Abstract
Polintons (also known as Mavericks) are large DNA transposons that are widespread in the genomes of eukaryotes. We have recently shown that Polintons encode virus capsid proteins, which suggests that these transposons might form virions, at least under some conditions. In this Opinion article, we delineate the evolutionary relationships among bacterial tectiviruses, Polintons, adenoviruses, virophages, large and giant DNA viruses of eukaryotes of the proposed order ‘Megavirales’, and linear mitochondrial and cytoplasmic plasmids. We hypothesize that Polintons were the first group of eukaryotic double-stranded DNA viruses to evolve from bacteriophages and that they gave rise to most large DNA viruses of eukaryotes and various other selfish genetic elements.
All cellular life forms, with the possible exception of some extremely reduced intra-cellular parasites, are hosts to viruses and/or other selfish elements, such as transposons and plasmids1. This ‘greater virus world’ is enormously diverse in terms of replication mechanisms, gene-expression mechanisms, virion structure, and genome structure and size — viral genomes consist of single-stranded (ss) or double-stranded (ds) RNA or DNA and range in size from <2 kb to >2 Mb2–4.
Viruses and other selfish elements do not share a single common ancestor; indeed, not a single gene is conserved in all or even most of these genomes5,6. However, selfish elements form a complex, dense network of evolutionary relationships in which genomes are linked through different shared genes3. This type of evolutionary relationship is most likely to have emerged from extensive gene exchange, sometimes between widely different elements. Viruses with large genomes possess many genes that have been acquired from the hosts at different stages of evolution. However, the hallmark genes of viruses, those that encode key proteins involved in genome replication and virion formation, are represented in a broad range of elements that form many edges in the evolutionary network3,5,6.
The viruses present in archaea, bacteria and eukaryotes are fundamentally different3,4. In archaea and bacteria, most viruses possess dsDNA genomes. The second-most-common class includes small ssDNA viruses. Retrotranscribing elements comprise a small minority (no retroviruses are known) and RNA viruses are rare. Many dsDNA viruses as well as ssDNA viruses of archaea and bacteria alternate between lytic and temperate modes of reproduction. In the temperate mode, a viral genome integrates into a bacterial or archaeal chromosome and is transmitted vertically in the form of a provirus or prophage4; a typical bacterial or archaeal genome carries multiple prophages.
In contrast to bacteria and archaea, eukaryotes host numerous, diverse RNA viruses, retrotranscribing elements and retroviruses that typically integrate into the host genome7,8. In comparison with RNA viruses and retroelements, ssDNA and dsDNA viruses and mobile elements are less diverse and less common in eukaryotes, although both of these classes of selfish elements are widespread3. By far the largest group of DNA viruses in eukaryotes (BOX 1) consists of seven families of large and giant viruses (including mimiviruses and pandoraviruses, which have genomes in the megabase range). These families seem to share a common ancestry, as indicated by the conservation of approximately 50 (putative) ancestral genes9–11. Together, this group is known as the nucleocytoplasmic large DNA viruses (NCLDVs), or more recently the proposed order ‘Megavirales’ (REF. 12). The giant viruses of the family Mimiviridae are associated with a distinct class of satellite viruses, the virophages, which reproduce within viral ‘factories’ inside protist cells infected by the Mimiviridae and which depend on the latter for their replication13–16.
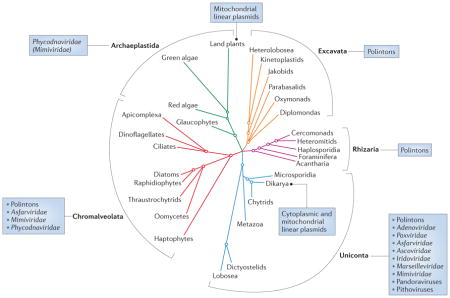
Box 1. Distribution of Polintons, linear plasmids and ‘Megavirales’ in Eukarya.
Viruses of the proposed order ‘Megavirales’ fall into seven well-established families12, but some recently isolated Megavirales-derived viruses, such as pandoraviruses71 and Pithovirus sibericum69, remain unclassified. Thus far, Megavirales have been shown to propagate in organisms from three of the five eukaryotic supergroups72, namely Archaeplastida, Chromalveolata and Uniconta (see the figure). The Megavirales associated with the Uniconta show the greatest diversity: within this supergroup, members of the families Marseilleviridae and Mimiviridae, as well as unclassified pandoraviruses and pithovirus, infect amoebae; members of the Ascoviridae infect insects; iridoviruses are associated with either insects or fish; poxviruses infect various animals, including insects, reptiles, molluscs, birds and mammals; and Asfarviridae infect mammals. In the Chromalveolata, members of the Phycodnaviridae infect brown algae (phylum Heterokontophyta) and coccolithophores (phylum Haptophyta); some members of the Mimiviridae prey on microflagellate grazers (Heterokontophyta) and bloom-forming microalgae (Haptophyta); and a putative member of Asfarviridae infects dinoflagellates. Finally, phycodnaviruses and some mimiviruses are known and thought, respectively, to infect green algae in the eukaryotic kingdom Archaeplastida.
Polintons and related large DNA transposons have an equally wide, if not wider, distribution in Eukarya20,21. Polintons are widespread in Uniconta, where they are found in diverse Amoebozoa, Metazoa and Dikarya (fungi). Among the Chromalveolata, Polintons are present in oomycetes, whereas a distinct type of Polinton, known as Tlr1 (Polinton-like transposable element from Tetrahymena thermophila) elements, is found in ciliates (phylum Ciliophora). Polintons have also been described in Excavata, in which they are extremely abundant in the parabasalid Trichomonas vaginalis, comprising up to 30% of the genome21. Polintons have not been previously described in Rhizaria. However, BLASTP (protein–protein BLAST) searches seeded with Polinton proteins showed that the partially sequenced genome of the foraminiferan Reticulomyxa filosa contains genes for key Polinton proteins, including protein-primed type B DNA polymerase, A32-like genome-packaging ATPase and RVE family integrase (Supplementary information S4 (table)). Thus, of the five major eukaryal kingdoms, only one (that is, Archaeplastida) does not contain identifiable Polintons.
In contrast to the wide distribution of Megavirales and Polintons, mitochondrial linear plasmids are found only in filamentous fungi (Dikarya) and plants (Viridiplantae), and so far the cytoplasmic plasmids are restricted to yeast (Dikarya)46,48.
Recently, an evolutionary connection has been shown to exist between the virophages and large eukaryotic dsDNA transposons of the Polinton family (also known as the Maverick family)17,18. The Polintons are widely distributed in diverse protists and animals (BOX 1), indicative of their ancient origin (which is perhaps concomitant with the origin of eukaryotes) and evolutionary success. We have recently shown that most Polintons encode two proteins that are homologous to viral capsid proteins, suggesting that at least under some conditions, Polintons can produce virions that could infect new hosts19. Thus, Polintons, which would perhaps be more properly named polintoviruses, seem to combine features of viruses and transposons. However, until production of viral particles is experimentally verified, we continue to refer to these virus-like elements as Polintons.
