ABSTRACT
Hippo signaling is regulated by biochemical and biomechanical cues that influence the cytoskeleton, but the mechanisms that mediate this have remained unclear. We show that all three mammalian Ajuba family proteins – AJUBA, LIMD1 and WTIP – exhibit tension-dependent localization to adherens junctions, and that both LATS family proteins, LATS1 and LATS2, exhibit an overlapping tension-dependent junctional localization. This localization of Ajuba and LATS family proteins is also influenced by cell density, and by Rho activation. We establish that junctional localization of LATS kinases requires LIMD1, and that LIMD1 is also specifically required for the regulation of LATS kinases and YAP1 by Rho. Our results identify a biomechanical pathway that contributes to regulation of mammalian Hippo signaling, establish that this occurs through tension-dependent LIMD1-mediated recruitment and inhibition of LATS kinases in junctional complexes, and identify roles for this pathway in both Rho-mediated and density-dependent regulation of Hippo signaling.
KEY WORDS: LIMD1, YAP, Junction, Hippo, Cytoskeleton, Tension
Highlighted Article: The mechanism through which YAP activity is regulated by tension at adherens junctions is demonstrated, which helps explain why cell proliferation is inhibited by high cell density and why Rho activity suppresses Hippo signaling.
INTRODUCTION
The Hippo signaling network integrates a wide range of biochemical and biomechanical cues to modulate organ growth and cell fate (Irvine and Harvey, 2015; Meng et al., 2016). This control of growth and cell fate is effected through regulation of two homologous transcriptional co-activator proteins, YAP (also known as YAP1) and TAZ. Many of the upstream cues that influence YAP and TAZ activity converge on cell junctions and the actin cytoskeleton (Dupont, 2016; Sun and Irvine, 2016). For example, YAP and TAZ can be affected by cell–cell contacts (Kim et al., 2011), cell–extracellular matrix contacts (Kim and Gumbiner, 2015; Serrano et al., 2013; Zhao et al., 2012), cell density (Zhao et al., 2007), F-actin levels (Aragona et al., 2013; Fernández et al., 2011; Sansores-Garcia et al., 2011), cytoskeletal tension (Dupont et al., 2011; Rauskolb et al., 2014), cell shape (Dupont et al., 2011; Wada et al., 2011), substrate stiffness (Dupont et al., 2011), cell stretching (Aragona et al., 2013; Benham-Pyle et al., 2015; Codelia et al., 2014) and Rho activity (Dupont et al., 2011; Yu et al., 2012). However, we have only a very limited understanding of the molecular processes that mediate this responsiveness to the cytoskeleton and mechanical stresses.
A major, though not exclusive, mechanism of YAP and TAZ regulation is through phosphorylation by the LATS kinases, LATS1 and LATS2 (Meng et al., 2016; Sun and Irvine, 2016). Phosphorylation of YAP and TAZ by LATS kinases inhibits their activity by promoting their degradation, and their exclusion from the nucleus. LATS kinases are activated through phosphorylation by upstream kinases, including Mst (also known as Stk) and Map4k family kinases. Hippo signaling was first identified in Drosophila based on the overgrowth phenotypes associated with mutations in warts and upstream pathway components that promote activation of Warts, which is the Drosophila orthologue of the LATS kinases (Reddy and Irvine, 2008). This overgrowth occurs in response to abnormally elevated levels of Yorkie activity (the orthologue of YAP and TAZ) (Huang et al., 2005). Similarly, elevated YAP and TAZ activity is observed in many cancers (Harvey et al., 2013).
Studies in Drosophila have identified a mechanism for biomechanical regulation of Hippo signaling involving tension-dependent recruitment of Warts into a complex at the adherens junctions with the Drosophila Ajuba family protein Jub (Rauskolb et al., 2014). Jub, which contributes to regulation of Hippo signaling during development and regeneration (Das Thakur et al., 2010; Meserve and Duronio, 2015; Sun and Irvine, 2011), is recruited to adherens junctions in a tension-dependent manner (Rauskolb et al., 2014). Jub is an inhibitor of Warts (Das Thakur et al., 2010), and recruitment of Warts to Jub complexes also prevents it from localizing to other junctional and apical complexes where Warts activation occurs (Su et al., 2017; Sun et al., 2015). Whether a comparable mechanism exists in mammalian cells has been disputed (Jagannathan et al., 2016), and studies of biomechanical regulation of Hippo signaling have focused on other potential cues, including tension at focal adhesions, actin levels and organization and mechanically gated channels (Dupont, 2016).
Mammals have three Ajuba family proteins: AJUBA, WTIP and LIMD1. They have been ascribed a variety of cellular localizations, including in the cytoplasm, nucleus, centrosomes, adherens junctions, focal adhesions and P bodies (Goyal et al., 1999; Hirota et al., 2003; Kanungo et al., 2000; Kim et al., 2012; Marie et al., 2003; Pratt et al., 2005; Spendlove et al., 2008; Srichai et al., 2004). They have also been ascribed a wide variety of biological functions, but one key function identified for Ajuba family proteins is physical interaction with, and inhibition of, LATS kinases (Abe et al., 2006; Das Thakur et al., 2010). The association of Ajuba family proteins with LATS kinases can be enhanced by JNK or ERK phosphorylation (Reddy and Irvine, 2013; Sun and Irvine, 2013). However, aside from one report implicating LIMD1 in a JNK-dependent activation of YAP after cyclic stretch (Codelia et al., 2014), it has not been shown that mammalian Ajuba family proteins contribute to biomechanical regulation of Hippo signaling. Indeed, a recent report has suggested that Ajuba proteins do not participate in biomechanical regulation of Hippo signaling, and that they interact with LATS kinases exclusively in the cytoplasm (Jagannathan et al., 2016). We also note that while association of LIMD1 with focal adhesions is reduced by blebbistatin treatment (Schiller et al., 2011), indicating that it depends upon myosin activity, whether LIMD1 localization to adherens junctions is also tension dependent has not been investigated, nor has any contribution of cytoskeletal tension to AJUBA, WTIP, LATS1 or LATS2 localization been reported.
Here, we describe investigations of the biomechanical regulation of Ajuba family proteins and their contribution to Hippo signaling. We find that AJUBA, WTIP and LIMD1 each exhibit a strong tension-dependent association to adherens junctions. We show that both LATS1 and LATS2 (henceforth collectively referred to as LATS) also exhibit a tension-dependent localization to adherens junctions. In MCF10A cells, one of the three Ajuba family proteins, LIMD1, is specifically required for the junctional localization of LATS. By undertaking pharmacological inhibition of cytoskeletal tension, cell density and Rho activation as models of cytoskeletal regulation of Hippo signaling, we show that LIMD1 is specifically required for cytoskeletal regulation of YAP, and that this regulation correlates with recruitment of LATS into complexes at adherens junctions. Our results indicate that LIMD1 is essential for certain modes of biomechanical regulation of Hippo signaling, and functions by recruiting LATS into an inhibitory complex at adherens junctions.
RESULTS
Tension-dependent localization of Ajuba family proteins in mammalian epithelial cells
To investigate regulation of Ajuba family protein localization in epithelial cells, we created constructs expressing GFP-tagged human cDNAs in a lentiviral vector with a doxycycline (Dox)-inducible promoter (CMV-TetON). We then used these to express Ajuba family proteins in the canine kidney epithelial cell line MDCKIIG, and in the human breast epithelial cell line MCF10A. Cells with relatively homogeneous expression of each construct were selected after viral transduction. In this system, proteins can be expressed at low levels by using low doses of Dox, thereby minimizing or eliminating mis-localization due to overexpression. Western blotting of cell lysates confirmed inducible expression for each protein, and in cases where we had antibodies that recognize endogenously expressed proteins on western blots (AJUBA and LIMD1), was used to identify conditions under which expression of GFP-tagged proteins is similar to endogenous expression levels (Fig. S1D,E). These doses were then used for all fluorescence imaging experiments.
