Abstract
CRISPR (clustered regularly interspaced short palindromic repeats)–Cas (CRISPR-associated) is an adaptive immunity system in bacteria and archaea that functions via a distinct self/non-self recognition mechanism that involves unique spacers homologous with viral or plasmid DNA and integrated into the CRISPR loci. Most of the Cas proteins evolve under relaxed purifying selection and some underwent dramatic structural rearrangements during evolution. In many cases, CRISPR–Cas system components are replaced either by homologous or by analogous proteins or domains in some bacterial and archaeal lineages. However, recent advances in comparative sequence analysis, structural studies and experimental data suggest that, despite this remarkable evolutionary plasticity, all CRISPR–Cas systems employ the same architectural and functional principles, and given the conservation of the principal building blocks, share a common ancestry. We review recent advances in the understanding of the evolution and organization of CRISPR–Cas systems. Among other developments, we describe for the first time a group of archaeal cas1 gene homologues that are not associated with CRISPR–Cas loci and are predicted to be involved in functions other than adaptive immunity.
Keywords: Cas1, clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated (Cas) system, evolution, phylogeny, repeat-associated mysterious protein (RAMP), toxin
Introduction
The molecular mechanism of CRISPR (clustered regularly interspaced short palindromic repeats)–Cas (CRISPR-associated) system, along with R-M (restriction-modification) and DNA phosphorothioation (DND) systems, is based on the self/non-self discrimination principle (see [1] and references therein). However, unlike R-M and DND systems that generally modify their own DNA and destroy unmodified DNA [2,3], the CRISPR–Cas system retains the memory of previous encounters with infectious agents. This memory is embodied in short nucleotide fragments matching the non-self DNA that are inserted into an array of CRISPRs and are then employed to attack and destroy the cognate virus or plasmid [4–7]. Therefore CRISPR–Cas is often referred to as a prokaryotic adaptive immunity system and its unique ability to inherit the acquired characteristics appears to be compatible with the Lamarckian mode of evolution [8].
The initial bioinformatic analysis of CRISPR, spacers and Cas proteins led to the prediction of the involvement of this system in antivirus defence [9–13]. This prediction was successfully confirmed experimentally [14] and, since then, CRISPR research has evolved into a dynamic field in microbiology with considerable biotechnology potential [15–17].
The classification and nomenclature of CRISPR–Cas systems is based on a combination of evidence from phylogenetic, comparative genomic and structural analysis [7]. The major challenge for this classification is to accommodate the remarkable diversity and fast evolution of Cas proteins (with the apparent single exception of the Cas1 protein which is involved in spacer integration and serves as a marker for the CRISPR–Cas systems). This challenge is not only technical, but also directly related to our ability to understand the basic mechanisms, principles of organization and the evolution of CRISPR–Cas.
The action of the CRISPR–Cas system is usually divided into three stages. The first stage is adaptation or insertion of alien DNA spacers into the CRISPR repeat cassettes. It has been demonstrated that two proteins, Cas1 and Cas2, are sufficient for this process [18]. The second stage is expression and processing of pre-crRNA (CRISPR RNA) into short guide crRNAs that is typically performed by a dedicated mechanism that involves one or several Cas proteins that form the Cascade (CRISPR-associated complex for antiviral defence) and in some cases also an RNA component. The third and final stage of the CRISPR-mediated immunity is interference, when the alien DNA or RNA is targeted by the Cascade with the bound crRNA guide [19–24]. The recent advances in the study of CRISPR–Cas systems have been covered in detail in many review articles (e.g. [5,25,26]).
Recently, a new aspect of CRISPR–Cas function has been introduced which has not yet been validated experimentally [1,27,28]. It is based on the fact that genes encoding homologues of known toxin domains involved in programmed cell death in bacteria and archaea are present in the neighbourhoods of the majority of CRISPR–Cas systems [27,28]. Remarkably, one of these components is the ribonuclease Cas2, the second gene present in most of the CRISPR–Cas systems and usually encoded in the same operon with Cas1. This feature is not unique to CRISPR–Cas systems; a variety of toxin domains is detected in association with R-M and DND systems [1,27,28]. These observations led to the hypothesis that in bacteria and archaea there exists a widespread coupling of antivirus immunity and programmed suicide or dormancy [1,27,28].
