Abstract
Here we present the genome of Methylovulum psychrotolerans strain HV10-M2, a methanotroph isolated from Hardangervidda national park (Norway). This strain represents the second of the two validly published species genus with a sequenced genome. The other is M. miyakonense HT12, which is the type strain of the species and the type species of the genus Methylovulum. We present the genome of M. psychrotolerants str. HV10-M2 and discuss the differences between M. psychrotolerans and M. miyakonense. The genome size of M. psychrotolerans str. HV10-M2 is 4,923,400 bp and contains 4415 protein-coding genes, 50 RNA genes and an average GC content of 50.88%.
Electronic supplementary material
The online version of this article (10.1186/s40793-018-0314-2) contains supplementary material, which is available to authorized users.
Keywords: Methylovulum psychrotolerans HV10-M2, Methylovulum, Gammaproteobacteria, Methanotroph, High-altitude
Introduction
Methanotrophs are a group of microorganisms that utilize methane as the sole energy and carbon source. They are important contributors to the global carbon budget and climate change, reducing methane emissions to the atmosphere as they represent the only known biological methane sink [1]. Aerobic methane oxidation can be performed by members of the phyla Proteobacteria (classes Alphaproteobacteria and Gammaproteobacteria) and Verrucomicrobia [2, 3]. The recently described genus Methylovulum [4] belongs to the family Methylococcaceae within the class Gammaproteobacteria. Their cells are described as aerobic, non-motile gram-negative bacteria with coccoid or rod-shape cells, they grow with methane and methanol as carbon sources and they have been isolated from cold environments [5]. So far, only one species within this genus, Methylovulum miyakonense HT12, has published available genome data. However, the 16S rRNA gene sequence of three isolates belonging to the Methylovulum genus have been recently reported [5].
Here we report the characteristics of M. psychrotolerans HV10-M2 (Fig. 1), isolated from plant material collected from a peat bog saturated with water at Hardangervidda, a high-altitude (> 1230 m above sea level) national park located in central Norway. We present the genome of M. psychrotolerans str. HV10-M2, and provide first insights into the genomic and physiological differences between M. psychrotolerans HV10-M2 and M. miyakonense HT12.
Fig. 1.
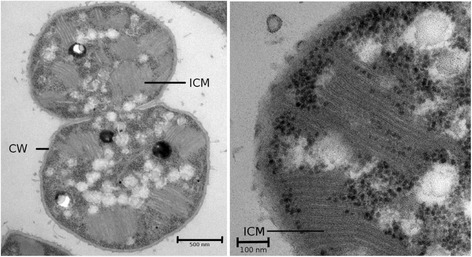
Transmission electron microscope picture of the strain Methylovulum psychrotolerans HV10-M2. Cell wall (CW) and intracytoplasmatic membrane (ICM) are labelled in the pictures. Scale bars represent 500 (left panel) and 100 (right panel) nm
Organism information
Classification and features
M. psychrotolerans HV10-M2 was isolated from wet plant material located in a peat bog at the high-altitude (1230 m above sea level) national park Hardangervidda (Norway) (GPS: 60.22 N, 7.25 E) on July 17, 2015. Air temperature during sampling was 9 °C. The strain was enriched from approximately 4 g of the plant material collected from the peat bog which was added directly to a 120 mL sterile serum flask containing 20 mL of LMM medium (Low-Salt-Methanotrophic medium; KNO3 0.1 g L− 1, MgSO4 0.1 g L− 1, CaCl2·2H2O 0.02 g L− 1, KBr 0.01 g L− 1 [6]), closed with a butyl rubber stopper and sealed with an aluminum crimp. Methane (purity 99.5%, Yara Praxair, Oslo, Norway) was amended with a syringe through a 0.2 μm DynaGard® filter (CA, USA) to a final ratio of 4:1 (methane/air; v/v). The flask was incubated under dark conditions at 16 °C for three weeks without shaking. The gas mixture was restored every seven days, and growth was monitored using phase contrast microscopy (Eclipse E400 microscope, Nikon Corporation, Tokyo, Japan).