These findings prompted us to revisit the evolutionary connections between the Polintons and other viruses and mobile elements of archaea, bacteria and eukaryotes. Here, we provide evidence to support an evolutionary scenario under which Polintons evolved directly from bacteriophages and became an evolutionary hotbed that gave rise to most of the large dsDNA viruses of eukaryotes, in addition to several groups of plasmids and transposons.
Conserved genome architecture
The Polinton genomes are 15–20 kb in size and encode several conserved proteins, including protein-primed type B DNA polymerase (pPolB), retroviral-like (RVE) family integrase, FtsK-like ATPase, adenovirus-type cysteine protease20–22 and two putative capsid proteins19. Polintons contain terminal inverted repeats (TIRs) and are thought to replicate by a protein-primed mechanism involving the element-encoded pPolBs. In some groups of Polintons, the putative capsid-protein-encoding genes have apparently been lost (BOX 1). Polintons from distant organisms display substantial variation in genome organization and gene content18,22. In addition to the conserved gene set outlined above, different Polintons encode less-conserved proteins, such as various helicases and primases, with homologues in viruses and transposons (BOX 2).
Box 2. Proteins shared between Polintons and ‘Megavirales’.
In addition to the highly conserved proteins involved in virion morphogenesis, at least 13 protein families encoded by different representatives of the proposed order ‘Megavirales’ have homologues in Polintons (see the table). Of particular note are the shared proteins that are potentially involved in genome replication and transcription. For example, Polintons from Glomus intraradices and Trichomonas vaginalis encode a divergent D5-like helicase–primase that is one of the hallmarks of the Megavirales. A Polinton of Dictyostelium fasciculatum encodes a superfamily 3 helicase, whereas elements of Tetrahymena thermophila and Hydra magnipapillata contain genes for PIF1-like superfamily 1 helicases. A related PIF1-like helicase is also encoded by transpovirons (FIG. 1), a group of recently described linear plasmids that replicate in association with mimiviruses73. Another element of H. magnipapillata encodes a dUTPase, an enzyme often found in Megavirales genomes10. A Polinton from Nematostella vectensis encodes a transcription initiation factor, TFIIB, that closely matches the corresponding proteins from marseilleviruses.
Some of the genes shared by Megavirales and Polintons might have been independently acquired by the respective viruses from distinct sources, as has been demonstrated for many of the Megavirales genes66. Nevertheless, it seems to be most likely that the core structural and morphogenetic proteins of the Megavirales — including two capsid proteins, the packaging ATPase and the maturation protease — have been derived from the Polinton ancestor.
| NCVOG | Protein* | Distribution among Polintons or plasmids | Best-match expectation (E) value |
|---|---|---|---|
| Megavirales proteins highly conserved in Polintons | |||
| NCVOG0022 | Double jelly-roll major capsid protein | Nearly universal | Structural modelling |
| NCVOG0249 | A32-like packaging ATPase | Nearly universal | 2.65E-09 |
| NCVOG0246 | Ulp1 protease (PF02902) | Nearly universal | 2.05E-06 |
| NCVOG0584 | Putative penton (mimiviruses and phycodnaviruses) | Nearly universal | 3.58E-03 |
| Megavirales proteins sporadically found in Polintons | |||
| NCVOG0045 | Highly derived D5-like helicase–primase | TV, GI and Df (S3H domain) | 1.02E-30 |
| NCVOG0248 | PIF1-like ATP-dependent DNA helicase (PF05970) | HM and Tt | 9.10E-43 |
| NCVOG1068 | dUTPase (PF00692) | HM | 1.20E-24 |
| NCVOG1127 | Transcription initiation factor TFIIB (PF00382) | NV | 3.27E-17 |
| NCVOG1086 | 259R of invertebrate iridescent virus 6 | NV and HM | 8.37E-09 |
| NCVOG0009 | BIR domain (PF00653) | TC | 1.81E-12 |
| NCVOG1360 | KilA domain (PF04383) | TV | 3.76E-12 |
| NCVOG0933 | MSV079 of Melanoplus sanguinipes entomopoxvirus | HM, TC, Nvi, NV, DBi, DGr and AP | 1.95E-08 |
| NCVOG0965 | Protein phosphatase 1 (PF10488) | TC, Nvi and Gf | 1.49E-07 |
| NCVOG1349 | Hypothetical Chlorovirus and Ostreococcus virus protein | Tt | 4.75E-17 |
| NCVOG0072 | HNH endonucleases (PF01844), found in marseille-, mimi- and phycodnaviruses | Tt and TV | 1.68E-09 |
| NCVOG0010 | Bro-N (PF02498) | NV | 9.19E-19 |
| No NCVOG | pB263R of African swine fever virus | GI | 5.49E-22 |
| Megavirales proteins highly conserved in cytoplasmic plasmids | |||
| NCVOG0031 | D6/D11 helicase (DEAD/SNF2-like helicases) | Universally present | 1.26E-31 |
| NCVOG0271 | DNA-dependent RNA polymerase, β-subunit | Universally present | 7.34E-23 |
| NCVOG1117 | mRNA-capping enzyme, large subunit | Universally present | 4.11E-05 |
When available, protein family (PF) identifiers from the Pfam database are indicated. AP, Acyrtosiphon pisum; DBi, Drosophila biarmipes; Df, Dictyostelium fasciculatum; DGr, Drosophila grimshawi; Gf, Glyptapanteles flavicoxis; GI, Glomus intraradices; HM, Hydra magnipapillata; NCVOG, nucleocytoplasmic virus orthologous group; NV, Nematostella vectensis; Nvi, Nasonia vitripennis; TC, Tribolium castaneum; Tt, Tetrahymena thermophila; TV, Trichomonas vaginalis.