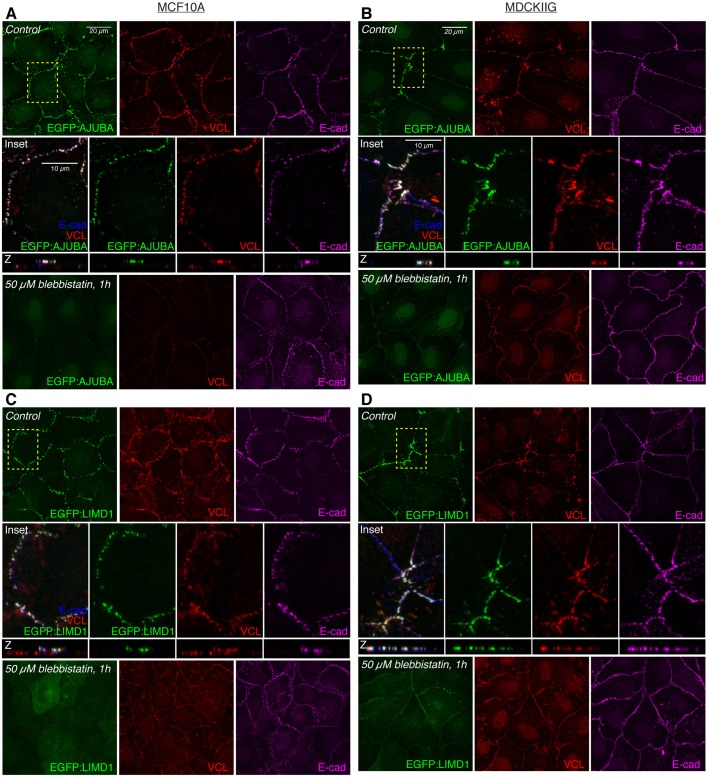
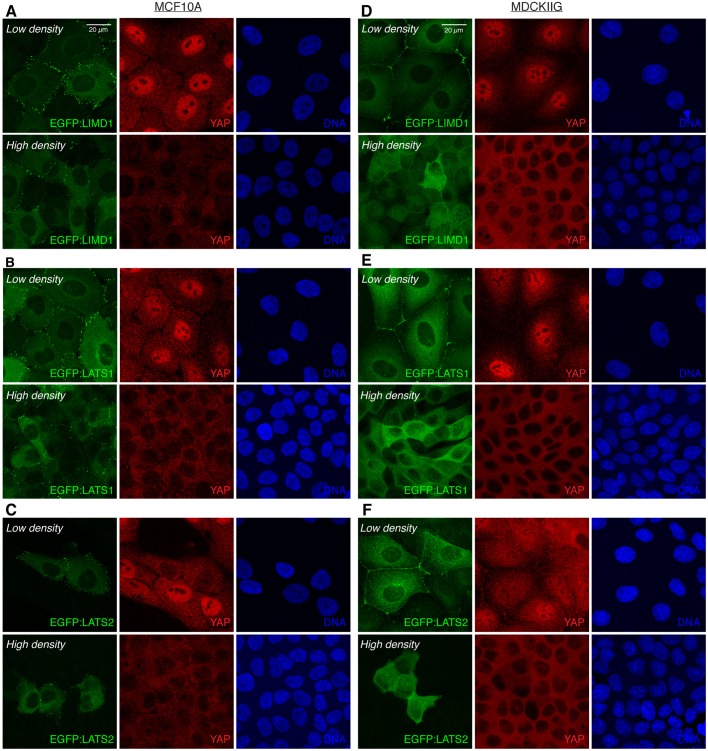
We found that in low-density cultures with cells in contact, each of the three Ajuba family proteins exhibit a prominent punctate localization to adherens junctions [identified by overlap with the apical circumferential localization of E-cadherin (E-cad)] (Fig. 1; Fig. S2A–D) and a faint localization to the cytoplasm. Some localization to focal adhesions, identified by overlap with basal puncta of vinculin (VCL), could also be detected, but at lower levels than at adherens junctions, and this focal adhesion localization was more evident in MDCKIIG cells than in MCF10A cells (Fig. S3). In MDCKIIG cells, which have linear adherens junctions, Ajuba family proteins preferentially accumulate in puncta near intercellular vertices (Fig. 1; Fig. S2B,D,F). This contrasts with the more even distribution of E-cad or ZO-1 (also known as TJP1) around the cell circumference (Fig. 1B,D, Fig. S2B,D), and is reminiscent of the distribution of Jub in Drosophila imaginal disc epithelia (Rauskolb et al., 2014). In MCF10A cells, which have punctate adherens junctions (Meng and Takeichi, 2009), Ajuba family proteins have a punctate distribution all around the apical cell periphery that matches the distribution of the junctional markers E-cad and ZO-1 (Fig. 1A,B; Fig. S2A,C). We also attempted to examine the localization of endogenous Ajuba family proteins using commercially available antisera. Antisera against AJUBA and LIMD1 that detect junctional staining in MCF10A and MDCKIIG cells similar to that of GFP-tagged proteins were identified (Fig. S1).
Fig. 1.
Tension-dependent localization of EGFP-tagged Ajuba family proteins in stable MCF10A or MDCKIIG cell lines. MCF10A (A,C) or MDCKIIG (B,D) cells from the indicated cell lines were plated at low density (15,000 cells/cm2) and cultured for 48 h in total. Transgene expression was induced with Dox for 24 h before treatments. Cells were treated with DMSO (control) or 50 µM blebbistatin for 1 h, fixed in the presence of 0.5% Triton X-100 and then stained for VCL (red) and E-cadherin (E-cad, magenta). Square images are apical slices for MCF10A and z-projections for MDCKIIG cells, and are representative of at least three biological replicates. Insets show higher magnification of the boxed regions (yellow dashes). Vertical confocal slices (z) are shown at the same magnification as the insets.
In addition to its localization to focal adhesions, VCL exhibits a tension-dependent localization to adherens junctions (le Duc et al., 2010; Yonemura et al., 2010). Notably, VCL localization overlaps with Ajuba protein family localization at adherens junctions, including both the localization to punctate adherens junctions in MCF10A cells and the localization to dispersed puncta in MDCKIIG cells (Fig. 1, Fig. S2B). This colocalization suggests that, like VCL, Ajuba family localization to adherens junctions is dependent upon cytoskeletal tension. To confirm this, we treated cells with either of two inhibitors of myosin activity: the Rho-associated protein kinase (ROCK) 1 and 2 inhibitor Y-27632, or the myosin II inhibitor blebbistatin. These compounds substantially reduce cytoskeletal tension, and treatment of cells with either compound led to a loss of the bright puncta of AJUBA, LIMD1 and WTIP, both at adherens junctions and at focal adhesions, and in both MDCKIIG and MCF10A cells (Fig. 1; Figs S2, S3). The cytoplasmic signal of the Ajuba family proteins appears to be slightly increased when tension is reduced, and total protein levels appear similar, suggesting that protein lost from junctions is now cytoplasmic (Fig. 1; Figs S2 and S3). In MDCKIIG cells, but not in MCF10A cells, we also detected a low level of Ajuba family protein distribution along cell junctions that is evenly distributed around the cell perimeter after inhibiting cytoskeletal tension (Fig. 1B,D, Fig. S2B,D,F). Thus each of the three Ajuba family proteins has a localization profile that is modulated by cytoskeletal tension, with strong punctate junctional localization, and also focal adhesion localization, dependent upon cytoskeletal tension.
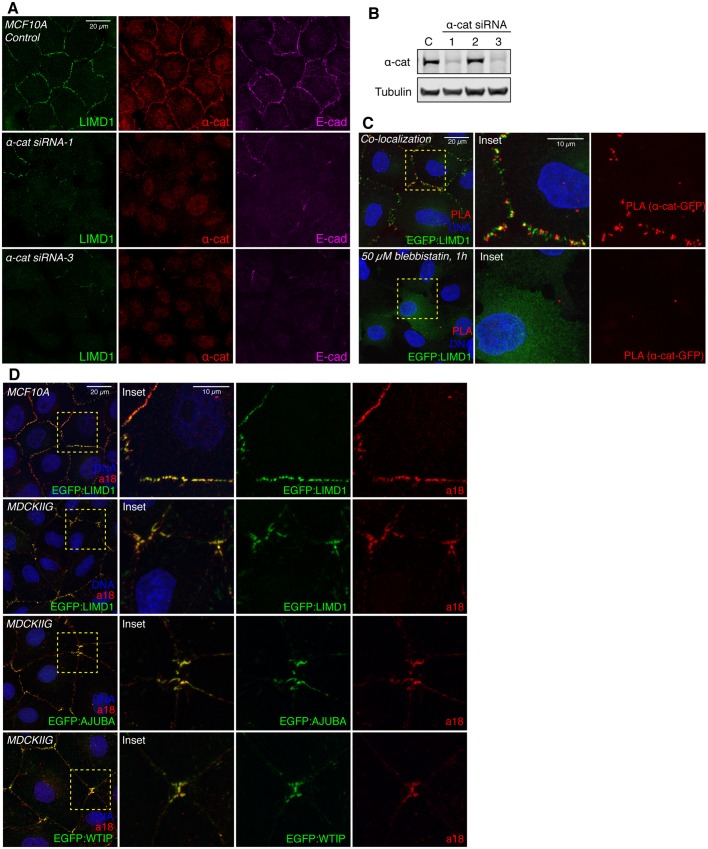
α-Catenin has been implicated in tension-dependent recruitment of VCL and Jub to adherens junctions, which is thought to occur through a tension-dependent change in α-catenin conformation (Yonemura et al., 2010). α-catenin has also been reported to bind to AJUBA, but the potential influence of tension on this interaction has not been investigated (Marie et al., 2003). To assess the requirement for α-catenin in tension-dependent recruitment of mammalian Ajuba family proteins, we examined LIMD1 localization in cells with an siRNA-mediated reduction in α-catenin expression. This substantially reduced junctional levels of LIMD1, however it also disrupts adherens junction formation, as E-cad staining is also lost (Fig. 2A,B). To further analyze potential interaction between LIMD1 and α-catenin, we used proximity ligation assays (PLAs) (Fredriksson et al., 2002) to probe their physical proximity in vivo. A robust and specific PLA signal was detected using antibodies against α-catenin and GFP in EGFP:LIMD1-expressing cells, and this signal was abolished by blebbistatin treatment (Fig. 2C). As PLA is thought to require proteins to be within 30–40 nm, this signal indicates that LIMD1 and α-catenin are closely associated at cell–cell junctions. The tension-dependent conformational change in α-catenin exposes the epitope recognized by the a18 monoclonal antibody (Yonemura et al., 2010). Junctional puncta of LIMD1, AJUBA and WTIP colocalize with a18 staining in both MCF10A and MDCKIIG cells (Fig. 2D). Taken together, these observations suggest that LIMD1 is recruited to adherens junctions by the tensed (open) conformation of α-catenin.
Fig. 2.
α-Catenin is required for LIMD1 recruitment to adherens junctions. (A) MCF10A cells from the indicated cell lines were plated at low density (15,000 cells/cm2) and cultured for 48 h in total. Cells were transfected with control or α-catenin siRNAs, fixed in the presence of 0.5% Triton X-100 and then stained for LIMD1 (Millipore, green), α-catenin (α-cat, red) and E-cadherin (E-cad, magenta). (B) Western blot showing the knockdown efficiency of the α-catenin siRNAs tested. C, control (non-specific siRNA). (C) PLA. MCF10A EGFP:LIMD1 cells were grown at low density, and treated with DMSO (control) or with 50 µM blebbistatin. Cells were induced with Dox for 24 h, fixed and incubated with mouse LIMD1 (Millipore) and mouse GFP antibodies, and signal was detected via anti-mouse MINUS and anti-rabbit PLUS probes. Insets show higher magnification of the boxed regions. (D) MCF10A and MDCKIIG from the indicated cell lines were grown at low density and stained with a18 (red) antibody and DNA (Hoechst 33342, blue). Insets show higher magnification of the boxed regions (yellow dashes).