In the present paper we combine the recent advances in comparative genomics, protein sequence and structure analysis of CRISPR–Cas systems to describe a scheme of their basic structural and functional blocks, origin and evolution.
Basic structural and functional blocks of CRISPR–Cas systems
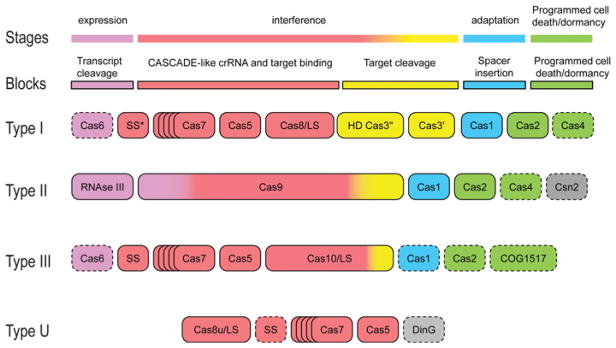
We have recently proposed a unifying scheme of the architectures of CRISPR–Cas systems on the basis of experimentally established functions of the Cas proteins and domains and recent in silico predictions [28] (Figure 1). Largely, the delineated building blocks correspond to the three functional stages of the CRISPR–Cas immune response and also encompass the putative associated immunity components that are predicted to be involved in dormancy or programmed cell suicide coupled with the CRISPR response [1,27,28].
Figure 1. The principal building blocks of CRISPR–Cas system types.
Gene names and other identifiers follow the current nomenclature and classification [7,30]. An asterisk indicates the putative small subunit that might be fused to the large subunit in several Type I subtypes [30]. Dispensable genes are indicated by broken outlines.
The block responsible for spacer integration, or the adaptation stage, consists, in the strictest sense, of the Cas1 protein only. Cas1 is an endonuclease that adopts a unique α-helical fold [29]. The cas1 gene is usually present in operons with other cas genes, and is associated with all Type I and II systems, many systems of subtype III-A and a few subtype III-B systems. As Cas1 is essential for adaptation, those Type III systems that lack the cas1 gene apparently employ the protein provided in trans by another CRISPR–Cas system within the same organism [10,19,30]. However, Cas1 was not detected within the highly reduced Type U CRISPR–Cas systems, and the functional mode of this derived CRISPR–Cas variant remains unclear [30]. The status of the Cas2 protein is somewhat ambiguous. There are controversial data on the enzymatic activity of Cas2 proteins from different bacteria and archaea. Initially, Cas2 proteins from six diverse organisms were characterized as ribonucleases [31]. However, Cas2 from Desulfovibrio vulgaris does not appear to be an active nuclease [31a] (PMID 21139194), whereas for Cas2 from Bacillus halodurans, DNase activity has been reported [31b] (PMID 22942283). Although both Casl and Cas2 are required for spacer acquisition [18], it appears unlikely that the enzymatic activity of Cas2 is involved. To our knowledge, the critical experiment with Cas2 proteins mutated in the predicted nuclease catalytic site so far has not been reported. Therefore Cas2 is probably best classified as one of the associated immunity components (see below).
Two distinct mechanisms are employed for the processing of the pre-crRNA transcript, the second stage of the CRISPR response that is associated with the second essential building block of CRISPR–Cas. In Type I and Type III systems, this process requires a dedicated ribonuclease. In most cases, this protein belongs to the Cas6 family of RAMPs (repeat-associated mysterious proteins) [30,32–34]. In some cases, however, Cas6 is apparently substituted for by other catalytically active RAMPs, such as Cas5 in subtype IC [35]. In the subtype I-E CRISPR–Cas, for which the processing mechanism was first characterized in detail, the Cas6 endonuclease is a subunit of the Cascade complex which also includes Cas5 family RAMPs as well as a large and a small subunit [36]. However, Type II systems use a seemingly unrelated mechanism that involves cellular RNAse III that is not encoded within the CRISPR–Cas loci and performs unrelated functions in RNA processing [37], a separately encoded small tracrRNA (transactivating crRNA) that is homologous with a cognate CRISPR repeat, and an unknown domain of Cas9, the multidomain protein that is the signature of the Type II CRISPR–Cas [37].