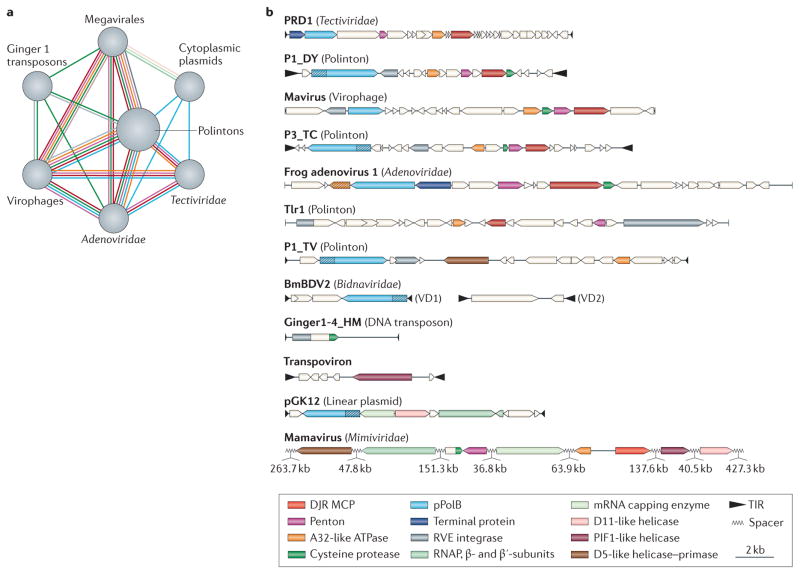
FIGURE 1 shows the homologous genes and gene blocks that are shared between the Polintons and other viruses, transposons and plasmids. All of these elements form a network of connections in which the edges are homologous genes (FIG. 1a). Polintons share the largest number of genes with mobile elements from the other nodes and hence represent the central hub of the network (FIG. 1a). Most notable are the multiple connections between bacteriophages of the family Tectiviridae, Polintons and the Mavirus virophage. All of these elements share four genes that encode two capsid proteins, a DNA-packaging ATPase and pPolB (FIG. 1b) (see below). Polintons share two additional genes with the Mavirus virophage, namely those for the capsid-maturation protease and the RVE integrase. The other known virophages lack pPolB and RVE integrase but share the capsid proteins, ATPase and protease. The adenoviruses join the network (FIG. 1) through pPolB, the two capsid proteins and the protease, whereas the much larger Megavirales possess the capsid proteins, the ATPase and the protease. The linear cytoplasmic plasmids isolated from yeast form a bridge between the mobile elements with protein-primed replication and the much more complex Megavirales; these plasmids encode pPolB along with four key proteins that are conserved in most of the Megavirales (FIG. 1). Below, we examine these homologous relationships in greater detail before proposing an evolutionary scenario in which diverse eukaryotic viruses and other mobile elements are derived from the Polintons.
Figure 1. Evolutionary relationships between Polintons and other mobile genetic elements.
a | Evolutionary network showing shared gene content between Polintons and other mobile genetic elements in archaea, bacteria and eukaryotes. Edges correspond to homologous genes. The colour key is provided in part b. Only those elements that are directly linked to Polintons are shown. b | Genome maps of various viruses, plasmids and transposons in archaea, bacteria and eukaryotes. Homologous genes are colour-coded. Hatched regions in the protein-primed type B DNA polymerase (pPolB) genes indicate the position of the (predicted) terminal protein domains. Hatching is also used to indicate the gene encoding the distinct adenoviral genome packaging ATPase IVa2. VD1 and VD2 are the two genomic segments of the Bombyx mori bidensovirus 2 (BmBDV2). DJR, double jelly-roll; Ginger1-4_HM, transposon 4 of Hydra magnipapillata; MCP, major capsid protein; P1_DY, Polinton 1 of Drosophila yakuba; P1_TV, Polinton 1 of Trichomonas vaginalis; P3_TC, Polinton 3 of Tribolium castaneum; RNAP, RNA polymerase; TIR, terminal inverted repeat; Tlr1, Polinton-like transposable element from Tetrahymena thermophila.
Virion morphogenesis proteins
Most Polintons encode two putative capsid proteins, one of which is predicted to adopt the double jelly-roll (DJR) topology19 found in the major capsid proteins (MCPs) of diverse dsDNA viruses that infect bacteria, archaea and eukaryotes23–25. The other predicted capsid protein has a single jelly-roll fold and corresponds to the minor capsid protein (mCP), often called the penton protein, which is also conserved in viruses with DJR MCPs26–30. In principle, the two proteins — the MCP and the mCP — are sufficient (and necessary) to construct the entire icosahedral capsid (FIG. 2), although viruses often encode additional minor structural proteins. The conservation of the MCP and mCP in Polintons closely follows the presence of the genome-packaging ATPase and the cysteine protease (that is, all of these proteins are either present or absent), which are also predicted to be involved in virion morphogenesis19.
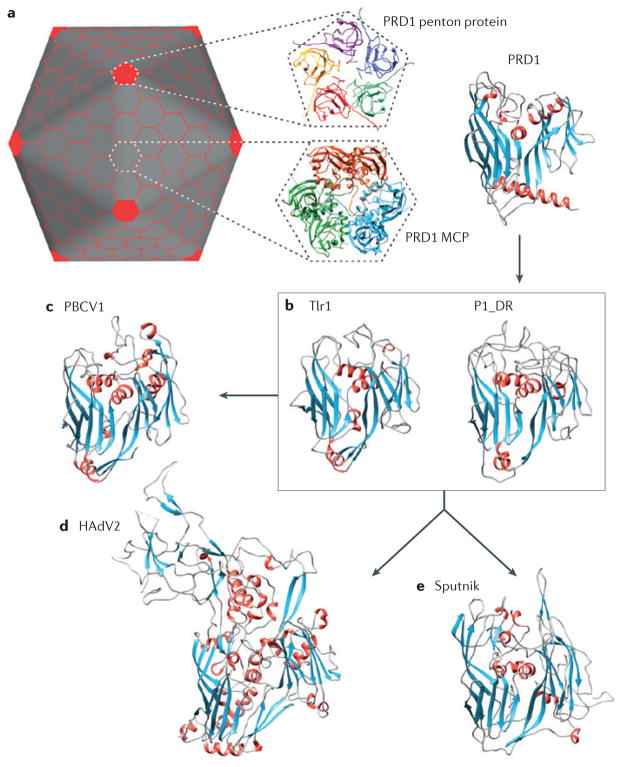
Figure 2. Structural features of some viruses with double jelly-roll major capsid proteins.
a | Representation of the icosahedral virion with triangulation number (T)=25; this organization is found in tectiviruses and adenoviruses. The pentagonal capsomers (shown in red) are composed of five copies of the penton protein, which has a single jelly-roll fold, whereas the hexagonal capsomers are trimers of the major capsid protein (MCP) with the double jelly-roll (DJR) topology. X-ray structures of the tectivirus PRD1 penton and MCP are shown as examples (Protein Data Bank (PDB) identifier: 1W8X)26. b | Structural models of the MCPs of Tlr1 (Polinton-like transposable element from Tetrahymena thermophila) and P1_DR (Polinton 1 from Danio rerio)19. c–e | X-ray structures of the DJR MCPs from the eukaryotic viruses PBCV1 (Paramecium bursaria Chlorella virus 1; PDB identifier: 1M4X)74 (c), HAdV2 (human adenovirus type 2; PDB identifier: 1P2Z)75 (d) and Sputnik (PDB identifier: 3J26)29 (e).
Major capsid proteins
As discussed above, Polinton genomes have a similar layout to those of bacterial tectiviruses, eukaryotic adenoviruses and virophages (FIG. 1), suggesting a specific evolutionary connection among all of these elements. However, the MCP of Polintons is most closely related to the MCPs of phycodnaviruses19, which are members of the proposed order Megavirales12. FIGURE 2 illustrates the similarity between the more compact MCPs of Polintons and the phycodnavirus Paramecium bursaria Chlorella virus 1 (PBCV1), and highlights the differences between these proteins and the larger MCPs of the Sputnik virophage and adenoviruses. The evolutionary scenario underlying this relationship remains uncertain, but it might involve fast evolution of the MCP in the virophages and adenoviruses. Thus, the presence of phycodnavirus-like MCPs in Polintons establishes an evolutionary link between the giant Megavirales and dsDNA viruses with smaller genomes.
