Abstract
Transient receptor potential (TRP) melastatin 4 (TRPM4) is a widely expressed cation channel associated with a variety of cardiovascular disorders. TRPM4 is activated by increased intracellular calcium in a voltage dependent manner, but unlike many other TRP channels is permeable to monovalent cations only. Here we present two structures of full-length human TRPM4 embedded in lipid nanodiscs at ~3Å resolution as determined by single particle electron cryo-microscopy. These structures, with and without calcium bound, reveal a general architecture for this major subfamily of TRP channels and a well-defined calcium binding site within the intracellular side of the S1–S4 domain. The structures correspond to two distinct closed states. Calcium binding induces conformational changes that likely prime the channel for voltage-dependent opening.
Transient receptor potential (TRP) channels comprise an extended superfamily of membrane ion channels that mediate diverse cellular and physiological functions. Permeability to both mono- and divalent cations is a defining feature for the majority of TRP channels (1). However, the TRP melastatin subfamily member 4 (TRPM4) channel is impermeable to Ca2+, yet activated by intracellular Ca2+ (2). Activation of TRPM4 depolarizes the plasma membrane through Na+-entry, which in turn enhances Ca2+-influx through Ca2+-permeable channels or otherwise modulates Ca2+ oscillations (2, 3). Like many other TRP channel subtypes, TRPM4 displays voltage sensitivity as evidenced by pronounced outward rectification of Ca2+-activated TRPM4 currents in response to voltage ramps (4). Binding of Ca2+ to TRPM4 is hypothesized to precede voltage-dependent opening, thereby giving rise to two closed states: Ca2+-unbound and Ca2+-bound (5). However, it is unclear where the Ca2+-binding site is located in the channel and a structural basis for voltage sensitivity and ion selectivity remains elusive. Here, we present two single particle electron cryo-microscopy (cryo-EM) structures of the human TRPM4 channel in nanodiscs, in both Ca2+-bound and unbound closed states. Comparison of the two structures reveals a Ca2+ binding site in the transmembrane domain, and conformational changes induced by Ca2+. Lipid molecules, including cholesteryl hemisuccinate (CHS), form tight interactions with the channel, likely stabilizing it in the lipid bilayer.
Recombinant full-length human TRPM4 (hTRPM4b) was purified in detergent, and reconstituted into lipid nanodiscs as described for other TRP ion channels (6) (fig. S1). Three-dimensional (3D) reconstructions with a C4 symmetry were obtained in the presence of either 5 mM EDTA or CaCl2 to overall resolutions of 3.2Å and 3.1Å (Fig. 1A, B, fig. S2A–F and S3A–F). In both structures, the transmembrane domain and part of the soluble domain are well resolved. However, the soluble regions distal to the membrane and the symmetry axis are of lower resolution, indicating conformational flexibility (fig. S2G, S3G). Focused refinement of the transmembrane domain with the central coiled-coil improved local resolution and enabled de novo atomic model building of these domains for both samples (fig. S2F, H; S3F, H; S4B, D and S5). For the rest of the cytoplasmic domains, further classification and focused refinement of individual monomers without symmetry improved the resolution to a level sufficient for de novo model building (Fig. 1C–F, fig. S2I–K, S3I-K, S4C, E and S5). The complete data processing scheme is described in Supplementary Methods and Figure S4.
Figure 1. Cryo-EM structure of hTRPM4.

(A and B) Unsharpened (in transparent light blue) and sharpened cryo-EM density map of hTRPM4 in nanodiscs in side (A) and top (B) views with each subunit colored differently. (C) Atomic model of the tetrameric hTRPM4 in ribbons, in the same orientation and colors as the density map in A. (D) Atomic model of the hTRPM4b monomer in ribbons with domains labeled. (E) bottom view of the hTRPM4 structure showing interactions between neighboring subunits. (F) A schematic representation of the major structural components in hTPPM4b. Dashed lines denote regions where density was insufficient for model building. Each domain is labeled and color-coded to match the domain representation in (D).