Two other distinct blocks are related to interference. One of these blocks encompasses the multiprotein Cascade complex which binds the crRNA and the target DNA that is then cleaved. After the seminal discovery of the subtype I-E Cascade in Escherichia coli [36], Cascade-like complexes were identified for the subtypes I-F, I-C and III-B [22,23,34,35,38]. Sequence and structure comparisons indicate that the Cascade-like complexes of Type I, Type III and Type U systems are homologous and generally consist of four components: large subunit, small subunit, Cas5 group RAMPs and Cas7 group RAMPs [30]. The large subunit is directly involved in target recognition, several identical or different Cas7 subunits bind crRNA and Cas5 probably interacts with both the large subunit and one of the Cas7 subunits, although the role of Cas5 has not been characterized in detail [22,23,34,35,38]. The function of the small subunit remains obscure. In Type II systems some unknown domains of Cas9 are responsible for both target recognition and crRNA binding [20]. Obviously, a DNase or a RNase is required to cleave the target. In Type I systems, an HD family nuclease has been shown to cleave DNA, and a homologous domain, present in many, but not all, Cas10 proteins is predicted to cleave DNA in DNA targeting Type III systems [30,39–41]. However, RNA cleavage by subtype III-B CRISPR–Cas is predicted to be catalysed by an enzymatically active RAMP [30]. In Type I systems, the HD domain is often fused to the Cas3 helicase which is also required for the target DNA cleavage [39]. In many Type III and subtype I-D systems, the HD domain is part of the Cas10 protein, the large subunit of the Cascade-like complex [30]. In contrast, Type II systems employ an unrelated target DNA cleavage mechanism that employs the RuvC-like and the HNH (His-Asn-His) nuclease domains of Cas9 [20].
Finally, the CRISPR–Cas systems contain the associated immunity block that is described in detail in a separate section below. In addition, several Cas proteins currently cannot be assigned to any of these major blocks with confidence. These, for example, include Csn2, the marker gene for subtype II-A and the DinG-like helicase associated with some systems of Type U (Figure 1).
Comparative analysis of the modular organization of the CRISPR–Cas systems reveals remarkable plasticity whereby the blocks are either dispensable or can be replaced by homologous or analogous components. As mentioned above, such replacements are especially pronounced in Type II systems. Subtype III-B systems often lack Cas1 and/or Cas6 that are often encoded in Type I systems in the same genome. Such subtype III-B systems apparently depend on crRNAs produced by other systems. Cas1, CRISPR arrays or modules responsible for transcript processing, or DNA target cleavage are not associated with Type U systems whose mechanism and function remain to be characterized. Thus the only module that is invariably present in all CRISPR–Cas systems is involved in crRNA binding (Figure 1). Below we briefly discuss the evolution of the components of three of these building blocks: Cas1, Cascade-like complexes and programmed cell death/dormancy machinery associated with CRISPR–Cas systems.
Evolution of Cas1
The endonuclease Cas1 and the CRISPR array embody the unique feature of CRISPR-systems, the ability to keep memory of encounters with infectious agents. Cas1 is closely associated with the ribonuclease Cas2. Both genes are present in all fully functional CRISPR–Cas systems. It has been shown that both proteins and a single CRISPR repeat are required and sufficient for spacer integration [18]. It appears, however, that Cas1 possesses all the required enzymatic activities, especially given that only DNA, and not RNA, is involved in spacer acquisition, whereas Cas2 might perform a distinct function (see discussion below).
