Abstract
Divergent long noncoding RNAs (lncRNAs) represent a major lncRNA biotype in mouse and human genomes. The biological and molecular functions of the divergent lncRNAs remain largely unknown. Here, we show that lncKdm2b, a divergent lncRNA for Kdm2b gene, is conserved among five mammalian species and highly expressed in embryonic stem cells (ESCs) and early embryos. LncKdm2b knockout impairs ESC self‐renewal and causes early embryonic lethality. LncKdm2b can activate Zbtb3 by promoting the assembly and ATPase activity of Snf2‐related CREBBP activator protein (SRCAP) complex in trans. Zbtb3 potentiates the ESC self‐renewal in a Nanog‐dependent manner. Finally, Zbtb3 deficiency impairs the ESC self‐renewal and early embryonic development. Therefore, our findings reveal that lncRNAs may represent an additional layer of the regulation of ESC self‐renewal and early embryogenesis.
Keywords: embryogenesis, ESC pluripotency, LncKdm2b, SRCAP, Zbtb3
Subject Categories: Development & Differentiation, RNA Biology, Stem Cells
Introduction
In mammals, embryonic reprogramming, the acquisition and loss of totipotency, and the first cell fate decision all occur within a 3‐day window after fertilization from a one‐cell zygote to formation of the blastocyst. Upon fertilization, the zygote first undergoes a series of divisions, without overt increase in cell volume, resulting in the formation of the blastocyst (Burton & Torres‐Padilla, 2014; Wu & Izpisua Belmonte, 2015). By this point, the first differentiation event has appeared, segregating the outer trophectoderm, which is developmentally limited to extra‐embryonic tissues, from the inner cell mass (ICM), which encompasses pluripotent embryonic stem cells (ESCs) that will give rise to the embryo. Mouse and human pluripotent ESCs have been established to investigate the epigenetic reprogramming and self‐renewal maintenance during early embryonic development (Borsos & Torres‐Padilla, 2016). However, how the pluripotent ESCs sustain their self‐renewal and undergo differentiation still remains largely unknown.
Long noncoding RNAs (lncRNAs) are recently identified as transcripts longer than 200 nucleotides (nt) that lack protein‐coding potential (Batista & Chang, 2013). Genome‐scale deep‐sequencing and transcriptome analysis have identified tens of thousands of lncRNAs (Derrien et al, 2012). Accumulating evidence shows that lncRNAs function in a wide range of biological processes and can be modulate gene expression in cis or trans by various mechanisms (Djebali et al, 2012; Kretz et al, 2013; Yildirim et al, 2013; Wang et al, 2015). Divergent lncRNAs are transcribed in the opposite direction to nearby protein‐coding genes. It has been reported that Evx1as, a divergent lncRNA against EVX1 gene, promotes transcription of its nearby gene EVX1 to modulate mesendodermal differentiation (Luo et al, 2016). Another divergent lncRNA Uph against Hand2 gene suppresses the expression of its nearby gene Hand2 to regulate heart development (Anderson et al, 2016). We recently showed that lncKdm2b, a divergent lncRNA against Kdm2b gene, is highly expressed in intestinal group 3 innate lymphoid cells (ILC3s) and initiates Zfp292 expression to promote ILC3 proliferation (Liu et al, 2017). We also found that lncKdm2b is highly expressed in mouse embryos and lncKdm2b deletion causes early embryonic lethality (Liu et al, 2017). However, how lncKdm2b regulates the ESC self‐renewal and embryonic development has been not defined yet.
Zinc finger and BTB/POZ domain containing (ZBTB) proteins are an emerging family of transcription factors, commonly containing a DNA binding zinc finger and a transcription‐repressing BTB/POZ domain. These ZBTB members have important roles in development, differentiation, and oncogenesis (Lee & Maeda, 2012). In humans, over 40 members of the ZBTB family have been identified, some of which are closely linked to cancer development and progression. ZBTB4 and ZBTB7 are implicated in the development of breast cancer (Qu et al, 2010; Kim et al, 2012). ZBTB27, also known as BCL6, is involved in the oncogenesis of B‐cell lymphoma (Polo et al, 2004). ZBTB29, also called as HIC1, regulates cancer growth, angiogenesis, and tumor invasion (Janeckova et al, 2015). However, the physiological roles of many of the other ZBTB family members have not been defined yet. Here, we show that lncKdm2b is required for the self‐renewal maintenance of mouse and human ESCs. Mechanistically, lncKdm2b initiates Zbtb3 transcription in ESCs by promoting SRCAP activity in trans and Zbtb3 activates Nanog expression to maintain ESC self‐renewal.
Results
LncKdm2b is highly expressed in ESCs and early embryos
Pluripotent ESCs were established as an ideal system to study the self‐renewal maintenance and differentiation during cell fate switches. Withdrawal of leukemia inhibitory factor (LIF) causes ESCs to differentiate into three germ layers. In order to identify key lncRNAs in the ESC self‐renewal maintenance, we performed RNA‐seq analyses in mouse ESCs, embryoid body (EB), and ESCs differentiated by removal of LIF; 556 lncRNAs (183 upregulated and 373 downregulated) were differentially expressed in mouse ESCs versus EB and differentiated ESCs (Fig 1A). Among the top differentially expressed lncRNAs in mouse ESCs, we focused on lncKdm2b (gene symbol A930024E05Rik) that was a divergent lncRNA for its nearby coding gene Kdm2b (Appendix Fig S1A). We previously demonstrated that lncKdm2b is highly expressed in mouse embryos and is involved in the regulation of embryonic development (Liu et al, 2017). We verified that lncKdm2b was actually highly expressed in mouse ESCs and early embryos (Fig 1B and C). High expression of lncKdm2b in mouse ESCs and early embryo cells was further validated by RNA fluorescence in situ hybridization (RNA‐FISH) (Fig 1D and Appendix Fig S1B). In addition, we detected about 800 copies of lncKdm2b transcripts per mouse ESC cell (Appendix Fig S1C). Moreover, lncKdm2b was transcribed with one transcript in mouse ESCs with 1,892 nt long identified by 5′‐RACE and 3′‐RACE (Appendix Fig S1A and D). Of note, lncKdm2b was distributed in the nucleus and cytoplasm of mouse ESCs through cellular fractionation assays (Appendix Fig S1E). Finally, lncKdm2b did not produce any detectable peptides by in vitro translation assays (Appendix Fig S1F–H).
Figure 1. LncKdm2b is highly expressed in ESCs and early embryos.
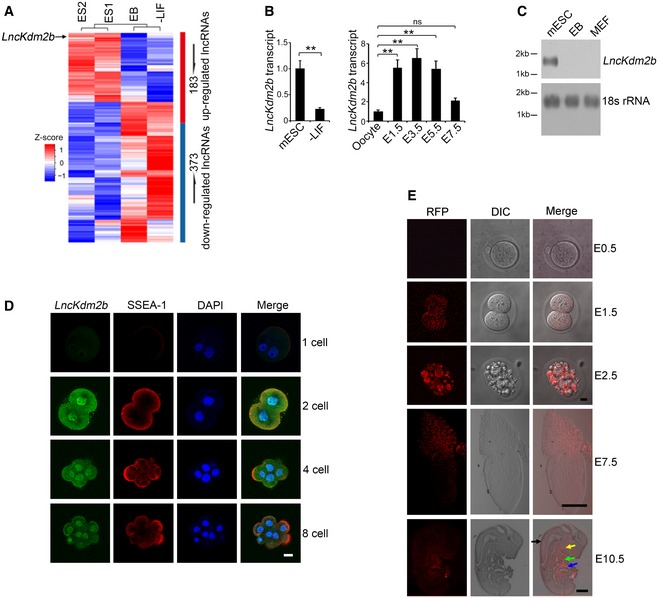
- Differentially expressed lncRNAs by RNA‐seq analyses of mouse ESCs versus embryoid body (EB) (culturing with LIF withdrawal for 5 days) and differentiated ESCs with LIF withdrawal (LIF withdrawal with differentiation for 10 days). ES1 and ES2 indicate two biological replicates of ESC samples in RNA‐seq assay.
- LncKdm2b transcript was analyzed in ESCs and embryo cells by real‐time qPCR. Primers are listed in Appendix Table S1. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. **P = 0.0059 (left panel), **P = 0.0091, **P = 0.0087, **P = 0.0081, ns (no significance), P = 0.0617 (right panel) by unpaired Student's t‐test. Mouse ESC line D3 cells were treated with LIF withdrawal (−LIF) for 3 days. Mice were stimulated for superovulation and naturally mated. Embryos were isolated in the indicated embryonic days.
- LncKdm2b expression in mouse ESCs and differentiated ESCs were examined by northern blot. A 277‐nt probe of lncKdm2b (81–358 nt) was labeled for northern blot analysis. RNAs were extracted from indicated cells; 18S RNA was used as a loading control. EB, embryoid body; MEF, mouse embryonic fibroblast.
- LncKdm2b was visualized in the indicated embryo stages by RNA‐FISH assays followed with immunofluorescence staining. Green: lncKdm2b probe, Red: SSEA‐1, nuclei were counterstained by DAPI. Scale bar, 10 μm. Sequences of probes are listed in Appendix Table S1. For one‐cell stage embryos, n = 51; for two‐cell stage embryos, n = 66; for four‐cell stage embryos, n = 56; and for eight‐cell stage embryos, n = 69.
