Abstract
In contrast to the cyanobacterial ancestor, chloroplast gene expression is predominantly governed on the post-transcriptional level such as modifications of the RNA sequence, decay rates, exo- and endonucleolytic processing as well as translational events. The concerted function of numerous chloroplast RNA-binding proteins plays a fundamental and often essential role in all these processes but our understanding of their impact in regulation of RNA degradation is only at the beginning. Moreover, metabolic processes and post-translational modifications are thought to affect the function of RNA protectors. These protectors contain a variety of different RNA-recognition motifs, which often appear as multiple repeats. They are required for normal plant growth and development as well as diverse stress responses and acclimation processes. Interestingly, most of the protectors are plant specific which reflects a fast-evolving RNA metabolism in chloroplasts congruent with the diverging RNA targets. Here, we mainly focused on the characteristics of known chloroplast RNA-binding proteins that protect exonuclease-sensitive sites in chloroplasts of vascular plants.
Keywords: chloroplast gene expression, plastid RNA metabolism, RNA binding proteins, RNA stability, RNA processing, RNases
Introduction
The plastid is the result of an endosymbiotic event that occurred billion years ago and comprised the incorporation of a cyanobacterium into a eukaryotic, mitochondria-possessing cell. The main benefit of the cyanobacterium for the host cell was the supply of additional energy and photoautotrophic growth, granting an enormous selective advantage. The genome of the endosymbiont underwent dramatic transformations, whereby the majority of the cyanobacterial genes were transferred into the nucleus or lost [1]. Meanwhile, about half of the plastid proteome consists of members, which are not of cyanobacterial origin. The genome of the model plant Arabidopsis thaliana contains a total of 87 genes encoding mostly ribosomal and photosynthetic subunits, 4 rRNA and 37 tRNA genes [2–4].
Similar to the operons of the cyanobacterial ancestor, the chloroplast genes are primarily organized in newly composed transcriptional units and often contain genes that do not belong to the same functional cluster. These units are transcribed as polycistronic pre-mRNAs by two types of RNA polymerases: the plastid-encoded bacterial type RNA polymerase (PEP) and at least one phage-type nuclear-encoded RNA polymerase (NEP). The PEP is composed of four basic plastid-encoded core subunits (rpo genes) and requires additional nucleus-encoded sigma factors (six in Arabidopsis) for promoter recognition and transcription initiation as well as further factors, which associate to the core during chloroplast development. In contrast, NEP is a one-subunit-enzyme which seems to require additional accessory factors as well. It was assumed that photosynthetic genes are predominantly transcribed by the PEP, while genes for the translational machinery and for PEP subunits are targets of the NEP [5]. However, most plastid genes contain promoter elements that are recognized by both polymerases. NEP and PEP promoters are not restricted to the transcriptional units, but are also found in intergenic regions and opposite to annotated genes, where they are responsible for the synthesis of the recently identified chloroplast noncoding RNAs [6], whose role is poorly understood. In contrast with cyanobacteria, the transcriptional regulation in plastids is global and large-scale rather than gene-specific. Both polymerase types are present and active during all stages of chloroplast development as well as in nongreen tissues. Upon light exposure and formation of the photosynthetic thylakoid membrane both, PEP and NEP, exhibit highest activity and remain active during the entire development of the leaf while in mature chloroplasts genes are preferentially transcribed by the PEP [7]. In most mutants generally affected in the expression of plastid genes including rpo genes and genes for the translational machinery, transcripts preferentially generated by the NEP predominate [8–10].
Even though there are examples for gene-specific transcriptional regulation, such as the psbD–psbC promoter switch in response to blue light in barley, gene-specific transcriptional regulation seems to be an exception rather than the rule [11,12]. Unlike in bacteria, little is known about processes of plastid transcription termination, which seems to be assisted by additional gene-specific factors [13]. Most obviously, the most pronounced difference between cyanobacteria and chloroplasts in terms of regulation of gene expression is the level of control. In bacteria, gene expression is mainly regulated via transcription initiation, while in chloroplasts post-transcriptional control predominates. The chloroplast retained some of the ancestral attributes such as a similar translational machinery, the operon-like gene organization and bacteria-related RNA degradation processes. However, over the course of evolution the chloroplast recruited many new features including the acquisition of introns, the ability to post-transcriptionally modify the RNA sequence, a process called RNA editing, and extensive processing events of the polycistronic pre-mRNAs leading to complex transcript patterns [14]. These new features together with the coevolution of numerous mostly newly acquired, nucleus-encoded RNA-binding proteins involved in the expression of chloroplast genes opened up new possibilities for targeted and more precise regulation of gene products at many different steps that enable adaptive and developmentally flexible chloroplast biogenesis and plant viability [15]. The processing of plastid polycistronic transcripts into complex mRNA isoforms is one of the most obvious differences between bacteria and chloroplasts and offers a platform for gene-specific regulation through RNA lifetime and translational control [14,16–18].
The conversion of polycistronic transcripts into oligocistronic or monocistronic isoforms, the post-transcriptional generation of 5′ and 3′ transcript termini as well as RNA decay in general is accomplished by plastid ribonucleases [19]. In chloroplasts, like in bacteria, two types of ribonucleases exist, endonucleases that execute internal RNA cleavage, and exonucleases that progressively remove nucleotides from the end of the RNA in the either 5′→3′ or 3′→5′ direction. In Arabidopsis, 17 gene products assigned to exhibit ribonucleolytic activity or to contain domains characteristic for ribonucleases were predicted to be localized in the chloroplast [19]. Among the endonucleases, the RNase J and RNase E are thought to be the most relevant enzymes for intercistronic mRNA cleavage in chloroplasts [16,17], while RNase Z (TRZ2) and PRORP are important for the maturation of tRNAs [19]. CSP41(a/b) have been shown to exhibit endonucleolytic activity and may serve to stabilize a large set of nontranslated target mRNAs and precursor rRNAs during the night [17,20,21]. Two mini-RNase III proteins (RNC3 and RNC4) have been demonstrated to be important for rRNA maturation and intron recycling [22]. Three exonucleases have been reported to be active in Arabidopsis chloroplasts, RNR1 (RNase R) and PNPase with 3′→5′ specificity, and RNase J with 5′→3′ activity. As in other organisms, plant ribonucleases are thought to be rather unspecific and certain substrate preferences might be determined by the RNA structure or chemical features and less by the RNA sequence [17,19].
Based on the current knowledge, it has been proposed that both RNA decay and intercistronic processing of polycistronic units are initiated by housekeeping endonucleases (RNase E and J, CSP41) that cleave the RNA in unstructured, unprotected regions [16]. Although accessibility of the RNA target is hypothesized to be determined by the extent of the association with RNA-binding proteins or ribosomes, the precise parameters for initiating the endonucleolytic attack and a regulatory impact of this activity remain unknown [16,19]. In a second step, the endonucleolytically cleaved RNA products are either stabilized to form translationally competent RNAs or rapidly degraded by the PNPase stimulated by polyadenylation that is accomplished either by the PNPase itself or by another yet unidentified plastid poly(A) polymerase [23]. The activity of the PNPase is impeded by secondary structures that are often found in the 3′-untranslated region of mRNAs. These structures are formed by short inverted repeat sequences (IP) that are also present in bacterial RNAs where they perform a different function, namely the termination of transcription. Thus, one of the parameters that determines RNA lifetime are stable secondary structures that could be found in several plastid mature RNA 3′ ends. The transcript 5′ ends are thought to be predominantly protected by tightly bound RNA-binding proteins, which also determine the mature 5′ transcript termini by serving as barriers for 5′→3′ exonucleases. In addition, protecting stem–loop structures also found in chloroplast transcript 5′ ends can be relevant for regulation of mRNA stability and translation as they might hinder ribosomal binding, i.e. access to the Shine–Dalgarno sequence. RNA-binding proteins also protect and define the 3′ termini in transcripts lacking prominent inverted repeat sequences [24]. It seems that the lifetime of chloroplast transcripts is mainly governed by protective RNA-binding proteins as well as by the structure of the RNA and presumably to a certain extent the regulation and/or expression of unspecific RNases.