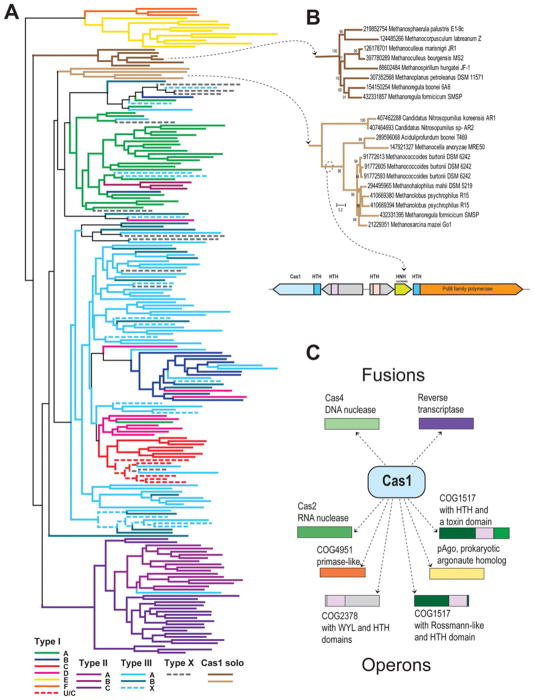
Cas1 phylogeny and operon organization are important for the classification of the subtypes of CRISPR–Cas systems [7]. The number of Cas1 proteins in the database of completely sequenced genomes more than doubled since 2011, so it is interesting to update the analysis of this family. At least one Cas1 protein is present in 953 of the 2262 available (as of 20 March 2013) archaeal and bacterial genomes. All Cas1 proteins detected were clustered by sequence similarity and 205 representatives (one from each cluster) were selected to represent the family in a further in-depth analysis (Figure 2). We also reconstructed a phylogenetic tree for the entire set of Cas1 proteins with a few fragments discarded (the Newick notation for both trees are available with the Supplementary Online data at http://www.biochemsoctrans.org/bst/041/bst0411392add.htm) and analysed their genomic neighbourhoods (Supplementary Table S1 at http://www.biochemsoctrans.org/bst/041/bst0411392add.htm). Figure 2(A) shows the phylogeny of the Cas1 family. Consistent with previous observations [7], most of the known Type I and Type II subtypes of the CRISPR–Cas systems are monophyletic, with a few exceptions in subtypes I-D, I-B and I-C. In contrast, Cas1 proteins associated with both Type III subtypes do not form monophyletic groups, especially in the case of subtype III-B. This observation is consistent with the previous conclusion that the majority of subtype III-B CRSIPR–Cas systems are not independent and have to be associated with other subtypes. Apparently, the same is partly true for subtype III-A, although there are many genomes that possess only this system, and even if they encode other systems, usually subtype III-A encompasses its own cas1–cas2 gene pair. Notably, a large fraction of Cas1 diversity is concentrated within subtype III-A, suggesting that this is a fast-evolving system.
Figure 2. The Cas1 family evolution and functional associations.
(A) The Cas1 family phylogeny. The PSIBLAST program [50] was used to retrieve homologues of the Cas1 family from 2262 completely sequenced genomes in the NCBI nr database. The BLASTCLUST program [51] (length coverage cut-off, 0.8; score density threshold, 1.0) was used to select 205 representative sequences. The multiple alignment was built using the MUSCLE program [52] with default parameters. The FastTree program [53] (JTT evolutionary model, discrete gamma model for site rates with 20 rate categories) was used for tree reconstruction. The branches are coloured according to the assignment of Cas1 genes to system subtypes based on the analysis of ten upstream and ten downstream genes. X denotes systems of unknown type or those that are predicted to be derivatives of the respective system (when coloured). (B) Phylogeny of the two solo Cas1 families. The conserved gene neighbourhood of the second group is shown underneath the trees. (C) Cas1 fusions and operonic associations.
The new tree reveals several unexpected results. The first one is the non-monophyly of the Type II system, whereby Cas1 sequences of subtype II-B form a clade within the subtype I-A branch. Interestingly, in this subtype, the cas1 gene seems to form an operon with the cas4 gene, suggesting that subtype II-B is a remnant of an ancestral fusion between a subtype I-A-like system and an element possessing RuvC and HNH nuclease domains [30].
