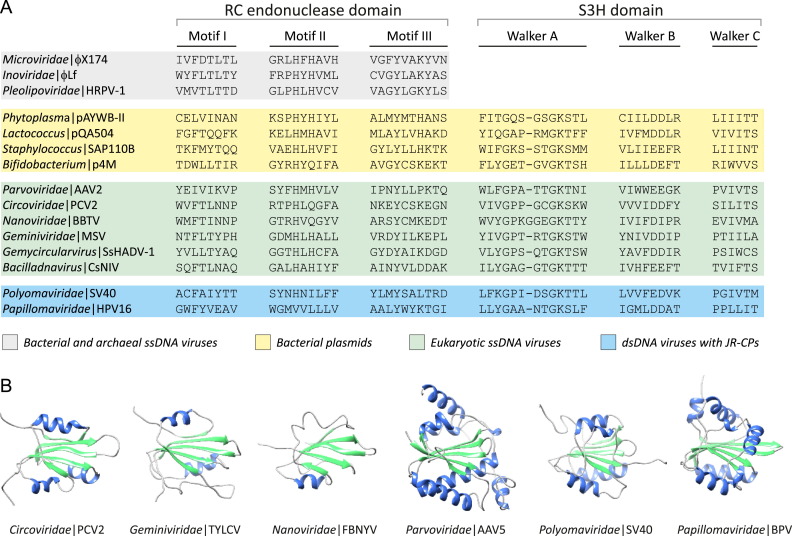
Abstract
Viruses and other selfish genetic elements are dominant entities in the biosphere, with respect to both physical abundance and genetic diversity. Various selfish elements parasitize on all cellular life forms. The relative abundances of different classes of viruses are dramatically different between prokaryotes and eukaryotes. In prokaryotes, the great majority of viruses possess double-stranded (ds) DNA genomes, with a substantial minority of single-stranded (ss) DNA viruses and only limited presence of RNA viruses. In contrast, in eukaryotes, RNA viruses account for the majority of the virome diversity although ssDNA and dsDNA viruses are common as well. Phylogenomic analysis yields tangible clues for the origins of major classes of eukaryotic viruses and in particular their likely roots in prokaryotes. Specifically, the ancestral genome of positive-strand RNA viruses of eukaryotes might have been assembled de novo from genes derived from prokaryotic retroelements and bacteria although a primordial origin of this class of viruses cannot be ruled out. Different groups of double-stranded RNA viruses derive either from dsRNA bacteriophages or from positive-strand RNA viruses. The eukaryotic ssDNA viruses apparently evolved via a fusion of genes from prokaryotic rolling circle-replicating plasmids and positive-strand RNA viruses. Different families of eukaryotic dsDNA viruses appear to have originated from specific groups of bacteriophages on at least two independent occasions. Polintons, the largest known eukaryotic transposons, predicted to also form virus particles, most likely, were the evolutionary intermediates between bacterial tectiviruses and several groups of eukaryotic dsDNA viruses including the proposed order “Megavirales” that unites diverse families of large and giant viruses. Strikingly, evolution of all classes of eukaryotic viruses appears to have involved fusion between structural and replicative gene modules derived from different sources along with additional acquisitions of diverse genes.
Keywords: Evolution of viruses, Transposable elements, Polintons, Bacteriophages, Recombination, Functional gene modules
Highlights
-
•
Eukaryotic virome dramatically differs from the viromes of bacteria and archaea.
-
•
Eukaryotic virome is dominated by RNA viruses and retroelements.
-
•
All classes of eukaryotic viruses evolved by gene module exchange.
-
•
Prokaryotic ancestry is traceable for core gene modules of most eukaryotic viruses.
-
•
Evolutionary histories of viruses and transposable elements are tightly linked.