Minor capsid proteins
The mCPs of Polintons correspond to the penton proteins that are found at the fivefold vertices of icosahedral capsids (FIG. 2). Penton proteins have been observed in all viruses with DJR MCPs for which high-resolution structural information is available, including tectivirus PRD1 (REF. 26), corticovirus PM2 (REF. 27), archaeal turrivirus STIV (Sulfolobus turreted icosahedral virus)28, virophage Sputnik29 and adenoviruses30. Thus far, an equivalent of the penton protein has not been identified among the Megavirales; however, considering the importance of this protein for the formation of icosahedral virions, one could expect that these giant viruses also encode a penton homologue31. Using the predicted mCP of Tlr1 (Polinton-like transposable element from Tetrahymena thermophila) as a seed in PSI-BLAST (position-specific iterative basic local alignment search tool) sequence similarity searches, we detected penton-like proteins in mimiviruses and phycodnaviruses (Supplementary information S1 (figure)). Consistent with this, in PBCV1 the predicted penton (A533R) has been detected in the virion32. In mimiviruses, the predicted penton-protein gene is adjacent to the gene encoding the maturation protease (FIG. 1), an arrangement that is also found in Mavirus and some Polintons. The finding that not only the MCP but also the mCP of Polintons has counterparts in Megavirales further strengthens the link between these genetic elements.
Cysteine proteases
Polintons encode a conserved cysteine protease that is homologous to the adenovirus maturation protease and eukaryotic Ulp1-like proteases, both classified as members of the CE (cysteine endopeptidase) clan of proteases33. In adenoviruses, the protease is the principal player responsible for transforming immature virions into fully infectious, mature virus particles34. In addition to adenoviruses, homologous proteases are encoded by virophages and most of the Megavirales18,35. In adenoviruses, poxviruses and asfarviruses, the proteases are responsible for proteolytic processing of immature virions36,37. The wide distribution and importance of these proteases in the maturation of both small and large eukaryotic viruses with DJR MCPs suggests that acquisition of the protease antedates the radiation of these viruses. By contrast, none of the viruses in archaea and bacteria with a DJR MCP encodes, or is known to require, a protease for virion maturation. Indeed, the key difference between Polintons and bacterial tectiviruses is that the latter lack genes for the cysteine protease and RVE family integrase. Although some tectiviruses form plasmidial prophages, none of these viruses has been found to integrate into the host genome38,39. Notably, some eukaryotic transposons carry genes for both integrase and cysteine protease22. For example, the DNA transposons of the Ginger 1 family encode Polinton-like RVE integrases containing carboxy-terminal Ulp1-like cysteine protease domains40 (FIG. 1). Thus, the proteases and integrases could have been incorporated into an ancestral viral genome through the integration and subsequent domestication of a DNA transposon.
Genome-packaging ATPases
All icosahedral viruses with DJR MCPs in archaea, bacteria and eukaryotes, with the exception of adenoviruses, encode genome-packaging ATPases of the FtsK/HerA superfamily41,42. The vaccinia virus protein A32 and the P9 protein of tectivirus PRD1 are essential for pumping the respective viral genomes into preformed immature virions42,43. However, adenoviruses encode a distinct ATPase (IVa2) that has the same role34,44. Although most Polintons encode A32-like ATPases19, some contain a gene encoding an ATPase that is more closely related to adenovirus IVa2 (Supplementary information S2 (figure)). Given the greater genomic diversity and broader taxonomic distribution of Polintons compared with adenoviruses, which are restricted to Metazoa (BOX 1), the diversification of packaging ATPases probably occurred during the evolution of Polintons, and IVa2 was subsequently inherited by adenoviruses from a specific Polinton lineage. This directionality is consistent with the phylogenetic analysis of the pPolBs, as described below.
Protein-priming mobile elements
Protein-primed DNA replication is an exclusive feature of viruses and plasmids that encode pPolBs. To initiate genome replication, these elements use a terminal protein that remains covalently attached to the 5′ termini of their linear genomes instead of the nucleic-acid primers that are used by cellular organisms and most DNA viruses. Terminal proteins are often encoded by separate genes45, but in some plasmids they are fused to the amino termini of their cognate pPolBs46. A similar fusion has been detected in Polintons and bidnaviruses47 (FIG. 1).
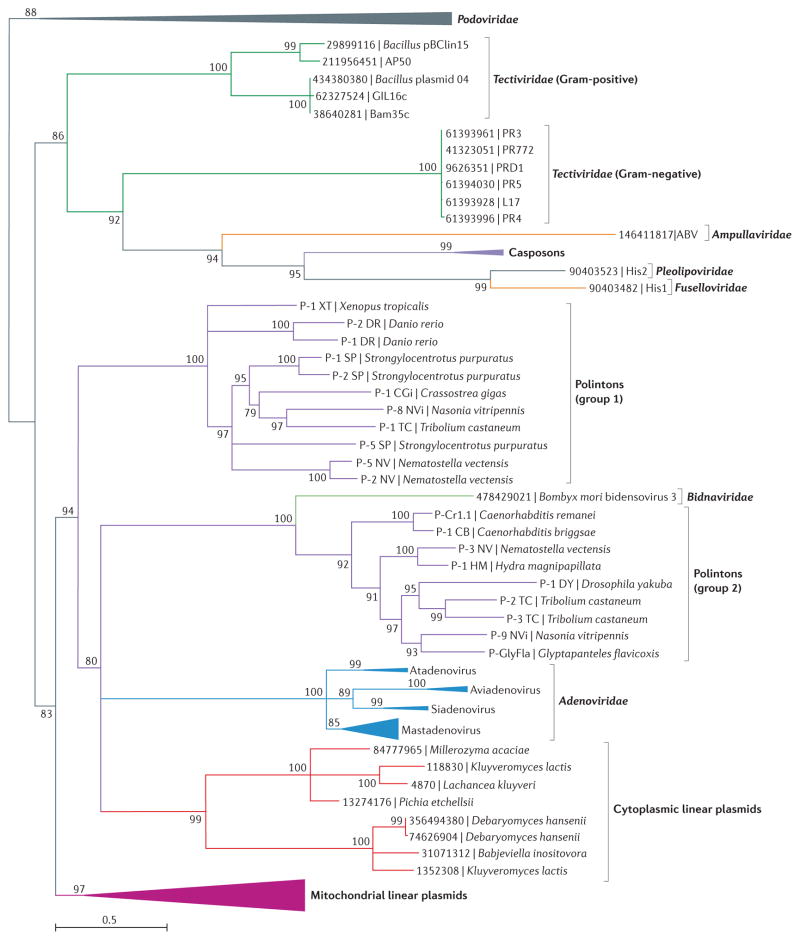
FIGURE 3 shows a phylogenetic tree of pPolBs from a wide range of viruses and plasmids in archaea, bacteria and eukaryotes. In this phylogeny, pPolBs are segregated into two major groups: bacterial and archaeal mobile elements, and eukaryotic plasmids and viruses. Among the pPolB-encoding elements in archaea and bacteria, only tectiviruses share additional genes with the eukaryotic pPolB-encoding viruses and plasmids (FIG. 1).