The enrichment culture was transferred five times into serum flasks with LMM medium as described above. Serial dilutions (10− 5 to 10− 8) were then prepared and aliquots of 0.1 mL of each dilution were spread onto LMM agar plates. Plates were incubated at 16 °C in jars filled with a methane:air gas mixture (4:1, v/v). Single pink colonies (previously isolated M. psychrotolerans strains show pink pigmentation [5]) were picked and re-streaked onto new agar plates. Finally, one single colony was transferred into a serum flask with LMM medium (see above) and incubated for one week at 16 °C. Purity of the isolate was confirmed by PCR and transmission electron microscopy (TEM, at 60 kV, Jeol JEM-1230, Tokyo, Japan). Contamination was assessed as reported previously [6, 7].
M. psychrotolerans HV10-M2 grows between 4 °C and 25 °C, with optimal growth between 13 °C to 20 °C. Strain HV10-M2 grows using methane and methanol as the carbon and energy source. The optimal pH for growth 6.8. Cells of HV10-M2 are aerobic, non-motile, coccoid- to rod-shaped and form light pink colonies when checked on LMM agar plates. Average cell size is 2 μm. The characteristics of M. psychrotolerans HV10-M2 are summarized in Table 1.
Table 1.
Classification and general features of Methylovulum psychrotolerans strain HV10_M2T
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [34] | |
| Phylum Proteobacteria | TAS [35] | ||
| Class Gammaproteobacteria | TAS [36] | ||
| Order Methylococcales | TAS [37] | ||
| Family Methylococcaceae | TAS [38] | ||
| Genus Methylovulum | TAS [4] | ||
| Species psychrotolerans | TAS [5] | ||
| Strain: Sph1 | TAS [5] | ||
| Gram stain | Negative | IDA | |
| Cell shape | Coccoid / Rod-shape | IDA | |
| Motility | Non-motile | IDA | |
| Sporulation | Non-sporulating | IDA | |
| Temperature range | 4–25 °C | IDA | |
| Optimum temperature | 13–20 °C | IDA | |
| pH range; Optimum | 5–7; 6.8 | IDA | |
| Carbon source | Methane | IDA | |
| MIGS-6 | Habitat | Peat bog | IDA |
| MIGS-6.3 | Salinity | ND | – |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living | IDA |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Hardangervidda, Norway | IDA |
| MIGS-5 | Sample collection | 2015 | IDA |
| MIGS-4.1 | Latitude | 60.22 N | IDA |
| MIGS-4.2 | Longitude | 7.25 E | IDA |
| MIGS-4.4 | Altitude | 1230 m a.s.l. | IDA |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [39]
The 16S rRNA gene of M. psychrotolerans HV10-M2 shows more than 99% sequence identity with M. psychrotolerans Sph1, Sph2 and Oz2 (GenBank accession numbers KT381578, KT381580 and KT381582, respectively; Fig. 2). However, none of those strains has genome data publicly available. The highest sequence identity with a strain with genome data was M. miyakonense HT12. The two strains M. psychrotolerans HV10-M2 and M. miyakonense HT12 show 95% sequence identity in the 16S rRNA gene (Fig. 2).
Fig. 2.
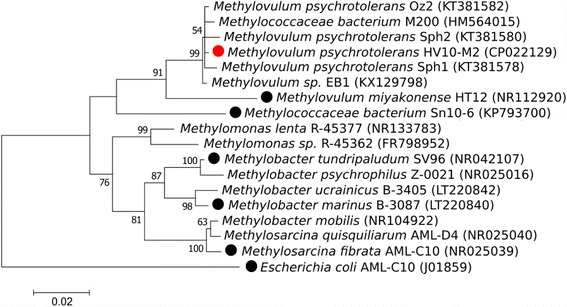
Phylogenetic tree. The tree is based on the 16S rRNA gene sequences of the best hits of cultured strains in the nucleotide database from NCBI (retrieved on June 5, 2017). The tree was reconstructed by using maximum likelihood analysis and the Jukes-Cantor nucleotide substitution model as implemented in MEGA v7 [40]. Robustness of the tree was determined using 1000 bootstrap replicates. Sequences were aligned with MUSCLE [41] in MEGA v7. The tree was rooted against the 16S rRNA gene sequence of Escherichia coli. Accession numbers are provided within parenthesis. The strain presented in this study is marked with a red dot. Strains with available genome data are marked with black dots. Bootstrap values less than 50 are not shown
Genome sequencing information
Genome project history
M. psychrotolerans HV10-M2 was whole genome sequenced at the Department of Bioscience at Aarhus University, Denmark in September 2016. The genome project was deposited in GOLD under the project Ga0185950. The Whole Genome Shotgun project was deposited at GenBank with the accession number CP022129. Summarized information about the project and the sequencing platform details are included in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | High-quality-draft |