Tension-dependent localization of LATS proteins in mammalian epithelial cells
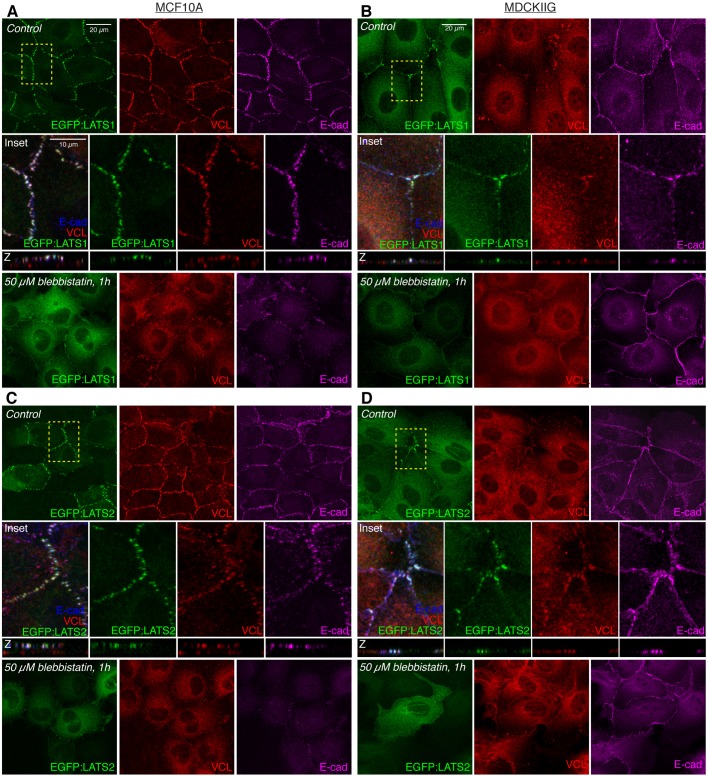
Published studies have described a variety of localizations for LATS, including nuclear, cytoplasmic and junctional (Abe et al., 2006; Jagannathan et al., 2016; Li et al., 2014; Paramasivam et al., 2011; Szymaniak et al., 2015; Yin et al., 2013). To define their localization in mammalian epithelial cells, we created constructs for inducible expression of EGFP-tagged human LATS1 and LATS2 in lentiviral vectors. MCF10A and MDCKIIG cell lines with relatively homogeneous expression of EGFP:LATS1 were selected, but cells transduced with EGFP:LATS2 constructs tend to be lost during culture, even without addition of Dox, and we were only able to achieve patchy and transient expression of EGFP:LATS2 in both MDCKIIG and MCF10A cell lines. Moreover, we found that much higher levels of Dox were needed to induce visible EGFP:LATS2 expression than for any of our other cell lines; these observations suggest that there is a strong selection against LATS2 expression, likely due to inhibition of YAP/TAZ activity. We also assayed several commercially available anti-LATS serum and confirmed that one of them has high specificity for LATS proteins. Western blotting of cells transduced to express EGFP:LATS1 or EGFP:LATS2 revealed that this antibody specifically recognizes human LATS1 (Fig. S4C). Moreover, the single band identified by this antibody in MCF10A cells was substantially reduced by siRNA-mediated knockdown of LATS1 (Fig. S4D). Immunostaining of MCF10A cells with this antibody revealed a localization pattern similar to that of EGFP:LATS1, and this pattern is lost when cells were transfected with LATS1 siRNAs, but not with LATS2 siRNAs (Fig. S4F). By using canine-specific siRNAs, we found that this antibody appears to recognize both endogenous LATS1 and LATS2 on western blots of MDCKIIG cell lysates (Fig. S4E), and reveals a localization pattern in MDCKIIG cells similar to that of GFP-tagged human LATS proteins (Figs 3 and 4). By using LATS2 siRNA knockdown, we also found an antibody that specifically recognized endogenous human LATS2 in western blots, although not in immunostainings (Fig. S4D,G). Western blotting lysates of stably transduced cells identified conditions under which induced expression of EGFP:LATS1 was similar to that of endogenous LATS1 expression (Fig. S4H,I), and these conditions were used for analysis of LATS protein localization. Direct comparisons to endogenous LATS2 levels were not possible due to the small fraction of EGFP:LATS2-expressing cells.
Fig. 3.
Tension-dependent localization of LATS proteins in MDCKIIG and MCF10A cell lines. (A,C) MCF10A or (B,D) MDCKIIG cells from the indicated cell lines were plated at low density (15,000 cells/cm2) and cultured for 48 h in total. Transgene expression was induced by adding Dox for 24 h before treatment. Cells were treated with DMSO (control) or 50 µM blebbistatin for 1 h and then fixed in the presence of 0.5% Triton X-100 (A,B; MCF10A) or without detergent (C,D; MDCKIIG) and stained for VCL (red) and E-cad (blue/magenta). Square images are z-projections (MDCKIIG) or apical slices (MCF10A) and are representative of at least three biological replicates. Insets show higher magnification of the boxed regions (yellow dashes). Vertical slices (z) are shown as the same magnification as the boxed regions.
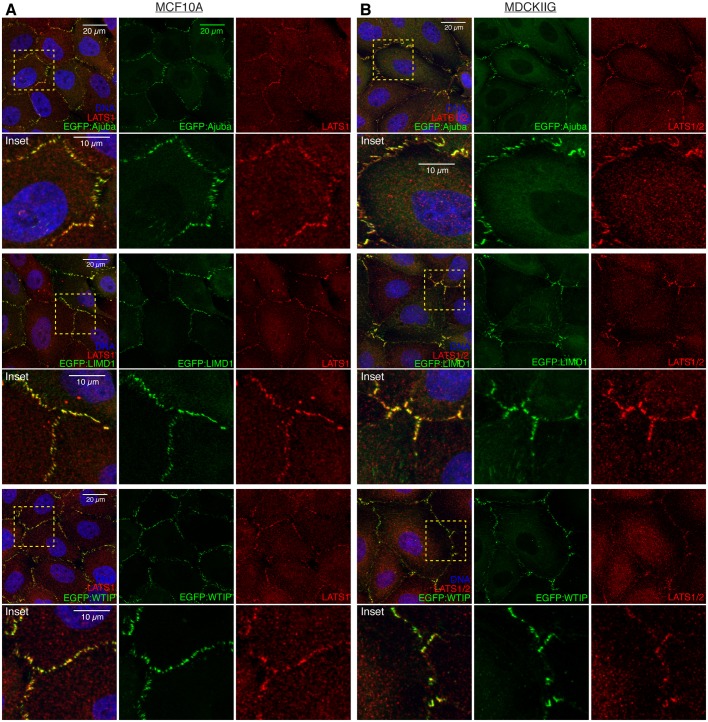
Fig. 4.
EGFP-tagged Ajuba family proteins colocalize with LATS1. (A) MCF10A cells from the indicated stable cell lines were plated at low density (15,000 cells/cm2) and cultured for 48 h in total. Transgene expression was induced with Dox for 24 h before fixation and then cells were stained for LATS1 (red) and DNA (Hoechst 33342, blue). (B) MDCKIIG cells from the indicated stable cell lines were plated at low density and treated as in A. Insets show higher magnification of the boxed regions (yellow dashes).
LATS1 and LATS2 both accumulate in puncta overlapping adherens junctions in the same manner as the Ajuba family proteins (Fig. 3; Fig. S4A,B). That is, in MDCKIIG cells they accumulate in puncta near intercellular vertices, whereas in MCF10A cells they accumulate in puncta around the circumference of the cell. However, in contrast to the Ajuba family proteins, accumulation of LATS proteins was never detected at focal adhesions (Fig. S5A–D). LATS puncta at adherens junctions overlap puncta of junctional VCL accumulation, implying that their localization is dependent on tension (Fig. 3). This was confirmed by observations that treatment of cells with either blebbistatin or Y-27632 leads to loss of the bright puncta of junctional LATS proteins, both in MCF10A and in MDCKIIG cells (Fig. 3, Fig. S5). Under these low-tension conditions, total LATS protein levels appear similar, and LATS proteins accumulate in the cytoplasm, but as for Ajuba family proteins, we could also detect some low-level uniform localization around the cell circumference in MDCKIIG cells (Fig. 3B,D; Fig. S5). Thus, junctional localization of both LATS proteins in mammalian epithelial cells is modulated by cytoskeletal tension.
LIMD1 recruits LATS to adherens junctions
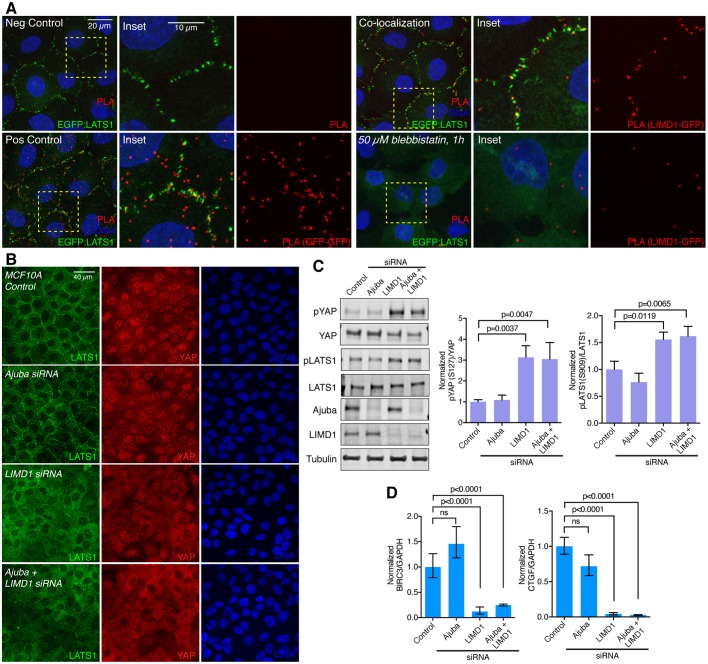
The similar localization of Ajuba and LATS family proteins to puncta overlapping VCL at adherens junctions implies that they colocalize. To examine this directly, we used anti-LATS1 serum to stain cells expressing GFP-tagged Ajuba family proteins. This revealed extensive colocalization of LATS1 with each of the GFP-tagged Ajuba family proteins in puncta at cell junctions, both in MDCKIIG and MCF10A cells (Fig. 4). To further analyze this colocalization, we used PLA to probe the physical proximity of Ajuba and LATS family proteins. In a PLA using an anti-LIMD1 antibody in combination with an anti-GFP antibody on EGFP:LATS1-expressing cells, a clear and consistent signal was detected in puncta along cell–cell junctions (Fig. 5A), which indicates that LIMD1 and LATS1 are closely associated on cell–cell junctions, and most likely in direct contact. This PLA signal is lost upon blebbistatin treatment (Fig. 5A). No determination could be made regarding AJUBA or WTIP proximity to LATS1 because these antisera were not of sufficiently high quality to give a clear PLA signal.