Figure 3. Phylogenetic analysis of protein-primed type B DNA polymerases from mobile elements in archaea, bacteria and eukaryotes.
The maximum likelihood tree was calculated using PhyML76, with the WAG (Whelan and Goldman) model of amino-acid substitution, including a gamma law (four categories) and an estimated proportion of invariable sites. Numbers at the branch points represent SH (Shimodaira–Hasegawa)-like local support values. The tree is rooted with bacterial phi29-like podoviruses. Branches with support values below 75% were collapsed. Branches are coloured according to the classification of the corresponding taxa. The branching of the major taxonomic groups is consistent with that obtained previously using different phylogenetic analysis methods and different taxonomic sampling20,46,47,77. The scale bar represents the number of substitutions per site.
The pPolBs from eukaryotic mobile elements form two clades. One clade exclusively contains linear plasmids that replicate in the mitochondria of filamentous fungi and some plants46,48. All of these plasmids contain TIRs and commonly encode two proteins: a pPolB fused to the terminal protein and a single-subunit DNA-dependent RNA polymerase (RNAP) related to the mitochondrial RNAPs that are encoded in the nucleus and derive from T7-like bacteriophages49,50. The second eukaryotic pPolB clade includes cytoplasmic linear plasmids from yeast and all eukaryotic viruses that use protein priming (FIG. 3). Notably, Polintons are at the base of this group, whereas cytoplasmic plasmids, adenoviruses and bidnaviruses emerge from within the Polinton clade (FIG. 3).
Adenoviruses
All known adenoviruses infect vertebrates and possess capsids that are geometrically identical to those of tectiviruses; that is, the number and arrangement of hexagonal capsomers (DJR MCP trimers) and pentagonal capsomers (penton pentamers) is exactly the same in these two groups of viruses26,51 (FIG. 2). Despite the strong evidence of the evolutionary relationship between tectiviruses and adenoviruses23,52,53, the exact evolutionary trajectory linking them has remained obscure. In our phylogenetic analysis, viruses of the four major genera of the family Adenoviridae form a well-supported clade that emerges from within Polintons (FIG. 3), in agreement with previous findings18. Given the comparatively low divergence of known adenoviruses and lack of known representatives outside vertebrates52, we suggest that adenoviruses probably evolved from Polintons relatively late in eukaryotic evolution.
Bidnaviruses
Members of the recently established family Bidnaviridae are small ssDNA viruses that infect insect hosts54. They have bipartite linear genomes with TIRs and, unlike other ssDNA viruses, encode pPolBs55. A recent analysis of the complex evolutionary history of bidnaviruses has shown that the ancestor of this viral group evolved from an arthropod-infecting ssDNA virus of the Parvoviridae family via multiple gene exchanges with diverse RNA and DNA viruses47. A key point in bidnavirus evolution was the loss of the rolling-circle replication initiation protein gene and acquisition of the pPolB-encoding gene from an insect Polinton.
Virophages
Mavirus-like virophages represent the fourth group of eukaryotic pPolB-encoding viruses17. Although virophages were excluded from the phylogenetic tree in FIG. 3 owing to the high divergence of their pPolBs, resulting in distortion to the tree topology, separate analyses place the virophages inside the Polinton branch of pPolB22 (Supplementary information S3 (figure)). Multiple lines of evidence have recently indicated that the genes shared between virophages and Polintons originated during the evolution of Polintons18, in contrast to the originally proposed scenario in which Polintons were derived from virophages17. In our view, the identification of Polinton DJR MCPs and mCPs leaves little doubt that virophages are direct descendants of Polintons19. Virophages show notable variation in gene content. In Sputnik-like virophages, the pPolB gene has been replaced with a gene encoding a distinct polymerase primase18,56. Although we remained uncertain about the genomic layout of the ancestral virophage in our previous analysis18, the evolutionary relationship between Polintons and virophages seems to point to a Mavirus-like virophage as the most likely ancestral state.
Cytoplasmic plasmids
A group of linear plasmids, distinct from those found in mitochondria of filamentous fungi and plants, is present in various yeast species46,57. These plasmids reside exclusively in the cytoplasm of their hosts and encode key components of their own replication and transcription machineries (FIG. 1). In phylogeny, pPolBs of the cytoplasmic plasmids emerge from within the Polintons as a sister group to adenoviruses and show no affinity to the mitochondrial plasmids (FIG. 3). As in Polintons, bidnaviruses and mitochondrial plasmids, the terminal proteins of linear cytoplasmic plasmids are fused to the N termini of the pPolBs46,47, suggesting that these plasmids share the most recent common ancestor with Polintons rather than adenoviruses. These plasmids do not encode any viral structural proteins that could have been lost during evolution. Notably, certain linear bacterial plasmids (for example, pBClin15) evolved from tectiviruses39 (FIG. 3), suggesting that a similar evolutionary transition could have occurred in the case of Polintons.
The RNAPs encoded by the cytoplasmic plasmids are unrelated to the single-subunit RNAPs of mitochondrial plasmids; instead, they are homologous to the two largest subunits (the β- and β′-subunits) of multisubunit RNAPs encoded by cellular organisms and members of the Megavirales10,46,57,58. The plasmid RNAPs are encoded by two genes (FIG. 1); the larger one encodes a subunit containing all characteristic motifs of the β-subunit and some of the motifs of the β′-subunit, whereas the second, shorter gene encodes a protein bearing additional motifs of the β′-subunit57,59. The cytoplasmic plasmids encode two additional proteins, the D11-like helicase and the mRNA-capping enzyme, which show specific evolutionary relationship to the homologues from Megavirales (FIG. 1) and are likely to be essential for the cytoplasmic replication of DNA genomes in eukaryotes, given the ubiquity of the respective genes in all known cytoplasmic DNA elements. In vaccinia virus (Poxviridae), the D11 protein (a DExH helicase family member) has an important role during transcription elongation and mRNA release on termination60. The mRNA-capping enzymes encoded by cytoplasmic plasmids possess a unique domain architecture that is not found in cellular organisms but only in diverse members of the Megavirales61–63. These viral and plasmid enzymes consist of three functional domains, namely RNA 5′-triphosphatase, RNA guanylyltransferase and RNA (guanine-N7)-methyltransferase, which in a series of consecutive reactions synthesize the 7-methylguanosine RNA cap on the nascent transcripts62–65.