The transmembrane core contains six helices (S1-S6) arranged in a domain-swapped architecture (Fig. 1C, D) reminiscent of many tetrameric voltage-gated cation channels, and all other TRP channels to date (7–13). hTRPM4 has additional membrane embedded fragments that surround the S1–S4 domain from the inner leaflet of the membrane (Fig. 1D, F), a feature that might be distinct to the TRPM subfamily. The C-terminus of each subunit joins to form a parallel coiled-coil resembling that of TRPA1 channel (11). The coiled-coil is surrounded by a large, intertwined cytoplasmic domain, comprised of four N-terminal TRPM Homology Regions (MHR1-4), which are highly conserved within the TRPM subfamily (14). MHR1-2 contains an eight-stranded β-sheet surrounded by eight α-helices. MHR3-4 is composed of stacked α-helices and linked to the transmembrane domain through MHR4, which clasps the TRP domain, thereby forming an interaction between the cytoplasmic domain and the transmembrane core (Fig. 1D). Between neighboring monomers, MHR1 of one subunit interacts with the MHR3 of the neighboring subunit forming a ring that does not interact with the central coiled-coil (Fig. 1E).
Comparison of Ca2+-free and Ca2+-bound structures reveals an extra density in a hydrophilic pocket within the cytoplasmic side of the S1–S4 domain (Fig. 2A and fig. S6). Several lines of analysis suggest that this density represents a bona fide bound Ca2+-ion: 1) Direct comparison between both half maps and the combined maps of the CaCl2 and EDTA samples confirms the existence and precise location of the additional density in the CaCl2 map (fig. S6A–C, E–G); 2) the omit densities calculated as the difference between the experimental maps and map calculated from the refined atomic models without Ca2+ show a strong density corresponding to a bound Ca2+-ion (fig. S6D) in the CaCl2 structure, but not in the EDTA structure; 3) the difference map between the CaCl2 and EDTA maps reveals a clear density with a high signal-to-noise ratio (Fig. 2A and fig. S6H); 4) side chain densities of surrounding negatively charged residues are stronger in the CaCl2 structure than in the EDTA structure (fig. S6A, E), consistent with the observation that negatively charged side chains are generally weaker in a cryo-EM density map unless they are engaged in specific interactions (15).
Figure 2. The Ca2+ binding site.
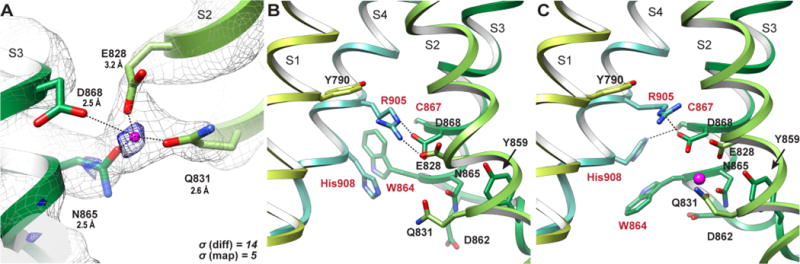
(A) Coordination of the bound Ca2+-ion by Glu828, Gln831, Asn865 and Asp868, with distances labelled. The density of the CaCl2 structure (gray mesh) is contoured at σ = 5 and overlaid with the difference density (blue mesh) between the CaCl2 structure and the EDTA structure, contoured at σ = 14. (B) Ca2+ binding site within the S1–S4 domain in the absence of bound ion. (C) The same site with a bound ion. Side chains are labeled. The bound Ca2+ ion is shown in magenta.