Most of the plastid RNA protectors present in vascular plants are not found in green algae, such as Chlamydomonas, and vice versa, reflecting the diverged genome–plastome interaction and the tight coevolution of the cellular genetic compartments in the respective lineages during endosymbiosis. Comparably dissimilar are post-transcriptional processes in plastids of land plants and green algae. For example, RNA editing requires a plethora of trans-acting factors and is widespread in vascular plants but entirely absent in green algae [25]. Often, chloroplast RNA-binding proteins required for different post-transcriptional processing steps were found to form either homomultimeric or heteromeric proteinaceous and/or RNA-containing high molecular complexes mostly of unknown composition (e.g. [10,13,26–32]).
In this review, we provide an overview about the protective role of mostly vascular plant-specific RNA-binding proteins in chloroplasts and highlight the progress made over two decades. We outline 18 known nucleus-encoded RNA-stabilizing factors according to the protein class they belong to: (1) pentatricopeptide repeat (PPR) proteins, (2) chloroplast ribonucleoproteins (cpRNPs), and 3) additional RNA protectors, which do not belong to a larger protein family. Our review article mainly focuses on vascular plant chloroplasts as gene expression in Chlamydomonas reinhardtii differs in many respects from vascular plants and extensive reports deserve an independent comprehensive review article. The reader is referred to recent articles reporting on plastid mRNA stability in this unicellular algae (e.g. [17,33–41] ). An overview of the described plastid factors, including their preferential targets and classification of their domains is given in Table 1. Figure 1 summarizes the main mechanisms how binding of RNA protectors confers transcript stability in chloroplasts.
Table 1. List of known chloroplast RNA protectors, their targets, and domains.
| Protein | Gene ID | Targets/Binding Site(s) | Protein Family/Domains | Organism | References |
|---|---|---|---|---|---|
| PPRs | |||||
| PPR10 | GRMZM2G177169 | atpH 5′ UTR, psaJ 3′ UTR | PPR | Zea mays | [24,52] |
| HCF152 | AT3G09650 | psbH-petB IGR, petB 5′ UTR, psbH 3′ UTR | PPR | Arabidopsis thaliana | [57,28] |
| CRP1 | GRMZM2G083950 | psaC 5′ UTR, petA 5′ UTR (role in translation) | PPR | Zea mays | [26,60,62] |
| AT5G42310 | petB-petD IGR (shown in Arabidopsis) | Arabidopsis thaliana | |||
| SOT1 | AT5G46580 | rrn23 5′ end | PPR-SMR | Arabidopsis thaliana | [63–65] |
| PPR53 | GRMZM2G438524 | Zea mays | |||
| PGR3 | AT4G31850 | petL, ndhA 5′ UTRs | PPR | Arabidopsis thaliana | [66–69] |
| GRMZM2G372632 | Zea mays | ||||
| PPR5 | GRMZM2G025409 | trnG(UCC) intron | PPR | Zea mays | [61,70] |
| AT4G39620 | Arabidopsis thaliana | ||||
| EMB175 | AT5G03800 | rpl16 5′ UTR | PPR-PLS-DYW | Arabidopsis thaliana | [72,73] |
| PPR103 | GRMZM2G170896 | Zea mays | |||
| SVR7 | AT4G16390 | atpF-atpA IGR, psaJ-rpl33 IGR, atpB/E 5′ UTR | PPR | Arabidopsis thaliana | [75,76] |
| ATP4 | GRMZM2G128665 | Zea mays | |||
| CRR2 | At3g46790 | rps7-ndhB IGR | PPR-PLS-DYW | Arabidopsis thaliana | [77,78] |
| PpPPR_38 | BAF02672.1 | clpP-rps12 IGR | PPR | Physcomitrella patens | [79,80] |
| cpRNPs | |||||
| 28RNP | X57955.1 | psbA, rbcL, petD, rps14 3′ UTRs | cpRNPs, RRM motif | Spinacea oleracea | [85] |
| CP31A | At4G24770 | 3′ ends of psbB, psbD, psaA/B, atpB, ndhB, ndhF | cpRNPs, RRM motif | Arabidopsis thaliana | [88,89] |
| CP29A | AT3G53460 | 3′ ends of psbB, psbD, psaA/B, atpB, ndhB | cpRNPs, RRM motif | Arabidopsis thaliana | [98] |
| CP33A | AT3G52380 | Multiple transcripts | cpRNPs, RRM motif | Arabidopsis thaliana | [90] |
| Additional chloroplast RNA-stabilizing RNA-binding proteins | |||||
| HCF145 | AT5G08720 | psaA 5′ UTR | 2 TMR domains and 2 SRPBCC domains | Arabidopsis thaliana | [91,92] |
| XM_001783694 | Physcomitrella patens | ||||
| PrfB1/HCF109 | AT5G36170 | Transcripts with UGA stop codons | ribosomal release factor, SPF and GGQ motifs | Arabidopsis thaliana | [94,95] |
| PrfB3 | AT3G57190 | petB 3′ UTR | homologous to PrfB1 (HCF109) | Arabidopsis thaliana | [31] |
| HCF107 | AT3G17040 | psbH 5′ UTR | HAT (Half a TPR) | Arabidopsis thaliana | [29,56,97–99] |
| Zm-HCF107 | GRMZM2G121960 | Zea mays | |||
Abbreviation: IGR, intergenic region. Homologs are shown in the same row.
Figure 1. Model for the function of nuclear-encoded PPR and other RNA-binding proteins in protecting plastid RNAs from ribonucleolytic attack.
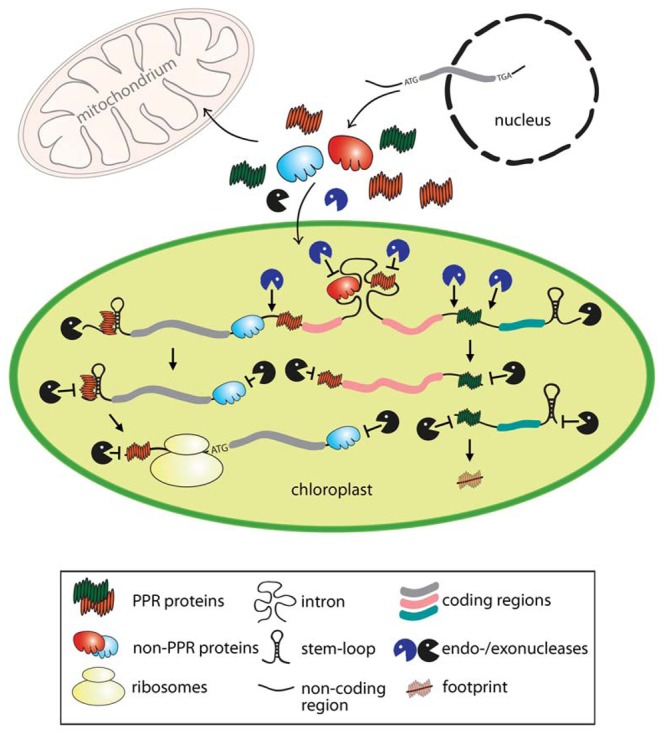
The mode of action shown is representative also for mitochondria. The two colors of protectors (PPR and non-PPR proteins) point to the specificity for particular targets. The scheme shows the endo- and exonucleolytic processing of a representative tricistronic precursor transcript resulting in three stable and translational competent monocistronic products. In chloroplasts, unprotected sites of precursor mRNAs are cleaved by site-specific endonucleases (blue) often in intergenic regions, giving rise to new 5′ and 3′ transcript ends. These extremities are protected against attacks of exonucleases (black) either by the specific binding of RNA protectors and/or by stabilizing secondary RNA stem–loop structures at both ends. For example, PPR10, HCF152, and CRP1 delineated in green show that binding to intergenic regions and subsequent endonucleolytic processing of both adjacent regions results in RNAs with overlapping 5′- and 3′-UTRs, which are protected by the same PPR protein. Binding of protectors to endonuclease-sensitive sites (e.g. intron regions) prevents the cleavage and subsequent unspecific exonucleolytic degradation of the processed RNAs. Furthermore, binding of these factors can stimulate the restructuring of the 5′ region that in turn promotes translation initiation. Short noncoding RNA fragments resulting from the protective role of tightly bound PPR proteins are shown as footprints.