Introduction
A major discovery of environmental genomics over the last decade is that the most common and abundant biological entities on earth are viruses, in particular bacteriophages (Edwards and Rohwer, 2005, Rohwer, 2003, Rohwer and Thurber, 2009, Suttle, 2005, Suttle, 2007). In marine, soil and animal-associated environments, virus particles consistently outnumber cells by one to two orders of magnitude. Viruses are major ecological and even geological agents that in large part shape such processes as energy conversion in the biosphere and sediment formation in water bodies by killing off populations of abundant, ecologically important organisms such as cyanobacteria or eukaryotic algae (Fuhrman, 1999, Rohwer and Thurber, 2009, Suttle, 2007). With the possible exception of some highly degraded intracellular parasitic bacteria, viruses and/or other selfish elements, such as transposons and plasmids, parasitize on all cellular organisms. Complementary to their physical dominance in the biosphere, viruses collectively appear to encompass the bulk of the genetic diversity on earth (Hendrix, 2003, Kristensen et al., 2010, Kristensen et al., 2013). The ubiquity of viruses in the extant biosphere and the results of theoretical modeling indicating that emergence of selfish genetic elements is intrinsic to any evolving system of replicators together imply that virus-host coevolution had been the mode of the evolution of life ever since its origin (Szathmary and Demeter, 1987, Takeuchi and Hogeweg, 2007, Takeuchi and Hogeweg, 2012, Takeuchi et al., 2011).
All cellular life forms possess genomes consisting of double-stranded (ds) DNA and employ the same, standard scheme for replication and expression. In contrast, viruses and other selfish elements exploit all theoretically conceivable inter-conversions of nucleic acids, with the genome represented by either RNA or DNA that can be either single-stranded or double-stranded, either circular or linear, and consists of either a single or multiple molecules (Agol, 1974, Baltimore, 1971, Koonin, 1991a). Typical viral genomes are small compared to genomes of cellular life forms but over the past few years the discovery of several groups of giant viruses has dramatically expanded the viral genome size range that now spans 3 orders of magnitude, from about 2 kilobases (kb) to over 2 megabases (Mb). The genomes of giant viruses are larger than the genomes of numerous bacteria and archaea, obliterating the gulf between cells and viruses in terms of genome size and complexity (Claverie and Abergel, 2009, Claverie et al., 2006, Legendre et al., 2014, Philippe et al., 2013, Raoult et al., 2004).
Given the fundamental differences in the reproduction strategy between viruses and cellular organisms, along with the prominence of viruses in the biosphere, it has been proposed that all organisms be classified into two primary “empires”, the ribosome-encoding (cellular) organisms and the capsid-encoding organisms (viruses) (Raoult and Forterre, 2008). This division captures some of the essential distinctions between cells and viruses but, due to the focus on capsids as a positive, defining trait of the virus empire, fails to reflect the full complexity of the evolutionary relationships among selfish genetic elements. Indeed, comparative genomic analyses make it increasingly clear that the evolutionary connections between viruses and various capsid-less elements are multifarious, involve all major groups of viruses and encompass multiple transitions from capsid-less elements to bona fide viruses and vice versa (Koonin and Dolja, 2013, Koonin and Dolja, 2014). Thus, any reconstruction of virus evolution that fails to take into account the evolutionary relationships with non-viral selfish elements is bound to be substantially incomplete. The capsid-less elements as well as many viruses differ in their extent of integration with the host cells: some insert into the cell genome and are transmitted mainly vertically through the host generations, others are largely autonomous, and many combine both strategies mixed in different proportions.
Viruses and other selfish elements certainly have not evolved from a single common ancestor: indeed, not a single gene is conserved across the entire “greater virus world” or even in the majority of selfish elements (Holmes, 2011, Koonin et al., 2006). However, these elements form a dense evolutionary network in which genomes are linked through different shared genes (Koonin and Dolja, 2014, Krupovic and Koonin, 2015, Yutin et al., 2013). This type of evolutionary relationship results from extensive exchange of genes and gene modules, in some cases between widely different elements, as well as parallel capture of homologous genes from the hosts by distinct elements. Viruses with large genomes possess numerous genes that were acquired from the hosts at different stages of evolution; such genes typically are restricted in their spread to a narrow group of viruses. However, a small group of viral hallmark genes that encode key proteins involved in genome replication and virion formation and are shared by overlapping sets of diverse viruses ensures the connectivity of the evolutionary network in the virus world (Holmes, 2011, Koonin and Dolja, 2014, Koonin et al., 2006). Virus hallmark genes have no obvious ancestors in cellular life forms, suggesting that virus-like elements evolved at a pre-cellular stage of the evolution of life.