- Endogenous lncKdm2b expression with RFP‐reporter fluorescence was visualized by live cell imaging (E0.5–E7.5) or fixed sagittal section imaging (E10.5). Scale bar, 10 μm for E0.5‐ to E2.5‐stage embryos, 100 μm for E7.5‐stage embryo, and 2 mm for E10.5‐stage embryo. Black arrow: cranial/spinal accessory nerve; yellow arrow: first branchial arch; green arrow: heart; blue arrow: liver. For E0.5‐stage embryos, n = 105; for E1.5‐stage embryos, n = 112; for E2.5‐stage embryos, n = 109; for E7.5‐stage embryos, n = 115; and for E10.5‐stage embryos, n = 101.
We noticed that lncKdm2b transcript was highly conserved among five mammalian species we analyzed, sharing ~69% RNA sequence homology with human lncKDM2B (Appendix Fig S1A and I). Surely, human lncKDM2B on chromosome 12 was also transcribed bidirectionally against its neighbor gene KDM2B as a divergent lncRNA (Appendix Fig S1I). Furthermore, human lncKDM2B was also highly expressed in human ESCs (Appendix Fig S1J and K). Using lncKdm2b RFP‐reporter mice (Liu et al, 2017), we observed that RFP signals appeared from the two‐cell stage of embryos and were elevated during early embryonic development (Fig 1E). At embryonic day 10.5 (E10.5), lncKdm2b was restrictedly expressed in the cranial/spinal accessory nerve, first branchial arch, heart, and liver (Fig 1E). The specificity of RFP fluorescence was confirmed with anti‐RFP antibody staining (Appendix Fig S1L). Altogether, we reveal that lncKdm2b is highly expressed in the pluripotent ESCs and early embryos.
LncKdm2b knockout impairs ESC self‐renewal and causes early embryonic lethality
To explore the physiological role of lncKdm2b in the regulation of ESCs, we generated lncKdm2b knockout (KO) mice by deletion of its exon 2 via CRISPR/Cas9 technology (Yin et al, 2015; Liu et al, 2017). Given that deletion of exon 2 led to a drastic change of lncKdm2b structure predicted by RNA folding analysis (Appendix Fig S2A), we thus deleted the exon 2 as our KO strategy. Deletion of 838‐bp fragment containing the exon 2 was confirmed by genotyping and real‐time qPCR in blastocyst‐derived ESCs (Fig 2A, and Appendix Fig S2B and C). As expected, lncKdm2b was completely deleted in lncKdm2b KO blastocyst‐derived ESCs (Fig 2A). At E3.5, lncKdm2b deficiency impaired blastocyst development, leading to early embryonic lethality (Liu et al, 2017; Fig 2B). Consistently, lncKdm2b KO mice did not produce homozygous pups after heterozygotes crossing (Fig 2C and Appendix Fig S2D). In addition, lncKdm2b‐deficient blastocysts formed decreased numbers of ICM‐derived ESC colonies with abnormal morphological features compared with WT blastocysts (Fig 2D and Appendix Fig S2E). Of note, we used serum‐free 2i culture condition and demonstrated that lncKdm2b KO impaired self‐renewal of ESCs (Appendix Fig S2F). We also deleted the exon 2 of lncKdm2b in mouse ESC cell lines and tested their self‐renewal capacities. Of note, lncKdm2b‐deficient ESCs drastically reduced expression levels of pluripotency genes (Fig 2E). Moreover, lncKdm2b deficiency failed to form three embryonic layers via an in vivo teratoma formation assay (Fig 2F).
Figure 2. LncKdm2b knockout impairs ESC self‐renewal and causes early embryonic lethality.
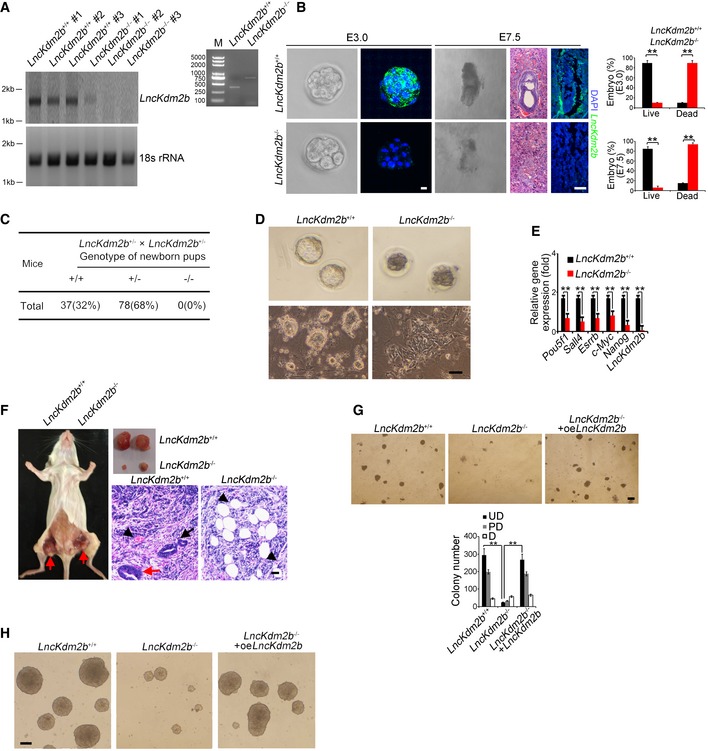
- LncKdm2b knockout mice were generated by CRISPR/Cas9 system. Blastocysts from lncKdm2b +/+ and lncKdm2b −/− were cultured in mouse ESC media. ESC clones were picked up and further cultured, followed by northern blot as in Fig 1C and genotyping.
- LncKdm2b‐deficient embryos were isolated at E3.0 and E7.5 stages. LncKdm2b transcript in embryos was examined by RNA‐FISH assays. Sequences of probes are listed in Appendix Table S1. Scale bar, 10 μm (left panel) and 50 μm (right panel). Percentages of live embryos and dead embryos were counted as mean ± SD (right panel). **P = 0.0055, **P = 0.0067 (upper panel), **P = 0.0046, **P = 0.0057 (lower panel) by unpaired Student's t‐test. For lncKdm2b +/+ E3.0 embryos, n = 125. For lncKdm2b +/+ E7.5 embryos, n = 101. For lncKdm2b −/− E3.0 embryos, n = 108. For lncKdm2b −/− E7.5 embryos, n = 102. Green, lncKdm2b; blue, DAPI.
- LncKdm2b‐deficient pups were genotyped after heterozygotes crossing.
- Blastocysts from lncKdm2b +/+ and lncKdm2b −/− mice were isolated and visualized as upper panel. Blastocysts were continuously cultured in mouse ESC media till embryonic stem cell clones formed as in (A) (lower panel). Scale bar, 50 μm.
- LncKdm2b knockout mouse ESC cell lines were generated by CRISPR/Cas9 system, followed by detection of mRNA levels of indicated genes with real‐time qPCR. Primers were listed in Appendix Table S1. Relative gene expression fold changes were normalized to endogenous β‐actin and counted as means ± SD. **P = 0.0058, **P = 0.0012, **P = 0.0049, **P = 0.0019, **P = 0.0032, **P = 0.0002 by unpaired Student's t‐test.
- LncKdm2b +/+ or lncKdm2b −/− ESCs (1 × 106 cells per point) were subcutaneously injected into NOD/SCID mice. One month later, teratomas were collected for H&E staining. Red arrow denotes teratomas in the left panel. Red arrow: endoderm; black arrow: ectoderm; black arrowhead: mesoderm for the right panels. Scale bar, 50 μm. n = 5 mice per group.
- ES cells were dissociated and plated at 4.5 × 104 per well of 24‐well plates and were infected by lncKdm2b overexpression (oe) lentivirus for 5 days, followed by fixation and AP staining. Colony numbers for undifferentiated (UD), partially differentiated (PD), or differentiated (D) clones were calculated as means ± SD. **P = 0.0004, **P = 0.0003 by unpaired Student's t‐test. Scale bar, 50 μm.
- LncKdm2b KO ESC cells were infected by lncKdm2b overexpression (oe) lentivirus followed by induction of EB differentiation. Morphology of EB bodies was imaged. Scale bar, 100 μm.
Pluripotent ESCs can be induced to generate EB containing three embryonic layers. We next tested the capacity for EB differentiation in lncKdm2b deleted mouse ESC cell lines. We observed that lncKdm2b deletion remarkably inhibited expression of multiple‐layer genes (Appendix Fig S2G). Notably, restoration of lncKdm2b in lncKdm2b −/− ESCs was able to rescue ESC self‐renewal (Fig 2G, and Appendix Fig S2H and I) and restored the capacity of EB formation (Fig 2H). We next depleted lncKdm2b expression with shRNA‐mediated knockdown approaches. Expectedly, lncKdm2b‐depleted mouse D3 ESC lines exhibited reduced ESC colony numbers (Appendix Fig S2J and K).