The second unexpected finding is the discovery of two distinct Cas1 groups, mostly from Methanomicrobia, that are not associated with any CRISPR–Cas systems (Figures 2A and 2B). Previously, cas1 genes have been described exclusively as CRISPR–Cas components. The Cas1 sequences from the first stand-alone group are encoded in all known Methanomicrobiales and do not show any indications of HGT (horizontal gene transfer). In contrast, the second group shows a patchy distribution in Methanomicrobia (with three copies in Methanoregula boonei) and several representatives are found in Thaumarchaeota and Aciduliprofundum boonei (an archaeon affiliated with Thermoplasmatales in the ribosomal protein tree [42]), suggestive of multiple HGT events. There is no apparent conserved gene context for the first group of solo cas1 genes, whereas most of the cas1 genes of the second group belong to a conserved neighbourhood that includes a diverged PolB-like polymerase, an HNH nuclease and two HTH (helix–turn–helix) domain-containing proteins. Moreover, both Cas1 and PolB-like proteins are fused to related Xre family HTH domains (Figure 2B). The functions of the non-CRISPR-associated cas1 genes remain unknown, although the observation of DNA repair phenotypes in cas1-knockout mutants of E. coli might provide a clue [43].
Finally, an examination of cas1 gene fusions and operonic associations emphasizes previously under-appreciated connections to genes involved in programmed cell death/dormancy and transcription regulation (Figure 2C), suggestive of functionally important interactions between Cas1 and the respective systems (see below).
Evolution of Cascade-like complexes
We have previously proposed a parsimonious evolutionary scenario whereby a small number of evolutionary events could explain the emergence of the CRISPR–Cas system types and subtypes [28,30]. The apparent homology of Cascade-like complexes of Type I, III and U systems implies that the majority of these complexes encompass four classes of components: large subunit, small subunit and two distinct RAMPs of the Cas5 and Cas7 families [30]. However, owing to the high sequence divergence, the evidence of the presence of some of these components in all CRISPR–Cas systems was weak, being based often only on secondary structure predictions and analysis of gene context. At this time, several key structures of Cas proteins and Cascade complexes have become available, allowing us to put the previous predictions to test. In particular, the structures or stoichiometry of four Cascade-like complexes have been determined, namely the subtypes I-E, I-F, I-A and III-B [22,23,34,35,38].
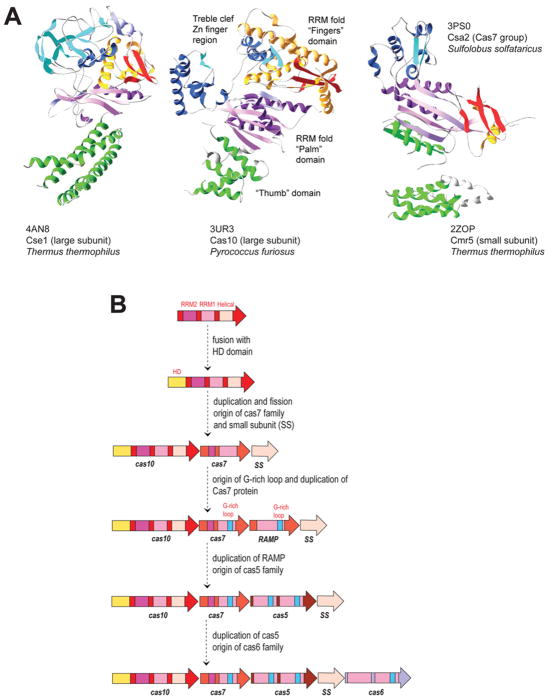
In all cases, either all four or three (subtype I-F in which the small subunit is either missing or fused to the large subunit) classes of subunits were detected. As predicted, there is only one Cas5 group RAMP in each complex and several subunits that belong to the Cas7 family. As predicted, the structure of the Cas5 group RAMP from subtype III-B system (Cmr3) encompasses two RRMs (RNA-recognition motifs) and is strikingly similar to Cas6 [44]. In all Cascade complexes that have been studied in detail, the Cas5-like RAMP interacts with the large subunit, whereas Cas7 binds crRNA [22,23,38,44], which is also compatible with our prediction. We further proposed that in subtype I-C systems Cas5 is catalytically active and could functionally replace Cas6, which is now validated experimentally [35]. Structures of the small subunits have been solved for subtype I-E (PDB code 2ZCA, Cse2), I-A (PDB code 3ZC4, Csa5) and subtype III-B systems (PDB code 2ZOP, Cmr5), and structural similarity has been clearly established between the N-terminal domains of Cse1 and Cmr5 and the C-terminal domains of Cse1 and Csa5 [45], in strong support of the homology between the Cascade-like complexes of Type I and Type III systems. Moreover, the Cmr5 structure also seems to resemble the C-terminal α-helical domain of Cas10 (PDB code 4DOZ) [46] which is compatible with our hypothesis on the origin of Cascade from a Cas10-like protein [28,30].