The Ca2+ ion is directly coordinated by side chains of Glu828 and Gln831 from S2, and Asn865 from S3 (Fig. 2A). Asp868 is also near the ion and may participate in its coordination. Together, these side chains contribute at least four oxygens that can coordinate the Ca2+. The Ca2+ binding site is located within a hydrophilic pocket that is large enough to accommodate additional water molecules for further coordination of the bound ion. This pocket is connected to the cytoplasmic space through a narrow path between the TRP domain and the S2–S3 linker (fig. S6I, J). Within the pocket, Arg905 from a single 310 helical turn in S4 and Tyr790 from the S1 are located above the Ca2+ binding site. In the absence of Ca2+, Arg905 is coordinated by Glu828 and Asp868 (Fig. 2B). Upon Ca2+ binding, the S2–S3 linker translates ~1.5 Å such that Tyr859 supports a tighter conformation for coordination of Ca2+ by Glu828, Gln831, Asn865 and Asp868 (fig. S6L). Furthermore, the side chain of His908 in S4 transitions from a π-π stacking arrangement with Trp864 in S3 to form a new interaction with Cys867 (Fig. 2B, C and fig. S6K–N). Without coordination of Glu828 and with rearrangement in S3 and S4, Arg905 moves slightly upward (fig. S6N). The configuration of Arg905 and Tyr790 is reminiscent of a positive gating charge (arginine) and charge transfer center (tyrosine) seen in voltage-gated potassium channels (16). We speculate that Ca2+ binding moves Arg905 up towards Tyr790 to prime the channel for voltage-dependent opening.
A previous mutagenesis study showed that the Glu1068Gln mutation significantly reduces TRPM4 Ca2+-sensitivity (17). Glu1068 is located in the pathway leading to the Ca2+-binding site from the cytoplasmic space (fig. S6I, J), and this mutation likely reduces the Ca2+ accessibility to the site. Of the four residues that constitute the Ca2+ binding site, Asn865 and the negative charge of Asp868 are conserved throughout the TRPM subfamily, while Glu828 and Gln831 as well as Tyr859 and Glu1068 are conserved only in subfamily members that are shown to be Ca2+ dependent: TRPM2, TRPM4, TRPM5 and TRPM8(18–20) (fig. S7). Therefore, it is likely that there is an analogous binding site in these channels. Electrophysiological and mutagenesis studies of TRPM8 suggest that Ca2+ binds directly to an analogous site in that channel to confer sensitivity to icilin, a synthetic cooling agent. Specifically, substituting the residues corresponding to Asn865 and Asp868 resulted in icilin-insensitive channels that retain robust Ca2+-independent responses to cold and menthol (21).
The central pore of hTRPM4 is formed by S5 and S6, and the intervening re-entrant pore helix and pore loop. Together, these elements form an ion permeation pathway with two restriction sites, similar to other TRP channels (Fig. 3A, B). As seen in TRPV1(10), the outer pore has a funnel shape with a negatively charged residue facing the funnel, attracting cations (fig. S8A, B). Two highly conserved residues, Phe975 and Gly976, located in the bottom of the pore loop, form the narrowest restriction point of the upper pore. The pore diameter at this site is very similar to that of the open gate in TRPV1 (22) (Fig. 3B), which is sufficient to accommodate partially dehydrated monovalent cations. However, no density corresponding to coordinated ions is found within the pore (fig. S8C, D). The location of a previously identified putative selectivity filter (981EDMDVA986) (23) does not coincide with this restriction site, but is located further towards the extracellular face (Fig. 3C). At the bottom of the ion permeation pathway, side chains of opposing Ile1040 residues in S6 form a tight seal at the lower restriction site, signifying that both structures are in closed conformations (Fig. 3A, B).
Figure 3. Ion permeation pore.
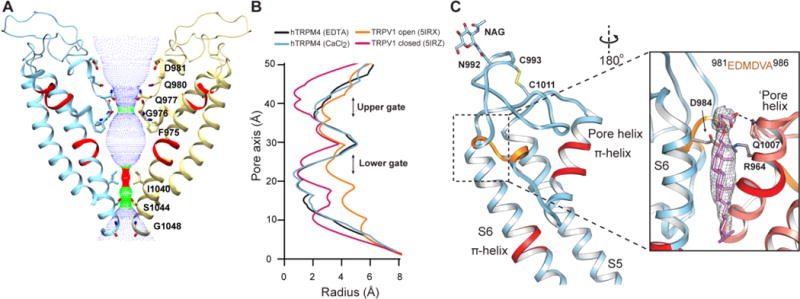
(A) The solvent-accessible pathway along the ion permeation pore represented by dots between two opposing monomers shown in ribbons colored yellow and blue. Residues aligning the pathway are shown in sticks and labelled. (B) The pore radius of the hTRM4 EDTA (black) and CaCl2 (blue) structures overlaid with the pore radius of TRPV1 in its closed (purple) and open (orange) states. (C) A close-up view of the pore helix and pore loop. The two single-turn π-helices are marked with red in the middle of S6 and the pore helix. The putative selectivity filter is marked with orange. The insert represents the density of a bound lipid, fitted with the atomic model of CHS, which forms a tripartite complex with S1 from one monomer and the pore helix of the neighboring monomer (‘Pore helix).