| MIGS-28 | Libraries used | Paired-end NexteraXT DNA |
| MIGS 29 | Sequencing platforms | Illumina MiSeq |
| MIGS 31.2 | Fold coverage | 303.7X |
| MIGS 30 | Assemblers | SPAdes 3.9.0 |
| MIGS 32 | Gene calling method | Prodigal v2.6.2 |
| Locus Tag | CEK71 | |
| Genbank ID | CP022129 | |
| GenBank Date of Release | 2017–06-27 | |
| GOLD ID | Gp029646 | |
| BIOPROJECT | PRJNA391059 | |
| MIGS 13 | Source Material Identifier | HV10-M2 |
| Project relevance | Environmental |
Growth conditions and genomic DNA preparation
M. psychrotolerans HV10-M2 was cultivated in a 120 ml serum flask at 16 °C containing LMM medium with methane addition. After turbidity was observed (approximately 2 months), it was transferred onto LMM agar plates and incubated with methane as headspace gas (4:1, v/v). A single colony was transferred into a flask containing LMM medium and finally, 1.5 mL of the culture was harvested by centrifugation and genomic DNA was isolated from the pellet using the GenElute Bacterial Genomic DNA kit (Sigma Aldrich, USA) following the manufacturer recommendations and purified using the DNA Clean and Concentrator kit (Zymo Research, USA).
Genome sequencing and assembly
The genomic DNA of strain HV10-M2 was sequenced with the Illumina MiSeq Reagent Kit V3 (Illumina, CA, USA) and the sequencing libraries were prepared using the Nextera XT Library Preparation Kit (Illumina). A total of 4,933,624 sequence reads were generated. FastQC [8] was used for quality control. Reads were adaptor- and quality trimmed using Trimmomatic v0.36 [9] when bases were at the end of the reads and when the average quality was below the quality threshold (Phred score < 33) with the parameters: CROP:289, HEADCROP:19, SLIDINGWINDOW:4:20, MINLEN:100, resulting in 4,019,650 paired-end reads and 4.92 Mb with an overall coverage estimate of 303.7×. Assembly of the data was performed using SPAdes v3.9.0 [10] with the “--careful” option and the k-mer values 21, 33, 55, 77, 99, 127. The assembly was evaluated with QUAST v4.3 [11]. The assembly yielded 186 contigs with a total length of 4,923,400 bp, and an N50 value of 71,358 bp.
Genome annotation
Gene prediction and annotation was performed using the standard operating procedure of the Integrated Microbial Genomes platform developed by the Joint Genome Institute [12]. In addition, the genome of M. psychrotolerans HV10-M2 was submitted to BlastKOALA [13] and Pathway tools [14] to be compared against the KEGG [15] and MetaCyc [16] databases, respectively, for pathway predictions.
Genome properties
The properties of the draft genome of M. psychrotolerans HV10-M2 are shown in Table 3 and the genes associated with COG functional categories in Table 4. The genome consists of 4,923,400 bp with a GC content of 50.88%. The genome is estimated to be 99% complete as determined by CheckM v1.0.7 [17] compared to the family Methylococcaceae. In total 4465 genes were predicted: 50 RNA genes and 4415 protein-coding genes. In addition, 2344 genes were assigned in COG functional categories. The PHAST program, used to detect prophages sequences in bacterial genomes [18], determined no evidence of completed prophages in the genome of Methylovulum psychrotolerans HV10-M2.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,923,400 | 100 |
| DNA coding (bp) | 4,194,869 | 85.20 |
| DNA G + C (bp) | 2,504,955 | 50.88 |
| DNA scaffolds | 186 | 100 |
| Total genes | 4465 | 100 |
| Protein coding genes | 4415 | 98.88 |
| RNA genes | 50 | 1.12 |
| Pseudo genes | 0 | 0 |
| Genes in internal clusters | 804 | 18.01 |
| Genes with function prediction | 2984 | 66.83 |
| Genes assigned to COGs | 2344 | 52.50 |
| Genes with Pfam domains | 3159 | 70.75 |
| Genes with signal peptides | 438 | 9.81 |
| Genes with transmembrane helices | 897 | 20.09 |
| CRISPR repeats | 0 | 0 |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | %age | Description |
|---|---|---|---|
| J | 194 | 7.48 | Translation, ribosomal structure and biogenesis |
| A | 2 | 0.08 | RNA processing and modification |
| K | 124 | 4.78 | Transcription |
| L | 121 | 4.66 | Replication, recombination and repair |