Specificity factors for plastid RNA stabilization
Pentatricopeptide repeat proteins
The family of plant PPR proteins presumably evolved from tetratricopeptide repeat (TPR) proteins and consists of more than 450 up to approximately 1000 conserved members, shared among the eukaryotic kingdom [42,43]. PPR proteins are characterized by their modular and superhelical structure, consisting of 2 to approximately 30 PPR motifs ranging in length from 31 to 36 amino acids [43,44], which were shown to be essential for sequence-specific one-repeat:one-nucleotide RNA recognition including combinatorial effects of two amino acids in each repeat [45,46]. PPR proteins fulfill essential functions in both, chloroplast and mitochondrial RNA metabolism, including endonucleolytic processing, splicing, editing, and translation initiation [44,47]. A subset of PPR proteins in both plant organelles was shown to be particularly important for the stabilization of mRNAs, rRNAs, and tRNAs [44]. The underlying mechanisms involve specific binding of the PPR proteins to the 5′-UTR, 3′-UTR, or other exo-/endonuclease-sensitive sites of certain transcripts that block their degradation by ribonucleases [48]. In contrast with the predominant PPR proteins in plant organelles, the 38–40 amino acid degenerate repeats of the octatricopeptide repeat (OPR) family expanded in Chlamydomonas [38,39,49]. According to mutant phenotypes and the architectural similarities to PPRs, OPRs are assumed to form α-helical RNA-binding domains and are implicated in a variety of post-transcriptional steps. This model of action is further supported by the discovery of short RNA footprints that accumulate as sRNAs, caused by association of RNA-binding proteins to their targets in monocots, dicots, and Chlamydomonas [24,48,50,51]. Several of these PPR proteins were reported to not only stabilize a certain transcript but also hold an additional function in stimulating its editing, splicing, and/or translation.
PPR10 protects the 5′ and 3′ ends of atpH and psaJ respectively, and facilitates the translation of atpH mRNA
The best understood PPR protein is PPR10 in maize. PPR10 is essential for the stabilization of two different RNA termini: the 5′-UTR of atpH and rpl33 as well as the 3′-UTR of atpI and psaJ, whose intergenic regions (atpI–aptH and psaJ–rpl33) share consensus sequences to which PPR10 specifically binds [24] (Figure 1). In concert with exoribonucleases, PPR10 is able to define the 5′ and 3′ termini of processed transcripts which overlap in the intergenic region and contributes thereby to the maturation of their RNA termini [52]. Studies of the PPR10 crystal structure showed that it forms homodimers when bound to psaJ, offering new RNA-binding regulation perspectives via different dimerization states [53]. However, the solution structure as well as more detailed studies on the mode of RNA recognition came to the conclusion that PPR10 binds to its target RNAs in a monomeric form [45,54]. Finally, PPR10 was shown to enhance the translation of atpH by restructuring its 5′ region and exposing its otherwise masked ribosome-binding site [52]. A similar mechanism could also be shown for at least two other PPR proteins, which provide sequence specificity for other 5′ and 3′ transcript termini: HCF152 and CRP1.
HCF152 binds to the psbH–petB intergenic region and is involved in the processing and stabilization of petB transcripts
In a first step to identify novel factors in Arabidopsis involved in chloroplast gene expression and the establishment of the photosynthetic thylakoid membrane, a large number of high chlorophyll fluorescence (hcf) mutants were screened [55]. The hcf152 mutant showed defects in the processing and accumulation of the psbB–psbT–psbH–petB–petD transcript cluster accompanied by severe defects in PSII and cytochrome b6f complex formation [27,56]. Further analysis revealed that the corresponding HCF152 protein forms an RNA-binding homodimer [57]. A screen for small noncoding RNAs in chloroplasts identified an abundant and conserved candidate in the psbH–petB intergenic region in monocots and dicots [48,50,58], supporting the preferential binding of HCF152 to this region, which also corresponded with the immediate 5′ or 3′ termini of petB and psbH RNAs respectively, as revealed by precise transcript mapping [24]. In vitro RNA-binding experiments validated that this sRNA represents the direct target of HCF152, suggesting that HCF152 protects the cleaved 5′ petB and 3′ psbH RNAs with identical sequences in their termini against exonucleolytic degradation, which resembles the function of PPR10 [48,24].
CRP1 stabilizes petB 3′ and petD 5′ ends and regulates translation of additional targets in maize and Arabidopsis
The maize mutant crp1 (chloroplast RNA processing 1) was identified in a screen for mutants with distinct defects in the expression of plastid-encoded genes. It was shown to lack the cytochrome b6f complex, which could be traced back to defects in the accumulation of monocistronic petB and petD transcripts [59]. In addition, CRP1 was shown to be important for translation initiation of petA, petD, and psaC mRNAs [26]. In vivo RIP-chip experiments with the 5′-untranslated regions of psaC and petA mRNAs further contributed to the understanding of CRP1 function as a translational regulator [60]. Electrophoretic mobility shift assays with recombinant ZmCRP1 verified the preferential binding to the petA 5′-UTR [61]. Further analysis of the Arabidopsis homolog AtCRP1 revealed an association with the 5′-UTR of psaC, the petB–petD intergenic region and to a much weaker extent the petA 5′-UTR [62]. This is further supported by the identification of footprints in those three RNA targets [51]. Taken together, although the strength of binding to the targets differs between Arabidopsis and maize the overall analyses argue for similar functions of both CRP1 proteins in stabilization of petD 5′ and petB 3′ RNA termini and in translational regulation of psaC and petA mRNAs.
PPR53 (Zea mays) and SOT1 (Arabidopsis thaliana) stabilize the 5′ end of the 23S rRNA precursor and stimulate the translation of ndhA
Analysis of the Zea mays mutant ppr53 revealed that 5′ transcript ends of rrn23 and ndhA RNAs were absent. PPR53 was able to bind to the 5′ end of the rrn23 RNA, which was also predicted to be the PPR53 target site, but failed to do so in case of the 5′ end of ndhA RNA. Thus, the authors concluded that PPR53 stabilizes the 23S rRNA by binding to its 5′ region and stimulates the translation of ndhA via another yet unknown factor(s) [63]. Experiments on the Arabidopsis ortholog SOT1 suggest that the binding of SOT1 to the 5′ end of the dicistronic 23S-4.5S rRNA prevents its exonucleolytic degradation [64]. Moreover, the SOT1 binding site was narrowed down to an RNA segment of 14 nucleotides and an endonuclease activity was attributed to its small MutS-related (SMR) domain, pointing to a role in stabilization and maturation of 23S-4.5S rRNA precursors [65].
Proton gradient regulation3 is responsible for the stability of petL and ndhA whose translation it activates
Proton gradient regulation3 (PGR3) is an example for another PPR protein that harbors two functions: transcript stabilization and translation initiation. Initial experiments including different pgr3 alleles revealed defects in cytochrome b6f and NDH complex accumulation. These defects were traced back to impaired stabilization of the tricistronic petL–petG–psaJ RNA precursor and putative defects in the translation of petL and presumably a member of the ndh transcripts [66]. Further in vitro studies revealed an association of recombinant PGR3 with the 5′-UTRs of petL and ndhA, arguing for a role of PGR3 in the stabilization and translational activation of these two RNA targets [67]. Finally, analysis of several alleles harboring point mutations in different PPR motifs could decipher which parts of PGR3 are responsible for the RNA-binding activity and the translational activation of petL and ndhA [68]. Comparing the phenotypes of maize and Arabidopsis pgr3 mutants revealed that the defects in petL stabilization and translation are conserved. However, as pgr3 mutants in maize showed more dramatic effects than the Arabidopsis ones, it was concluded that ZmPGR3 has acquired additional functions compared with the Arabidopsis ortholog [69].