The pore helix of hTRPM4 has an additional turn compared to that in other TRP channels (Fig. 3C) (9–13). In the middle of the pore helix there is a single-turn π-helix (Arg964-Arg969) followed by a proline residue, Pro970, conserved in all human TRPM channels except TRPM2. Likewise, similar to what was first identified in TRPV1 (10) and subsequently observed in other TRP channels (7, 11), there is a one-turn π-helix (Val1030-Leu1035) in the middle of the S6 helix (Fig. 3C). While the exact role of these single-turn π-helices is unclear, they could potentially facilitate helix bending under different functional states, leading to movement of the lower gate or modulation of the upper pore.
Outside of the pore, hTRPM4 has an extensive extracellular loop comprising more than 30 residues that connect the pore loop to the S6 helix. The loop is clearly resolved, likely stabilized by a disulfide bond between two cysteines (Cys993 and Cys1011) (Fig. 3C and fig. S8F, G), a feature predicted to exist in the majority of TRPM channels. This disulfide bond is not seen in the CaCl2 structure (fig. S7H), where disulfide bond breakage may have been caused by radiation damage (24). The loop also contains a glycosylation site (Asn992) with the attached glycan pointing towards the extracellular space (Fig. 3C and fig. S7G, H).
In addition to the transmembrane S1–S6 domain, hTRPM4 has distinct membrane embedded α-helical segments (Fig. 4A). These segments surround the exterior of the S1–S4 domain within the inner leaflet of the membrane and mediate extensive interactions with the soluble domain. Preceding the S1 helix, there are two short helices shaped as an inverted ‘V’ embedded within the inner leaflet of the membrane (Fig. 4B). We term it the “pre-S1 elbow”, similar to that observed in the mechanotransduction channel NOMPC (9). A disordered loop unresolved in both structures, connects the pre-S1 elbow to an amphipathic helix termed as the “pre-S1 shoulder” positioned at the inner surface of the membrane (Fig. 4C). The pre-S1 shoulder helix contains large hydrophobic residues buried in the membrane and a number of charged residues facing the cytosol. Among these charged residues, Arg767 was identified as important for interactions with the phosphatidylinositol lipids, PIP2 and PIP3 (25).
Figure 4. Membrane embedded helical segments surround the S1–S4 domain.

(A) Side view of the hTRPM4 monomer with membrane embedded helical segments labeled and in solid color. (B) The pre-S1 elbow (gold) and pre-S1 shoulder (yellow). Lipid density between the pre-S1 elbow and S1 is modeled with CHS. Side chains interacting with the CHS are shown as sticks. (C) Same as (B), viewed from the opposite direction. (D) Cytoplasmic domain MHR3 interacts with the S2–S3 linker. (E) The CH1 helix after the TRP domain. Charged residues in the pre-S1 shoulder (C) and CH1 (E) are shown in sticks. Missing links are shown in dashed lines in (A) and (D).
An extended S2–S3 linker distinct to the TRPM subfamily forms a short amphipathic helix, positioned at the membrane surface, near the Ca2+-binding site (Fig. 2B). The loop that connects S2 with the S2–S3 linker is ~12 residues longer in hTRPM4 compared to other human TRPM channels and extends beyond the membrane bilayer, interacting with MHR4. Ser839 in the linker is a predicted phosphorylation site important for trafficking TRPM4 to plasma membrane (26), however, this linker is only weakly resolved in the CaCl2 structure. In addition, the S2–S3 linker also interacts loosely with the cytoplasmic domain MHR3 (Fig. 4D).