| B | 1 | 0.04 | Chromatin structure and dynamics |
| D | 40 | 1.54 | Cell cycle control, Cell division, chromosome partitioning |
| V | 88 | 3.39 | Defense mechanisms |
| T | 182 | 7.01 | Signal transduction mechanisms |
| M | 231 | 8.9 | Cell wall/membrane biogenesis |
| N | 56 | 2.16 | Cell motility |
| U | 56 | 2.16 | Intracellular trafficking and secretion |
| O | 137 | 5.28 | Posttranslational modification, protein turnover, chaperones |
| C | 161 | 6.2 | Energy production and conversion |
| G | 97 | 3.74 | Carbohydrate transport and metabolism |
| E | 160 | 6.17 | Amino acid transport and metabolism |
| F | 63 | 2.43 | Nucleotide transport and metabolism |
| H | 156 | 6.01 | Coenzyme transport and metabolism |
| I | 68 | 2.62 | Lipid transport and metabolism |
| P | 154 | 5.93 | Inorganic ion transport and metabolism |
| Q | 45 | 1.73 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 243 | 9.36 | General function prediction only |
| S | 148 | 5.7 | Function unknown |
| – | 2121 | 47.5 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
Here we present the draft genome sequencing and annotation of M. psychrotolerans HV10-M2. The 16S rRNA gene of Methylovulum psychrotolerans HV10-M2 shows a 99% sequence identity with M. psychrotolerans strains Sph1, Sph2 and Oz2 as well as Methylovulum sp. Eb1 and Methylococcaceae bacterium M200 (see also Fig. 2). The highest identity to a strain with an available genome sequence was 95% with Methylovulum miyakonense HT12, which is the only genome sequenced species within the Methylovulum genus.
Extended insights
Methanotrophic microorganisms oxidize methane to carbon dioxide and water. In a first step, methane is converted into methanol in a process catalyzed by the methane monooxigenase enzyme (MMO). There are two types of MMO, the membrane-bound particulate MMO (pMMO), which is found in all methanotrophs except for some members of the genera Methylocella and Methyloferula [19, 20], and the cytoplasmatic soluble MMO (sMMO), which is limited to very few species [21]. In the genome of M. psychrotolerans HV10-M2, the presence of a single copy of the gene cluster pmoABC was observed (locus tags RS17435, RS17440, RS17440). The pmoABC cluster contains the genes encoding for the pMMO. However, the mmoX gene encoding for the sMMO was absent in the genome of M. psychrotolerans HV10-M2. The mmoX gene is reported to be present in M. miyakonense HT12. To confirm the absence of this gene in M. psychrotolerans HV10-M2, the mmoX gene sequence was blasted against the genome of M. psychrotolerans HV10-M2 using the sequence of M. miyakonense HT12 as query (Genbank accession number AB501287). In addition, a mmoX-specific PCR with the primer set 882F/1403R [22] was performed. Both approaches confirmed the absence of the mmoX gene in M. psychrotolerans HV10-M2. Additionally, the pxmABC operon described in some gammaproteobacterial methanotrophs [23], was not observed in the genome of M. psychrotolerans HV10-M2.
The following step in the oxidation of methane is the conversion of methanol into formaldehyde. This reaction is catalyzed by a methanol dehydrogenase that contains a pyrroloquinoline quinone as a cofactor and requires a cytochrome c as electron acceptor [24]. The gene clusters associated with this step present in the genome of M. psychrotolerans HV10-M2 are: (i) the gene cluster mxaFJ that encodes the components active in methanol oxidation (locus tags RS12435, RS12440); (ii) the cluster mxaACKL required for MDH synthesis and PQQ insertion into the MDH (locus tags RS12465, RS12475, RS12480); and (iii) the gene cluster pqqABCDE involved in the PQQ biosynthesis (locus tags RS20845, RS01900, RS01880, RS05860, RS12150). The MDH enzyme is also present in M. miyakonense HT12. The gene xoxF that encodes for a polypeptide with similar sequence to the MxaF protein, has been suggested as an alternative to the MDH [25]. High identity (89%) between a gene encoding for a PQQ-dependent dehydrogenase in M. psychrotolerans HV10-M2 (locus tag RS12435) and a methanotrophic XoxF protein (Accesion number: SAJ59293), suggesting that this protein could be also present in M. psychrotolerans HV10-M2.
