volume of 20 µl. Samples were analyzed by western blotting. Guinea pig anti-Mbt (Walther et al., 2016) and mouse anti-α-Tubulin antibodies (AA4.3, Developmental Studies Hybridoma Bank) (Walsh, 1984) were used for protein detection at concentrations of 1:1000 and 1:200, respectively.
Data analysis
For length and pixel intensity measurements, a threshold was applied to define the ZA domain and a line was drawn along the apical-basal axis of the cell, running in the middle of the ZA to measure the length of the Arm, Baz and Mbt domains. Mean pixel intensity was measured by using the wand (tracing) tool in Fiji (Schindelin et al., 2012). In all cases, at least four independent mosaic retinas were used for each genotype. The intensity profiles of Rap1::GFP, dzy::GFP and Cno relative to Arm were measured in Fiji. A 1 µm line was drawn along the apical membrane and continued for 4 µm along the stalk membrane and ZA. For each profile, pixel intensities were subjected to unity-based normalization and adjusted such that the normalized maximum value of Arm was placed at 2 µm. Statistical analysis was performed in Prism 7.0. Data sets were tested for normality (D'Agostino and Pearson normality test) and P-values were calculated using a Student's t-test or the Mann–Whitney test as appropriate.
Fluorescence recovery after photobleaching
FRAP analysis was performed as previously described (Walther et al., 2016). At 40% APF, the pupal cuticle was removed to expose the retina and the animal was mounted in Voltalef oil. Live imaging was performed on a Leica SP5 confocal microscope with a 63×1.4 NA oil immersion objective at the following settings: pixel resolution 512×512, speed 400 Hz, 10% 488 nm laser power at 20% argon laser intensity and 5× zoom. FRAP analysis of ubi-ECad::GFP was performed by marking the basal tip of the AJ with a 5 pixel diameter circlular region of interest (ROI) followed by photo-bleaching with a single pulse using 90% 488 nm laser power at 20% argon laser intensity. AJ recovery was recorded every 1.293 s with the previously mentioned settings for ∼1000 s. FRAP data were drift corrected in Fiji (Schindelin et al., 2012) using the StackReg plugin. Three different z-axis profiles were analyzed: (1) from the photo-bleached area; (2) from an equivalent area of a neighboring non-photo-bleached AJ; and (3) from an equivalent area of a background region. The data were normalized with easyFRAP. ECad::GFP data were fitted to a two-phase association curve in GraphPad Prism. The P-values were calculated with an unpaired two-tailed Student's t-test with Welch's correction.
Scanning electron microscopy
Flies were fixed in 2% paraformaldehyde, 2% glutaraldehyde and 0.1 M cacodylate for 2 h and then dehydrated in ethanol, as previously described (Richardson and Pichaud, 2010). The samples were then critical-point dried and mounted on aluminum stubs before gold coating. Imaging was carried out on a JEOL Variable Pressure scanning electron microscope (SEM).
Supplementary Material
Acknowledgements
The authors wish to thank all members of the Pichaud laboratory for helpful discussion, in particular Francisca Nunes de Almeida for help with the FRAP assay. We are grateful to Van Aelst and Andreas Wodarz for sharing reagents. The N2 A71 anti-Armadillo and AA4.3 anti-α-tubulin monoclonal antibodies, developed by Eric Weischaus and Charles Walsh, respectively, were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and the KYOTO Stock Center (DGRC) at Kyoto Institute of Technology were used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.F.W., F.P.; Methodology: R.F.W., F.P.; Validation: M.B., C.R.; Formal analysis: R.F.W.; Investigation: R.F.W., M.B., N.P.; Data curation: R.F.W., M.B.; Writing - original draft: R.F.W., F.P.; Writing - review & editing: R.F.W., F.P.; Visualization: R.F.W.; Supervision: F.P.; Project administration: F.P.; Funding acquisition: F.P.