PPR5 stabilizes the trnG–UCC RNA precursor
The stabilizing function of PPR proteins is not restricted to protein coding mRNAs and rRNAs but extends to tRNAs, as demonstrated for the maize PPR5 protein that associates with the trnG–UCC precursor [70]. In vitro binding assays with recombinant ZmPPR5 could narrow down its binding site to the central region of the trnG–UCC intron, suggesting that PPR5 protects an endonuclease-sensitive site through its binding and might even facilitate the splicing of this group II intron [61] (Figure 1).
PPR103 stabilizes the 5′ end of rpl16 in maize chloroplasts
Reverse-genetic approaches led to the identification of several PPR mutants exhibiting an embryo-defective phenotype indicating that they are indispensable for early plant development [71]. One of the identified mutants, emb175, exhibited a morphological arrest in early embryo development, pointing to a general house-keeping function of the respective PPR protein [72]. Analysis of the corresponding ppr103 mutant in maize unveiled that the PPR103 protein stabilizes the processed rpl16 mRNA by specific binding to its 5′-UTR [73]. Another interesting feature of PPR103 is its membership to the class of PLS-type proteins (PLS-class proteins contain alternating canonical P-type motifs and variant long (L)- and short (S)-type motifs) that are typically involved in RNA editing; however, a role of PPR103 in editing could be ruled out [73].
ATP4, a multifunctional PPR protein, involved in translation and stabilization of several chloroplast RNAs
Analysis of Arabidopsis suppressor of variegation7 (svr7) mutant showed that rRNA processing and accumulation are affected causing a prevalent reduction of plastid proteins [74]. A later study of the maize ortholog ATP4 unveiled a function of ATP4 in promoting atpA and atpB translation [75]. Additionally, it was hypothesized that ATP4 plays a role in stabilizing the 3′ end of the atpF–atpA and psaJ–rpl33 intergenic regions, thus acting on similar RNA targets as PPR10. Differences in the mutant phenotype in monocot and dicot species argue for evolutionary diverged functions of ATP4 and SVR7 [76].
CRR2 is required for the accumulation of processed ndhB transcripts
The rps12–rps7–ndhB precursor RNA is likely to be processed at not less than two sites in the rps7–ndhB intercistronic region. RNase protection assays revealed that the PPR protein CRR2 is required for intergenic processing of the site close to ndhB [77]. Additionally, CRR2 could stabilize the 5′ end of processed ndhB transcripts by binding to this region, thus hindering 5′-3′ exonucleolytic cleavage. This is reflected by the lack of 5′ processed ndhB transcripts whereas the precursor RNA accumulates at normal levels in crr2 mutants in Arabidopsis, which are unable to accumulate stable and functional NADH dehydrogenase-like complexes mediating cyclic electron flow around photosystem I [77]. A further role in translation could not be excluded. In accordance with the proposed function of CRR2, its essential and C-terminal tripeptide motif DYW, consisting of aspartate (D), tyrosine (Y) and tryptophan (W), has been shown to be capable of cleaving RNA endonucleolytically in vitro even if the domain was considered to be related to RNA editing [78]. Interestingly, although there are no obvious significant differences between the DYW motifs, characteristic for many PPR proteins, the CRR2 DYW domain could not be replaced functionally by the apparently equivalent motifs of other PPR proteins. Thus, CRR2 is a DYW-dependent endoribonuclease with N-terminal PPR repeats likely to confer sequence-specificity to the protein [78]. Although binding of CRR2 to the ndhB 5′-UTR has never been investigated, its potential binding site is presumably represented by an abundant RNA footprint downstream of the 5′ end of the monocistronic ndhB mRNA implying that CRR2 could also function in the stabilization of processed RNAs; however, future research is needed to confirm this interaction [6,50].
PpPPR_38 is involved in splicing and stabilization of the clpP mRNA
The PPR531-11 protein from Physcomitrella patens, later renamed to PpPPR_38, was initially shown to be involved in splicing of clpP and processing of the clpP–rps12 intergenic region resulting in elevated levels of precursor mRNAs [79]. In a following study, it was demonstrated that recombinant PpPPR_38 binds to the first intron of the clpP mRNA and to the clpP–rps12 intergenic region in vitro. Studies with isolated chloroplast lysates suggested that binding of PpPPR_38 to the clpP–rps12 intergenic region stabilizes a predicted stem–loop structure that first provides a correct intergenic endonucleolytic processing site and then protects the cleaved clpP mRNA against 3′→5′ exoribonuclease activity [80].
CpRNPs are general RNA stabilization factors in chloroplasts
In general cpRNPs are divided into three different groups according to predicted structural features and phylogenetic considerations: I (cp28 and cp31), II (cp29A and cp29B), and III (cp33) [81]. CpRNPs are RNA-Recognition Motifs (RRM)-containing, abundant stroma-localized proteins that associate with various ribosome-free mRNAs and pre-tRNAs [82,83]. Interestingly, depletion of the cpRNPs in stromal extracts was followed by rapid RNA degradation while addition of recombinant cpRNPs could restore RNA-stability, suggesting that cpRNPs act as general mediators for RNA stabilization [84]. Meanwhile, in-depth analyses of some of the cpRNP members are available, confirming their involvement in post-transcriptional processes. Spinach 28RNP was suggested to be required for processing and stabilization of several 3′ ends of chloroplast mRNAs including psbA, rbcL, petD, and rps14 as revealed by in vitro studies [85]. In addition, recombinant 28RNP was assumed to be regulated via phosphorylation as its RNA-binding affinity decreased 3- to 4-fold upon phosphorylation [86]. In contrast, 24RNP exhibits an opposite mode of regulation, as phosphorylation enhances the affinity toward its targets [87]. CP31A in Arabidopsis has a more specific function and was shown to be important for editing of different sites and stabilization of several but mainly ndhF mRNAs, whereas CP31B was assumed to support editing of specific CP31A-dependent sites [88]. Further transcriptome-wide analysis revealed that two cpRNPs with overlapping function, CP31A and CP29A, associate with the 3′ end of numerous sense and antisense transcripts including psbB, psbD, psaA/B, atpB, and ndhB to protect them from degradation under cold stress conditions [89]. The Arabidopsis CP33A protein is another example for the global role of cpRNPs in processing and stabilizing of multiple chloroplast RNAs [90].
Additional chloroplast RNA-stabilizing RNA-binding proteins
HCF145 stabilizes the polycistronic psaA–psaB–rps14 mRNA at the 5′ end
The seedling-lethal hcf145 mutant was initially found to specifically lack PSI [91]. This coincided with a severe destabilization of the tricistronic psaA–psaB–rps14 transcript encoding the two photosystem I core proteins PsaA and PsaB. Binding of HCF145 to the psaA 5′-UTR probably protects it from exoribonucleotytic attack [92]. The HCF145 protein contains two highly homologous transcript-binding motif repeat (TMR) domains of approximately 70 amino acids which are responsible for specific binding to the psaA 5′-UTR. The TMR motifs are present rarely as single domains but often as multiple (up to ten) repeats in quite diverse proteins of unknown function in photosynthetic organisms including moss, red and green algae, and cyanobacteria [92]. The predicted helical TMR motifs probably add a new example to the repertoire of RNA-binding domains in photosynthetic organisms.
Two additional N-terminal repeated regions of unknown function were found in the HCF145 protein that showed homology to the ubiquitously distributed SRPBCC (START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC) binding domains superfamily consisting of 11 Pfam families, including START domains, phosphatidylinositol transfer proteins, Bet_v_1, CalC related, CoxG, and polyketide cyclase related families [93]. The structural similarity of the large ligand binding domains to already crystallized members of known function and ligands suggests that they either bind hydrophobic ligands like lipids, phytohormones, steroids, coenzyme Q, alkaloids, polyketides, and/or represent a catalytic pocket for the production of complex polyaromatic substances of the secondary metabolism, such as aromatic hydrocarbon hydroxylating enzymes. Structurally related SRPBCC domains are present in 37 Arabidopsis proteins with unknown function and localization [92].