The next step is the formaldehyde oxidation. This step is crucial, as formaldehyde is a cytotoxic compound. The methanotrophs can use different pathways to perform oxidation of formaldehyde to formate. In the tetrahydromethanopterin (H4MPT)-linked C1 transfer pathway, presence of the genes encoding for the tetrahydromethanopterin protein and processes such as fae, mch and mtdB, were observed in the genome of M. psychrotolerans HV10-M2. Most likely the H4MPT-linked pathway will act as a secondary pathway involved in formaldehyde assimilation as the Ribulose MonoPhosphate will act as the principal pathway [24]. In addition, like in most of the methylotrophs, presence of the genes encoding for the tetrahydrofolate pathway including the methylene-H4F dehydrogenase and methenyl-H4F cyclohydrolase enzymes of the FoID [24], are also present in the genome of M. psychrotolerans HV10-M2 (locus tags RS10730, RS15610).
Methanotrophic microorganisms can be divided into type I (Gammaproteobacteria) and type II (Alphaproteobacteria) based on the cyclic pathway followed to perform C1 assimilation. Recently, type X methanotrophs have also been described [26]. Type I methanotrophs, such as M. psychrotolerans HV10-M2, use the RuMP cycle, whereas type II methanotrophs use the serine cycle. Briefly, in the first step of the RuMP pathway D-arabino-3-hexulose-6-phosphate is formed from ribulose-5-phosphate, which will be later converted into fructose-6-phosphate. Then, fructose-6-phosphate is converted into Fructose-1,6-bisphosphate through the 6-phosphofructokinase using PPi as the donor in a reversible reaction. Finally, an aldolase will form glyceraldehyde-3-phosphate.
Several genes encoding for enzymes used in the Serine cycle such as hydroxytransmethylase, serine-glyoxylate aminotransferase and hydroxypyruvate reductase were also present in the genome of M. psychrotolerans HV10-M2. However, the serine cycle is not completed, as the genes encoding for the malyl-CoA lyase enzyme could not be found in the genome. This is not surprising as other type I methanotrophs such as M. miyakonense HT12, Methylobacter tundripaludum SV96 [27] or Methylomicrobium album BG8 [28] also contain the genes encoding for most of the serine pathway. However, in the latter two, the phosphoenolpyruvate carboxylase enzyme is absent in the genome while in the Methylovulum strains is present. So far, there is no knowledge about any methanotrophic strain encoding for all the enzymes in both, the RuMP and the serine cycle.
The oxidation of formate to CO2 is performed by the formate dehydrogenase enzyme. The genes encoding for the FDH enzyme were also present in the genome of M. psychrotolerans HV10-M2 (locus tag RS07700). It has been previously reported that this step is less demanding in organisms using the RuMP pathway for formaldehyde assimilation, such as M. psychrotolerans HV10-M2, and therefore FDH activities are very low [24].
It is known that some methanotrophs such as members of the genera Methylococcus and Methylocaldum (Type X methanotrophs), have genes encoding for enzymes involved in the Calvin-Benson-Basham cycle responsible for carbon dioxide fixation [29]. However, no evidence of the genes encoding for the ribulose-1,5-biphosphate carboxylase/oxygenase (RuBisCO), cbbL, cbbS and cbbQ, was found in the genome of M. psychrotolerans HV10-M2.