To exclude the possibility that the deleted exon 2 contains functional enhancers, we used a published dataset GSE98063 to analyze modifications such as H3K27ac (marking transcriptional active and enhancer regions) and H3K4me1 (marking active enhancer) on the deleted 838‐bp region. We noticed that H3K4me1 displayed no peaks, while H3K27ac had some peaks in this region (Appendix Fig S3A). However, we observed that the 838‐bp region did not promote GFP transcription through an enhancer assay (Appendix Fig S3B). This result was further validated by an enhancer‐reporter assay (Appendix Fig S3C). These results suggest that the deleted 838‐bp region does not contain functional enhancer potential. Finally, we generated polyA insertion ESC lines and noticed that transcription of lncKdm2b was disrupted in polyA insertion ESC lines (Appendix Fig S3D). PolyA insertion ESC lines displayed a similar phenotype to that of exon 2‐deletion ESCs (Appendix Fig S3E) and inhibited expression of pluripotency factors as well (Appendix Fig S3F). These data indicate that the deletion of exon 2 successfully deletes the lncKdm2b transcript, but does not affect the cis regulatory motifs. In parallel, lncKDM2B silencing in human ESC H1 cell lines also disrupted the self‐renewal of ESCs (Appendix Fig S3G and H). Collectively, we conclude that lncKdm2b is required for the ESC self‐renewal maintenance and early embryonic development.
LncKdm2b associates with SRCAP to promote its ATPase activity
Divergent lncRNAs are often positively correlated with regulation of their nearby protein‐coding genes (Anderson et al, 2016; Luo et al, 2016). We previously showed that deletion of lncKdm2b does not affect the expression level of Kdm2b in ILC3s (Liu et al, 2017). Similarly, lncKdm2b KO did not affect the expression levels of Kdm2b and its neighboring genes in mouse ESCs (Appendix Fig S4A and B). Therefore, in ESCs lncKdm2b also exerts its regulatory functions in trans as well. We then performed a biotin‐labeled RNA pull‐down assay to identify potential lncKdm2b‐associated proteins from mouse ESC lysates. SRCAP (also called as SWR1 in Yeast), the core subunit of SRCAP remodeling complex, was identified to bind lncKdm2b in ESCs (Fig 3A, and Appendix Fig S4C and D). The interaction of lncKdm2b with SRCAP was further validated by immunoblotting and CHIRP assays (Fig 3B and C). Moreover, lncKdm2b was co‐localized with SRCAP in the nuclei of ESCs and E3.5 embryos (Fig 3D). The interaction of SRCAP with lncKdm2b was further confirmed by RNA immunoprecipitation (RIP) (Fig 3E). Of note, the fragment (nt 450–700) of lncKdm2b was necessary to bind SRCAP protein through fragment mapping analysis (Fig 3F and Appendix Fig S4E). This region of lncKdm2b is also required to associate with Satb1 protein (Liu et al, 2017). Additionally, this region (nt 450–700) harbored a stable stem‐loop structure predicted by RNA folding analysis (Appendix Fig S4F). Of note, we found that Satb1 was not expressed in mouse ESCs (Appendix Fig S4G), suggesting that the interaction of lncKdm2b with Satb1 does not take place in mouse ESCs. The binding of lncKdm2b with SRCAP was further validated by RNA electrophoretic mobility shift assay (RNA‐EMSA) (Fig 3G). These results indicate that lncKdm2b directly binds to the SRCAP protein in mouse ESCs.
Figure 3. LncKdm2b promotes the ATPase activity of SRCAP.
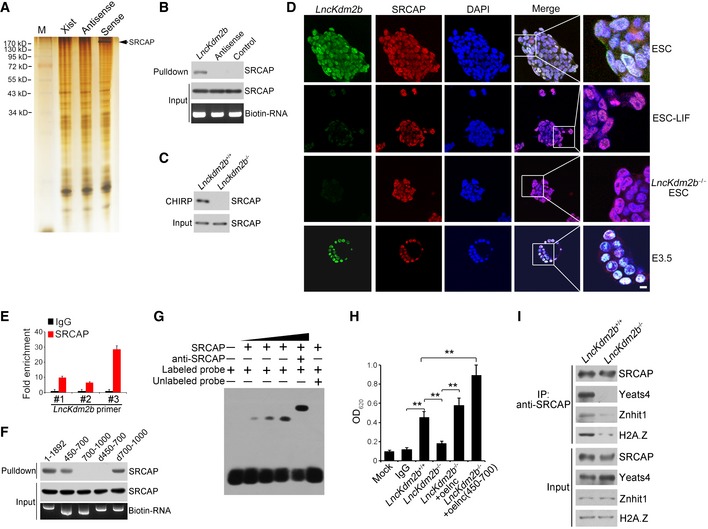
-
ABiotin RNA pull‐downs were performed with nuclear extracts of mouse ESCs using full‐length lncKdm2b transcript (Sense), antisense, and Xist A repeats sequence control followed with mass spectrometry.
-
B, CThe interaction of SRCAP with lncKdm2b was confirmed by immunoblotting (B) and CHIRP assay (C). Biotinylated probes were hybridized to lncKdm2b, and chromatin complex was purified by magnetic streptavidin beads, followed by elution and immunoblotting. CHIRP probe sequences are listed in Appendix Table S1.
-
DPluripotent ESCs and E3.5 blastocysts were probed with lncKdm2b by RNA‐FISH, followed by immunofluorescence staining for SRCAP. Green: lncKdm2b probe; red: SRCAP; nuclei were counterstained by DAPI. Scale bar, 10 μm. For normal ESC clone, n = 240; for LIF‐withdrawal ESC clone, n = 130; for lncKdm2b −/− ESC clone, n = 105; for E3.5 embryos, n = 119.
-
EInteraction of lncKdm2b with SRCAP was verified by RIP assay. ESC lysates were incubated with anti‐SRCAP antibody, followed by RNA immunoprecipitation (RIP) assay. RNA was extracted and reversely transcribed. LncKdm2b transcript was analyzed by real‐time qPCR. Fold changes were shown as means ± SD. Primers are listed in Appendix Table S1.
-
FFull‐length and truncated fragments of lncKdm2b were in vitro‐transcribed to biotin‐labeled RNA followed with RNA pull‐down and immunoblotting. d450–700 denotes the truncated fragment of lncKdm2b deleting nt 450–700. d700–1,000 denotes the truncated fragment of lncKdm2b deleting nt 700–1,000.
-
GNuclear extracts of ESCs and biotin‐labeled lncKdm2b (450–700 nt) probes were incubated for EMSA assays. Anti‐SRCAP antibody was preincubated with nuclear extracts that caused supershift.
-
HESC lysates were immunoprecipitated with anti‐SRCAP antibody, followed by detection of ATPase activities. Biotin‐labeled lncKdm2b and lncKdm2b (nt 450–700) fragments were generated by in vitro transcription by T7 RNA polymerase. Mouse IgG IP was used as a background control. Relative OD values were normalized to IgG background control and shown as fold changes as means ± SD. lnc, lncKdm2b; oe, overexpression. **P = 0.0034, **P = 0.0089, **P = 0.0023, **P = 0.0011 by unpaired Student's t‐test.
-
IlncKdm2b +/+ and lncKdm2b −/− ESC cell lysates were immunoprecipitated with anti‐SRCAP antibody, followed by immunoblotting with the indicated antibodies.
The SRCAP protein is a major subunit of the SRCAP remodeling complex, which confers its ATPase activity (Kobor et al, 2004). We observed that lncKdm2b deletion in ESCs remarkably suppressed the ATPase activity of SRCAP subunit (Fig 3H), whereas lncKdm2b overexpression in lncKdm2b −/− ESCs could restore the ATPase activity of SRCAP protein (Fig 3H). Notably, overexpression of lncKdm2b RNA and the binding fragment (nt 450–700) could enhance the activity of SRCAP complex (Fig 3H). More importantly, lncKdm2b deletion abrogated the assembly of the SRCAP complex in ESCs as well as its ATPase activity (Fig 3I), but does not affect the NuRD complex integrity (Appendix Fig S4H). Finally, we purified the SRCAP complex with anti‐SRCAP antibody from ESC lysates and treated with RNAse, followed by size fractionation assay. We found that RNAse treatment impaired the SRCAP complex integrity (Appendix Fig S4I). Taken together, lncKdm2b interacts with SRCAP to modulate the assembly and activity of SRCAP‐contained remodeling complex in ESCs.
LncKdm2b activates Zbtb3 transcription in ESCs
We next performed transcriptome microarray analysis between lncKdm2b −/− ESCs and WT ESCs. Totally, 11,302 genes (5,933 upregulated and 5,369 downregulated, fold change > 2.0, FDR < 0.05) were differentially expressed in WT ESCs versus lncKdm2b −/− ESCs. Gene ontology (GO) enrichment analysis demonstrated that transcription‐related factors were mainly differentially expressed genes in lncKdm2b −/− ESCs (Appendix Fig S5A). Among the top 10 downregulated transcription factors (TFs) in lncKdm2b −/− ESCs (Fig 4A), Pbx3, Zim1, Taf1c, Tcf4, and Nr2c1 have been defined to participate in the ESC self‐renewal maintenance (Ibrahim et al, 2012; Baker et al, 2016). However, the other 5 TFs were uncharacterized in the regulation of ESC self‐renewal. In order to determine their roles in the ESC self‐renewal maintenance, we depleted these genes in mouse ESC line D3 cells by transducing lentivirus‐mediated shRNAs. We observed that Zbtb3 depletion significantly decreased ESC colony numbers (Appendix Fig S5B). Pbx3 depletion was used as a positive control (Appendix Fig S5B). Intriguingly, of the top 10 downregulated TFs, Zbtb3 was the most highly expressed TF in mouse ESCs (Appendix Fig S5C). We carried out chromatin immunoprecipitation (ChIP) from ESC lysates with anti‐SRCAP antibody. We found that SRCAP was enriched at the transcription start site (TSS) region of Zbtb3 promoter in ESCs (Fig 4B). Indeed, lncKdm2b KO abolished the expression of Zbtb3 in early embryo cells (Fig 4C).