Interestingly, the SRPBCC-related ligand-binding motifs support association to the 5′ psaA mRNA, although they do not contribute to the binding on their own. This points to a regulatory role of the SRPBCC domains in adjusting PSI levels [92] and provides a control of metabolic processes in fine-tuning RNA binding and thus stabilization of the psaA RNA. Further studies will show whether a metabolism-dependent regulation of psaA–psaB–rps14 mRNA levels is important during development, greening and/or acclimation processes.
PrfB1 is required for stabilization of UGA stop codon-containing transcripts
Different from eukaryotes harboring only one release factor (eRF) for ribosomal release at all three stop codons, eubacteria and organelles contain two release factors, PrfA (RF1) and PrfB (RF2), for termination of translation at UAG and UGA stop codons respectively. The UAA codon is recognized by both factors. PrfB1 and PrfB2 encode the only functional plastid and mitochondrial ribosomal RF2-related proteins in land plants respectively [94]. In contrast with numerous eubacterial prfB mutants described so far, in Arabidopsis lines lacking PrfB1 most plastid UGA stop codon-containing transcripts are unstable [94,95]. The essential PrfB1 protein typically harbors two most important tripeptide motifs characteristic for all organellar and eubacterial PrfB homologs: the “SPF” stops codon recognition motif and the “GGQ” reaction center for peptidyl-tRNA hydrolysis. In some algae, such as C. reinhardtii, a PrfB-related gene is lacking in the nuclear genome and accordingly TGA stop codons are absent in their plastid genomes [94]. It is likely that similar to eubacterial systems lack of mutations of PrfB causes frame shifting at UGA stop codons which produces altered C-termini of different length depending on the position of the next in-frame UAG or UAA stop codons. Either these undesirable reading mistakes or read-through into the 3′-UTR are responsible for destabilization of plastid transcripts harboring UGA stop codons in prfB1 mutants. In either case, a hitherto unrecognized surveillance system efficiently removes all these nonfunctional transcripts. This demonstrates an advanced and more sophisticated regulatory level of plastid gene expression in comparison with known eubacterial systems.
Interestingly, a possible regulatory function of translational termination at UGA stop codons is reflected by a significant increase in the TGA content in land plant plastid genomes in comparison with those in algae [94]. Presumably, evolutionary constraints keep the number of plastid TGA stop codon high in land plants in order to deal with tissue specificity, developmental programs, and/or changing environmental conditions during acclimation processes on the molecular level.
PrfB3 stabilizes the petB mRNAs with cleaved 3′ ends
Land plant genomes surprisingly encode an additional eubacterial RF2-related protein, named PrfB3, which lost its two most important “SPF” and “GGQ” motifs of a release factor and thus the capability to terminate translation at UGA and UAA stop codons [31]. Accordingly, PrfB3 mutants in Arabidopsis are able to terminate translation at UGA stop codons but are specifically affected in the accumulation of the cytochrome b6f complex due to the lack of petB transcripts encoding cytochrome b6. The petB gene is part of the primary pentacistronic psbB–psbT–psbH–petB–petD transcript encoding three proteins of PSII (PsbB, PsbT, and PsbH) and two proteins of the cytochrome b6f complex (PetB and PetD). Similar to most plastid polycistronic transcripts, numerous endonucleolytic processing events generate a wealth of overlapping RNA species, which are subjected to individual life times and translational control. Data revealed that petB transcripts that are cleaved at the 3′ end are efficiently generated but rapidly degraded in prfB3 mutants indicating that PrfB3 protects these RNAs from 3′→5′ exonucleolytic attacks. This is in agreement with preferential binding of PrfB3 to the petB 3′-UTR with respect to control transcripts [31]. Surprisingly, spliced but 3′ unprocessed petB-containing precursor transcripts accumulate in prfB3 but petB is not translated. This indicates that binding of PrfB3 to the 3′ end could have an impact on elongation and/or initiation of translation at the 5′ end or that only processed monocistronic petB transcript are translational competent. Alternatively, a potential weak binding of PrfB3 also to the petB 5′-UTR is required for efficient initiation of translation [31].
A drop in the expression of PrfB3 in partially complemented mutant lines correlated with a decrease in petB mRNA levels [31]. Most importantly, a remarkable decrease in the accumulation of PrfB3 upon changing light conditions and in photoreceptor mutants was also accompanied to a certain extent by a loss of cytochrome b6f amounts. Therefore, PrfB3 could well serve a regulatory function in vivo during acclimation processes by rate limiting levels of petB transcripts when changes in the amount of the cytochrome b6f complex are required. A similar regulatory role was assigned to the plastid PPR protein MCA1 in Chlamydomonas in determining levels of petA mRNA and accordingly its product cytochrome f [96].
HCF107 is required for accumulation of the psbH mRNA
The HCF107 gene was initially identified by high resolution mapping of the corresponding Arabidopsis mutation, which was characterized by a lack of some but not all oligo- and monocistronic psbH transcripts generated by processing of the pentacistronic psbB–psbT–psbH–petB–petD precursor message [55,56]. Determination of transcript termini demonstrated that only transcripts that are processed at position -45 in the 5′-UTR are lacking in the mutant [56]. All other transcripts of this operon accumulate at normal levels and size already indicating that stability of the processed psbH messages rather than processing is affected. PsbH encodes an essential 8 kDa phosphoprotein of PSII whose absence leads to a pronounced deficiency of PSII activity and protein levels in the hcf107 mutant [55,56]. Ectopic expression of psbH in the nuclear genome, which targets the protein back to the chloroplast, revealed that HCF107 is exclusively required for the expression of psbH [97]. This is in contrast with the function of the evolutionary ortholog MBB1 in C. reinhardtii that affects expression of psbB [37].
HCF107 encodes a half-a-tetratricopeptide (HAT) protein containing 11 repeats and no additional domains [98]. It is mostly associated with thylakoid membranes and forms high molecular weight complexes of 60–190 and 600–800 kDa [29]. Crystal structure analysis of HAT proteins revealed that they form helical α-solenoid hairpins similar to TPR proteins and that they are capable of binding RNA. Accordingly, HAT proteins are involved in splicing, polyadenylation, and the maturation of ribosomal RNAs [99]. It appeared that HCF107 binds specifically 11 nucleotides in the 5′ leader 44 nucleotides upstream of the psbH start codon in agreement with the one repeat—one nucleotide model of PPR proteins [98] and according to the 5′ mapped psbH transcripts lacking in hcf107. Expectedly, binding to its target protects the psbH RNA from 5′→3′ exonucleolytic attack in vitro [98]. RNA structure probing revealed that binding of HCF107 to the psbH 5′-UTR remodels the RNA structure, which may also allow ribosome binding and subsequent enhancement of translation, hinting to a dual role of the HCF107 protein. Obviously, PPRs and HATs can act in a similar and presumably analogous way to the recently identified family containing predicted superhelical and repeated RNA-binding TMR motifs in photosynthetic organisms [92].
Conclusion and outlook
Here, we summarized the characteristics of plastid RNA-binding proteins with a protective role in vascular plants. Whether the regulation of endonucleolytic activities, which were assumed to initiate degradation, is rate-limiting for mRNA decay still remains elusive. The RNA-binding proteins addressed here evolved mostly to hinder exonucleolytic attacks of tRNAs and translationally competent mRNAs, some of which were also shown to impact additional processing events and/or translation. Ribosome profiling was shown to partially overcome the challenge to investigate the role of the protectors in translation in particular using genetic approaches with low abundant RNA targets in the respective mutants [63,100]. Nevertheless, questions need to be addressed regarding a direct involvement of the RNA protectors in translational stimulation, e.g. by recruiting additional proteins or restructuring the mRNA to render it accessible for ribosomes. Thus, chloroplast RNA-binding protectors could also act as RNA chaperones, which resolve kinetically trapped misfolded RNA structures to produce functional RNAs, e.g. translationally competent RNAs, which are accessible for ribosomes. In addition, the function of the recently identified chloroplast noncoding RNAs in the overall context of RNA stability still remains to be determined [6,58]. Furthermore, the relationship between mRNA abundance, translational rates of individual mRNA species, and accumulation of the respective gene product in correlation to RNA protectors needs to be investigated quantitatively and under changing environmental conditions to identify rate-limiting steps in plastid gene expression.