Fig. 5.
LIMD1 recruits LATS1 to adherens junctions and promotes YAP activity. (A) PLA. MCF10A EGFP:LATS1 cells were grown at low density, induced with Dox and treated with or without 50 µM blebbistatin. Cells were fixed and incubated with only secondary antibodies (negative control), mouse GFP antibody and both anti-mouse MINUS and PLUS probes (positive control), and rabbit LIMD1 and mouse GFP antibodies, with signal was detected via detected with anti-mouse MINUS and anti-rabbit PLUS probes (colocalization and 50 µM blebbistatin, 1 h). Insets show higher magnification of the boxed regions (yellow dashes). (B) MCF10A cells grown at low density transfected with control, Ajuba, LIMD1 or Ajuba+LIMD1 siRNAs, and stained for LATS1 (green), YAP (red) and DNA (Hoechst 33342, blue). Images are z-projections and representative of at least three biological replicates. (C) Western blots of lysates of MCF10A cells transfected with control, AJUBA, LIMD1 and Ajuba+LIMD1 siRNA, blotted using the indicated antisera. All the blots are from the same experiment and a representative loading control (tubulin) is shown. Histograms show quantification of the pYAP (S127) to total YAP ratio and pLATS1 (S909) to total LATS1 ratio from three biological replicates normalized to the ratio in the control siRNA transfection. Error bars indicate s.e.m. and P-values less than 0.05 are shown. (D) Quantification of BIRC3 and CTGF mRNA abundance (n=3 biological replicates) by means of qPCR on MCF10A cells transfected with control, Ajuba, LIMD1 and Ajuba+LIMD1 siRNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a standard. The BIRC3 or CTGF to GAPDH ratio was normalized to the ratio in the control siRNA transfection. Error bars indicate the 95% c.i. and P-values less than 0.05 are shown. ns, not significant.
To determine whether the colocalization of Ajuba and LATS family proteins reflects a functional requirement for Ajuba proteins in recruiting LATS proteins to junctions, we examined LATS1 and LATS2 localization in MCF10A cells treated with at least two independent siRNAs targeting AJUBA, LIMD1 and/or WTIP. Western blotting and immunostaining confirmed that these siRNAs could effectively reduce Ajuba family protein levels, and also could reduce the levels of the EGFP-tagged Ajuba family proteins (Figs S1B,C, S6A). Knockdown of AJUBA or WTIP had no visible effect on LATS1 localization (Fig. 5B; Fig. S6B). In contrast, knockdown of LIMD1 clearly suppressed LATS1 and LATS2 recruitment to junctions (Fig. 5B; Fig. S6B,C). Thus, junctional localization of LATS proteins in MCF10A cells specifically requires LIMD1.
When overexpressed, each of the three Ajuba family proteins has been reported to be able to co-immunoprecipitate with LATS proteins (Das Thakur et al., 2010; Sun and Irvine, 2013). Our observation that localization of endogenous LATS to junctional puncta in MCF10A cells specifically requires LIMD1 provided an opportunity to investigate the relationship between LATS protein localization and YAP activity. YAP activity was examined by assaying YAP localization, YAP phosphorylation and expression of YAP target genes. Based on all three criteria, we found that YAP activity was reduced in MCF10A cells by knockdown of LIMD1, as LIMD1 siRNA reduced nuclear localization of YAP, increased phosphorylation of YAP at the key LATS phosphorylation site (S127), and reduced the mRNA levels of the YAP targets CTGF and BIRC3 (Fig. 5B–D; Fig. S6B). Conversely, knockdown of AJUBA or WTIP had no effect on YAP activity (Fig. 5B–D; Fig. S6B). We also assayed for potential additive effects of knockdowns of two Ajuba family genes, in all three pairwise combinations, but no additive effects were detected (Fig. 5B–D; Fig. S6B). Staining western blots with anti-phospho-LATS serum revealed that knockdown of LIMD1, but not of AJUBA, increases LATS activation (Fig. 5C), which could account for the influence of LIMD1 on YAP activity. Taken together, these observations imply that among the three Ajuba family proteins, LIMD1 specifically regulates YAP activity in MCF10A cells, and that it does so by recruiting LATS to adherens junctions to suppress their activation.
Regulation of Ajuba and LATS family proteins by cell density
Hippo signaling is influenced by cell density, and plays a key role in the density-dependent regulation of cell proliferation (contact inhibition) (Zhao et al., 2007). To investigate the potential for junctional localization of Ajuba and LATS family proteins to contribute to density-dependent regulation of Hippo signaling, we investigated whether their localization is affected by cell density. At low densities (15,000 cells/cm2, corresponding to the conditions used in all of the experiments described above), Ajuba and LATS family proteins localize in bright puncta overlapping adherens junctions (Figs 1–4). In contrast, at high densities (150,000 cells/cm2) bright puncta of junctional localization are rare or absent (Fig. 6; Fig. S7).
Fig. 6.
Regulation of LIMD1 and LATS proteins localization and YAP activity by cell density. (A-C) The indicated MCF10A or (D–F) MDCKIIG cell lines were plated at low (15,000 cells/cm2) and high density (150,000 cells/cm2) and grown for 48 h in total. Transgene expression was induced by adding Dox 24 h before fixation. Cells were stained for YAP (red) and DNA (Hoechst 33342, blue). Images are projections through z-stacks.
Examination of YAP activity in MCF10A cells confirmed that changes in YAP correlate with these changes in LIMD1 and LATS protein localization. Thus, under our low-cell density conditions, YAP is predominantly nuclear, whereas under our high-cell density conditions, YAP is predominantly cytoplasmic (Fig. 6; Fig. S7). (Note that expression of EGFP:LATS2 at detectable levels reduced YAP nuclear localization at low-cell density, but further reductions are nonetheless evident at higher cell density; Fig. 6.) The correlation between YAP localization and LIMD1 and LATS localization suggests that density-dependent regulation of their localization could contribute to density-dependent regulation of YAP. Consistent with this hypothesis, we observed that LIMD1 knockdown decreases YAP activity at low-cell density (Fig. 5B–D), but has no significant effect at high-cell density (see Fig. 8E,F, compare control and siRNA-treated cells without Rho activator).
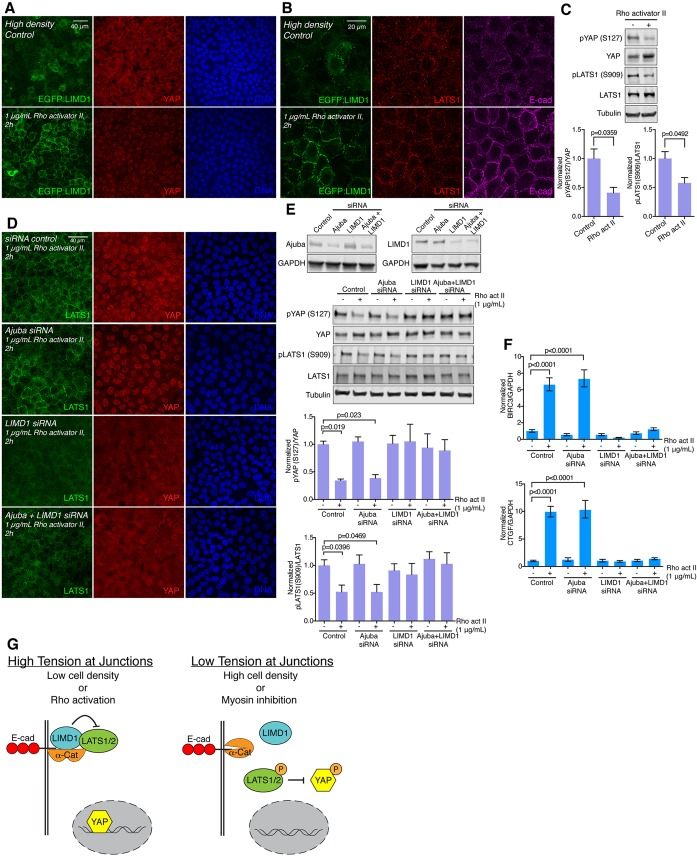
Fig. 8.