The major surprise came from the structure and biochemical analysis of the large subunit of the I-E system (PDB codes 4AN8 and 4H3T, Cse1) that shows no structural similarity with Cas10 (PDB code 3UR3), the large subunit of subtype III-B, at least when analysed with conventional structure comparison methods [21]. However, visual inspection of both structures suggests that they nevertheless share a common architectural layout of domains and subdomains (Figure 3A). Although we could not identify a palm-like RRM in this protein, it seems that the characteristic β2-β3 hairpin is conserved and is located exactly where it has been predicted [30]. Dramatic structural rearrangements are common among Cas proteins. For example, extensively reorganized C-terminal RRMs are present in Cas5 and Cas6f [30]. We also detected several distant homologues of large subunits that appear to have lost more than half of the protein, e.g. Csf1 in Type U systems or CsaX in subtype I-A [30]. Thus the most parsimonious interpretation of the available data still might be that Cse1 is homologous with Cas10, but severely rearranged.
Figure 3. Putative common architectural organization of central Cascade components and the evolutionary scenario for the Cascade origin.
(A) The subdomain architectures of Cse1, Cas10 and Cas7. The similarly coloured domains could be homologous but dramatically rearranged. The small subunit of subtype III-B that could be homologous with C-terminal helical domain of Cas10 is also shown. (B) Evolutionary scenario for the origin of the Cascade-like complex. Homologous domains or subdomains are colour-coded and identified by a family name, which follow the modified classification [30]. This scenario is a modification of the previously described one [28].
Given these apparent extraordinary structural rearrangements of Cas proteins, it is tempting to speculate on some even deeper relationships. Thus it cannot be excluded that Cas7, the only RAMP that contains additional subdomains and, in several cases, a zinc-finger module, could be a rearranged homologue of Cas10-like proteins (Figure 3A). Then a more detailed scenario of early evolution of the Cascade-like complex can be proposed whereby a series of duplications of an ancestral protein containing two RRMs and an α-helical domain, followed by several rearrangements, gave rise to all Cascade subunits (Figure 3B).
Programmed cell death/dormancy systems associated with CRISPR–Cas
On the basis of the analysis of gene neighbourhoods and domain composition of the numerous extremely diverse defence systems of prokaryotes, we have recently proposed a hypothesis on the coupling of antivirus immunity and programmed suicide/dormancy [1,27,28]. Under this hypothesis, a toxin associated with an immunity system, such as CRISPR–Cas, could act either as an inducer of dormancy, which prevents virus reproduction and could ‘buy the time’ for the immune system to jump into action, or as a ‘dead man’s switch’ that is released when immunity fails, by analogy with many abortive infection systems [27]. As pointed out above, the cas1 gene often associates with genes encoding (predicted) toxins, most commonly Cas2, but also Cas4 and others (Figure 2C). These associations seem to suggest that, in addition to its key roles in adaptation, Cas1 might function as an antitoxin to different toxins, thereby providing the connection between immunity and dormancy/cell suicide.