Figure 4. LncKdm2b activates Zbtb3 transcription.
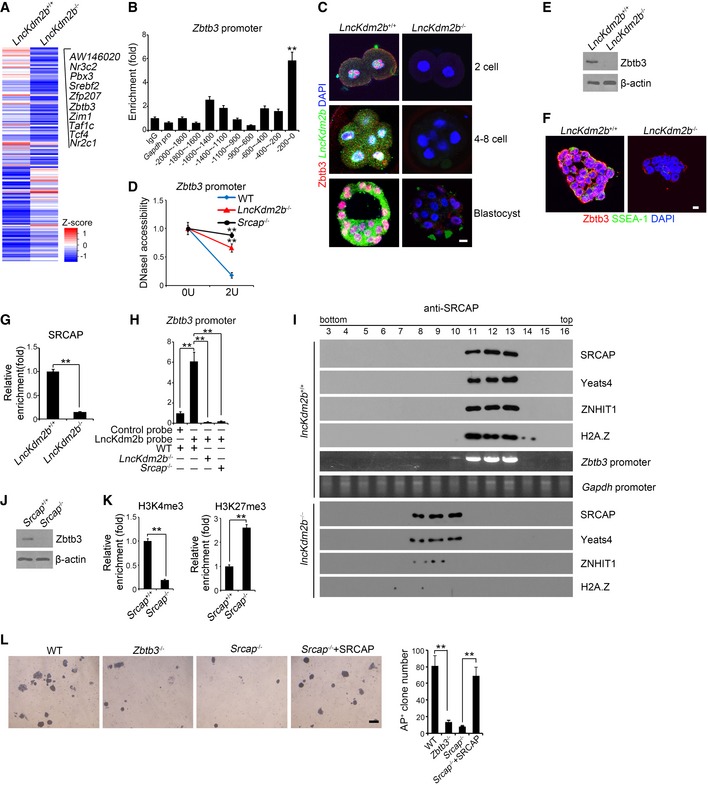
- Heat map of transcriptome microarray analysis between lncKdm2b −/− ESCs and WT ESCs. Top 10 downregulated TFs in lncKdm2b −/− ESCs were listed.
- SRCAP was recruited near the transcription start site (TSS) region of Zbtb3 promoter in ESCs. ESCs were lysed for ChIP assays by anti‐SRCAP antibody. SRCAP protein enriched to Zbtb3 promoter was examined by real‐time qPCR. Signals were normalized to input DNA. Primers are listed in Appendix Table S1. IgG and SRCAP proteins enriched on Gapdh promoter were used as negative controls. Data are from three independent lncKdm2b −/− clones. Relative fold changes were counted as means ± SD. **P = 0.0001 by unpaired Student's t‐test.
- Zbtb3 expression was visualized in indicated embryo stages by immunofluorescence staining. Green: lncKdm2b probe; red: Zbtb3; nuclei were counterstained by DAPI. Scale bar, 10 μm. For lncKdm2b +/+ one‐cell stage embryos, n = 111. For lncKdm2b +/+ four‐ to eight‐cell stage embryos, n = 128. For lncKdm2b +/+ blastocysts, n = 101. For lncKdm2b −/− one‐cell stage embryos, n = 104. For lncKdm2b −/− four‐ to eight‐cell stage embryos, n = 110. For lncKdm2b −/− blastocysts, n = 101.
- Nuclei were extracted from indicated ESCs followed by DNase I digestion. Total DNA was extracted and quantitated by qPCR with Zbtb3 promoter‐specific primers. Primers are listed in Appendix Table S1. Data are from three independent lncKdm2b −/− clones. Relative fold changes were counted as means ± SD. **P = 0.0012, **P = 0.0026 by unpaired Student's t‐test.
- Zbtb3 expression was analyzed by Western blotting in lncKdm2b +/+ and lncKdm2b −/− ESCs.
- Zbtb3 expression was analyzed in lncKdm2b +/+ and lncKdm2b −/− ESCs by immunofluorescence staining. For lncKdm2b +/+ ESC clone, n = 180. For lncKdm2b −/− ESC clone, n = 121. Scale bar, 10 μm.
- LncKdm2b +/+ and lncKdm2b −/− ESCs were lysed for ChIP assays. SRCAP protein enriched to Zbtb3 promoter was examined by real‐time qPCR. Signals were normalized to input DNA. Relative fold changes were counted as means ± SD. **P = 0.0005 by unpaired Student's t‐test.
- Interaction of lncKdm2b with Zbtb3 promoter was analyzed by CHIRP assay as in Fig 3C. Relative fold changes were counted as means ± SD. **P = 0.0006, **P = 0.0001, **P = 0.0001 by unpaired Student's t‐test.
- Indicated ESCs were lysed and treated with 1% formaldehyde for cross‐linking and sonicated. Then anti‐SRCAP antibody was incubated with treated lysates for ChIP assays, followed by size fractionation with sucrose gradient ultracentrifugation. Eluate gradients were examined by Western blot and PCR assays. Pro: promoter.
- Zbtb3 expression was tested in Srcap +/+ and Srcap −/− ESCs by Western blotting.
- ESCs were lysed for ChIP assays. Indicated proteins enriched to Zbtb3 promoter were examined by real‐time qPCR. Signals were normalized to input DNA. Enrichment fold changes were counted as means ± SD. **P = 0.0024, **P = 0.0052 by unpaired Student's t‐test.
- Indicated ESC cells were cultured for AP staining as in Fig 2G. AP‐positive ES clone numbers were counted as means ± SD. **P = 0.0048, **P = 0.0052 by unpaired Student's t‐test. Scale bar, 50 μm.
We noticed that lncKdm2b deficiency in mouse ESCs dramatically repressed the expression of Zbtb3 (Fig 4D–F). Of note, lncKdm2b deficiency almost abolished recruitment of SRCAP to the promoter region of Zbtb3 gene (Fig 4G). In addition, SRCAP deletion abrogated the binding of lncKdm2b to Zbtb3 promoter (Fig 4H). Importantly, lncKdm2b deficiency completely lost the recruitment of SRCAP complex to Zbtb3 promoter, whereas the SRCAP complex bound Zbtb3 promoter in lncKdm2b +/+ ESCs (Fig 4I). These data suggest that lncKdm2b is required for the binding of SRCAP complex to Zbtb3 promoter.
We next deleted Srcap and Zbtb3 in mouse ESC cell lines via CRISPR/Cas9 approaches (Appendix Fig S5D and E) and established their respective Srcap −/− and Zbtb3 −/− ESCs. We observed that SRCAP deletion failed to activate Zbtb3 transcription (Fig 4J and K). Notably, deletion of Zbtb3 or SRCAP impaired ESC self‐renewal (Fig 4L). By contrast, restoration of SRCAP in Srcap −/− ESCs could sustain ESC self‐renewal (Fig 4L). Therefore, these data suggest that SRCAP activates Zbtb3 expression in a lncKdm2b‐dependent manner.
Zbtb3 initiates Nanog expression to potentiate ESC self‐renewal
To further explore the mechanism by which Zbtb3 regulated ESC self‐renewal, we compared expression levels of major pluripotency factors in lncKdm2b −/−, Srcap −/−, and Zbtb3 −/− ESCs versus WT ESCs. We found that several master pluripotency factors, including Pou5f1, Nanog, Sox2, c‐Myc, and Klf4, were downregulated in the three ESC lines with deletion of lncKdm2b, Srcap, and Zbtb3 genes (Fig 5A). Of note, Zbtb3 was only enriched onto Nanog promoter among these master pluripotency factors (Fig 5B and C). However, neither SRCAP nor lncKdm2b bound the Nanog promoter, suggesting that Nanog is a direct target gene of Zbtb3 (Appendix Fig S5F and G). Of note, lncKdm2b restoration in lncKdm2b KO or silenced ESCs could restore Zbtb3 and Nanog expression to its WT levels (Appendix Fig S5H). Deletion of lncKdm2b, SRCAP, or Zbtb3 surely suppressed the expression of Nanog in ESCs (Fig 5D and E). By contrast, restoration of Zbtb3 in lncKdm2b −/−, Srcap −/−, and Zbtb3 −/− ESCs augmented expression of Nanog gene (Fig 5D and E), indicating the Nanog gene is a downstream target for Zbtb3.
Figure 5. Zbtb3 initiates Nanog expression to potentiate ESC self‐renewal.