Ajuba and LATS family proteins contribute to Rho-mediated regulation of Hippo signaling. (A) MCF10A EGFP:LIMD1 cells grown at high density, induced with Dox for 24 h before treatments and mock-treated with water (control) or treated with 1 µg/mL Rho activator II for 2 h. After treatment, cells were stained for YAP (red) and DNA (Hoechst 33342, blue). (B) Same cell line and treatments as in A, but cells were stained for LATS1 (red) and E-cad (magenta). Images are z-projections and representative of at least three biological replicates. (C) Western blots of lysates of MCF10A cells mock-treated with water (control) or treated with 1 µg/ml Rho activator II for 2 h, blotted using the indicated antisera. All the blots are from the same experiment and a representative loading control (Tubulin) is shown. Histograms show quantification of the pYAP (S127) to total YAP ratio and pLATS1 (S909) to total LATS1 ratio from three biological replicates normalized to the ratio in the control. Error bars indicate s.e.m. and P-values less than 0.05 are shown. (D) MCF10A cells grown at high density and transfected with control, Ajuba, LIMD1 or Ajuba+LIMD1 siRNA. After 48 h, cells were treated with 1 µg/ml Rho activator II for 2 h and stained for LATS1 (green), YAP (red) and DNA (Hoechst 33342, blue). Images are z-projections and representative of at least three experiments. (E) Western blots of lysates of MCF10A cells grown at high density and transfected with control, Ajuba, LIMD1 or Ajuba+LIMD1 siRNA, and blotted using the indicated antisera. Efficiency of the knockdown is reduced when the cells are transfected at high density as compared to low density (compare with Fig. 5). All the blots are from the same experiment and a representative loading control (tubulin or GAPDH) is shown. Histograms show the quantification of the pYAP (S127) to total YAP ratio and pLATS1 (S909) to total LATS1 ratio from three biological replicates normalized to the ratio in the control siRNA transfection. Error bars indicate the s.e.m. and P-values less than 0.05 are shown. (F) Quantification of BIRC3 and CTGF mRNA abundance (n=3 biological replicates) as determined by qPCR on MCF10A cells transfected with control, Ajuba, LIMD1 or Ajuba+LIMD1 siRNA and treated with 1 µg/ml Rho activator II. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a standard. The BIRC3 or CTGF to GAPDH ratio was normalized to the ratio in the control siRNA transfection. Error bars indicate 95% c.i. and P-values less than 0.05 are shown. (G) Summary model for regulation of YAP by LIMD1. When the cells are under high tension (left, e.g. cell low density or Rho activation), LIMD1 is associated with adherens junctions through α-catenin, where it recruits and inhibits LATS. This allows YAP to go to the nucleus and activate transcription. When cells are under low tension (right, e.g. high density or myosin inhibition), the α-catenin conformation is altered, LIMD1 and LATS are released from junctions, and LATS can be activated. Activated (phosphorylated) LATS phosphorylates and inhibits YAP by promoting its cytoplasmic localization and degradation.
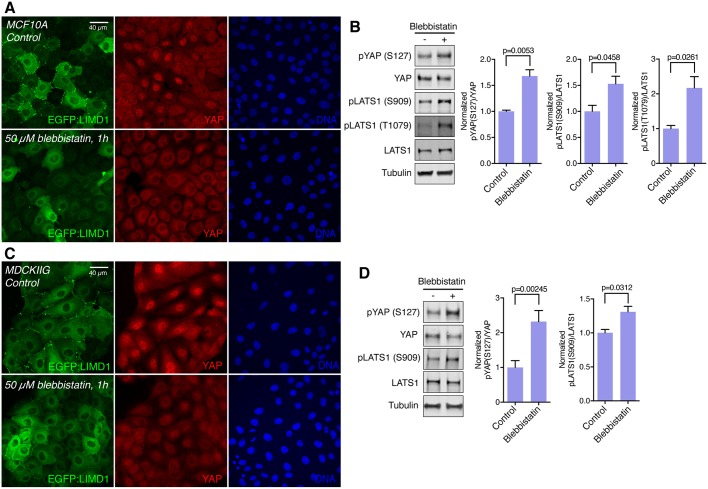
The localization of Ajuba and LATS family proteins at high density becomes similar to that observed when myosin activity is inhibited at low-cell density; junctional localization is reduced in MCF10A cells, and a weak uniform junctional localization is observed in MDCKIIG cells (Fig. 6; Fig. S7). Indeed, examination of phosphorylated myosin light chain (pMLC; the activated form) and VCL localization indicate that cytoskeletal tension is lower in cells at high-cell density (Fig. S7C–F), which could account for the observed changes in Ajuba and LATS family protein localization. Moreover, the tension dependence of Ajuba and LATS family protein localization also correlates with influences of cytoskeletal tension on YAP activity. Thus, under the same doses of blebbistatin that causes Ajuba and LATS family proteins to be lost from puncta at adherens junctions despite low-cell density, LATS phosphorylation is increased and YAP activity is reduced, as revealed by western blotting with anti-phospho-YAP and immunostaining for YAP (Fig. 7).
Fig. 7.
Tension-dependence of Ajuba and LATS family protein localization correlates with influences of cytoskeletal tension on YAP activity. (A) MCF10A EGFP:LIMD1 cells were plated at low density and grown for 48 h in total. Transgene expression was induced by adding Dox 24 h before treatments. Cells were treated with DMSO (control) or treated with 50 µM blebbistatin for 1 h. After the treatment, cells were stained for YAP (red) and DNA (Hoechst 33342, blue) (B) Western blots of lysates of MCF10A cells treated with DMSO (control) or 50 µM blebbistatin for 1 h, blotted using the indicated antisera. All the blots are from the same experiment and a representative loading control (tubulin) is shown. Histograms show a quantification of the pYAP (S127) to total YAP ratio and pLATS1 (S909 and T1079) to total LATS1 ratio from three biological replicates normalized to the ratio in the control. (C) MDCKIIG EGFP:LIMD1 cells were grown and stained as in A. Images are z-projections and are representative of at least three biological replicates. (D) Western blots of lysates of MDCKIIG cells treated with DMSO (control) or 50 µM blebbistatin for 1 h, blotted using the indicated antisera. Tubulin was used as a loading control. Histograms show a quantification of the pYAP (S127) to total YAP ratio and pLATS1 (S909) to total LATS1 ratio from three biological replicates normalized to the ratio in the control. In all histograms, error bars indicate s.e.m. and P-values less than 0.05 are shown.
Ajuba and LATS family proteins contribute to Rho-mediated regulation of Hippo signaling
Multiple upstream inputs into Hippo signaling appear to act through, or are dependent upon, the small GTPase Rho. For example, YAP and TAZ activation associated with cell attachment to substrates, large cell area, stiff substrates, or activation of certain G protein-coupled receptor (GPCR) signaling pathways is suppressed by Rho inhibitors (Dupont et al., 2011; Yu et al., 2012; Zhao et al., 2012), whereas expression of activated Rho can promote YAP activation (Zhao et al., 2012). However, the mechanism(s) by which Rho activity ultimately impinges upon YAP activity have remained unclear. Based upon the well-established ability of Rho to influence F-actin and myosin activity (Fig. S8E; Etienne-Manneville and Hall, 2002), we asked whether Rho-mediated regulation of Hippo signaling involves Ajuba family proteins.
This was examined using MCF10A cells plated at high density, which have low recruitment of Ajuba and LATS family proteins to adherens junctions, and correspondingly low YAP activity (Fig. 8; Fig. S8). Endogenous Rho proteins were activated either by treating cells with lysophosphatidic acid (LPA), which activates Rho through GPCRs (Yu et al., 2012), or Rho activator II [cytotoxic necrotizing factor-1 (CNF1) from E. coli], which deamidates glutamine-63 of RhoA and its homologs, preventing GTP hydrolysis and thereby keeping Rho proteins in their active, GTP-bound state (Schmidt et al., 1997). Each of these treatments substantially decreased LATS activity and increased YAP activity (Fig. 8; Fig. S8). At the same time, Rho activation substantially increased localization of Ajuba and LATS family proteins to adherens junctions (Fig. 8; Fig. S8). Thus, Rho-mediated YAP activation correlates with Rho-mediated Ajuba and LATS protein localization to cell–cell junctions.
To directly test the requirement for Ajuba family proteins, and the significance of the correlation between LATS recruitment to junctions and activation of YAP in response to Rho activation, we examined the consequences of siRNA-mediated knockdown AJUBA and/or LIMD1 on cells plated at high density and then treated with Rho activator II. These experiments revealed that knockdown of AJUBA did not visibly decrease Rho-mediated LATS1 recruitment or YAP activation (Fig. 8). Conversely, knockdown of LIMD1 suppressed Rho-mediated LATS1 recruitment to junctions, YAP activation and expression of YAP target genes (Fig. 8). Thus, LIMD1 is specifically required for Rho-mediated LATS re-localization and YAP activation.
DISCUSSION
Hippo signaling has emerged as a key conduit for transduction of the responses of cells to their biomechanical environment. Nonetheless, the molecular mechanisms by which biomechanical signals are perceived and transduced to influence YAP and TAZ activity in mammalian cells have remained poorly understood. Our observations showing that LIMD1 localizes to puncta at adherens junctions under conditions of low-cell density, Rho activation and cytoskeletal tension, that this LIMD1 recruits LATS kinases to adherens junctions and inhibits LATS activity, and that LIMD1 is required for YAP activation under conditions of low-cell density, Rho activation and cytoskeletal tension, all support a model in which tension-dependent recruitment and inhibition of LATS kinases in complexes with LIMD1 at adherens junctions contributes to biomechanical (and biochemical) regulation of Hippo signaling (Fig. 8G).
The mechanism by which LIMD1 inhibits LATS activation is not clear, but studies of Jub inhibition of Warts in Drosophila suggested a model in which recruitment of Warts to Jub sequesters it away from upstream complexes that promote Warts activation (Sun et al., 2015). In Drosophila, these include junctional Expanded complexes, where Hippo, Salvador and Expanded can be seen to overlap with activated Warts (Sun et al., 2015), and apical Merlin complexes (Su et al., 2017). Junctional or apical localization of proteins that could scaffold assembly of LATS-activating complexes has also been reported in mammalian cells (Sun and Irvine, 2016), and observations that at least in some contexts the key upstream regulators merlin (also known as NF2) and angiomotins associate with tight junctions (Yi et al., 2011) suggests that sequestration of LATS into adherens junction complexes might prevent its activation. It is also possible that association with LIMD1 physically precludes association of LATS with one or more upstream activators. Conversely, under low-tension conditions, such as upon blebbistatin treatment or high-cell density, LATS could be free to associate with and be activated by apical complexes including upstream factors like angiomotin and merlin that can bind to LATS and promote its activation. The reduced junctional accumulation of LATS under activating (e.g. low tension) conditions suggests that LATS normally only associates with activating complexes transiently and/or at low levels; a similar situation occurs in Drosophila wing discs, where phospho-Warts staining is normally undetectable even though some Warts is active (Sun et al., 2015).