The last segment is positioned between the pre-S1 shoulder and S2–S3 linker helix (Fig. 4A, F). Here, a loop fills the gap between S1 and S2 and connects the conserved TRP domain to the C-terminal Helix 1 (CH1), which is located at a steep angle relative to S1, extending downwards from the middle of the membrane bilayer to the cytoplasmic surface more than 20Å away from the end of the TRP domain (Fig. 4E). The C-terminus of CH1 contains five arginine residues, similar to the pre-S1 shoulder mentioned above, which likely constitute interaction partners for negatively charged lipid headgroups at the membrane interface. CH1 connects to CH2 through a 19-residue linker which was only partially resolved in the structures and observed to associate with MHR4 and the C-terminal end of the TRP-domain.
A number of lipids were observed in our structures (fig. S9A). In addition to annular phospholipids, we identified three densities as CHS based on its characteristic shape (Fig. 3C, 4B and fig. S9B). CHS is often used as a cholesterol analog (27), thus a cholesterol molecule is likely to bind at the same sites. One CHS molecule fills a cleft at the backside of the pore, interacting with the S6 helix and the pore loop or the proposed selectivity filter of one subunit and the pore helix of the neighboring subunit. This arrangement, together with the relative rigidity of CHS over annular phospholipids, may stabilize the conformation of the pore (Fig. 3C and fig. S8E). A second CHS molecule is positioned at a location equivalent to the vanilloid binding pocket in TRPV1 (6) (fig. S9C). A third CHS is located at the pre-S1 elbow, which together with S1 and S4 creates a cavity (Fig. 4B).
While there is no noticeable change of the pore between the two structures (Fig. 3B, fig. S10A, C), we observe subtle yet well-defined changes in the region surrounding the Ca2+ binding site (fig. S10). In addition, CH2 is horizontally displaced by ~2.5 Å around the central coiled-coil, measured from the Cα of Glu1116, in the distal end of CH2 (fig. S10D). The rotation results in a slight tightening of the central coiled-coil (fig. S10E), likely stabilizing it as evident by the observation that more residues in the C-terminal end of the coiled-coil were resolved in the CaCl2 structure (fig. S2G, 3G). Given that the lower gate remains closed in both structures, such conformational changes are insufficient for channel opening. However, as the extensive soluble domains are implicated in interactions with various co-factors (28), such conformational changes may be important functional features that enable the channel to better detect or response to its co-factors (28).
Supplementary Material
One sentence summary.
Structures of the human TRPM4 channel reveal the architecture of the TRPM channel subfamily and its calcium binding site.
Acknowledgments
We thank M. Diver, P. Dominik, A. Kintzer, R. Stroud for valuable discussions, M. Diver and E. Green for reading the manuscript. We thank Y. Jiang for coordinating co-submission of our studies. H.E.A. is supported by a postdoctoral fellowship from the Danish Council of Independent Research (Grant No. DFF-5051-00085). This work was supported by grants from National Institute of Health (R01NS047723 to D.J., R01GM098672, S10OD020054, S10OD021741 to Y.C). Y.C. is an Investigator of the Howard Hughes Medical Institute. Accession numbers of the human TRPM4 structures in EDTA and CaCl2 are: XXXX and XXXX (coordinates of atomic models), EMD-XXXX and EMD-XXXX (density maps).