Based on the unique shared domain architecture of the capping enzymes, Shuman has proposed that poxviruses and asfarviruses evolved from linear cytoplasmic plasmids63. However, neither the origin of the cytoplasmic plasmids themselves nor the source of the virion morphogenetic unit of the Megavirales has been clarified. By contrast, Klassen and Meinhardt have proposed that eukaryotic linear plasmids evolved from an adenovirus-like or tectivirus-like ancestor through the loss of genes required for virion formation46. Inclusion of the Polintons provides a better-supported scenario for the origin of both linear plasmids and Megavirales. Indeed, the pPolB phylogeny implies that cytoplasmic plasmids evolved from a Polinton ancestor rather than from a tectivirus or an adenovirus (FIG. 3). We hypothesize that a Polinton escaped from the nucleus by acquiring the RNAP, capping apparatus and D11-like helicase from the host66, and subsequently followed two opposite evolutionary pathways, one leading to the origin of cytoplasmic plasmids and the other to the emergence of the ancestor of Megavirales (see below).
Polintons: a hotbed of virus evolution
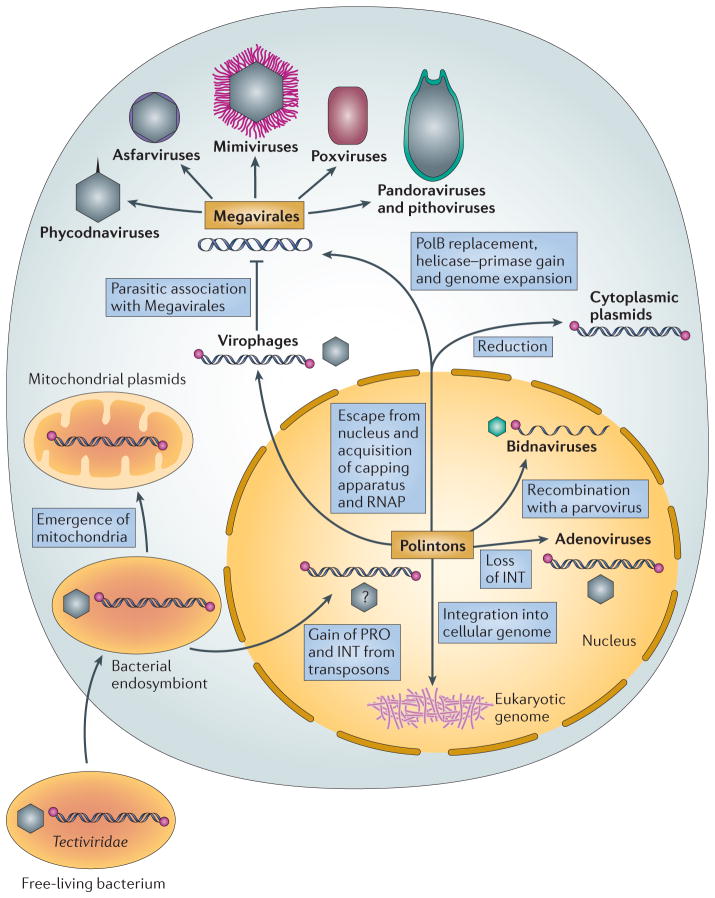
The comparative analysis of the various viruses, plasmids and transposons in archaea, bacteria and eukaryotes presented here places the Polintons at the root of many families of eukaryotic DNA viruses. Integrating different lines of evidence, we propose a unifying scenario in which Polintons were a hotbed of eukaryotic virus, transposon and plasmid evolution (FIG. 4). In this scenario, Polintons evolved at the onset of eukaryogenesis from a tectivirus-like ancestor. This virus could have entered the proto-eukaryotic host along with the bacterial endosymbiont that subsequently gave rise to the mitochondria (FIG. 4). This route of evolution is consistent with the presence of linear plasmids in the mitochondria of fungi and plants and the primary split between the pPolBs of mitochondrial plasmids and the rest of the eukaryotic plasmids and viruses (FIG. 3).
Figure 4. A hypothetical scenario for the evolution of various eukaryotic viruses and plasmids.
In this scenario, the Polintons evolved from a tectivirus-like ancestor, which entered the proto-eukaryotic host within a bacterial endosymbiont that subsequently gave rise to the mitochondria. Polintons reside in the nucleus, and the key event in the emergence of both cytoplasmic plasmids and the proposed order ‘Megavirales’ was escape from the nucleus, which was associated with the acquisition of RNA polymerase (RNAP) and the capping apparatus from the host. The essential events in the evolution of Megavirales from the cytoplasmic Polinton-like ancestor were the replacement of protein-primed type B DNA polymerase (pPolB) with the RNA/DNA-primed PolB, the acquisition of the D5-like helicase–primase and genome expansion. Polintons are also implicated in the evolution of virophages, bidnaviruses and adenoviruses. The hexagons represent icosahedral capsids; in the case of Polintons, the capsids have been predicted to exist but have not yet been observed (indicated by a question mark). Double strands represent double-stranded DNA, whereas the single-stranded DNA genome of bidnaviruses is shown as a single strand. INT, retroviral-like (RVE) family integrase; PRO, adenovirus-type cysteine protease.
The major distinction between tectiviruses and Polintons is the presence in the latter of the genes encoding the Ulp1-like cysteine protease and RVE family integrase. Both of these genes could have been introduced into the genome of the tectiviral ancestor of the Polintons in a single recombination event with a eukaryotic DNA transposon. Notably, Ulp1-like cysteine proteases are characteristic of eukaryotes, whereas bacteria encode only distantly related cysteine proteases, suggesting that the Polinton ancestor had already acquired the protease and integrase genes in the (proto-)eukaryotic host. The protease was adopted for virion maturation and retained in all major virus lineages emerging from Polintons, including virophages, adenoviruses and Megavirales, although it was subsequently lost from some members of the Megavirales.
The acquisition of the RVE integrase gene granted the emerging Polintons the ability to follow two alternative lifestyles, one that is typical of transposable elements and one that is typical of bona fide viruses. Such duality is also embraced by Mu-like bacteriophages and eukaryotic metaviruses (Ty3-gypsy retrotransposons), and pseudoviruses (Ty1-copia retrotransposons)3,4,67. The ability to persist in the integrated form in the host genome could have played a key part in the further diversification and successful spread of Polintons and their derivatives in eukaryotes. Integration into the host chromosome provides a ‘safe haven’ for viral genomes, where they have the opportunity to accumulate changes by genetic drift or recombination without the immediacy of producing infectious virus progeny. After a viable and sufficiently fit individual evolves, it can ‘spring out’ of the host chromosome and revert to the viral way of life.
Some Polintons seem to have abandoned the virus-like reproduction strategy altogether and have lost the genes implicated in virion formation19. The loss of the genes for DJR MCPs might be linked to the extraordinary expansion of Polintons in Trichomonas vaginalis, where they constitute up to 30% of the host genome21. The ancestor of adenoviruses seems to have followed the opposite evolutionary route, whereby the integrase gene was lost along with the transposition ability (FIG. 4), thus committing adenoviruses to the strict viral strategy. Polintons are also implicated in the evolution of ssDNA viruses of the Bidnaviridae family; this family emerged as a result of extensive gene shuffle between four groups of selfish elements, including Polintons, which contributed pPolB47.