The absence of visible recruitment of LATS proteins to focal adhesions, despite the presence of Ajuba family proteins indicates that there must be additional cues at adherens junctions that promote the recruitment of LATS kinases. The absence of LATS kinases at focal adhesions also implies that distinct mechanisms are involved in biomechanical signaling mediated through focal adhesions. This is consistent with observations that some biomechanical signals that impinge on focal adhesions, such as substrate stiffness, substrate stretching, or increasing cell shape through modulation of cell attachment sites, can influence YAP/TAZ activity through mechanisms that are independent of LATS activity (Aragona et al., 2013; Das et al., 2016; Dupont et al., 2011). Similarly, a reported lack of requirement for Ajuba family proteins for ‘mechanical signals’ is not inconsistent with our results, because that study (Jagannathan et al., 2016) examined potential requirements for AJUBA and LIMD1 in YAP activation under conditions where cells were on stiff substrates or undergoing spreading, which are expected to exert tension on focal adhesions rather than adherens junctions. Jagannathan et al. (2016) also reported observing increased LATS activity only when both AJUBA and LIMD1 were knocked down, and not when LIMD1 alone was knocked down. However, this was under low-density conditions that differed from ours, as they examined isolated cells lacking cell–cell junctions, which consequently could not have the LIMD1-specific junctional recruitment of LATS that we identified. We also note that while Jagannathan et al. (2016) reported that LIMD1 and LATS1 could be detected at adherens junctions in cells cultured at ‘high-cell density’, based on cell area some of their high density images appear to correspond conditions that we would classify as low-cell density.
Our observations implicate contact inhibition of cell proliferation [more accurately described as cell density-dependent regulation of cell proliferation (Puliafito et al., 2012)] as a biological context where tension-dependent recruitment of LIMD1 and LATS to adherens junctions plays a role in modulating Hippo signaling. Hippo signaling has been shown to mediate density-dependent regulation of cell proliferation (Zhao et al., 2007), but the mechanism through which this occurs was unclear. The reduction in junctional localization and LATS inhibition as cell density increases appears to stem from decreased cytoskeletal tension, as it correlates with reduced phospho-myosin staining and VCL localization, and it can be reversed by activation of Rho. The observation that suppression of YAP activity at high-cell density can be reversed by Rho activation, and that this reversal is dependent upon LIMD1, implicates tension-dependent regulation of LIMD1 and LATS as a key contributor to contact inhibition. Conversely, pharmacological reduction of cytoskeletal tension can abolish junctional localization of LIMD1 and LATS even under low-cell density conditions. The observation that cytoskeletal tension is reduced as cells become more crowded is also consistent both with modeling of epithelial mechanics (Noll et al., 2017; Pan et al., 2016; Puliafito et al., 2012), and with experimental studies in Drosophila, where growth-induced cell crowding has been shown to reduce cytoskeletal tension (Pan et al., 2016).
Our observations also imply that formation of junctional complexes between LATS and LIMD1 contributes to Hippo pathway regulation by GPCR signaling. Regulation of Hippo signaling by GPCRs is known to depend upon Rho, but the molecular mechanism by which Rho affects Hippo signaling has been unclear. GPCR signaling is activated by LPA, and mediated through Rho activation (Yu et al., 2012), both of which we found to promote junctional recruitment of LIMD1 and LATS, and LIMD1-dependent activation of YAP.
Our studies identified all three Ajuba family proteins as exhibiting tension-dependent recruitment to adherens junctions. The distinct localization profiles observed in MCF10A versus MDCKIIG cells are consistent with tension-dependent localization, as the distinct organization of the cytoskeleton in cells with punctate versus linear adherens junctions is expected to correlate with differences in the spatial pattern of tension along junctions (Takeichi, 2014). This tension-dependent recruitment is likely mediated through conformational changes in α-catenin structure (Yonemura et al., 2010), as both AJUBA and Jub have been reported to associate with α-catenin (Marie et al., 2003; Rauskolb et al., 2014), and the puncta of Ajuba family localization at adherens junctions colocalize both with VCL, which associates with stretched α-catenin, and with a monoclonal antibody (a18) that specifically recognizes stretched α-catenin. Junctional localization of LIMD1 also requires α-catenin, and we confirmed that LIMD1 and α-catenin are closely associated at junctions through PLAs. As Ajuba family proteins have many other functions besides their contribution to Hippo signaling (Schimizzi and Longmore, 2015), our observation that the localization of all three proteins is regulated by tension raises the possibility that some of their other functions could also be modulated by cytoskeletal tension.
Our observations identify a role for α-catenin in promoting YAP activation, but earlier studies have revealed that, in some contexts, loss of α-catenin is associated with activation of YAP (Schlegelmilch et al., 2011). This presumably reflects the existence of positive and negative effects of apical junctions, and specifically α-catenin, on YAP activity. For example, while our results implicate α-catenin in recruiting an inhibitor of LATS (LIMD1), earlier studies have reported that α-catenin can interact with an activator of LATS (merlin) (Gladden et al., 2010). Whether YAP-activating or YAP-inhibiting functions of α-catenin are predominant could depend on a variety of factors, including cytoskeletal tension and the cell type-specific character of cell–cell junctions. We also note that activation of YAP in cells lacking α-catenin has been attributed to Src activation, rather than a reduction in Hippo pathway activity (Li et al., 2016).
Given their similar tension-dependent recruitment, and the reported ability of all three Ajuba family proteins to associate with LATS family proteins (Das Thakur et al., 2010), the specific requirement for LIMD1 in LATS recruitment was unexpected. Although there could be in vivo differences in requirements due to different expression patterns, both AJUBA and LIMD1 are clearly expressed in the MCF10A cells we examined. It is conceivable that there are differences in binding affinity, differences in association with partner proteins or differences in post-translational modifications that account for the specific requirement for LIMD1 as opposed to other Ajuba family proteins, and this will be an important question for future studies.
MATERIALS AND METHODS
Plasmids
To generate Dox-inducible lentiviral vector, the coding sequence of TurboRFP in pTRIPZ (GE Healthcare) was replaced with the GFP fusion proteins. EGFP-tagged human AJUBA, LIMD1, LATS1 and WTIP were generated by PCR from pcDNA3.1-V5:His containing AJUBA, LIMD1, LATS1 or WTIP (Reddy and Irvine, 2013; Sun and Irvine, 2013) and cloned into pEGFP-C3. Human EGFP-LATS2 was a gift of Norikazu Yabuta and Hiroshi Nojima, Osaka University, Japan (Yabuta et al., 2011).
Generation of stable cell lines
Lentiviral Dox-inducible vectors were co-transfected with Trans-Lentiviral Packaging Mix into HEK-293T (Dharmacon) cells by using calcium phosphate. After 16 h of transfection, medium was replaced with UltraCULTURE medium (Lonza) supplemented with 1× Gluta-MAX (Life Technologies) and antibiotics. Starting the next day, the lentivirus-containing supernatants were collected every 24 h for 2 days and stored at 4°C. After centrifugation at 800 g for 10 min to remove cell debris, supernatants were filtered using a 0.45-μm-pore size filter. About 30 ml of filtered supernatants were then transferred into ultracentrifuge tubes containing 5 ml of 20% sucrose on a PBS cushion. Lentiviral particles were concentrated by centrifugation at 100,000 g for 2 h at 4°C and resuspended in 1× Hank's balanced salt solution (HBSS) buffer. MCF-10A or MDCKIIG cells were incubated overnight with lentiviral particles and 8 µg/ml polybrene (Sigma) in DMEM/F12 for MCF10A or low-glucose DMEM for MDCKIIG, which was replaced with DMEM/F12 supplemented with 5% horse serum, EGF (20 μg/ml), insulin (10 μg/ml), cholera toxin (0.1 μg/ml) hydrocortisone (0.5 μg/ml) and antibiotic-antimycotic (MCF10A complete medium), or low-glucose DMEM supplemented with 10% fetal bovine serum (MDCKIIG complete medium) the next morning. Antibiotics selection was started 2 days after transduction with 2 μg/ml puromycin and maintained on complete medium with 2 μg/ml puromycin for an additional 7 days. After selection, cells were isolated to establish clonal populations. Induction of the transgenes was performed with Dox 24 h before fixation or harvesting. Dox doses used for induction of EGFP-labeled transgenes for confocal imaging experiments in all figures shown for the different cell lines were: MCF10A EGFP:AJUBA, 0.025 µg/ml; MCF10A EGFP:LIMD1, 0.05 µg/ml; MCF10A EGFP:WTIP, 0.05 µg/ml; MCF10A EGFP:LATS1, 0.25 µg/ml; MCF10A EGFP:LATS2, 1.0 µg/ml; MDCKIIG EGFP:AJUBA, 0.01 µg/ml; MDCKIIG EGFP:LIMD1, 0.05 µg/ml; MDCKIIG EGFP:WTIP, 0.05 µg/ml; MDCKIIG EGFP:LATS1, 0.25 µg/ml; MDCKIIG EGFP:LATS2, 5 µg/ml.
Cell culture, transfections and treatments
MDCKIIG (a gift from W. James Nelson, Stanford University) cells were cultured in low-glucose Dubecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) and antibiotic-antimycotic, and MCF10A (a gift from Jay Debnath, UCSF, CA) were cultured in DMEM/F-12 (Life Technologies) supplemented with 5% horse serum, epidermal growth factor (20 µg/ml), insulin (10 µg/ml), cholera toxin (0.1 µg/ml), hydrocortisone (0.5 µg/ml) and antibiotic-antimycotic at 37°C and 5% CO2. Cell lines were obtained within the past 3 years, used at low passage number, and checked regularly for contamination by cell morphology and mycoplasma testing.
For immunostainings, cells were grown on coverslips coated with 0.6 mg/ml of collagen for 15 min at room temperature and washed with PBS. For the cytoskeletal inhibitor treatments, cells were grown at low density (15,000 cells/cm2) for 48 h and blebbistatin (50 µM) (Sigma) or Y-27632 (10 µM for MCF10A and 20 µM for MDCKIIG cells) (Cytoskeleton) were applied to the cells for 1 h. For cytoskeletal activator experiments, cells were grown at high density (150,000 cells/cm2) for 48 h and Rho activator II (1 µg/ml; Cytoskeleton) or lysophosphatidic acid (LPA; 1 µM; Sigma) was added for 2 or 1 h, respectively.