Methanotrophic bacteria also play a major role in the nitrogen cycle. The MMO can oxidize ammonia into nitrite and nitrous oxide as they are evolutionary related [30]. The prevailing view was that only type II and type X methanotrophs could have the ability to fix nitrogen, and although recently it has been suggested that some type I methanotrophs (Methylomonas and Methylobacter) could also perform nitrogen assimilation. Although in the genome of M. psychrotolerans HV10-M2 the nitrogenase gene cluster nifDKH was present (locus tag RS1055, RS01060, RS01050), the anfG gene encoding for the nitrogenase delta subunit was absent, therefore M. psychrotolerans HV10-M2 cannot fix nitrogen. This result matches with other members of the genus Methylovulum where no growth in nitrogen-free medium was reported [4]. The nitrite reductase genes nirK and nirS involved in the formation of nitric oxide were not found in the genome of M. psychrotolerans HV10-M2, furthermore the genes norBC, involved in the following reaction (formation of nitrous oxide from nitric oxide), were not detected in the genome of M. psychrotolerans HV10-M2. Although the nitrite reductase genes are common in Gammaproteobacteria methanotrophs, they have not been found in others such as M. miyakonense HT12 or Methylomicrobium alcaliphilum or Methylomonas denitrificans [4, 31]. The absence of the genes norBC, present in the latter methanotrophs, in M. psychrotolerans HV10-M2 could be due to the completeness of the genome. Additionally the presence of the haoAB genes, encoding for the hydroxylamine dehydrogenase, that have been observed to be variable in Gammaproteobacteria methanotrophs, are absent in the genome of M. miyakonense HT12 and M. psychrotolerans HV10-M2.
Compared to the reference strain of the Methylovulum genus, Methylovulum miyakonense HT12, there are genetic and morphological differences with M. psychrotolerans HV10-M2. The former has a pink color while the latter exhibited brown color. Additionally, the sMMO enzyme has been found only in one strain within the Methylovulum genus (M. miyakonense HT12). Further, the optimal growth temperature range between M. psychrotolerans and M. miyakonense is different. M. psychrotolerans is psychrotolerant with an optimal growth temperature between 13 and 20 °C, whereas Methylovulum miyakonense HT12 is mesophilic with an optimal temperature between 24 and 32 °C. Interestingly, most of the characteristics not shared between those strains such as, the pink color and the absence of the sMMO enzyme, are common within the M. psychrotolerans strains including the strain M200 in the family Methylococcaceae [32] (Information about Methylovulum sp. Eb1 is not available).
Those differences together with the 95% identity in the 16S rRNA gene between M. miyakonense HT12 and M. psychrotolerans HV10-M2 could suggest that the former belong to a different genus. To investigate this, the average nucleotide identity using BLAST were performed with the draft genomes. The strains shared ANIb values of 79.2%, being the highest amongst the closest strains with genome data available (Additional file 1: Table S1). Furthermore, recently it has been proposed that a prokaryotic genus can be defined as a group of species with pairwise values in the percentage of conserved proteins higher than 50% [33]. The POCP value between M. miyakonense HT12 and M. psychrotolerans HV10-M2 was 62.9%, therefore suggesting that M. miyakonense HT12 and M. psychrotolerans HV10-M2 belong to the same genus.
Conclusions
In the present study, we present the high-quality draft genome of Methylovulum psychrotolerans HV10-M2. The genome consists of 4,923,400 bp in 4415 protein-coding genes, 50 RNA genes with and an average 50.88% GC content. As the Methylovulum genus has been recently described [4] only one genome has been available so far and this is from Methylovulum miyakonense HT12. M. psychrotolerans HV10-M2 has a 95% sequence identity with M. miyakonense HT12. In addition, there are some differences between both species, such as the mmoX gene, which encodes for the sMMO enzyme. The mmoX gene is present only in M. miyakonense whereas in M. psychrotolerans is absent. The other differences are the colour, as M. miyakonense showed a brown colour and M. psychrotolerans is pink, and the optimal growth temperature. M. miyakonense is mesophilic and M. psychrotolerans is psychrotolerant.
Additional file
Table S1. ANIb analysis results with the similarities between the draft genomes of the four closest strains to M. psychrotolerans HV10-M2. (DOCX 15 kb)
Acknowledgements
We thank Britta Poulsen and Hilde R. Armo for technical assistance and John Colin Murrell for valuable comments and discussions.
Funding
AMR was funded by a PhD research grant from Western Norway University of Applied Sciences. This study is part of the project “Microorganisms in the Arctic: Major drivers of biogeochemical cycles and climate change” (RCN 227062) funded by the Norwegian Research Council. Additional funding was provided by Aarhus University Graduate School of Science and Technology, the Danish National Research Foundation (grant no. DNRF104), ERC Advanced Grant MICROENERGY [grant no. 294200] (European Union 7th Framework Program) and the Marie Curie IIF fellowship “ATP_adapt_low_energy” (European Union 7th Framework Program).