The major legacy of Polintons is the central role they are likely to have had in the emergence of the Megavirales, the most widespread and diverse group of eukaryotic dsDNA viruses. The Megavirales apparently inherited from the Polintons the key proteins involved in virion morphogenesis, including the DJR MCP, the genome-packaging ATPase, the maturation protease and possibly the penton protein (FIG. 1). The Polinton MCP is more closely related to those of phycodnaviruses than it is to the MCPs of any other viral groups, including tectiviruses19 (FIG. 2), which is suggestive of an evolutionary link between Polintons and Megavirales. Although the packaging ATPases and maturation proteases both show high levels of sequence divergence, the topologies of the respective phylogenetic trees are compatible with the existence of such a link18.
Polintons reside in the nucleus and accordingly rely on host enzymes for transcription. The major event underlying the emergence of the Megavirales was apparently the escape from the nucleus, which was associated with the acquisition of RNAP and the capping apparatus from the host. The putative escaped element that would replicate in the cytoplasm using the ancestral Polinton pPolB gave rise to two groups of mobile elements (FIG. 4); namely, the cytoplasmic plasmids and Megavirales that share the unique trifunctional capping enzyme, RNAP and D11-like helicase (FIG. 1). The cytoplasmic plasmids retained pPolB but lost the genes implicated in virion morphogenesis, succumbing to the exclusive intra-cellular lifestyle. By contrast, the ancestor of the Megavirales evolved via the route of increasing complexity.
The essential events in the evolution of Megavirales from the cytoplasmic Polinton-like ancestor were the replacement of the pPolB with the RNA/DNA-primed PolB and the acquisition of the D5-like helicase–primase (FIG. 4). The pPolBs might be unable to ensure efficient replication of genomes larger than a certain threshold size (~45 kb, as in adenoviruses), conceivably owing to the lack of a dedicated primase that would provide multiple internal primers along the genome. Notably, some Polintons encode divergent D5-like helicase–primases (FIG. 1; BOX 2), which tend to cluster with the homologues from Megavirales in phylogenetic trees18. This suggests that Megavirales inherited this key enzyme from Polintons. Several other genes that are common and probably ancestral in the Megavirales are also shared with various Polintons (BOX 2). The PolB of Megavirales was probably acquired from the eukaryotic host66, replacing the ancestral pPolB and, together with the helicase–primase, opening the route to genome expansion. The extent of this expansion, which involved massive acquisition of genes not only from eukaryotes but also from bacteria68, was such that the genes apparently inherited from Polintons comprise but a tiny proportion of the gene complement of these large and giant viruses. Crucially, however, a few losses notwithstanding, the proteins of Polinton descent form the core of the virion morphogenesis machinery, as well as important parts of the replication apparatus, in most members of the Megavirales (BOX 2).
The evolution of the Megavirales was marked by genome expansion that has been pushed to the extreme in at least three independently evolved groups of giant viruses: Mimiviridae, which are distantly related to Phycodnaviridae; pandoraviruses, which evolved from a common ancestor with coccolithoviruses within the Phycodnaviridae; and pithoviruses, which are related to Iridoviridae and Marseilleviridae68,69. The ancestral icosahedral capsids constructed from DJR MCPs were substituted with less-regular, ovoid or brick-shaped virion morphologies in several groups within the Megavirales — namely, in ascoviruses, poxviruses, pandoraviruses and pithoviruses — indicating that all facets of the viral life cycle are susceptible to dramatic changes. Unlike in the case of adenoviruses, the wide distribution of Megavirales in eukaryotes (BOX 1) implies that these viruses diverged from the Polinton ancestor early in eukaryal evolution.
The virophages followed a different strategy to escape from the nucleus. Instead of encoding their own factors that would enable their cytoplasmic propagation, these viruses evolved to parasitize on their giant relatives by hijacking the necessary molecular machinery14–17.
Conclusions
The extended virus world, which includes both bona fide viruses and related mobile elements, is a complex network of genomes that share partially overlapping sets of genes3. Discerning specific evolutionary scenarios in this maze is a major challenge. In particular, the origins of the diverse groups of eukaryotic viruses remain obscure. Clearly, the evolution of these viruses involved contributions from bacteriophages, as particularly indicated by the relationships between the respective capsid proteins and packaging enzymes25,41,70. It is equally clear that some of the key viral genes were acquired from the hosts at different stages of evolution, but otherwise the routes of viral evolution remain hazy. The synthesis of phylogenomic analyses presented here pushes the evolutionary study of eukaryotic DNA viruses beyond general considerations and into the territory of concrete, evidence-based reconstruction. The central role in our emerging scenario belongs to the Polintons, a remarkable group of selfish elements that are nearly ubiquitous in eukaryotes and combine features of integrating mobile elements with those of bona fide viruses. Polintons show strong evolutionary connections with bacteriophages of the family Tectiviridae, suggesting that Polintons were the first group of eukaryotic dsDNA viruses to evolve from archaeal and bacterial ancestors. The apparent dual lifestyle of the Polintons seems to have entailed outstanding evolutionary resilience and versatility, making them a hotbed of evolution of eukaryotic selfish elements that spawned diverse groups of viruses in a broad range of sizes, along with related plasmids. This evolutionary scenario parallels the apparent course of evolution of retrotranscribing viruses from retroelements with a dual lifestyle3. In even more general terms, the emerging story of virus evolution is compatible with the concept of viral hallmark genes that form different combinations in diverse selfish elements and on multiple occasions serve as ‘kernels’ for gene accretion and genome expansion.
Acknowledgments
E.V.K. is supported by the intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
Protein Data Bank: http://www.rcsb.org/pdb/home/home.do
SUPPLEMENTARY INFORMATION
See online article: S1 (figure) | S2 (figure) | S3 (figure) | S4 (table)
This article is dedicated to the memory of Jerzy Jurka (1950–2014), a co-discoverer of Polintons and a pioneer in the field of mobile genetic elements.
Contributor Information
Mart Krupovic, Institut Pasteur, Unité Biologie Moléculaire du Gène chez les Extrêmophiles, Department of Microbiology, Paris 75015, France.