For siRNA delivery, cells were transfected with Lipofectamine RNAi Max (Life Technologies) according with the manufacturer's protocols, and fixed or harvested 48 h after transfection. Sequences (5′ to 3′) and sources of siRNAs used were: control NC1, CGUUAAUCGCGUAUAAUACGCGUAT (IDT) (Figs 2A; 5B–D; 8D–F; Figs S1C S6A–C); human AJUBA-1, GAGUACUUCUGAGCUAUUAUCAGCA (IDT) (Fig. S1C); human AJUBA-2, GGACUUCUCCAACCAAGUAUACUGT (IDT) (Figs S1B,C, S6B); human AJUBA-3, AUAUCCAUGGACCGGGAUUAUCACT (IDT) (Figs 5B–D; 8D–F; Fig. S1B,C; S6A,B);
human LIMD1-1, GCCAAGAAUCAGUCCUAAUUUCUTT (IDT) (Figs S1B,C, S6B); human LIMD1-2, AGCAUGGAUAAGUAUGACGACCUGG (IDT) (Figs 5B,C, 8D–F; Figs S1B,C, S6A,C); human LIMD1-3, GGUUUCAGGUGUGAUGUCCAAACCC (IDT) (Fig. S1C); human WTIP-1, CUACUUCGGCAUUUGCAUCAAGUGT (IDT) (Fig. S1C); human WTIP-2, AUCUACUGCGUGCGAGACUAUCACA (IDT) (Figs S1C, S6A,B); human WTIP-3, AAAAGGACAAGAUUUGACUUAAATT (IDT) (Figs S1C, S6B); human LATS1-1, UAGCAUGGAUUUCAGUAAUUU (Dharmacon) (Fig. S4D,F); human LATS1-2, CUAACAACAGAAGUAUAGAUU (Dharmacon) (Fig. S4D,F); human LATS2-1, GGUUCUCUAUAGGAACUACUU (Dharmacon) (Fig. S4D,F); human LATS2-2, UCAACGUGGACCUGUAUGAUU (Dharmacon) (Fig. S4D,F); human α-catenin-1, CGUGAACAUGCCAACAAAUUGAUTG (IDT) (Fig. 2A); human α-catenin-2, GCUAAGGCAGUAAUUUAGACUUUAC (IDT) (Fig. 2A); human α-catenin-3, CCAAUGUUCCACUUUUGGUAUUGAT (IDT) (Fig. 2A); canine LATS1-1, CGGCAGAUGUUGCAAGAAAUU (Dharmacon) (Fig. S4E); canine LATS1-2, UAGUAUGGAUUUCAGUAAUUU (Dharmacon) (Fig. S4E); canine LATS2-1, GGUUCUCUAUAGGAACUACUU (Dharmacon) (Fig. S4E); canine LATS2-2, UCAACGUGGACCUAUAUGAUU (Dharmacon) (Fig. S4E).
Immunostaining and imaging
Cells were fixed with 4% paraformaldehyde in PBS++ (phosphate-buffered saline supplemented with 100 mM MgCl2 and 50 mM CaCl2) for 10 min at room temperature. For the detection of VCL, α-catenin or a18, cells were fixed in 1% paraformaldehyde with 0.5% Triton X-100 in PBS++ for 3 min, rinsed and then fixed in 1% PFA in PBS++ for 10 min. Then cells were washed three times for 5 min each with 200 mM glycine containing PBS, followed by permeabilization with 0.5% Triton X-100 in PBS for 20 min.
After blocking with 5% bovine serum albumin (BSA) in PBS for 1 h, cells were incubated with primary antibody diluted in a 5% BSA in PBS solution overnight at 4°C. After washing with PBS, cells were incubated with Alexa Fluor 488- (Life Technologies), Cy3- or Alexa Fluor 647-conjugated (Jackson ImmunoResearch) secondary antibodies for 2 h and washed four times with PBS. Cell nuclei were counterstained with Hoechst 33342 (1 µg/ml; Invitrogen) and mounted with mounting medium (Dako). Antibodies used for immunostaining include mouse anti-Yap (1:100; Santa Cruz Biotechnology, sc-101199), rabbit anti-Yap (1:100, Cell Signaling Technology, #14074), rabbit anti-LATS1 (1:600; Cell Signaling Technology, #3477), rabbit anti-α-catenin (1:500; Sigma, C8114), mouse anti-vinculin (1:2000; Sigma, V9131), rat anti-a18 [1:50; a gift from Akira Nagafuchi (Nara Medical University, Japan)], mouse anti-ZO-1 (1:1000; Life Technology, #33-9100), mouse anti-phospho-myosin light chain (S19) (1:200; Cell Signaling Technology, #3675), rat anti-E-cadherin (1:500; Life Technology, #13-1900), rabbit anti-Ajuba (1:500; Cell Signaling Technology, #4897), rabbit anti-LIMD1 (1:500; Bethyl, A303-182A) and mouse anti-LIMD1 (1:500; EMD Millipore, MABD85). F-actin was stained with Alexa Fluor 647-conjugated phalloidin (1:50; Life Technologies). Images were acquired using LAS X software on a Leica TCS SP8 confocal microscope system using a HC PL APO 63×/1.40 objective. Images were processed by using ImageJ and Adobe Photoshop CS6, and figures were assembled in Adobe Illustrator CS6.
Proximity ligation assay
The PLA was performed with the Duolink® Proximity Ligation Assay kit according to the manufacturer's instructions (Sigma). Fixation and permeabilization steps were performed as in the normal immunostaining procedure. Antibodies used for PLA include mouse anti-GFP (1:250; Life Technologies, A11120), rabbit anti-LIMD1 (1:250; Bethyl, A303-182A) and rabbit anti-α-catenin (1:250; Sigma, C8114) antibodies and secondary anti-mouse MINUS and anti-rabbit PLUS probes were used. As a negative control for the PLA signal, only secondary antibodies were used and as a positive control, EGFP:LATS1 was recognized by using only GFP antibody and secondary anti-mouse MINUS and anti-mouse PLUS probes.
Immunoblotting
Cells were lysed in lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% CHAPS, 0.1% NP-40, 1 mM EDTA, 5% glycerol) or in 2× Laemmli Sample Buffer (Bio-Rad) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Calbiochem). Protein samples were loaded in 4% to 15% gradient gels (Bio-Rad). Antibodies used for immunoblotting include rabbit anti-LATS1 (1:1000; Cell Signaling Technology, #3477), rabbit anti-LATS2 (1:1000; Bethyl, A300-479A), rabbit anti-phospho-LATS1 (T1079) (1:1000; Cell Signaling Technology, #8654), rabbit anti-phospho-LATS1 (S909) (1:1000; Cell Signaling Technology, #9157), rabbit anti-phospho-Yap (S127) (1:1000; Cell Signaling Technology, #4911), rabbit anti-Yap (1:1000; Abcam, ab52771), mouse anti-LIMD1 (1:1000; EMD Millipore, MABD85), rabbit anti-Ajuba (1:1000; Cell Signaling Technology, #4897), goat anti-WTIP (1:1000; Santa Cruz Biotechnology, sc-241738), rabbit α-catenin (1:1000; Sigma, C8114), mouse anti-GFP (1:1000; Cell Signaling Technology, #2955) and rabbit anti-GFP (1:1000; Cell Signaling Technology, #2555). As loading controls, mouse anti-α-tubulin (1:10,000; Sigma, T6199), rabbit anti-GAPDH (1:5000; Santa Cruz Biotechnology, sc-25778) or mouse anti-GAPDH (1:10,000; Novus Biologicals, NBP2-27103) were used. Blots were visualized and quantified by means of staining with fluorescent-conjugated secondary antibodies (LI-COR Biosciences) and the Odyssey Imaging System (LI-COR Biosciences). All the blots from the same experiment were loaded equally and run on multiple gels in parallel, then blotted with the indicated antibodies and included loading controls to be able to make comparisons between the phosphorylated protein and total protein level.
Quantitative PCR protocol
Total RNA from MCF10A cells was extracted using Trizol reagent (Life Technologies) according to the manufacturer's instructions. Reverse transcription PCR (RT-PCR) was carried out on 2 µg of total RNA by using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer's instructions. First-strand cDNA was subjected to quantitative (q)PCR by using SYBR Select Master Mix and specific BIRC3, CTGF and GAPDH (as housekeeping gene) primers (sequences available upon request).
Statistical analysis
Statistical significance was determined with Graphpad Prism software by performing a paired two-tailed t-test for two sample comparisons or analysis of variance (ANOVA) for more than two samples, with P<0.05 set as the criteria for significance. The Tukey test was used to derive adjusted P-values for multiple comparisons. Error bars on figure panels show the s.e.m., except for the qPCR data, which show the 95% c.i.
Supplementary Material
Acknowledgements
We thank W. J. Nelson, J. Debnath, G. Sun, A. Nagafuchi, N. Yabuta and H. Nojima for reagents.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.I., K.D.I.; Methodology: C.I.; Formal analysis: C.I., K.D.I.; Investigation: C.I., E.K., B.K., E.E., K.F.; Resources: K.D.I.; Writing - original draft: C.I., K.D.I.; Writing - review & editing: C.I., K.D.I.; Supervision: C.I., K.D.I.; Project administration: K.D.I.; Funding acquisition: K.D.I.