Differential expression of RNA protector genes under changing environmental conditions and the effects on their downstream targets speaks in favor of the idea that RNA degradation is mostly regulated at the RNA extremities. One example is provided by a comparable differential expression of AtprfB3 and its target petB in Arabidopsis [31]. Whether PrfB3 is indeed rate limiting for the amounts of the cytochrome b6f complex during stress and/or acclimation processes still needs to be shown.
The progression in mass spectrometric methods will prompt future studies on the metabolic impact and post-translational modifications of RNA protectors on the regulation of their activity, interaction with other hitherto unidentified association partners, and/or RNA affinity in a quantitative manner. Generally, these analyses remained poor, nevertheless the potential integrator HCF145 seems to be a promising candidate for such experiments. The two SRPBCC ligand binding domains of HCF145 offer an excellent platform for their metabolic control by the integration of external stimuli to finally fine-tune photosynthesis by regulating the effect of HCF145 on psaA stability and thus photosystem I amounts [91]. Also, cpRNPs deserve a closer look as they were reported to be of particular importance under cold stress conditions and influence a wide range of target transcripts [88]. Studies on chloroplast gene expression is important not only for understanding regulatory aspects but also for guiding transplastomic approaches for scientific and biotechnological purposes in model as well as agriculturally important plants [3,4].
Most protectors are plant specific congruent with the fast divergence of their targets at the 5′- and 3′-UTRs of plastid transcripts [101]. Even if counterparts of the RNA-binding proteins are occasionally found in algae or even rarely in cyanobacteria, they exert a different function on transcripts or have different targets. This demonstrates that the RNA metabolism represents a fast-evolving process, which presumably reflects a combined adaptation to multicellularity, developmental programs, and changes in light, temperature, humidity, and water supply. Specificity factors, such as plastid RNA protectors, may integrate external signals on the molecular level to rapidly cope with changes of environmental conditions allowing to perform stress responses and acclimation processes. This is reflected by changes in plastid RNA patterns and abundance during development and greening, in different tissues and under changing environmental conditions [9].
Summary
Chloroplast gene expression is predominantly governed on the level of post-transcriptional processes including RNA editing, methylation, polyadenylation, splicing, endo- and exonucleolytic digestion as well as translation.
Generally, newly evolved and vascular plant-specific RNA-binding factors encoded in the nuclear genome are required for protecting specific chloroplast RNAs at exo- and endoribonuclease-sensitive sites reflecting a fast-evolving RNA metabolism congruent with the diverging RNA targets.
Chloroplast RNA-binding protectors, usually found in high molecular weight complexes with largely unknown components, can act as RNA chaperones, occasionally affect other post-transcriptional processes, such as splicing, editing and translation, and contain a variety of different RNA recognition motifs, which often appear as multiple repeats.
Interactions with other proteins, post-translational modifications of protectors, metabolic pathways, and presumably noncoding RNAs may have an important impact on the characteristics of RNA protectors, which are required for normal plant growth and development as well as diverse stress responses and acclimation processes.
Abbreviations
- cpRNP
chloroplast ribonucleoprotein
- HAT
half-a-tetratricopeptide repeat
- NEP
nuclear-encoded RNA polymerase
- OPR
octatricopeptide repeat
- PEP
plastid-encoded bacterial type RNA polymerase
- PPR
pentatricopeptide repeat
- RRM
RNA-recognition motifs
- TMR
transcript-binding motif repeat
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors wish to thank the Deutsche Forschungsgemeinschaft for financial support to J.M. [TRR 175].
References
- 1.Allen J.F. (2015) Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. U.S.A. 112, 10231–10238 10.1073/pnas.1500012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green B.R. (2011) Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66, 34–44 10.1111/j.1365-313X.2011.04541.x [DOI] [PubMed] [Google Scholar]
- 3.Bock R. (2015) Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66, 211–241 10.1146/annurev-arplant-050213-040212 [DOI] [PubMed] [Google Scholar]
- 4.Daniell H., Lin C.S., Yu M. and Chang W.J. (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134 10.1186/s13059-016-1004-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Börner T., Aleynikova A.Y., Zubo Y.O. and Kusnetsov V.V. (2015) Chloroplast RNA polymerases:role in chloroplast biogenesis. Biochim. Biophys. Acta 1847, 761–769 10.1016/j.bbabio.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 6.Zhelyazkova P., Sharma C.M., Forstner K.U., Liere K., Vogel J. and Börner T. (2012) The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24, 123–136 10.1105/tpc.111.089441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Börner T., Aleynikova A.Y., Zubo Y.O. and Kusnetsov V.V. (2015) Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 1847, 761–769 10.1016/j.bbabio.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 8.Hajdukiewicz P.T., Allison L.A. and Maliga P. (1997) The two RNA polymerases encoded by the nuclear and plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048 10.1093/emboj/16.13.4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho W.K., Geimer S. and Meurer J. (2009) Cluster analysis and comparison of various chloroplast transcriptomes and genes in Arabidopsis thaliana. DNA Res. 16, 31–44 10.1093/dnares/dsn031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meurer J., Schmid L.M., Stoppel R., Leister D., Brachmann A. and Manavski N. (2017) PALE CRESS binds to plastid RNAs and facilitates the biogenesis of the 50S ribosomal subunit. Plant J. 92, 400–413 10.1111/tpj.13662 [DOI] [PubMed] [Google Scholar]
- 11.Gamble P.E. and Mullet J.E. (1989) Blue light regulates the accumulation of two psbD-psbC transcripts in barley chloroplasts. EMBO J. 8, 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sexton T.B., Christopher D.A. and Mullet J.E. (1990) Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J. 9, 4485–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi W., He B., Manavski N., Mao J., Ji D., Lu C. et al. (2014) RHON1 mediates a Rho-like activity for transcription termination in plastids of Arabidopsis thaliana. Plant Cell 26, 4918–4932 10.1105/tpc.114.132118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germain A., Hotto A.M., Barkan A. and Stern D.B. (2013) RNA processing and decay in plastids. Wiley Interdiscip Rev. RNA 4, 295–316 10.1002/wrna.1161 [DOI] [PubMed] [Google Scholar]
- 15.Shi X., Hanson M.R. and Bentolila S. (2017) Functional diversity of Arabidopsis organelle-localized RNA-recognition motif-containing proteins. Wiley Interdiscip Rev. RNA 8, 1–14, 10.1002/wrna.1420 [DOI] [PubMed] [Google Scholar]
- 16.Barkan A. (2011) Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155, 1520–1532 10.1104/pp.110.171231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern D.B., Goldschmidt-Clermont M. and Hanson M.R. (2010) Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155 10.1146/annurev-arplant-042809-112242 [DOI] [PubMed] [Google Scholar]
- 18.Stoppel R. and Meurer J. (2013) Complex RNA metabolism in the chloroplast: an update on the psbB operon. Planta 237, 441–449 10.1007/s00425-012-1782-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoppel R. and Meurer J. (2012) The cutting crew - ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 63, 1663–1673 10.1093/jxb/err401 [DOI] [PubMed] [Google Scholar]
- 20.Bollenbach T.J., Sharwood R.E., Gutierrez R., Lerbs-Mache S. and Stern D.B. (2009) The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol. Biol. 69, 541–552 10.1007/s11103-008-9436-z [DOI] [PubMed] [Google Scholar]