Funding
This work, including support to R.F.W., M.B. and N.P. was funded by a Medical Research Council grant to F.P. (award code MC_UU_12018/3). Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.207779.supplemental
References
- Aaku-Saraste E., Hellwig A. and Huttner W. B. (1996). Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure--remodeling of the neuroepithelium prior to neurogenesis. Dev. Biol. 180, 664-679. 10.1006/dbio.1996.0336 [DOI] [PubMed] [Google Scholar]
- Afonso C. and Henrique D. (2006). PAR3 acts as a molecular organizer to define the apical domain of chick neuroepithelial cells. J. Cell Sci. 119, 4293-4304. 10.1242/jcs.03170 [DOI] [PubMed] [Google Scholar]
- Berger S., Bulgakova N. A., Grawe F., Johnson K. and Knust E. (2007). Unraveling the genetic complexity of Drosophila stardust during photoreceptor morphogenesis and prevention of light-induced degeneration. Genetics 176, 2189-2200. 10.1534/genetics.107.071449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B. and Van Aelst L. (2007). The Rap GTPase activator Drosophila PDZ-GEF regulates cell shape in epithelial migration and morphogenesis. Mol. Cell. Biol. 27, 7966-7980. 10.1128/MCB.01275-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B., Harjes P., Ishimaru S., Heke M., Fan H. Q., Qin Y., Van Aelst L. and Gaul U. (2003). The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics 165, 159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello T. T., Perez-Vale K. Z., Sumigray K. D. and Peifer M. (2018). Rap1 acts via multiple mechanisms to position Canoe and adherens junctions and mediate apical-basal polarity establishment. Development 145, dev157941 10.1242/dev.157941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., de Rooij J. and Reedquist K. A. (2001). Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2, 369-377. 10.1038/35073073 [DOI] [PubMed] [Google Scholar]
- Bosch J. A., Tran N. H. and Hariharan I. K. (2015). CoinFLP: a system for efficient mosaic screening and for visualizing clonal boundaries in Drosophila. Development 142, 597-606. 10.1242/dev.114603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W., Harris N. J., Sumigray K. D. and Peifer M. (2013). Rap1 and Canoe/afadin are essential for establishment of apical-basal polarity in the Drosophila embryo. Mol. Biol. Cell 24, 945-963. 10.1091/mbc.E12-10-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J., Boenink N. M., van Triest M., Cool R. H., Wittinghofer A. and Bos J. L. (1999). PDZ-GEF1, a guanine nucleotide exchange factor specific for Rap1 and Rap2. J. Biol. Chem. 274, 38125-38130. 10.1074/jbc.274.53.38125 [DOI] [PubMed] [Google Scholar]
- Freeman M. (1996). Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87, 651-660. 10.1016/S0092-8674(00)81385-9 [DOI] [PubMed] [Google Scholar]
- Hayashi S., Ito K., Sado Y., Taniguchi M., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T. et al. (2002). GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34, 58-61. 10.1002/gene.10137 [DOI] [PubMed] [Google Scholar]
- Hogan C., Serpente N., Cogram P., Hosking C. R., Bialucha C. U., Feller S. M., Braga V. M., Birchmeier W. and Fujita Y. (2004). Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 24, 6690-6700. 10.1128/MCB.24.15.6690-6700.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Ackerman L., Jan L. Y. and Jan Y. N. (2003). Distinct roles of Bazooka and Stardust in the specification of Drosophila photoreceptor membrane architecture. Proc. Natl. Acad. Sci. USA 100, 12712-12717. 10.1073/pnas.2135347100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann S., Hepper C., Marchese D., Knoll C. and Reuter R. (2006). The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development 133, 2915-2924. 10.1242/dev.02449 [DOI] [PubMed] [Google Scholar]
- Izaddoost S., Nam S.-C., Bhat M. A., Bellen H. J. and Choi K.-W. (2002). Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature 416, 178-183. 10.1038/nature720 [DOI] [PubMed] [Google Scholar]