Funding
This research was supported by National Institutes of Health (grant R01 GM121537 to K.D.I.) and a New Jersey Commission on Cancer Research postdoctoral fellowship (DHFS16PPC035 to C.I.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.214700.supplemental
References
- Abe Y., Ohsugi M., Haraguchi K., Fujimoto J. and Yamamoto T. (2006). LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 580, 782-788. 10.1016/j.febslet.2005.12.096 [DOI] [PubMed] [Google Scholar]
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S. and Piccolo S. (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047-1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Benham-Pyle B. W., Pruitt B. L. and Nelson W. J. (2015). Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024-1027. 10.1126/science.aaa4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codelia V. A., Sun G. and Irvine K. D. (2014). Regulation of YAP by mechanical strain through Jnk and hippo signaling. Curr. Biol. 24, 2012-2017. 10.1016/j.cub.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Fischer R. S., Pan D. and Waterman C. M. (2016). YAP nuclear localization in the absence of cell-cell contact is mediated by a filamentous actin-dependent, myosin II- and phospho-YAP-independent pathway during extracellular matrix mechanosensing. J. Biol. Chem. 291, 6096-6110. 10.1074/jbc.M115.708313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M., Feng Y., Jagannathan R., Seppa M. J., Skeath J. B. and Longmore G. D. (2010). Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20, 657-662. 10.1016/j.cub.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S. (2016). Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp. Cell Res. 343, 42-53. 10.1016/j.yexcr.2015.10.034 [DOI] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179-183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. and Hall A. (2002). Rho GTPases in cell biology. Nature 420, 629-635. 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Fernández B. G., Gaspar P., Brás-Pereira C., Jezowska B., Rebelo S. R. and Janody F. (2011). Actin-capping protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337-2346. 10.1242/dev.063545 [DOI] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Östman A. and Landegren U. (2002). Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473-477. 10.1038/nbt0502-473 [DOI] [PubMed] [Google Scholar]
- Gladden A. B., Hebert A. M., Schneeberger E. E. and McClatchey A. I. (2010). The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev. Cell 19, 727-739. 10.1016/j.devcel.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal R. K., Lin P., Kanungo J., Payne A. S., Muslin A. J. and Longmore G. D. (1999). Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol. Cell. Biol. 19, 4379-4389. 10.1128/MCB.19.6.4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K. F., Zhang X. and Thomas D. M. (2013). The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246-257. 10.1038/nrc3458 [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K. and Saya H. (2003). Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585-598. 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K. and Pan D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421-434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Irvine K. D. and Harvey K. F. (2015). Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harbor Perspect. Biol. 7, a019224 10.1101/cshperspect.a019224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan R., Schimizzi G. V., Zhang K., Loza A. J., Yabuta N., Nojima H. and Longmore G. D. (2016). AJUBA LIM proteins limit hippo activity in proliferating cells by sequestering the hippo core kinase complex in the cytosol. Mol. Cell. Biol. 36, 2526-2542. 10.1128/MCB.00136-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanungo J., Pratt S. J., Marie H. and Longmore G. D. (2000). Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell 11, 3299-3313. 10.1091/mbc.11.10.3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.-G. and Gumbiner B. M. (2015). Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 210, 503-515. 10.1083/jcb.201501025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. G., Koh E., Chen X. and Gumbiner B. M. (2011). E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA 108, 11930-11935. 10.1073/pnas.1103345108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Mukherjee A., Madhavan S. M., Konieczkowski M. and Sedor J. R. (2012). WT1-interacting protein (Wtip) regulates podocyte phenotype by cell-cell and cell-matrix contact reorganization. Am. J. Physiol. Renal. Physiol. 302, F103-F115. 10.1152/ajprenal.00419.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Duc Q., Shi Q., Blonk I., Sonnenberg A., Wang N., Leckband D. and de Rooij J. (2010). Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II–dependent manner. J. Cell Biol. 189, 1107-1115. 10.1083/jcb.201001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Cooper J., Zhou L., Yang C., Erdjument-Bromage H., Zagzag D., Snuderl M., Ladanyi M., Hanemann C. O., Zhou P. et al. (2014). Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell 26, 48-60. 10.1016/j.ccr.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Silvis M. R., Honaker Y., Lien W.-H., Arron S. T. and Vasioukhin V. (2016). αE-catenin inhibits a Src-YAP1 oncogenic module that couples tyrosine kinases and the effector of Hippo signaling pathway. Genes. Dev. 30, 798-811. 10.1101/gad.274951.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie H., Pratt S. J., Betson M., Epple H., Kittler J. T., Meek L., Moss S. J., Troyanovsky S., Attwell D., Longmore G. D. et al. (2003). The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J. Biol. Chem. 278, 1220-1228. 10.1074/jbc.M205391200 [DOI] [PubMed] [Google Scholar]
- Meng W. and Takeichi M. (2009). Adherens junction: molecular architecture and regulation. Cold Spring Harb. Perspect Biol. 1, a002899 10.1101/cshperspect.a002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Moroishi T. and Guan K.-L. (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1-17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meserve J. H. and Duronio R. J. (2015). Scalloped and Yorkie are required for cell cycle re-entry of quiescent cells after tissue damage. Development (Cambridge, England) 142, 2740-2751. 10.1242/dev.119339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll N., Mani M., Heemskerk I., Streichan S. and Shraiman B. I. (2017). Active tension network model of epithelial mechanics. Nat. Phys. (in press) 13, 1221-1226. 10.1038/nphys4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Heemskerk I., Ibar C., Shraiman B. I. and Irvine K. D. (2016). Differential growth triggers mechanical feedback that elevates Hippo signaling. Proc. Natl. Acad. Sci. USA 113, E6974-E6983. 10.1073/pnas.1615012113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivam M., Sarkeshik A., Yates J. R., Fernandes M. J. G. and McCollum D. (2011). Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell 22, 3725-3733. 10.1091/mbc.E11-04-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt S. J., Epple H., Ward M., Feng Y., Braga V. M. and Longmore G. D. (2005). The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J. Cell Biol. 168, 813-824. 10.1083/jcb.200406083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puliafito A., Hufnagel L., Neveu P., Streichan S., Sigal A., Fygenson D. K. and Shraiman B. I. (2012). Collective and single cell behavior in epithelial contact inhibition. Proc. Natl. Acad. Sci. USA 109, 739-744. 10.1073/pnas.1007809109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C., Sun S., Sun G., Pan Y. and Irvine K. D. (2014). Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143-156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B. V. and Irvine K. D. (2008). The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development 135, 2827-2838. 10.1242/dev.020974 [DOI] [PubMed] [Google Scholar]
- Reddy B. V. and Irvine K. D. (2013). Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell 24, 459-471. 10.1016/j.devcel.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L., Bossuyt W., Wada K.-I., Yonemura S., Tao C., Sasaki H. and Halder G. (2011). Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30, 2325-2335. 10.1038/emboj.2011.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H. B., Friedel C. C., Boulegue C. and Fässler R. (2011). Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12, 259-266. 10.1038/embor.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimizzi G. V. and Longmore G. D. (2015). Ajuba proteins. Curr. Biol. 25, R445-R446. 10.1016/j.cub.2015.02.034 [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R. et al. (2011). Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144, 782-795. 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G., Sehr P., Wilm M., Selzer J., Mann M. and Aktories K. (1997). Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387, 725-729. 10.1038/42735 [DOI] [PubMed] [Google Scholar]
- Serrano I., McDonald P. C., Lock F., Muller W. J. and Dedhar S. (2013). Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat. Commun. 4, 2976 10.1038/ncomms3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spendlove I., Al-Attar A., Watherstone O., Webb T. M., Ellis I. O., Longmore G. D. and Sharp T. V. (2008). Differential subcellular localisation of the tumour suppressor protein LIMD1 in breast cancer correlates with patient survival. Int. J. Cancer 123, 2247-2253. 10.1002/ijc.23851 [DOI] [PubMed] [Google Scholar]
- Srichai M. B., Konieczkowski M., Padiyar A., Konieczkowski D. J., Mukherjee A., Hayden P. S., Kamat S., El-Meanawy M. A., Khan S., Mundel P. et al. (2004). A WT1 co-regulator controls podocyte phenotype by shuttling between adhesion structures and nucleus. J. Biol. Chem. 279, 14398-14408. 10.1074/jbc.M314155200 [DOI] [PubMed] [Google Scholar]
- Su T., Ludwig M. Z., Xu J. and Fehon R. G. (2017). Kibra and merlin activate the hippo pathway spatially distinct from and independent of expanded. Dev. Cell 40, 478-490.e3. 10.1016/j.devcel.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. and Irvine K. D. (2011). Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev. Biol. 350, 139-151. 10.1016/j.ydbio.2010.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. and Irvine K. D. (2013). Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 6, ra81 10.1126/scisignal.2004324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. and Irvine K. D. (2016). Cellular organization and cytoskeletal regulation of the hippo signaling network. Trends Cell Biol. 26, 694-704. 10.1016/j.tcb.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Reddy B. V. V. G. and Irvine K. D. (2015). Localization of Hippo Signaling complexes and Warts activation in vivo. Nat. Commun. 6, 8402 10.1038/ncomms9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymaniak A. D., Mahoney J. E., Cardoso W. V. and Varelas X. (2015). Crumbs3-mediated polarity directs airway epithelial cell fate through the hippo pathway effector yap. Dev. Cell 34, 283-296. 10.1016/j.devcel.2015.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. (2014). Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15, 397-410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Wada K.-I., Itoga K., Okano T., Yonemura S. and Sasaki H. (2011). Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907-3914. 10.1242/dev.070987 [DOI] [PubMed] [Google Scholar]
- Yabuta N., Mukai S., Okada N., Aylon Y. and Nojima H. (2011). The tumor suppressor Lats2 is pivotal in Aurora A and Aurora B signaling during mitosis. Cell Cycle 10, 2724-2736. 10.4161/cc.10.16.16873 [DOI] [PubMed] [Google Scholar]
- Yi C., Troutman S., Fera D., Stemmer-Rachamimov A., Avila J. L., Christian N., Luna Persson N., Shimono A., Speicher D. W., Marmorstein R. et al. (2011). A tight junction-associated merlin-angiomotin complex mediates merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell 19, 527-540. 10.1016/j.ccr.2011.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Yu J., Zheng Y., Chen Q., Zhang N. and Pan D. (2013). Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342-1355. 10.1016/j.cell.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A. and Shibata M. (2010). alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533-542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Yu F. X., Zhao B., Panupinthu N., Jewell J. L., Lian I., Wang L. H., Zhao J., Yuan H., Tumaneng K., Li H. et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780-791. 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L. et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747-2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Wang L., Wang C.-Y., Yu J. and Guan K.-L. (2012). Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 26, 54-68. 10.1101/gad.173435.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.