References
- 1.Julius D. TRP channels and pain. Annu Rev Cell Dev Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 2.Launay P, et al. TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell. 2002;109:397–407. doi: 10.1016/s0092-8674(02)00719-5. [DOI] [PubMed] [Google Scholar]
- 3.Launay P, et al. TRPM4 regulates calcium oscillations after T cell activation. Science. 2004;306:1374–1377. doi: 10.1126/science.1098845. [DOI] [PubMed] [Google Scholar]
- 4.Nilius B, et al. Voltage dependence of the Ca2+-activated cation channel TRPM4. J Biol Chem. 2003;278:30813–30820. doi: 10.1074/jbc.M305127200. [DOI] [PubMed] [Google Scholar]
- 5.Nilius B, Prenen J, Janssens A, Voets T, Droogmans G. Decavanadate modulates gating of TRPM4 cation channels. J Physiol. 2004;560:753–765. doi: 10.1113/jphysiol.2004.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, et al. Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature. 2017;550:415–418. doi: 10.1038/nature24035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirschi M, et al. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature. 2017;550:411–414. doi: 10.1038/nature24055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin P, et al. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature. 2017;547:118–122. doi: 10.1038/nature22981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmiege P, Fine M, Blobel G, Li X. Human TRPML1 channel structures in open and closed conformations. Nature. 2017;550:366–370. doi: 10.1038/nature24036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen PS, et al. The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell. 2016;167:763–773 e711. doi: 10.1016/j.cell.2016.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelps CB, Gaudet R. The role of the N terminus and transmembrane domain of TRPM8 in channel localization and tetramerization. J Biol Chem. 2007;282:36474–36480. doi: 10.1074/jbc.M707205200. [DOI] [PubMed] [Google Scholar]
- 15.Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci U S A. 2014;111:11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi S, Tanimoto A, Otsuguro K, Hibino H, Ito S. Negatively charged amino acids near and in transient receptor potential (TRP) domain of TRPM4 channel are one determinant of its Ca2+ sensitivity. J Biol Chem. 2014;289:35265–35282. doi: 10.1074/jbc.M114.606087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faouzi M, Penner R. Trpm2. Handb Exp Pharmacol. 2014;222:403–426. doi: 10.1007/978-3-642-54215-2_16. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann T, Chubanov V, Gudermann T, Montell C. TRPM5 is a voltage-modulated and Ca(2+)-activated monovalent selective cation channel. Curr Biol. 2003;13:1153–1158. doi: 10.1016/s0960-9822(03)00431-7. [DOI] [PubMed] [Google Scholar]
- 20.McKemy DD, Neuhausser WM, Julius D. Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature. 2002;416:52–58. doi: 10.1038/nature719. [DOI] [PubMed] [Google Scholar]
- 21.Chuang HH, Neuhausser WM, Julius D. The super-cooling agent icilin reveals a mechanism of coincidence detection by a temperature-sensitive TRP channel. Neuron. 2004;43:859–869. doi: 10.1016/j.neuron.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 22.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilius B, et al. The selectivity filter of the cation channel TRPM4. J Biol Chem. 2005;280:22899–22906. doi: 10.1074/jbc.M501686200. [DOI] [PubMed] [Google Scholar]
- 24.Weik M, et al. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci U S A. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bousova K, et al. PIP2 and PIP3 interact with N-terminus region of TRPM4 channel. Biophys Chem. 2015;205:24–32. doi: 10.1016/j.bpc.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Shin SH, Lee EJ, Chun J, Hyun S, Kang SS. Phosphorylation on TRPV4 Serine 824 Regulates Interaction with STIM1. Open Biochem J. 2015;9:24–33. doi: 10.2174/1874091X01509010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zocher M, Zhang C, Rasmussen SG, Kobilka BK, Muller DJ. Cholesterol increases kinetic, energetic, and mechanical stability of the human beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2012;109:E3463–3472. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilius B, et al. Regulation of the Ca2+ sensitivity of the nonselective cation channel TRPM4. J Biol Chem. 2005;280:6423–6433. doi: 10.1074/jbc.M411089200. [DOI] [PubMed] [Google Scholar]
- 29.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie TK, et al. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A, Brubaker MA, Fleet DJ. Building Proteins in a Day: Efficient 3D Molecular Structure Estimation with Electron Cryomicroscopy. IEEE Trans Pattern Anal Mach Intell. 2017;39:706–718. doi: 10.1109/TPAMI.2016.2627573. [DOI] [PubMed] [Google Scholar]
- 35.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