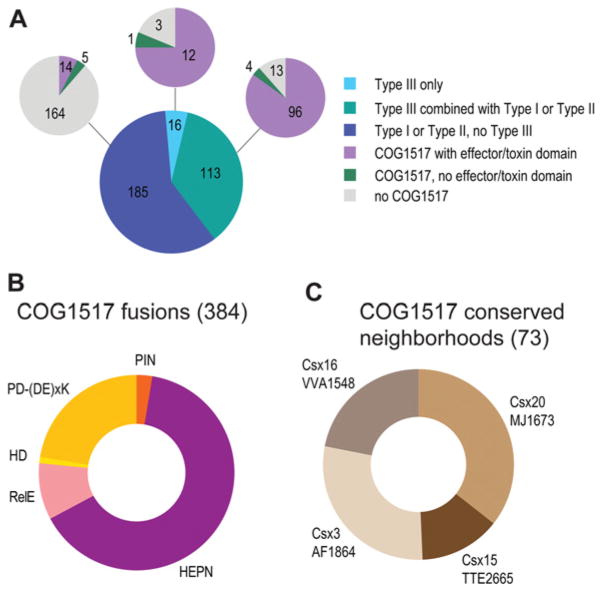
Additional observations along the same lines involved the specific link between the COG1517 genes and Type III CRISPR–Cas systems. The COG1517 family of proteins typically consist of a Rossmann-like domain and an HTH domain [47,48] and are associated, although not strictly, with CRISPR–Cas loci [7,13]. A more detailed analysis of genomic neighbourhoods (K.S. Makarova and E.V. Koonin, unpublished work) suggests a strong link between COG1517 and Type III CRISPR–Cas systems (Figure 4A). Furthermore, Type III systems are specifically associated with COG1517 family proteins that possess a third ‘effector’ domain. In most cases, the effector domains are homologous with known toxins of several families or PD-(D/E)XK nucleases which belong to the same superfamily as Cas4 (Figure 4B) [1]. More than half of the ‘effector’ domains identified belong to the HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domain superfamily (Figure 4B) and are predicted to be active ribonucleases [49]. In particular, this group includes the large Csm6 family that is strongly associated with subtype III-A systems [7,10]. COG1517-related genes are also linked to genes of several uncharacterized families, in particular Csx20 and Csx19 (Figure 4C) that also might possess toxin activity.
Figure 4. COG1517, a dormancy/programmed cell death system component associated with CRISPR–Cas systems of Type III.
(A) CRISPR–Cas-associated ‘effector’ domains. The bottom pie chart shows the breakdown of 314 CRISPR–Cas-positive genomes by the system types. The pie charts at the top show the breakdown of the respective sectors with respect to the presence of at least one COG1517 family protein and the presence or absence of an ‘effector/toxin’ domain in these proteins. (B) Effector/toxin domain fusions. HD and PD-(D/E)xK are DNA nucleases, and PIN, RelE and HEPN are ribonucleases. The number of fusions identified is indicated in parentheses. (C) Uncharacterized gene families associated with COG1517 genes in putative operons. The number of operons predicted is indicated in parentheses.
Taken together, these observations seem to suggest that a separate ‘associated immunity’ module is present in the vast majority of the CRISPR–Cas systems. Most likely, this module is not directly involved in the CRISPR–Cas molecular machinery, but rather mediates the functional coupling of the CRISPR–Cas immunity with dormancy induction and programmed cell suicide. Some of the proteins of this module might physically interact with Cas proteins that would function as antitoxins, as could be the case for Cas1 and Cas2.
Conclusion
Sequencing of numerous diverse archaeal and bacterial genomes together with the determination of the structures of many Cas proteins provides for increasingly detailed reconstructions of the modular organization and evolutionary relationships of the CRISPR–Cas systems. The emerging overarching theme seems to be the combination of the extreme diversity of the sequence and structures of Cas proteins, as well as the compositions and genomic organization of the CRISPR–Cas loci, with the remarkable structural unity that provides for relatively simple evolutionary scenarios. Indeed, the entire diversity of the Cas proteins appears to be based on diversification and embellishment of only a few basic domain types, above all the RRM that comes in a striking variety of incarnations and combinations with other domains. The second key generalization is the tight link between the CRISPR–Cas adaptive immunity systems and various toxin/antitoxin systems, which implies tight co-ordination of immune response with dormancy and programmed cell suicide in archaea and bacteria.