Abbreviations
- CBB
Calvin-Benson-Basham
- FDH
Formate dehydrogenase
- H4F
Tetrahydrofolate
- H4MPT
Tetrahydromethanopterin
- LMM
Low-salt methanotrophic medium
- MDH
Methanol dehydrogenase
- MMO
Methane monooxigenase
- pMMO
Membrane-bound particulate MMO
- POCP
Percentage of conserved protein
- PQQ
Pyrroloquinoline quinone
- RuBisCO
Ribulose-1,5-biphosphate carboxylase/oxygenase
- RuMP
Ribulose MonoPhosphate
- sMMO
cytoplasmatic soluble MMO
Authors’ contributions
LØ and TI designed the experiment. TI collected the samples. TI and AMR isolated and characterized the strain. AMR performed the bioinformatics analyses with significant help from IM and LS. AMR wrote the manuscript with significant help from LØ, TI, IM and LS. All authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40793-018-0314-2) contains supplementary material, which is available to authorized users.
References
- 1.Aronson E, Allison S, Helliker BR. Environmental impacts on the diversity of methane-cycling microbes and their resultant function. Front Microbiol. 2013;4:225. doi: 10.3389/fmicb.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes AJ, Tujula NA, Holley M, Contos A, James JM, Rogers P, Gillings MR. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ Microbiol. 2001;3:256–264. doi: 10.1046/j.1462-2920.2001.00187.x. [DOI] [PubMed] [Google Scholar]
- 3.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PL, Liesack W, Feng L, Wang L, Alam M. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature. 2007;450:879–882. doi: 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 4.Iguchi H, Yurimoto H, Sakai Y. Methylovulum miyakonense gen. nov., sp. nov., a type I methanotroph isolated from forest soil. Int J Syst Evol Microbiol. 2011;61:810–815. doi: 10.1099/ijs.0.019604-0. [DOI] [PubMed] [Google Scholar]
- 5.Oshkin I, Belova S, Danilova O, Miroshnikov K, Rijpstra W, Sinninghe Damsté J, Liesack W, Dedysh S. Methylovulum psychrotolerants sp. nov., a cold-adapted methanotroph from low-temperature terrestrial environments, and emended description of the genus Methylovulum. Int J Syst Evol Microbiol. 2016;66:2417–2423. doi: 10.1099/ijsem.0.001046. [DOI] [PubMed] [Google Scholar]
- 6.Islam T, Larsen Ø, Torsvik V, Øvreås L, Panosyan H, Murrell JC, Birkeland NK, Bodrossy L. Novel methanotrophs of the family Methylococcaceae from different geographical regions and habitats. Microorganisms. 2015;3:484–499. doi: 10.3390/microorganisms3030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam T, Torsvik V, Larsen Ø, Bodrossy L, Øvreås L, Birkeland NK. Acid-tolerant moderately thermophilic methanotrophs of the class Gammaproteobacteria isolated from tropical topsoil with methane seeps. Front Microbiol. 2016;7:851. doi: 10.3389/fmicb.2016.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. [Google Scholar]
- 9.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Palaniappan K, Szeto E, Pillay M, Chen IA, Pati A, Nielsen T, Markowitz VM, Kyrpides NC. The standard operating procedure of the DOE-JGI microbial genome annotation pipeline (MGAP v.4) Stand Genomic Sci. 2015;10:86. doi: 10.1186/s40793-015-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Karp PD, Paley S, Romero P. The pathway tools software. Bioinformatics. 2002;18:S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.S225. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Liang Y, Lynch K, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dedysh SN, Dunfield PF. Facultative methane oxidizers. In: Timmis KN, editor. Handbook of hydrocarbon and lipid microbiology. Springer: Berlin; 2010. pp. 1967–1976. [Google Scholar]
- 20.Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh S. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int J Syst Evol Microbiol. 2011;61:2465–2463. doi: 10.1099/ijs.0.028118-0. [DOI] [PubMed] [Google Scholar]
- 21.Miguez CB, Bourque D, Sealy JA, Greer CW, Groleau D. Detection and isolation of methanotrophic bacteria possessing soluble methane monooxygenase (sMMO) genes using the polymerase chain reaction (PCR) Microb Ecol. 1997;33:21–31. doi: 10.1007/s002489900004. [DOI] [PubMed] [Google Scholar]