The viromes and mobilomes (i.e. the supersets of viruses and other selfish elements) of the three domains of cellular life (bacteria, archaea and eukaryotes) are fundamentally different. Although several families of dsDNA viruses are represented in both bacteria and archaea, no viruses are known to be shared by eukaryotes with any of the other two cellular domains, even at the family or order level (King et al., 2011). The evolutionary connections between viruses of eukaryotes and those that infect bacteria and archaea are distant and complex. In this review article, we quantify the differences between the prokaryotic and eukaryotic viromes, summarize the existing evidence on putative prokaryotic ancestry of the major classes of eukaryotic viruses and virus-like elements, and delineate the likely key events in the evolution of each class.
The contrasting viromes of prokaryotes and eukaryotes
The high level classification of viruses that was introduced by Baltimore in 1971 (largely inspired by his co-discovery, with Temin, of reverse transcription in animal tumor viruses) is based on the replication-expression strategies and in particular on the form of nucleic acid that is incorporated into virions (obviously, this criterion is only applicable to bona fide viruses) (Baltimore, 1971). The following 7 classes have been delineated under this approach (Koonin, 1991a): (i) positive-strand RNA viruses (virions contain RNA of the same polarity as mRNA), (ii) negative-strand RNA viruses (virions contain RNA molecules complementary to the mRNA), (iii) dsRNA viruses, (iv) reverse-transcribing viruses with positive-strand RNA genomes, (v) reverse-transcribing viruses with dsDNA genomes (these were characterized subsequent to the seminal publication of Baltimore), (vi) ssDNA viruses, (vii) dsDNA viruses.
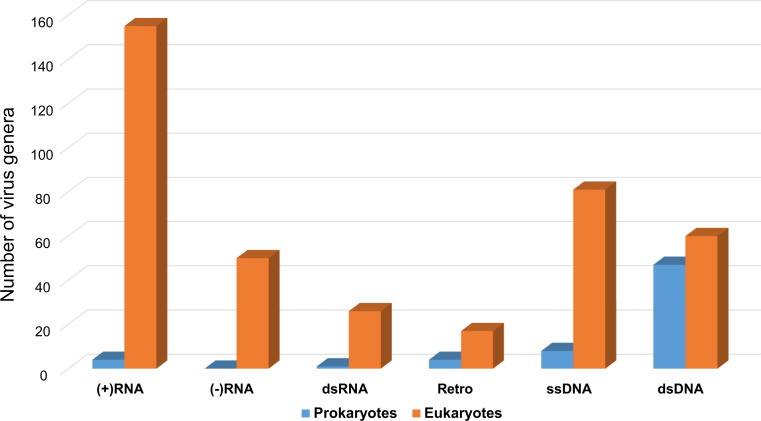
The viromes of prokaryotes and eukaryotes dramatically differ with respect to the contribution of the different Baltimore classes to the overall viral diversity ( Fig. 1). In both bacteria and archaea, the vast majority of the viruses possess dsDNA genomes, mostly within the range of 10 to 100 kb. The second most common class includes small ssDNA viruses. Positive-strand RNA and dsRNA viruses are extremely rare, and no retroviruses are known (reverse-transcribing elements exist but are not highly abundant) (Fig. 1).
Fig. 1.
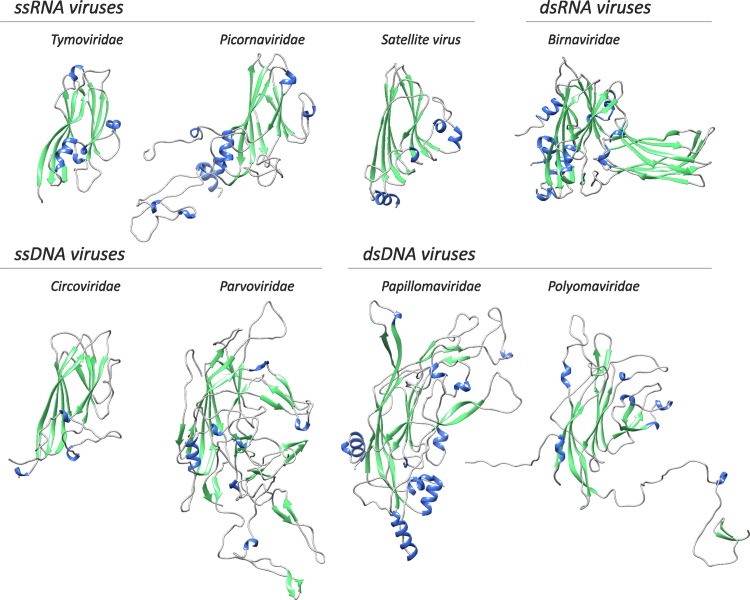
Representation of different “Baltimore classes” of viruses in prokaryotes and eukaryotes. The bars show the number of genera in the respective classes according to the latest ICTV report (King et al., 2011). Unclassified viruses are disregarded. The numbers for ssDNA viruses also include those for papillomaviruses and polyomaviruses.