- Jin D., Durgan J. and Hall A. (2015). Functional cross-talk between Cdc42 and two downstream targets, Par6B and PAK4. Biochem. J. 467, 293-302. 10.1042/BJ20141352 [DOI] [PubMed] [Google Scholar]
- KawAJiri A., Itoh N., Fukata M., Nakagawa M., Yamaga M., Iwamatsu A. and Kaibuchi K. (2000). Identification of a novel beta-catenin-interacting protein. Biochem. Biophys. Res. Commun. 273, 712-717. 10.1006/bbrc.2000.3002 [DOI] [PubMed] [Google Scholar]
- Knox A. L. and Brown N. H. (2002). Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295, 1285-1288. 10.1126/science.1067549 [DOI] [PubMed] [Google Scholar]
- Krahn M. P., Bückers J., Kastrup L. and Wodarz A. (2010). Formation of a Bazooka-Stardust complex is essential for plasma membrane polarity in epithelia. J. Cell Biol. 190, 751-760. 10.1083/jcb.201006029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law S. H. W. and Sargent T. D. (2014). The serine-threonine protein kinase PAK4 is dispensable in zebrafish: identification of a morpholino-generated pseudophenotype. PLoS One 9, e100268 10.1371/journal.pone.0100268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K., Rikitake Y., Shimono Y. and Takai Y. (2013). Afadin/AF-6 and canoe: roles in cell adhesion and beyond. Prog. Mol. Biol. Transl. Sci. 116, 433-454. 10.1016/B978-0-12-394311-8.00019-4 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Takahashi K., Kondo S., Kaibuchi K. and Yamamoto D. (1997). Regulation of cone cell formation by Canoe and Ras in the developing Drosophila eye. Development 124, 2671-2680. [DOI] [PubMed] [Google Scholar]
- McGill M. A., McKinley R. F. A. and Harris T. J. C. (2009). Independent cadherin-catenin and Bazooka clusters interact to assemble adherens junctions. J. Cell Biol. 185, 787-796. 10.1083/jcb.200812146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel N., Schneeberger D. and Raabe T. (2007). The Drosophila p21 activated kinase Mbt regulates the actin cytoskeleton and adherens junctions to control photoreceptor cell morphogenesis. Mech. Dev. 124, 78-90. 10.1016/j.mod.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Menzel N., Melzer J., Waschke J., Lenz C., Wecklein H., Lochnit G., Drenckhahn D. and Raabe T. (2008). The Drosophila p21-activated kinase Mbt modulates DE-cadherin-mediated cell adhesion by phosphorylation of Armadillo. Biochem. J. 416, 231-241. 10.1042/BJ20080465 [DOI] [PubMed] [Google Scholar]
- Morais-de-Sá E., Mirouse V. and St Johnston D. (2010). aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell 141, 509-523. 10.1016/j.cell.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai-Tamai Y., Mizuno K., Hirose T., Suzuki A. and Ohno S. (2002). Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells 7, 1161-1171. 10.1046/j.1365-2443.2002.00590.x [DOI] [PubMed] [Google Scholar]
- Nam S.-C. and Choi K. W. (2003). Interaction of Par-6 and Crumbs complexes is essential for photoreceptor morphogenesis in Drosophila. Development 130, 4363-4372. 10.1242/dev.00648 [DOI] [PubMed] [Google Scholar]
- Newsome T. P., Asling B. and Dickson B. J. (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851-860. [DOI] [PubMed] [Google Scholar]
- Niessen C. M. and Gottardi C. J. (2008). Molecular components of the adherens junction. Biochim. Biophys. Acta 1778, 562-571. 10.1016/j.bbamem.2007.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Frohnhofer H. G. and Lehmann R. (1987). Determination of anteroposterior polarity in Drosophila. Science 238, 1675-1681. 10.1126/science.3686007 [DOI] [PubMed] [Google Scholar]
- O'Keefe D. D., Gonzalez-Nino E., Burnett M., Dylla L., Lambeth S. M., Licon E., Amesoli C., Edgar B. A. and Curtiss J. (2009). Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila. Dev. Biol. 333, 143-160. 10.1016/j.ydbio.2009.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H. and Tsukita S. (2001). Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J. Cell Sci. 114, 493-501. [DOI] [PubMed] [Google Scholar]