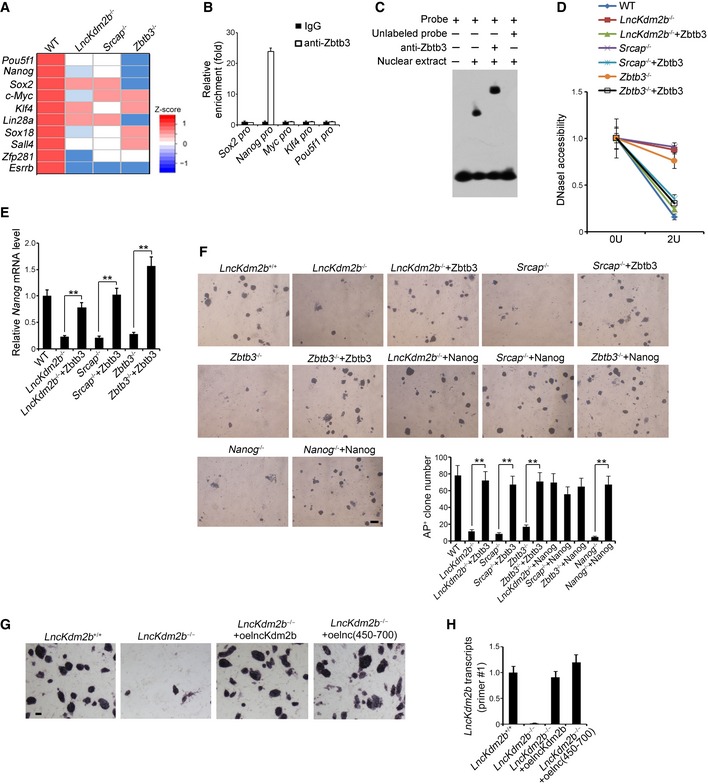
- mRNAs levels of 10 major pluripotency TFs were analyzed by real‐time qPCR. Relative gene expression folds were normalized to endogenous β‐actin. Primers are listed in Appendix Table S1.
- Zbtb3 was highly enriched on Nanog promoter compared with other pluripotent factors by ChIP assays. Enrichment fold changes were counted as means ± SD.
- Zbtb3 protein was incubated with labeled and unlabeled probes against Nanog promoter, followed by EMSA assays. Anti‐Zbtb3 antibody was preincubated that caused supershift. Sequence of Nanog promoter probe is listed in Appendix Table S1.
- Nuclei were extracted from indicated ESCs followed by DNase I digestion. Total DNA was extracted and quantitated by qPCR with Zbtb3 promoter‐specific primers. Relative fold changes were counted as means ± SD.
- ESC cells were infected with the indicated lentivirus, sand Nanog expression was assessed by qPCR. Relative gene expression fold changes were counted as means ± S .D. **P = 0.0027, **P = 0.0018, **P = 0.0012 by unpaired Student's t‐test.
- Indicated KO ESCs were generated by CRISPR/Cas9 technology and ESC cells were infected with the indicated lentivirus, followed by AP staining as in Fig 2G. AP‐positive ES clone numbers were counted as means ± SD. **P = 0.0025, **P = 0.0033, **P = 0.0019, **P = 0.0022 by unpaired Student's t‐test. Scale bar, 100 μm.
- SRCAP‐interacting region (nt 450–700) of lncKdm2b was cloned into pSicoR plasmid and infected lncKdm2b −/− ESCs for rescue assay. ESCs were stained with AP substrate after 5 days. Scale bar, 20 μm.
- Expression levels of lncKdm2b fragment (nt 450–700) in rescued lncKdm2b KO ESCs were tested by real‐time PCR. Primer#1 against the region nt 518–697 of lncKdm2b was used for probing the fragment (nt 450–700). Relative gene expression fold changes were normalized to endogenous β‐actin and counted as means ± SD.
Of note, lncKdm2b, Srcap, Zbtb3, or Nanog deleted ESC D3 cells impaired ESC self‐renewal (Fig 5F). In addition, restoration of either Zbtb3 or Nanog in lncKdm2b −/−, Srcap −/−, Zbtb3 −/−, or Nanog −/− ESCs could well rescue the self‐renewal capacity of ESC D3 cells (Fig 5F). Moreover, rescue of the SRCAP‐interacting region (nt 450–700) could restore the lost stemness phenotype in lncKdm2b KO ESCs akin to rescue of full‐length lncKdm2b (Fig 5G and H). Collectively, lncKdm2b maintains the ESC self‐renewal by initiating Zbtb3 expression in a Nanog‐dependent manner.
Zbtb3 deficiency abolishes ESC self‐renewal and early embryonic development
To further determine the physiological role of Zbtb3 in embryogenesis, we generated Zbtb3 KO mice via the same CRISPR/Cas9 approach. Zbtb3 heterozygotes crossing failed to generate homozygous pups (Fig 6A). At E3.5 stages, we isolated WT and Zbtb3 −/− embryos and stained with anti‐Zbtb3 antibody. We found that Zbtb3 deletion disrupted blastocyst development (Fig 6B). We then isolated fertilized eggs from WT and Zbtb3 +/− pregnant mice to assess ex vivo development of embryos. Zbtb3 deficiency could not form blastocyst in vitro (Fig 6C), which was reminiscent of lncKdm2b −/− mice. In addition, Zbtb3 deficiency also suppressed expression of pluripotency genes in ESCs (Fig 6D). Of note, ZBTB3 was also highly expressed in undifferentiated human ESCs based on transcriptome dataset (GSE23034). Consistently, depletion of ZBTB3 in human ESC H1 cells also abolished human ESC self‐renewal and inhibited expression of pluripotency factors (Fig 6E–G), suggesting ZBTB3 also exerts a pivotal role in human ESC self‐renewal maintenance. Altogether, Zbtb3 plays a critical role in ESC self‐renewal maintenance and early embryogenesis.
Figure 6. Zbtb3 deletion abolishes ESC self‐renewal and early embryonic development.
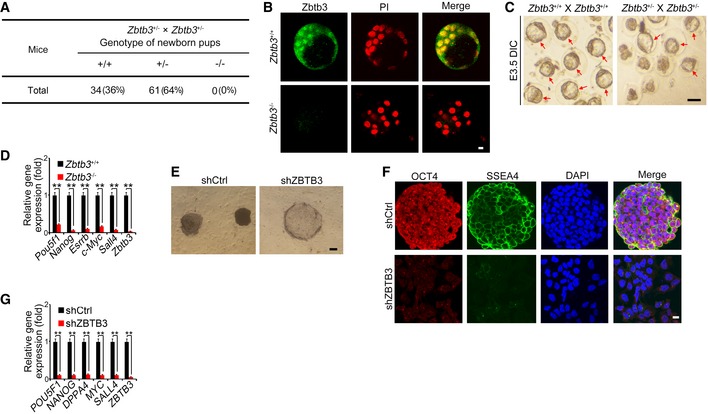
-
AZbtb3 knockout mice were generated by CRISPR/Cas9 technology. Zbtb3‐deficient pups were genotyped after heterozygotes crossing.
-
BE3.5 embryos were isolated and stained with anti‐Zbtb3 antibody and PI. Scale bar, 10 μm. For Zbtb3 +/+ embryos, n = 106. For Zbtb3 −/− embryos, n = 101.
-
CEmbryos were isolated at E3.0 stage and cultured in KSOM media supplied with 1 mg/ml BSA at 37°C for 8 h. Red arrows denote normal blastocysts. Scale bar, 100 μm. For Zbtb3 +/+ male mice, n = 5. For Zbtb3 +/+ female mice, n = 10. For Zbtb3 +/− male mice, n = 8. For Zbtb3 +/− female mice, n = 16.
-
DmRNAs of indicated genes were analyzed in embryos by real‐time qPCR. Relative gene expression folds were normalized to endogenous β‐actin and shown as means ± SD. **P = 0.0018, **P = 0.0002, **P = 0.0009, **P = 0.0015, **P = 0.0012, **P = 0.0001 by unpaired Student's t‐test. Primers are listed in Appendix Table S1.
-
E–GHuman ESC H1 cells were infected with lentivirus expressing shZBTB3 and cultured in human ESC media for 3 weeks, followed by AP staining. Scale bar, 20 μm. (E); fluorescence staining with anti‐SSEA4 (green) and anti‐OCT4 (red) antibodies, and nuclei were counterstained by DAPI (scale bar, 10 μm) (F); and mRNA expression analyses by real‐time qPCR (G). Relative gene expression fold changes were counted as means ± SD. **P = 0.0015, **P = 0.0017, **P = 0.0029, **P = 0.0012, **P = 0.0025, **P = 0.0005 by unpaired Student's t‐test.
Discussion
Long noncoding RNAs (lncRNAs), a recently recognized class of genes, have been reported to regulate gene expression and nuclear architecture (Ulitsky & Bartel, 2013; Engreitz et al, 2016). As a gene class, lncRNAs tend to be expressed at lower levels and are predominantly distributed in the nucleus, in contrast to mRNAs (Derrien et al, 2012). Moreover, lncRNA expression profiles are more cell type specific than those of mRNAs (Cabili et al, 2011). Divergent lncRNAs represent a major lncRNA biotype in mouse and human genomes. Some divergent lncRNAs exert their functions through regulation of their neighboring protein‐coding genes (Anderson et al, 2016; Luo et al, 2016). However, many of the other divergent lncRNAs are still uncharacterized. In this study, we identified lncKdm2b, a divergent lncRNA for Kdm2b gene, is highly conserved among five mammalian species and highly expressed in ESC cells and early embryos. LncKdm2b deletion impairs ESC self‐renewal and causes early embryonic lethality (Liu et al, 2017). LncKdm2b can activate Zbtb3 by promoting the ATPase activity of SRCAP subunit. Zbtb3 potentiates the ESC self‐renewal in a Nanog‐dependent manner. Finally, Zbtb3 deficiency disrupts ESC self‐renewal and early embryonic development.