- 21.Leister D. (2014) Complex(iti)es of the ubiquitous RNA-binding CSP41 proteins. Front. Plant Sci. 5, 255 10.3389/fpls.2014.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hotto A.M., Castandet B., Gilet L., Higdon A., Condon C. and Stern D.B. (2015) Arabidopsis chloroplast mini-ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell 27, 724–740 10.1105/tpc.114.134452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuster G. and Stern D. (2009) RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 85, 393–422 10.1016/S0079-6603(08)00810-6 [DOI] [PubMed] [Google Scholar]
- 24.Pfalz J., Bayraktar O.A., Prikryl J. and Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28, 2042–2052 10.1038/emboj.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shikanai T. (2015) RNA editing in plants: Machinery and flexibility of site recognition. Biochim. Biophys. Acta 1847, 779–785 10.1016/j.bbabio.2014.12.010 [DOI] [PubMed] [Google Scholar]
- 26.Fisk D.G., Walker M.B. and Barkan A. (1999) Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621–2630 10.1093/emboj/18.9.2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meierhoff K., Felder S., Nakamura T., Bechtold N. and Schuster G. (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15, 1480–1495 10.1105/tpc.010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura T., Schuster G., Sugiura M. and Sugita M. (2004) Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem. Soc. Trans. 32, 571–574 10.1042/BST0320571 [DOI] [PubMed] [Google Scholar]
- 29.Sane A.P., Stein B. and Westhoff P. (2005) The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 42, 720–730 10.1111/j.1365-313X.2005.02409.x [DOI] [PubMed] [Google Scholar]
- 30.Stoppel R., Manavski N., Schein A., Schuster G., Teubner M., Schmitz-Linneweber C. et al. (2012) RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res. 40, 8593–8606 10.1093/nar/gks613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoppel R., Lezhneva L., Schwenkert S., Torabi S., Felder S., Meierhoff K. et al. (2011) Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 23, 2680–2695 10.1105/tpc.111.085324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paieri F., Tadini L., Manavski N., Kleine T., Ferrari R., Morandini P.A. et al. (2017) The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrin D.L. and Nickelsen J. (2004) Chloroplast RNA processing and stability. Photosynth. Res. 82, 301–314 10.1007/s11120-004-2741-8 [DOI] [PubMed] [Google Scholar]
- 34.Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S. et al. (2010) MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22, 234–248 10.1105/tpc.109.066266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tourasse N.J., Choquet Y. and Vallon O. (2013) PPR proteins of green algae. RNA Biol. 10, 1526–1542 10.4161/rna.26127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefebvre-Legendre L., Merendino L., Rivier C. and Goldschmidt-Clermont M. (2014) On the complexity of chloroplast RNA metabolism: psaA trans-splicing can be bypassed in Chlamydomonas. Mol. Biol. Evol. 31, 2697–2707 10.1093/molbev/msu215 [DOI] [PubMed] [Google Scholar]
- 37.Loizeau K., Qu Y., Depp S., Fiechter V., Ruwe H., Lefebvre-Legendre L. et al. (2014) Small RNAs reveal two target sites of the RNA-maturation factor Mbb1 in the chloroplast of Chlamydomonas. Nucleic Acids Res. 42, 3286–3297 10.1093/nar/gkt1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lefebvre-Legendre L., Choquet Y., Kuras R., Loubéry S., Douchi D. and Goldschmidt-Clermont M. (2015) A nucleus-encoded chloroplast protein regulated by iron availability governs expression of the photosystem I subunit PsaA in Chlamydomonas reinhardtii. Plant Physiol. 167, 1527–1540 10.1104/pp.114.253906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., Johnson X., Cavaiuolo M., Bohne A.V., Nickelsen J. and Vallon O. (2015) Two Chlamydomonas OPR proteins stabilize chloroplast mRNAs encoding small subunits of photosystem II and cytochrome b6 f. Plant J. 82, 861–873 10.1111/tpj.12858 [DOI] [PubMed] [Google Scholar]
- 40.Douchi D., Qu Y., Longoni P., Legendre-Lefebvre L., Johnson X., Schmitz-Linneweber C. et al. (2016) A nucleus-encoded chloroplast phosphoprotein governs expression of the Photosystem I Subunit PsaC in Chlamydomonas reinhardtii. Plant Cell 28, 1182–1199 10.1105/tpc.15.00725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavaiuolo M., Kuras R., Wollman F.A., Choquet Y. and Vallon O. (2017) Small RNA profiling in Chlamydomonas: insights into chloroplast RNA metabolism. Nucleic Acids Res. 45, 10783–10799 10.1093/nar/gkx668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Toole N., Hattori M., Andrès C., Iida K., Lurin C., Schmitz-Linneweber C. et al. (2008) On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25, 1120–1128 [DOI] [PubMed] [Google Scholar]
- 43.Small I.D. and Peeters N. (2000) The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 10.1016/S0968-0004(99)01520-0 [DOI] [PubMed] [Google Scholar]
- 44.Barkan A. and Small I. (2014) Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 10.1146/annurev-arplant-050213-040159 [DOI] [PubMed] [Google Scholar]
- 45.Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S. et al. (2012) A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 10.1371/journal.pgen.1002910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yagi Y., Hayashi S., Kobayashi K., Hirayama T. and Nakamura T. (2013) Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8, e57286 10.1371/journal.pone.0057286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shikanai T. and Fujii S. (2013) Function of PPR proteins in plastid gene expression. RNA Biol. 10, 1446–1456 10.4161/rna.25207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suarez M., Börner T. et al. (2012) Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 40, 3092–3105 10.1093/nar/gkr1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahire M., Laroche F., Cerutti L. and Rochaix J.D. (2012) Identification of an OPR protein involved in the translation initiation of the PsaB subunit of photosystem I. Plant J. 72, 652–661 10.1111/j.1365-313X.2012.05111.x [DOI] [PubMed] [Google Scholar]
- 50.Ruwe H. and Schmitz-Linneweber C. (2012) Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic. Acids. Res. 40, 3106–3116 10.1093/nar/gkr1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruwe H., Wang G., Gusewski S. and Schmitz-Linneweber C. (2016) Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 44, 7406–7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prikryl J., Rojas M., Schuster G. and Barkan A. (2011) Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 108, 415–420 10.1073/pnas.1012076108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q., Yan C., Xu H., Wang Z., Long J., Li W. et al. (2014) Examination of the dimerization states of the single-stranded RNA recognition protein pentatricopeptide repeat 10 (PPR10). J. Biol. Chem. 289, 31503–31512 10.1074/jbc.M114.575472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gully B.S., Cowieson N., Stanley W.A., Shearston K., Small I.D., Barkan A. et al. (2015) The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 43, 1918–1926 10.1093/nar/gkv027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meurer J., Meierhoff K. and Westhoff P. (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198, 385–396 10.1007/BF00620055 [DOI] [PubMed] [Google Scholar]
- 56.Felder S., Meierhoff K., Sane A.P., Meurer J., Driemel C., Plücken H. et al. (2001) The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 13, 2127–2141 10.1105/tpc.13.9.2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura T., Meierhoff K., Westhoff P. and Schuster G. (2003) RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270, 4070–4081 10.1046/j.1432-1033.2003.03796.x [DOI] [PubMed] [Google Scholar]
- 58.Lung B., Zemann A., Madej M.J., Schuelke M., Techritz S., Ruf S. et al. (2006) Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 34, 3842–3852 10.1093/nar/gkl448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barkan A., Walker M., Nolasco M. and Johnson D. (1994) A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13, 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitz-Linneweber C., Williams-Carrier R. and Barkan A. (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17, 2791–2804 10.1105/tpc.105.034454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams-Carrier R., Kroeger T. and Barkan A. (2008) Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14, 1930–1941 10.1261/rna.1077708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferrari R., Tadini L., Moratti F., Lehniger M.K., Costa A., Rossi F. et al. (2017) CRP1 Protein: (dis)similarities between Arabidopsis thaliana and Zea mays. Front. Plant Sci. 8, 163 10.3389/fpls.2017.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zoschke R., Watkins K.P., Miranda R.G. and Barkan A. (2016) The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 85, 594–606 10.1111/tpj.13093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W., Liu S., Ruwe H., Zhang D., Melonek J., Zhu Y. et al. (2016) SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 85, 607–621 10.1111/tpj.13126 [DOI] [PubMed] [Google Scholar]