- 22.McDonald IR, Bodrossy L, Chen Y, Murrell JC. Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol. 2008;74:1305–1315. doi: 10.1128/AEM.02233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MSM, Klotz MG. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3:91–100. doi: 10.1111/j.1758-2229.2010.00192.x. [DOI] [PubMed] [Google Scholar]
- 24.Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson SM, Gleisten MP, Donohue TJ. Identification of proteins involved in formaldehyde metabolism by Rhodobacter sphaeroides. Microbiology. 2008;154:296–305. doi: 10.1099/mic.0.2007/011346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward N, Larsen O, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, Lewis M, Nelson KE, Methé BA, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, RT DB, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonasen I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR, Eisen JA. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (bath) PLoS Biol. 2004;2:1616–1628. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svenning MM, Hestnes AG, Wartiainen I, Stein LY, Klotz MG, Kalyuzhnaya MG, Spang A, Bringel F, Vuilleumier S, Lajus A, Médigue C, Bruce DC, Cheng JF, Goodwin L, Ivanova I, Han J, Han CS, Hauser L, Held B, Land ML, Lapidus A, Lucas S, Nolan M, Pitluck S, Woyke T. Genome sequenced of the Arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol. 2011;193:6418–6419. doi: 10.1128/JB.05380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kits KD, Kalyuzhnaya MG, Klotz MG, Jetten MSM, Op den Camp HJ, Vuilleumier S, Bringel F, Dispirito AA, Murrell JC, Bruce D, Cheng JF, Copeland A, Goodwin L, Hauser L, Lajus A, Land ML, Lapidus A, Lucas S, Médigue C, Pitluck S, Woyke T, Zeytun A, Stein LY. Genome sequence of the obligate gammaproteobacterial methanotroph Methylomicrobium album strain BG8. Genome Announc. 2013;1:e00170–e00113. doi: 10.1128/genomeA.00170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baxter NJ, Hirt RP, Bodrossy L, Kovacs KL, Embley TM, Prosser JI, Murrell JC. The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath) Arch Microbiol. 2002;177:279–289. doi: 10.1007/s00203-001-0387-x. [DOI] [PubMed] [Google Scholar]
- 30.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionary related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1111/j.1574-6968.1995.tb07834.x. [DOI] [PubMed] [Google Scholar]
- 31.Vuilleumier S, Khmelenina VN, Bringel F, Reshetnikov AS, Lajus A, Mangenot S, Rouy Z, Op Den Camp HJM, Jetten MSM, Dispirito AA, Dunfield P, Klotz MG, Semrau JD, Stein LY, Barbe V, Medigue C, Trotsenko YA, Kalyuzhnaya MG. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J Bacteriol. 2012;194:551–552. doi: 10.1128/JB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kip N, Ouyang W, van Winden J, Raghoebarsing A, Van Niftrik L, Pol A, Pan Y, Bodrossy L, Van Donselaar EG, Reichart GJ, Jetten MS, Damsté JS, Op den Camp HJ. Detection, isolation, and characterization of acidophilic Methanotrophs from Sphagnum mosses. Appl Environ Microbiol. 2011;77:5643–5654. doi: 10.1128/AEM.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains archaea, Bacteria, and Eukarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrity G, Bell J, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity G, Brenner D, Krieg N, Staley J, editors. Bergey’s manual of systematic bacteriology. New York: Springer; 2005. [Google Scholar]
- 36.Williams KP, Kelly DP. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63:2901–2906. doi: 10.1099/ijs.0.049270-0. [DOI] [PubMed] [Google Scholar]
- 37.Bowman JP. Order VII. Methylococcales ord. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergeys manual of systematic bacteriology, second edition, vol. 2 (the Proteobacteria), part B (the Gammaproteobacteria) New York: Springer; 2005. pp. 248–252. [Google Scholar]
- 38.Bowman JP, Sly LI, Nichols PD, Hayward AC. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int J Syst Bacteriol. 1993;43:735–753. doi: 10.1099/00207713-43-4-735. [DOI] [Google Scholar]
- 39.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Tamura K. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. ANIb analysis results with the similarities between the draft genomes of the four closest strains to M. psychrotolerans HV10-M2. (DOCX 15 kb)