LncRNAs are preferentially localized at the vicinity of gene promoters in antisense orientation (Guttman et al, 2010; Sigova et al, 2013). For instance, the divergent lncRNA Evx1as promotes transcription of its nearby gene EVX1 to modulate mesendodermal differentiation (Luo et al, 2016). We recently demonstrated that lncKdm2b knockout in the hematopoietic system impairs the maintenance and effector functions of ILC3s (Liu et al, 2017). In ILC3s, lncKdm2b fails to affect the regulation of its neighboring genes, which interacts with Satb1 to recruit the NURF remodeling complex for triggering Zfp292 expression in trans for the regulation of ILC maintenance. Consistently, here we show that lncKdm2b deletion does not impact the expression of Kdm2b and other neighboring genes in mouse ESCs. LncKdm2b associates with the SRCAP subunit of the SRCAP remodeling complex to activate Zbtb3 expression for the regulation of ESC self‐renewal in trans as well.
LncKdm2b was predicted to contain a stable stem‐loop structure at the exon 1 (nt 450–700). Of note, the fragment (nt 450–700) of lncKdm2b was required to bind SRCAP and Satb1 protein as well (Liu et al, 2017). However, we showed that Satb1 was not expressed in mouse ESCs, suggesting that the interaction of lncKdm2b with Satb1 does not take place in mouse ESCs. Based on our findings, lncKdm2b exerts different regulatory roles in various types of tissues with unique regulatory machineries. Through analyses of published datasets, when ESCs were induced for neuron differentiation treated with RA (GSE85061) or activin (GSE36114), lncKdm2b was actually upregulated during neuron differentiation. However, when ESCs were induced for spontaneous differentiation by LIF withdrawal (GSE48229), lncKdm2b was surely downregulated during spontaneous differentiation, which is consistent with our findings. Furthermore, we showed that lncKdm2b was restrictedly expressed in the cranial/spinal accessory nerve. These results suggest that lncKdm2b may also play an important role in the regulation of neuron differentiation. Given that lncKdm2b and Zbtb3 also express in both ICM and trophectoderm, lncKdm2b may also be involved in the regulation of cell proliferation or survival on embryonic lethality.
ATP‐dependent chromatin remodeling complexes (called as remodelers) exert important functions in the regulation of gene expression (Hartley & Madhani, 2009; Zhang et al, 2011). All remodelers encompass an ATPase domain, a member of the superfamily 2 (SF2) of translocases, within their core subunits (Clapier & Cairns, 2009). These remodelers also have domains located in cis to the ATPase that can modulate its ATPase activity and bind accessory subunits and/or histone modifications (Clapier & Cairns, 2012; Nguyen et al, 2013). The chromatin remodeling complexes are classified into four subfamilies: SWI/SNF, ISWI, CHD, and INO80 by their auxiliary domains (Tosi et al, 2013). The SNF2‐related CBP activator protein (SRCAP) complex, belonging to the INO80 subfamily, modulates the exchange of histone H2A to its variant H2A.Z of nucleosomes for gene expression (Watanabe et al, 2013). The SRCAP subunit contributes to its ATPase activity of the SRCAP complex. SRCAP mutations are related to human disease Floating‐Harbor syndrome (Hood et al, 2012). How the SRCAP complex regulates ESC self‐renewal remains unknown. Here, we show that lncKdm2b interacts with SRCAP and promotes the assembly and its ATPase activity of SRCAP complex, resulting in Zbtb3 transcription in ESCs. Finally, Zbtb3 initiates Nanog expression to sustain the ESC self‐renewal. However, how lncKdm2b assembles the SRCAP complex still needs to be further investigated.
Pluripotency and self‐renewal capacity are major characteristics of ESCs, and some pluripotency TFs such as Oct3/4, STAT3, and Nanog are implicated in the ESC properties (Niwa et al, 2000; Mitsui et al, 2003). In coordination with other factors, these TFs regulate downstream target genes collectively forming a gene regulatory network in ESCs (Young, 2011; Martello & Smith, 2014). Here, we identified Zbtb3 is required for the self‐renewal maintenance of ESCs, whose deletion impairs the ESC self‐renewal and causes early embryonic lethality. Zbtb3 binds to the promoter of Nanog gene and triggers Nanog expression to maintain the ESC self‐renewal. In this study, we demonstrate that Nanog is a downstream target of the TF Zbtb3 that is involved in the self‐renewal maintenance of ESCs, and Zbtb3 can be activated by a divergent lncRNA lncKdm2b. Therefore, our findings reveal that lncRNAs may represent an additional layer of the regulation of ESC self‐renewal and early embryogenesis.
Materials and Methods
Antibodies and reagents
Antibodies against human or mouse Oct4 (2750), H3K4me3 (9751), and H3K27me3 (9733) were purchased from Cell Signaling Technology (Danvers, USA). Antibodies against mouse SSEA‐1 (sc‐21702), human SSEA‐4 (sc‐21704), and mouse Fgf5 (sc‐7914) were purchased from Santa Cruz Biotechnology (Santa Cruz, USA). Antibody against mouse β‐actin (clone AC‐74, A2228) was from Sigma‐Aldrich (St. Louis, USA). Antibodies against mouse Gata6 (ab175349), mouse Brachyury (ab20680), and mouse Kdm2b (ab137547) were purchased from Abcam (Cambridge, USA). Anti‐mouse or human SRCAP antibody (ESL104) was from Kerafast (Boston, USA). Anti‐mouse or human Zbtb3 antibody (25242) was from Signalway Antibody (Baltimore, USA). Secondary antibodies conjugated with Alexa‐594, Alexa‐488, Alexa‐405, or Alexa‐649 were purchased from Molecular Probes Inc. (Eugene, USA). All trans‐retinoic acid (RA) was from Sigma‐Aldrich (St. Louis, USA). DNaseI was purchased from Roche Molecular Biochemicals (Basel, Switzerland). Streptavidin beads were from Sigma‐Aldrich (St. Louis, USA). Protein A/G beads were from Santa Cruz Biotechnology (Santa Cruz, USA). Recombinant LIF protein was from Millipore (Billerica, USA). Alkaline phosphatase detection kit was purchased from Millipore (Billerica, USA). SuperReal premix plus qPCR buffer was from TIANGEN Biotech (Beijing, China). ATPase/GTPase activity assay kit was purchased from Sigma‐Aldrich (St. Louis, USA). EZ prep nuclei isolation kit was from Sigma‐Aldrich (St. Louis, USA). LightShift Chemiluminescent RNA‐EMSA kit was from Thermo Scientific (Waltham, USA).
Cell culture
Mouse ESC D3 cells were obtained from the stem cell core facility of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). ESCs were grown on CF‐1 mouse MEFs treated with mitomycin C. mESCs were cultured in DMEM supplemented with 15% FBS (Invitrogen, USA), 2 mM l‐glutamine, 0.1 mM 2‐mercaptoethanol, 1 mM nucleotide, 0.1 mM non‐essential amino acids, and 103 units/ml mouse leukemia inhibitory factor (LIF). LncKdm2b knockout ES cells were generated either by ICM in vitro culture from KO mice blastocysts or CRISPR/Cas9‐mediated deletion in ES cells. ICM‐derived ESCs were isolated by E3.5 blastocysts and grown on CF‐1 mouse MEFs treated with mitomycin C in mESC media. ESC subclones were picked up after 5 day culture. Alternatively, sgRNA sequences were cloned into lentiCRISPRv2‐GFP plasmid, followed by Lipofectamine 2000 (Thermo Fisher) transfection to ESCs for generation of KO cells. Single GFP‐positive cell was sorted to 96‐well plates. Single cell‐derived ES clones were identified by PCR screening and DNA sequencing. At least three independent KO ESC clones were generated for each gene. gRNA sequences were listed in Appendix Table S2. For serum‐free 2i culture, mouse ESCs were cultured in ESGRO‐2i medium (EMD Millipore) with serum‐free and feeder‐free condition. Human ESC H1 cells was obtained from WiCell Research Institute (Madison, WI) and was grown on CF‐1 mouse MEFs treated with mitomycin C. Culture medium contained DMEM/F12 medium, 20% knockout serum replacement (KSR) (Invitrogen), and 2 mM l‐glutamine, 0.1 mM non‐essential amino acids, 0.1 mM 2‐mercaptoethanol, and 4 ng/ml hFGF2 (Sigma‐Aldrich, USA). Human 293T cells were cultured with DMEM supplemented with 10% FBS and 100 U/ml penicillin and 100 mg/ml streptomycin. Lentivirus was produced in 293T cells using the standard protocols. Transfection was performed using Lipofectin (Invitrogen). For shRNA knockdown and lncKdm2b overexpression experiments, shRNA, full‐length lncKdm2b, or fragment nt 450–700 of lncKdm2b was constructed into pSicoR plasmid. Full length of Zbtb3, Srcap, and Nanog was constructed into pSIN plasmid. Lentivirus was produced by 293T cells. Mouse ESCs and hESCs were infected by lentivirus. Most efficient shRNA among three shRNA constructs was screened for following experiments. Three independent KD cell lines were used for biological replicates in each assay. At least four independent experiments were performed as biological replicates. shRNA sequences were listed in Appendix Table S2. Experiments on embryonic stem cells were approved by the Institutional Medical Research Ethics Committee at Institute of Biophysics, Chinese Academy of Sciences.