In contrast to bacteria and archaea, eukaryotes host numerous, highly diverse RNA viruses (particularly of the positive-strand class) as well as reverse-transcribing elements and retroviruses that typically integrate into the host genome and are extremely abundant, comprising a substantial fraction of the genome in many groups of eukaryotes (Goodier and Kazazian, 2008, Kazazian, 2004). Collectively, the diversity and abundance of RNA viruses and retroviruses in eukaryotes exceeds the diversity and abundance of DNA viruses (Fig. 1; in this comparison, we refer to bona fide viruses because the prevalence of capsid-less elements is much more difficult to quantify).
The comparison in Fig. 1 that uses the number of recognized viral genera from each of the Baltimore classes infecting prokaryotes and eukaryotes as the measure of diversity most likely fails to pay full justice to the actual prevalence of the dominant classes, in particular dsDNA viruses, in the case of prokaryotes, and retroelements in eukaryotes. In the first instance, this appears to be the case given the existence of numerous unclassified bacteriophages and undoubtedly an even much greater number of phages that remain to be discovered. As a case in point, 39 new genera have been recently proposed within the bacteriophage family Siphoviridae (Adriaenssens et al., 2014). Despite the rapid accumulation of bacteriophage sequences, the diversity of phage genes does not show any signs of saturation, suggestive of a vast phage supergenome that so far has been barely tapped into (Kristensen et al., 2013). In the case of eukaryotes, the diversity of retroelements is not captured by the existing classification of viruses, resulting in a severe underestimate of the true impact of this class of genomic parasites. Thus, the actual discrepancy between the prokaryotic and eukaryotic viromes is likely to be even greater than suggested by the data in Fig. 1.
The biological causes of the dramatic difference in the composition of the virome between eukaryotes and prokaryotes remain unclear. It stands to reason that the emergence of the eukaryotic nucleus severely shrunk the niche for dsDNA virus reproduction by creating a barrier for the access of viral DNA to the sites of host genome replication and transcription, and complicating the process of virus maturation. Notably, the majority of dsDNA viruses of eukaryotes replicate in the cytoplasm (see below) suggesting that those few groups of dsDNA viruses that replicate in the nucleus have evolved specific adaptations to overcome the barriers. Conversely, the cytosolic compartment of eukaryotic cells, with its elaborate intracellular membrane system, might provide a fertile niche for the reproduction of RNA viruses (Belov, 2014, den Boon and Ahlquist, 2010, Greninger, 2015, Nagy and Pogany, 2012). With respect to the dramatic proliferation of retroelements, an accommodating niche could have been provided by the expanding genomes of eukaryotes and their greater tolerance to insertion of mobile elements compared to genomes of prokaryotes (Lynch, 2007, Lynch and Conery, 2003).
Regardless of the underlying causes, reconstruction of the evolution of the eukaryotic virome, with its dramatic differences from the viromes of bacteria and archaea and comparatively greater diversity, is a major challenge in the study of virus evolution. In the following sections of this article, we discuss the evolutionary scenarios that have been developed for different classes of eukaryotic viruses over the last few years and how the evolutionary relationships between viruses of prokaryotes and eukaryotes become apparent in these scenarios.