- Pannekoek W.-J., Kooistra M. R. H., Zwartkruis F. J. and Bos J. L. (2009). Cell-cell junction formation: the role of Rap1 and Rap1 guanine nucleotide exchange factors. Biochim. Biophys. Acta 1788, 790-796. 10.1016/j.bbamem.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Pellikka M., Tanentzapf G., Pinto M., Smith C., McGlade C. J., Ready D. F. and Tepass U. (2002). Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 416, 143-149. 10.1038/nature721 [DOI] [PubMed] [Google Scholar]
- Ready D. F. (2002). Drosophila compound eye morphogenesis: Blind mechanical engineers? In Results and Problems in Cell Differentiation, vol. Drosphila Eye Development (ed. Moses K.), pp. 191-204. Berlin Heildelderg New York: Springer-Verlag. [DOI] [PubMed] [Google Scholar]
- Richard M., Grawe F. and Knust E. (2006). DPATJ plays a role in retinal morphogenesis and protects against light-dependent degeneration of photoreceptor cells in the Drosophila eye. Dev. Dyn. 235, 895-907. 10.1002/dvdy.20595 [DOI] [PubMed] [Google Scholar]
- Richardson E. C. N. and Pichaud F. (2010). Crumbs is required to achieve proper organ size control during Drosophila head development. Development 137, 641-650. 10.1242/dev.041913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J. K., Harris N. J., Slep K. C., Gaul U. and Peifer M. (2009). The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186, 57-73. 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Lv Z. and Grosshans J. (2018). ELMO and Sponge specify subapical restriction of Canoe and formation of the subapical domain in early Drosophila embryos. Development 145, dev157909 10.1242/dev.157909 [DOI] [PubMed] [Google Scholar]
- Schneeberger D. and Raabe T. (2003). Mbt, a Drosophila PAK protein, combines with Cdc42 to regulate photoreceptor cell morphogenesis. Development 130, 427-437. 10.1242/dev.00248 [DOI] [PubMed] [Google Scholar]
- Spahn P., Ott A. and Reuter R. (2012). The PDZ-GEF protein Dizzy regulates the establishment of adherens junctions required for ventral furrow formation in Drosophila. J. Cell Sci. 125, 3801-3812. 10.1242/jcs.101196 [DOI] [PubMed] [Google Scholar]
- Tepass U. (2012). The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28, 655-685. 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- Wallace S. W., Durgan J., Jin D. and Hall A. (2010). Cdc42 regulates apical junction formation in human bronchial epithelial cells through PAK4 and Par6B. Mol. Biol. Cell 21, 2996-3006. 10.1091/mbc.E10-05-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. (1984). Synthesis and assembly of the cytoskeleton of Naegleria gruberi flagellates. J. Cell Biol. 98, 449-456. 10.1083/jcb.98.2.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther R. F. and Pichaud F. (2006). Immunofluorescent staining and imaging of the pupal and adult Drosophila visual system. Nat. Protoc. 1, 2635-2642. 10.1038/nprot.2006.379 [DOI] [PubMed] [Google Scholar]
- Walther R. F. and Pichaud F. (2010). Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 20, 1065-1074. 10.1016/j.cub.2010.04.049 [DOI] [PubMed] [Google Scholar]
- Walther R. F., Nunes de Almeida F., Vlassaks E., Burden J. J. and Pichaud F. (2016). Pak4 is required during epithelial polarity remodeling through regulating AJ stability and bazooka retention at the ZA. Cell Rep. 15, 45-53. 10.1016/j.celrep.2016.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-C., Khan Z. and Wieschaus E. F. (2013). Distinct Rap1 activity states control the extent of epithelial invagination via alpha-catenin. Dev. Cell 25, 299-309. 10.1016/j.devcel.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S.-Y., Escudero L. M., Yu F., Chang L. H., Chen L. Y., Ho Y. H., Lin C. M., Chou C. S., Chia W., Modolell J. et al. (2005). Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev. Cell 8, 493-504. 10.1016/j.devcel.2005.03.015 [DOI] [PubMed] [Google Scholar]
- Xu T. and Rubin G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.