Supplementary Material
Acknowledgments
Funding
This research was supported by the International Research Program of the National Institutes of Health, National Library of Medicine.
Abbreviations used
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas
CRISPR-associated
- Cascade
CRISPR-associated complex for antiviral defence
- crRNA
CRISPR RNA
- HEPN
higher eukaryotes and prokaryotes nucleotide-binding
- HGT
horizontal gene transfer
- HTH
helix–turn–helix
- R-M
restriction–modification
- RAMP
repeat-associated mysterious protein
- RRM
RNA recognition motif
References
- 1.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu T, Yao F, Zhou X, Deng Z, You D. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38:7133–7141. doi: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, Horvath P. CRISPR: new horizons in phage resistance and strain identification. Annu Rev Food Sci Technol. 2012;3:143–162. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- 5.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 6.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV, Wolf YI. Is evolution Darwinian or/and Lamarckian? Biol Direct. 2009;4:42. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 10.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 12.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 13.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 15.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR–Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll D. A CRISPR approach to gene targeting. Mol Ther. 2012;20:1658–1660. doi: 10.1038/mt.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat Biotechnol. 2012;30:1002–1006. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 18.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, Terns MP. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Duijn E, Barbu IM, Barendregt A, Jore MM, Wiedenheft B, Lundgren M, Westra ER, Brouns SJ, Doudna JA, van der Oost J, Heck AJ. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced short-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteomics. 2012;11:1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiedenheft B, van Duijn E, Bultema J, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck A, Boekema E, Dickman M, Doudna JA. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrangou R. CRISPR–Cas systems and RNA-guided interference. Wiley Interdiscip Rev: RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 26.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 27.Makarova KS, Anantharaman V, Aravind L, Koonin EV. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin EV, Makarova KS. CRISPR–Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–686. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 30.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Samai P, Smith P, Shuman S. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr, Sect F: Struct Biol Crystal Commun. 2010;66:1552–1556. doi: 10.1107/S1744309110039801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31b.Nam KH, Ding F, Haitjema C, Huang Q, DeLisa MP, Ke A. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J Biol Chem. 2012;287:35943–35952. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–2480. doi: 10.1093/nar/gkr1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR–Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulepati S, Bailey S. Structural and biochemical analysis of the nuclease domain of the clustered regularly interspaced short palindromic repeat (CRISPR) associated protein 3 (CAS3) J Biol Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yutin N, Puigbo P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS ONE. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, et al. A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao Y, Cocozaki AI, Ramia NF, Terns RM, Terns MP, Li H. Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex. Structure. 2013;21:376–384. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeks J, Graham S, ANderson L, Liu H, White MF, Naismith JH. Structure of the archaeal Cascade subunit Csa5: relating the small subunits of CRISPR effector complexes. RNA Biol. 2013;10:762–769. doi: 10.4161/rna.23854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu X, Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR–Cas systems. FEBS Lett. 2012;586:939–945. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 47.Kim YK, Kim YG, Oh BH. Crystal structure and nucleic acid-binding activity of the CRISPR-associated protein Csx1 of Pyrococcus furiosus. Proteins. 2013;81:261–270. doi: 10.1002/prot.24183. [DOI] [PubMed] [Google Scholar]
- 48.Lintner NG, Frankel KA, Tsutakawa SE, Alsbury DL, Copie V, Young MJ, Tainer JA, Lawrence CM. The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J Mol Biol. 2010;405:939–955. doi: 10.1016/j.jmb.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wheeler D, Bhagwat M. BLAST QuickStart: example-driven web-based BLAST tutorial. Methods Mol Biol. 2007;395:149–176. [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.