Generation of knockout mice
LncKdm2b KO and Zbtb3 KO mice were generated by CRISPR‐Cas9 approaches (Shen et al, 2014; Liu et al, 2017). Briefly, vector pST1374 expressing Cas9 and pUC57kan‐T7 expressing sgRNAs was constructed as previously described. Mixture of Cas9 mRNA (100 ng/μl) and sgRNA (50 ng/μl) was microinjected into the cytoplasm of C57BL/6 fertilized eggs. The injected zygotes were transferred into the uterus of pseudopregnant ICR females (Xia et al, 2016). Frameshift mutations or deletions were identified by PCR screening and DNA sequencing. Animal use and protocols were approved by the Institutional Animal Care and Use Committees at Institute of Biophysics, Chinese Academy of Sciences. We used littermates with the same age (8–12 weeks old) and gender for each group. We excluded the mice 5 g thinner than other littermates before any treatment or analysis. We did not use randomization in our animal studies. We were not blinded to the group in our animal studies.
Microarray and RNA‐seq analysis
RNAs from mouse lncKdm2b −/− ESCs and WT ESCs were prepared for NimbleGen microarray analysis according to the manufacturer's instruction. Briefly, total RNA was extracted with TRIzol reagent. Double‐strand cDNA was synthesized by an Invitrogen SuperScript cDNA synthesis kit from 5 μg total RNA. Double‐strand cDNA was labeled according to the NimbleGen Gene Expression Analysis protocol. Microarrays were hybridized, washed, and scanned according to the manufacturer's instruction. Differentially expressed genes were identified as fold change cutoff > 2.0, FDR < 0.05. Microarray data were deposited in GEO with an accession number (GSE92868). Data were from one biological replicate. RNA‐seq analyses of mouse ESCs, EB, and differentiated ESCs by LIF withdrawal were performed by library construction and sequenced on Illumina Hiseq2000. RNA‐seq data were deposited in GEO with an accession number (GSE93238).
Immunofluorescence assay
mESCs or hESCs with feeder cells were grown on 0.01% poly‐l‐lysine‐treated coverslips and fixed with 4% paraformaldehyde (PFA, Sigma‐Aldrich) for 20 min at room temperature. Cells were permeabilized with 1% Triton‐X 100 in PBS for 20 min and blocked with 10% donkey serum. Cells were then incubated with appropriate primary antibodies at 4°C overnight followed by incubation with corresponding fluorescence‐conjugated secondary antibodies. Nuclei were stained with DAPI. Mouse EBs and embryos were stained in M2 suspending buffer. Images were obtained with Olympus FV1000 laser scanning confocal microscopy (Olympus, Japan).
Northern blot
Total RNA was extracted from different mouse cells with TRIzol; 10 μg RNA from each sample was subjected to formaldehyde denaturing agarose electrophoresis followed by transferring to positively charged NC film with 20× SSC buffer (3.0 M NaCl and 0.3 M sodium citrate, pH 7.0). Membrane was UV‐cross‐linked and incubated with biotin‐labeled RNA probes (lncKdm2b 81–358 nt) generated by in vitro transcription. Biotin signals were detected with HRP‐conjugated streptavidin for the Chemiluminescent Nucleic Acid Detection Module according to the manufacturer's instruction (Han et al, 2014).
RNA‐FISH
Fluorescence‐conjugated lncKdm2b probes were generated according to protocols from Biosearch Technologies. LncKdm2b probe sets for RNA‐FISH are listed in Appendix Table S1. mESCs, hESCs, or mouse embryos were hybridized with DNA probe sets then stained with indicated antibodies (Wang et al, 2015). Images were obtained with Olympus FV1200 laser scanning confocal microscopy (Olympus, Japan).
In vivo assay of teratomas
Mouse ESCs were trypsinized, washed twice with PBS, and then subcutaneously injected into the bilateral inguens of male NOD/SCID mice (1 × 106 cells per injection). After 28 days, mice were sacrificed. Teratomas were fixed in 4% paraformaldehyde, sectioned, and stained with hematoxylin and eosin (H&E).
Electrophoretic mobility shift assay (EMSA)
For RNA‐EMSAs, biotin‐labeled lncKdm2b RNA probe (450–700 nt) was obtained by in vitro transcription with Biotin RNA Labeling Mix (Roche). LightShift Chemiluminescent RNA‐EMSA Kit (Thermo Scientific) was used for shift assay according to manufacturer's instructions. For Nanog promoter DNA EMSA, biotin‐labeled double‐stranded probes were generated by annealing complementary single‐stranded oligonucleotides. The probe sequence used for detection was listed in Appendix Table S1. LightShift Chemiluminescent EMSA Kit (Thermo Scientific) was used for shift assay according to manufacturer's instructions. 100× unlabeled Nanog promoter probe was used for competitive reaction (Wang et al, 2013).
RNA pull‐down assay
Biotin‐labeled lncKdm2b full‐length (sense), antisense, and control RNA (Xist A repeats) were obtained with Biotin RNA Labeling Mix (Roche) in vitro followed by incubation with nuclear extracts separated from ESCs. RNA‐binding proteins were pulled down by streptavidin beads. Pull‐down components were separated with SDS–PAGE followed by immunoblotting with anti‐SRCAP antibody or silver staining. Differential bands enriched by lncKdm2b were analyzed by LTQ Orbitrap XL mass spectrometry. Truncated fragments of lncKdm2b were in vitro‐transcribed to biotin‐labeled RNA followed with RNA pull‐down and immunoblotting (Zhu et al, 2016).
RIP assay
Embryonic stem cells were treated with 1% formaldehyde then dissolved with RNase‐free RIPA buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.5% sodium desoxycholate, 0.1% SDS, 5 mM EDTA, 2 mM PMSF, 20 mg/ml aprotinin, 20 mg/ml leupeptin, 10 mg/ml pepstatin A, 150 mM benzamidine, 1% Nonidet P‐40, and RNase inhibitors]. Samples were sonicated on ice and centrifuged. Supernatants were precleared and incubated with antibodies, followed by protein A/G bead immunoprecipitation. Total RNA was extracted from eluents. LncKdm2b enrichment was analyzed by qPCR.
CHIRP assay
Embryonic stem cells were treated with 1% formaldehyde and resuspended in swelling buffer (0.1 M Tris pH 7.0, 10 mM KOAc, 15 mM MgOAc, 1% NP‐40, 1 mM DTT, 1 mM PMSF, complete protease inhibitor (GE), and 0.1 units/microliter RNase inhibitor). Nuclei was further lysed in nuclear lysis buffer at 100 mg/ml (50 mM Tris 7.0, 10 mM EDTA, 1% SDS, 1 mM DTT, PMSF, protease inhibitor, and RNase inhibitor) and sonicated in the size range of 100–500 bp. Chromatin is diluted in hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris 7.0, 1 mM EDTA, 15% formamide). Probes of 100 pmol were added to 3 ml of diluted chromatin. Probe sequence is listed in Appendix Table S1. Streptavidin‐magnetic C1 beads were added, and beads:biotin probes:RNA:chromatin adducts were captured by magnets (Invitrogen) and washed with wash buffer (2× SSC, 0.5% SDS, add DTT and PMSF fresh). For protein detection, beads were resuspended in 3× original volume of DNase buffer (100 mM NaCl and 0.1% NP‐40), and protein was eluted with a cocktail of 100 μg/ml RNase A (Sigma‐Aldrich) and 0.1 units/microliter RNase H (Epicenter), and 100 U/ml DNase I (Invitrogen) followed with immunoblotting. For DNA detection, beads were resuspended in 3× original volume DNA elution buffer (50 mM NaHCO3, 1% SDS, 200 mM NaCl), and DNA was eluted with a cocktail of 100 μg/ml RNase A (Sigma‐Aldrich) and 0.1 units/microliter RNase H (Epicenter). Chromatin was reverse–cross‐linked at 65°C overnight. DNA was then extracted with equal volumes of phenol:chloroform:isoamyl (Invitrogen) and precipitated with ethanol. Eluted DNA was subject to QPCR (Chu et al, 2011).
Embryoid body (EB) formation
Mouse ESCs were cultured on 0.1% gelatin‐coated dishes with DMEM supplemented with 15% FBS (Invitrogen, USA), 2 mM l‐glutamine, 0.1 mM 2‐mercaptoethanol, 1 mM nucleotide, 0.1 mM non‐essential amino acids, and 103 units/ml mouse leukemia inhibitory factor (LIF). EB formation was induced by hanging drop method on bacteriological‐grade Petri dishes in medium without LIF for 6 days. EBs were collected for further analysis (Ye et al, 2014).
ATPase activity assay
mESCs cells were lysed with low‐salt buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA) followed by extraction of nuclear components in high‐salt buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 1.2 M KCl, 0.2 mM EDTA). Nucleosomes were purified from mESCs by Superdex 200 10/300 GL (GE Healthcare) according to the manufacturer's instruction. LncKdm2b RNA and its truncated fragments were generated by in vitro transcription. SRCAP was immunoprecipitated by the indicated antibody and protein A/G beads from nuclear extracts and measured ATPase activity using the ATPase/GTPase activity assay kit (Sigma‐Aldrich, St. Louis, USA) according to the manufacturer's instruction (Chen et al, 2013).