Evolutionary scenarios for the origin of eukaryotes and their impact on the reconstruction of virus evolution
The origin of eukaryotes is a major problem in evolutionary biology that is generally considered to be unresolved. It is now clear that nearly all extant eukaryotes possess membrane-bounded, energy-converting organelles, the mitochondria or partially degraded derivatives thereof (such as mitosomes or hydrogenosomes), and the few known cases of actual loss of mitochondria are secondary (Hjort et al., 2010, van der Giezen, 2009, van der Giezen and Tovar, 2005). Accordingly, the Last Eukaryotic Common Ancestor (LECA) is believed to have been a typical, mitochondriate eukaryotic cell (Embley and Martin, 2006, Lane and Martin, 2010, Lane and Martin, 2012). Another well established, key piece of information pertinent for the origin of eukaryotes is the sharp split of the evolutionarily conserved eukaryotic genes into the genes with an archaeal evolutionary affinity and those with a bacterial affinity (along with some with no detectable prokaryotic homologs) (Brown and Doolittle, 1997, Esser et al., 2004, Yutin et al., 2008). The archaeal ancestry is apparent primarily for genes encoding components of informational systems along with some key components of the cytoskeleton and the cell division machinery (Koonin and Yutin, 2014), whereas operational genes, such as metabolic enzymes, appear to be largely of bacterial origin.
Within the constraints set by these key observations, two distinct classes of scenarios for the origin of eukaryotes are currently considered; the scenarios within each class differ in detail but the classes are sharply differentiated by the postulated nature of the organism that played host to the protomitochondrial endosymbiont (Embley and Martin, 2006). The historically first scenario postulates a lineage of primary amitochondrial eukaryotes (sometimes called archaezoa) that are perceived to have evolved as a sister group of archaea or possibly as a sister group of one of the major archaeal branches, such as the ‘TACK (Thaumarchaeota–Aigarchaeota–Crenarchaeota–Korarchaeota) superphylum’ (Guy et al., 2014). Under this scenario, the hypothetical amitochondrial ancestor of eukaryotes possessed the principal features of the eukaryotic cellular architecture such as the advanced cytoskeleton and endomembrane system including the nucleus (Kurland et al., 2006, Poole et al., 1999, Poole and Penny, 2007). These features would facilitate engulfment of the protomitochondrial endosymbiont (and bacteria in general) which is conceivably the strongest aspect of the primary amitochondrial scenario (hereinafter protoeukaryote scenario). The obvious weakest point of this scenario is the lack of any evidence of the existence of primary amitochondrial eukaryotic forms despite intensive search. The proponents of the protoeukaryotic scenario thus have to postulate that such forms are either extinct or exceedingly rare. Furthermore, there is no precedent for the evolution of large, internally compartmentalized cells among prokaryotes, and it has been argued that emergence of such cells is unfeasible without highly efficient cellular energetics that is provided by the multiple mitochondria residing within a single cell (Lane and Martin, 2010, Lane and Martin, 2012).
The alternative, symbiogenetic scenario (Embley and Martin, 2006, Martin et al., 2007), obviously fueled by the ubiquity of mitochondria and related organelles in eukaryotes, postulates that the host of the proto-mitochondrial endosymbiont was not a protoeukaryote endowed with the key features of the eukaryotic cellular organization, including the nucleus, but rather a regular archaeon, most likely a mesophilic form that could comprise a deep branch within the TACK superphylum or possibly a sister group thereof (Koonin and Yutin, 2014). The symbiogenetic scenario implies a plausible succession of events leading to the key innovations of the eukaryotic cell such as the endomembrane system including the nucleus, the cytoskeleton, the ubiquitin-centered signaling system and pre-mRNA splicing (Koonin, 2006, Martin and Koonin, 2006). The weakness of the symbiogenetic scenario is the extreme rarity of endosymbiosis among prokaryotes (although bacteria living inside other bacteria have been described Husnik et al., 2013, von Dohlen et al., 2001) and the apparent absence of mechanisms, such as phagocytosis, that would facilitate engulfment of bacteria. The proponents of this scenario therefore are forced to postulate a (extremely) rare event at the root of eukaryogenesis. However, the recent discovery of archaeal homologs (and putative ancestors) of key elements of the eukaryotic cytoskeleton, cell division systems and ubiquitin machinery provide for an amended symbiogenetic scenario. Under this hypothesis, the archaeal ancestor of eukaryotes, the host of the protomitochondrial endosymbiont, could have possessed relatively complex intracellular organization that would facilitate engulfment of bacteria and evolution of the compartmentalized eukaryotic cell (Guy et al., 2014, Koonin and Yutin, 2014, Yutin et al., 2009).