- 65.Zhou W., Lu Q., Li Q., Wang L., Ding S., Zhang A. et al. (2017) PPR-SMR protein SOT1 has RNA endonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 114, E1554–E1563 10.1073/pnas.1612460114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamazaki H., Tasaka M. and Shikanai T. (2004) PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38, 152–163 10.1111/j.1365-313X.2004.02035.x [DOI] [PubMed] [Google Scholar]
- 67.Cai W., Okuda K., Peng L. and Shikanai T. (2011) PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 67, 318–327 10.1111/j.1365-313X.2011.04593.x [DOI] [PubMed] [Google Scholar]
- 68.Fujii S., Sato N. and Shikanai T. (2013) Mutagenesis of individual pentatricopeptide repeat motifs affects RNA binding activity and reveals functional partitioning of Arabidopsis PROTON gradient regulation3. Plant Cell 25, 3079–3088 10.1105/tpc.113.112193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belcher S., Williams-Carrier R., Stiffler N. and Barkan A. (2015) Large-scale genetic analysis of chloroplast biogenesis in maize. Biochim. Biophys. Acta 1847, 1004–1016 10.1016/j.bbabio.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 70.Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B. and Barkan A. (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28, 5337–5347 10.1128/MCB.00563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., Bruyere C. et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 10.1105/tpc.104.022236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cushing D.A., Forsthoefel N.R., Gestaut D.R. and Vernon D.M. (2005) Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221, 424–436 10.1007/s00425-004-1452-x [DOI] [PubMed] [Google Scholar]
- 73.Hammani K., Takenaka M., Miranda R. and Barkan A. (2016) A PPR protein in the PLS subfamily stabilizes the 5′-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res. 44, 4278–4288 10.1093/nar/gkw270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu X., Yu F. and Rodermel S. (2010) An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 154, 1588–1601 10.1104/pp.110.164111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zoschke R., Kroeger T., Belcher S., Schöttler M.A., Barkan A. and Schmitz-Linneweber C. (2012) The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 72, 547–558 10.1111/j.1365-313X.2012.05081.x [DOI] [PubMed] [Google Scholar]
- 76.Zoschke R., Qu Y., Zubo Y.O., Börner T. and Schmitz-Linneweber C. (2013) Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J. Plant Res. 126, 403–414 10.1007/s10265-012-0527-1 [DOI] [PubMed] [Google Scholar]
- 77.Hashimoto M., Endo T., Peltier G., Tasaka M. and Shikanai T. (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36, 541–549 10.1046/j.1365-313X.2003.01900.x [DOI] [PubMed] [Google Scholar]
- 78.Okuda K., Chateigner-Boutin A.L., Nakamura T., Delannoy E., Sugita M., Myouga F. et al. (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21, 146–156 10.1105/tpc.108.064667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hattori M., Miyake H. and Sugita M. (2007) A Pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. J. Biol. Chem. 282, 10773–10782 10.1074/jbc.M608034200 [DOI] [PubMed] [Google Scholar]
- 80.Hattori M. and Sugita M. (2009) A moss pentatricopeptide repeat protein binds to the 3′ end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 276, 5860–5869 10.1111/j.1742-4658.2009.07267.x [DOI] [PubMed] [Google Scholar]
- 81.Ohta M., Sugita M. and Sugiura M. (1995) Three types of nuclear genes encoding chloroplast RNA-binding proteins (cp29, cp31 and cp33) are present in Arabidopsis thaliana: presence of cp31 in chloroplasts and its homologue in nuclei/cytoplasms. Plant Mol. Biol. 27, 529–539 10.1007/BF00019319 [DOI] [PubMed] [Google Scholar]
- 82.Nakamura T., Ohta M., Sugiura M. and Sugita M. (1999) Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett. 460, 437–441 10.1016/S0014-5793(99)01390-3 [DOI] [PubMed] [Google Scholar]
- 83.Lorkovic Z.J. and Barta A. (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635 10.1093/nar/30.3.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakamura T., Ohta M., Sugiura M. and Sugita M. (2001) Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem. 276, 147–152 10.1074/jbc.M008817200 [DOI] [PubMed] [Google Scholar]
- 85.Schuster G. and Gruissem W. (1991) Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 10, 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lisitsky I. and Schuster G. (1995) Phosphorylation of a chloroplast RNA-binding protein changes its affinity to RNA. Nucleic Acids Res. 23, 2506–2511 10.1093/nar/23.13.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Loza-Tavera H., Vargas-Suarez M., Diaz-Mireles E., Torres-Marquez M.E., Gonzalez de la Vara L.E., Moreno-Sanchez R. et al. (2006) Phosphorylation of the spinach chloroplast 24 kDa RNA-binding protein (24RNP) increases its binding to petD and psbA 3′ untranslated regions. Biochimie 88, 1217–1228 10.1016/j.biochi.2006.03.009 [DOI] [PubMed] [Google Scholar]
- 88.Tillich M., Hardel S.L., Kupsch C., Armbruster U., Delannoy E., Gualberto J.M. et al. (2009) Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 6002–6007 10.1073/pnas.0808529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I. and Schmitz-Linneweber C. (2012) Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 24, 4266–4280 10.1105/tpc.112.103002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teubner M., Fuss J., Kühn K., Krause K. and Schmitz-Linneweber C. (2017) The RNA recognition motif protein CP33A is a global ligand of chloroplast mRNAs and is essential for plastid biogenesis and plant development. Plant J. 89, 472–485 10.1111/tpj.13396 [DOI] [PubMed] [Google Scholar]
- 91.Lezhneva L. and Meurer J. (2004) The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 38, 740–753 10.1111/j.1365-313X.2004.02081.x [DOI] [PubMed] [Google Scholar]
- 92.Manavski N., Torabi S., Lezhneva L., Arif M.A., Frank W. and Meurer J. (2015) HIGH CHLOROPHYLL FLUORESCENCE145 binds to and stabilizes the psaA 5′ UTR via a newly defined repeat motif in embryophyta. Plant Cell 27, 2600–2615 10.1105/tpc.15.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Radauer C., Lackner P. and Breiteneder H. (2008) The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8, 286 10.1186/1471-2148-8-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meurer J., Lezhneva L., Amann K., Gödel M., Bezhani S., Sherameti I. et al. (2002) A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell 14, 3255–3269 10.1105/tpc.006809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meurer J., Berger A. and Westhoff P. (1996) A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell 8, 1193–1207 10.1105/tpc.8.7.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.A. et al. (2007) Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 104, 9093–9098 10.1073/pnas.0703162104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levey T., Westhoff P. and Meierhoff K. (2014) Expression of a nuclear-encoded psbH gene complements the plastidic RNA processing defect in the PSII mutant hcf107 in Arabidopsis thaliana. Plant J. 80, 292–304 10.1111/tpj.12632 [DOI] [PubMed] [Google Scholar]
- 98.Hammani K., Cook W.B. and Barkan A. (2012) RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. U.S.A. 109, 5651–5656 10.1073/pnas.1200318109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Preker P.J. and Keller W. (1998) The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 23, 15–16 10.1016/S0968-0004(97)01156-0 [DOI] [PubMed] [Google Scholar]
- 100.Zoschke R., Watkins K.P. and Barkan A. (2013) A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25, 2265–2275 10.1105/tpc.113.111567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manavski N., Torabi S., Stoppel R. and Meurer J. (2012) Phylogenetic and ontogenetic integration of organelles into the compartmentalized genome of the eukaryotic cell. J. Endocytobiosis Cell Res. 25–31 [Google Scholar]