Chromatin immunoprecipitation (ChIP) assay
Quantitative ChIP was performed according to a standard protocol (Upstate). Sheared chromatin (sonicated to 200–500 bp) from mESCs (2 × 106) fixed in 1% formaldehyde was incubated with 5 μg antibody overnight at 4°C followed by immunoprecipitation with salmon sperm DNA/protein agarose beads. After washing, elution and cross‐link reversal, DNA from each ChIP sample and corresponding input sample were purified and analyzed using qPCR. Primers are listed in Appendix Table S1. For ChIP immunoblotting assay, ESCs were lysed and treated with 1% formaldehyde for cross‐linking. Chromatin was lysed with SDS lysis buffer and sonicated to 200–500 bp. Lysates were precleared with protein A agarose/salmon sperm DNA (50% slurry). Then, 4 μg anti‐SRCAP antibody was incubated with treated lysates for ChIP assays, followed by size fractionation with sucrose gradient ultracentrifugation; 500 μl eluate was put onto 30 ml 5–30% (V/V) sucrose gradient followed by ultracentrifugation at 55,000 g with S Beckman SW28 rotor. Eluent gradients were concentrated and reversely cross‐linked. Proteins were examined by Western blot. DNA was extracted and examined by PCR assays.
DNase accessibility assay
Nuclei were isolated from 1 × 106 mESCs using Nuclei Isolation Kit (Sigma‐Aldrich) according to the manufacturer's protocol. Nuclei were resuspended in 200 μl of DNase I digestion buffer (1 mM EDTA, 0.1 mM EGTA, 5% sucrose, 1 mM MgCl2, 0.5 mM CaCl2). Two equal aliquots of 100 μl nuclei were treated with the indicated units of DNase I (Sigma, USA) and incubated at 37°C for 5 min. Reactions were stopped by 2× DNase I stop buffer (20 mM Tris pH 8.0, 4 mM EDTA, 2 mM EGTA). DNA was extracted and analyzed by qPCR. Primers were listed in Appendix Table S1.
Statistical analysis
An unpaired Student's t‐test was used as statistical analysis in this study. Statistical calculation was performed by using Microsoft Excel or SPSS 13.
Author contributions
BY designed and performed experiments, analyzed data, and wrote the paper; BL, DZ, and XZ performed experiments and analyzed data; LY constructed plasmids and established mice tools. WW, PZ, YW, SW, PX, and RC analyzed data; YD, SM, GH, and JW performed some experiments; YT initiated the study, designed animal experiments, and analyzed data; ZF initiated the study, organized, designed, and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Source Data for Appendix
Review Process File
Acknowledgements
We thank Dr. James Thomson at University of Wisconsin‐Madison and Dr. Xiaoyan Ding at Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences for providing human ES cell line H1. We thank Peng Xue, Yihui Xu, Lei Sun, Junfeng Hao, Dongdong Fan, Yan Teng, Junying Jia, Xiaoyan Wang, Xudong Zhao, and Xiaofei Guo for technical support. We also thank Jing Li (Cnkingbio Company) for technical support. This work was supported by the National Natural Science Foundation of China (91640203, 31530093, 91419308, 31470864, 31670886, 81472413, 81672956, 31601189, 31401098, 31520103905, 81572433, 81772646, 31601189), Beijing Natural Science Foundation (7181006), and the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDB19030203).
The EMBO Journal (2018) 37: e97174
Contributor Information
Runsheng Chen, Email: crs@sun5.ibp.ac.cn.
Yong Tian, Email: ytian@ibp.ac.cn.
Zusen Fan, Email: fanz@moon.ibp.ac.cn.
References
- Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel‐Duby R, Olson EN (2016) Transcription of the non‐coding RNA upperhand controls Hand2 expression and heart development. Nature 539: 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Dunn KA, Mingrone J, Wood BA, Karpinski BA, Sherwood CC, Wildman DE, Maynard TM, Bielawski JP (2016) Functional divergence of the nuclear receptor NR2C1 as a modulator of pluripotentiality during hominid evolution. Genetics 203: 905–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152: 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsos M, Torres‐Padilla ME (2016) Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev 30: 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres‐Padilla ME (2014) Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 15: 723–734 [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon‐Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Conaway RC, Conaway JW (2013) Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin‐remodeling complex. Proc Natl Acad Sci USA 110: 20497–20502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA‐chromatin interactions. Mol Cell 44: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2012) Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T et al (2012) Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539: 452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A (2010) Ab initio reconstruction of cell type‐specific transcriptomes in mouse reveals the conserved multi‐exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien HC, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HS, Quertermous T, Chang CP (2014) A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514: 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJ, Allanson J, Kim CA, Wieczorek D, Moilanen JS, Lacombe D, Gillessen‐Kaesbach G, Whiteford ML, Quaio CR, Gomy I, Bertola DR, Albrecht B, Platzer K, McGillivray G, Zou R et al (2012) Mutations in SRCAP, encoding SNF2‐related CREBBP activator protein, cause Floating‐Harbor syndrome. Am J Hum Genet 90: 308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim EE, Babaei‐Jadidi R, Saadeddin A, Spencer‐Dene B, Hossaini S, Abuzinadah M, Li N, Fadhil W, Ilyas M, Bonnet D, Nateri AS (2012) Embryonic NANOG activity defines colorectal cancer stem cells and modulates through AP1‐ and TCF‐dependent mechanisms. Stem Cells 30: 2076–2087 [DOI] [PubMed] [Google Scholar]
- Janeckova L, Pospichalova V, Fafilek B, Vojtechova M, Tureckova J, Dobes J, Dubuissez M, Leprince D, Baloghova N, Horazna M, Hlavata A, Stancikova J, Sloncova E, Galuskova K, Strnad H, Korinek V (2015) HIC1 tumor suppressor loss potentiates TLR2/NF‐kappaB Signaling and promotes tissue damage‐associated tumorigenesis. Mol Cancer Res 13: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre‐Garau X, Defossez PA, Park YY, Lee JS, Safe S (2012) Identification of oncogenic microRNA‐17‐92/ZBTB4/specificity protein axis in breast cancer. Oncogene 31: 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J (2004) A protein complex containing the conserved Swi2/Snf2‐related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2: E131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA (2013) Control of somatic tissue differentiation by the long non‐coding RNA TINCR. Nature 493: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SU, Maeda T (2012) POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev 247: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, Du Y, Wu J, Qin X, Chen R, Tian Y, Fan Z (2017) Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol 18: 499–508 [DOI] [PubMed] [Google Scholar]
- Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, Shao W, Lu Z, Li H, Na J, Tang F, Wang J, Zhang YE, Shen X (2016) Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18: 637–652 [DOI] [PubMed] [Google Scholar]
- Martello G, Smith A (2014) The nature of embryonic stem cells. Annu Rev Cell Dev Biol 30: 647–675 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Ranjan A, Stengel F, Wei D, Aebersold R, Wu C, Leschziner AE (2013) Molecular architecture of the ATP‐dependent chromatin‐remodeling complex SWR1. Cell 154: 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG (2000) Quantitative expression of Oct‐3/4 defines differentiation, dedifferentiation or self‐renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Polo JM, Dell'Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, Prive GG, Licht JD, Melnick A (2004) Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B‐cell lymphoma cells. Nat Med 10: 1329–1335 [DOI] [PubMed] [Google Scholar]
- Qu H, Qu D, Chen F, Zhang Z, Liu B, Liu H (2010) ZBTB7 overexpression contributes to malignancy in breast cancer. Cancer Invest 28: 672–678 [DOI] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC (2014) Efficient genome modification by CRISPR‐Cas9 nickase with minimal off‐target effects. Nat Methods 11: 399–402 [DOI] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA (2013) Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci USA 110: 2876–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner KP (2013) Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154: 1207–1219 [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xia P, Ye B, Huang G, Liu J, Fan Z (2013) Transient activation of autophagy via Sox2‐mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell 13: 617–625 [DOI] [PubMed] [Google Scholar]
- Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, Zhang G, Tian Y, Chen R, Fan Z (2015) The long noncoding RNA lncTCF7 promotes self‐renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16: 413–425 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Radman‐Livaja M, Rando OJ, Peterson CL (2013) A histone acetylation switch regulates H2A.Z deposition by the SWR‐C remodeling enzyme. Science 340: 195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Izpisua Belmonte JC (2015) Dynamic pluripotent stem cell states and their applications. Cell Stem Cell 17: 509–525 [DOI] [PubMed] [Google Scholar]
- Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z, Tian Y, Fan Z (2016) Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol 17: 369–378 [DOI] [PubMed] [Google Scholar]
- Ye B, Dai Z, Liu B, Wang R, Li C, Huang G, Wang S, Xia P, Yang X, Kuwahara K, Sakaguchi N, Fan Z (2014) Pcid2 inactivates developmental genes in human and mouse embryonic stem cells to sustain their pluripotency by modulation of EID1 stability. Stem Cells 32: 623–635 [DOI] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT (2013) Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152: 727–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, Zhao Y, Shen B, Huang X, Zhu H, Orkin SH, Shen X (2015) Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16: 504–516 [DOI] [PubMed] [Google Scholar]
- Young RA (2011) Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF (2011) A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, Du Y, He L, Fan Z (2016) lnc‐beta‐Catm elicits EZH2‐dependent beta‐catenin stabilization and sustains liver CSC self‐renewal. Nat Struct Mol Biol 23: 631–639 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Source Data for Appendix
Review Process File