Eugene V. Koonin, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894, USA
References
- 1.Koonin EV, Dolja VV. A virocentric perspective on the evolution of life. Curr Opin Virol. 2013;3:546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupovic M, Bamford DH. Order to the viral universe. J Virol. 2010;84:12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV, Dolja VV. Virus world as an evolutionary network of viruses and capsid-less selfish elements. Microbiol Mol Biol Rev. 2014;78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev. 2011;75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes EC. What does virus evolution tell us about virus origins? J Virol. 2011;85:5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 8.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colson P, et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol. 2013;158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Scola B, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 14.Claverie JM, Abergel C. Mimivirus and its virophage. Annu Rev Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 15.Desnues C, Boyer M, Raoult D. Sputnik, a virophage infecting the viral domain of life. Adv Virus Res. 2012;82:63–89. doi: 10.1016/B978-0-12-394621-8.00013-3. [DOI] [PubMed] [Google Scholar]
- 16.Krupovic M, Cvirkaite-Krupovic V. Virophages or satellite viruses? Nature Rev Microbiol. 2011;9:762–763. doi: 10.1038/nrmicro2676. [DOI] [PubMed] [Google Scholar]
- 17.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 18.Yutin N, Raoult D, Koonin EV. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupovic M, Bamford DH, Koonin EV. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol Direct. 2014;9:6. doi: 10.1186/1745-6150-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci USA. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritham EJ, Putliwala T, Feschotte C. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene. 2007;390:3–17. doi: 10.1016/j.gene.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genom Hum Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 23.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Krupovic M, Bamford DH. Virus evolution: how far does the double β-barrel viral lineage extend? Nature Rev Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 25.Krupovic M, Bamford DH. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol. 2011;1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Abrescia NG, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 27.Abrescia NG, et al. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol Cell. 2008;31:749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 28.Veesler D, et al. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc Natl Acad Sci USA. 2013;110:5504–5509. doi: 10.1073/pnas.1300601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, et al. Structure of Sputnik, a virophage, at 3.5-Å resolution. Proc Natl Acad Sci USA. 2012;109:18431–18436. doi: 10.1073/pnas.1211702109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17:121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 31.Xiao C, Rossmann MG. Structures of giant icosahedral eukaryotic dsDNA viruses. Curr Opin Virol. 2011;1:101–109. doi: 10.1016/j.coviro.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunigan DD, et al. Paramecium bursaria Chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J Virol. 2012;86:8821–8834. doi: 10.1128/JVI.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett AJ, Rawlings ND. Evolutionary lines of cysteine peptidases. Biol Chem. 2001;382:727–733. doi: 10.1515/BC.2001.088. [DOI] [PubMed] [Google Scholar]
- 34.San Martín C. Latest insights on adenovirus structure and assembly. Viruses. 2012;4:847–877. doi: 10.3390/v4050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yutin N, Wolf YI, Raoult D, Koonin EV. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andres G, Alejo A, Simon-Mateo C, Salas ML. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J Biol Chem. 2001;276:780–787. doi: 10.1074/jbc.M006844200. [DOI] [PubMed] [Google Scholar]
- 37.Byrd CM, Hruby DE. A conditional-lethal vaccinia virus mutant demonstrates that the I7L gene product is required for virion morphogenesis. Virol J. 2005;2:4. doi: 10.1186/1743-422X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillis A, Mahillon J. Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future. Viruses. 2014;6:2623–2672. doi: 10.3390/v6072623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strömsten NJ, Benson SD, Burnett RM, Bamford DH, Bamford JK. The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J Bacteriol. 2003;185:6985–6989. doi: 10.1128/JB.185.23.6985-6989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao W, Kapitonov VV, Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA. 2010;1:3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strömsten NJ, Bamford DH, Bamford JK. In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J Mol Biol. 2005;348:617–629. doi: 10.1016/j.jmb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Cassetti MC, Merchlinsky M, Wolffe EJ, Weisberg AS, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 45.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 46.Klassen R, Meinhardt F. Linear protein-primed replicating plasmids in eukaryotic microbes. Microbiol Monogr. 2007;7:188–216. [Google Scholar]
- 47.Krupovic M, Koonin EV. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep. 2014;4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Handa H. Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions? Mitochondrion. 2008;8:15–25. doi: 10.1016/j.mito.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Filée J, Forterre P. Viral proteins functioning in organelles: a cryptic origin? Trends Microbiol. 2005;13:510–513. doi: 10.1016/j.tim.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22:90–95. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 52.Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J Gen Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 53.Merckel MC, Huiskonen JT, Bamford DH, Goldman A, Tuma R. The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture. Mol Cell. 2005;18:161–170. doi: 10.1016/j.molcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Hu ZY, Li GH, Li GT, Yao Q, Chen KP. Bombyx mori bidensovirus: the type species of the new genus Bidensovirus in the new family. Bidnaviridae Chin Sci Bull. 2013;58:4528–4532. [Google Scholar]
- 55.Krupovic M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr Opin Virol. 2013;3:578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 56.Iyer LM, Abhiman S, Aravind L. A new family of polymerases related to superfamily A DNA polymerases and T7-like DNA-dependent RNA polymerases. Biol Direct. 2008;3:39. doi: 10.1186/1745-6150-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeske S, Meinhardt F, Klassen R. In: Progress in Botany. Esser K, Lüttge U, Beyschlag W, Murata J, editors. Springer; 2007. pp. 98–129. [Google Scholar]
- 58.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson DW, Meacock PA. Extranuclear gene expression in yeast: evidence for a plasmid-encoded RNA polymerase of unique structure. Nucleic Acids Res. 1988;16:8097–8112. [PMC free article] [PubMed] [Google Scholar]
- 60.Deng L, Shuman S. Vaccinia NPH-I, a DExH-box ATPase, is the energy coupling factor for mRNA transcription termination. Genes Dev. 1998;12:538–546. doi: 10.1101/gad.12.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larsen M, Gunge N, Meinhardt F. Kluyveromyces lactis killer plasmid pGKL2: evidence for a viral-like capping enzyme encoded by ORF3. Plasmid. 1998;40:243–246. doi: 10.1006/plas.1998.1367. [DOI] [PubMed] [Google Scholar]
- 62.Kyrieleis OJ, Chang J, de la Pena M, Shuman S, Cusack S. Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus. Structure. 2014;22:452–465. doi: 10.1016/j.str.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nature Rev Mol Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 64.Shuman S. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harb Symp Quant Biol. 2001;66:301–312. doi: 10.1101/sqb.2001.66.301. [DOI] [PubMed] [Google Scholar]
- 65.Tiggemann M, Jeske S, Larsen M, Meinhardt F. Kluyveromyces lactis cytoplasmic plasmid pGKL2: heterologous expression of Orf3p and proof of guanylyltransferase and mRNA-triphosphatase activities. Yeast. 2001;18:815–825. doi: 10.1002/yea.728. [DOI] [PubMed] [Google Scholar]
- 66.Yutin N, Koonin EV. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J. 2012;9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sandmeyer SB, Menees TM. Morphogenesis at the retrotransposon-retrovirus interface: gypsy and copia families in yeast and Drosophila. Curr Top Microbiol Immunol. 1996;214:261–296. doi: 10.1007/978-3-642-80145-7_9. [DOI] [PubMed] [Google Scholar]
- 68.Yutin N, Wolf YI, Koonin EV. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology. 2014;466–467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Legendre M, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci USA. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rixon FJ, Schmid MF. Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr Opin Virol. 2014;5:105–110. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Philippe N, et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 72.Keeling PJ, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 73.Desnues C, et al. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci USA. 2012;109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rux JJ, Kuser PR, Burnett RM. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J Virol. 2003;77:9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 77.Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]