In the following sections, we examine the implications of each of these scenarios of the evolution of eukaryotes for the origin of different classes of eukaryotic viruses.
Origins of the major classes of eukaryotic viruses and evolutionary relationships between viruses of prokaryotes and eukaryotes
A general perspective on RNA virus evolution: Out of the primordial RNA world?
According to the widely accepted RNA world hypothesis, the RNA-only replication cycle antedates reverse transcription and DNA-based replication (Bernhardt, 2012, Gilbert, 1986, Neveu et al., 2013, Robertson and Joyce, 2012). Under this premise, the RNA viruses and related selfish elements whose replication relies on RNA-dependent RNA-polymerase (RdRp), are the only major group of organisms (apart from small, non-coding parasitic RNAs such as viroids Diener, 1989) that could be direct descendants of RNA world inhabitants. Because RdRp is the only viral hallmark protein that is universally conserved in RNA viruses (Kamer and Argos, 1984, Koonin and Dolja, 1993, Koonin et al., 2006), this enzyme is the key to reconstructing their evolutionary histories. Together with distantly related RNA-dependent DNA polymerases or reverse transcriptases (RT), viral RdRps represent a deeply branching lineage within the ancient superfamily of palm domain-containing polymerases and primases (Iyer et al., 2005). As is typical of viral hallmark genes (Koonin et al., 2006), cellular organisms encode no homologs of viral RdRps with the same enzymatic activity. The only known family of RdRps encoded in cellular genomes, those involved in the amplification of small interfering RNAs in eukaryotes, are homologs of the DNA-dependent RNA polymerases (Iyer et al., 2003, Salgado et al., 2006).
Based on the structure of the encapsidated genome and genome replication/expression cycles, the ‘RNA only’ viruses are divided into three Baltimore classes: positive-strand, double-strand and negative-strand (+RNA, dsRNA and −RNA, respectively). All non-defective viruses from each of these classes employ virus-encoded RdRps for genome replication and often for the distinct process of genome transcription to generate viral subgenomic mRNAs. Early comparative analyses identified 6 signature amino acid sequence motifs that are conserved in RdRps of diverse +RNA viruses infecting bacteria, plants and animals, suggesting their monophyletic origin (Kamer and Argos, 1984, Koonin, 1991b, Xiong and Eickbush, 1990). It has been further demonstrated that similar motifs were present in RdRps of dsRNA viruses and the RTs (Kamer and Argos, 1984, Koonin et al., 1989, Xiong and Eickbush, 1990). Although the RdRps of the –RNA viruses possess certain motifs resembling those conserved in +RNA and dsRNA viruses (Tordo et al., 1988, Xiong and Eickbush, 1990), the overall level of similarity is extremely low, making the evolutionary connection between the −RNA viruses and the rest of RNA viruses tenuous at best.
In addition to protein sequence analysis, reconstruction of the RdRp evolution is substantially aided by the comparisons of their atomic structures. It has been found that RdRps from diverse +RNA and dsRNA viruses of bacteria and animals possess a characteristic ‘right-handed’ fold, comprising palm, fingers, and thumb domains (Choi and Rossmann, 2009, Ferrer-Orta et al., 2006, Kidmose et al., 2010, Monttinen et al., 2014). A long-awaited first atomic structure of the RdRp of a −RNA virus, bat influenza A virus, helped to demystify the origins of these viruses by revealing a high level of structural similarity to RdRps of both +RNA and dsRNA viruses (Pflug et al., 2014). Thus, the three classes of RNA viruses share the homologous core enzyme that is responsible for their replication and, by implication, related origins.
Under the symbiogenetic scenario for the origin of eukaryotes, it seems natural to assume that RNA viruses of eukaryotes originate from either RNA bacteriophages or RNA viruses of Archaea. This assumption, however, is challenged by the striking scarcity of bacterial and archaeal RNA viruses compared to the flourishing genomic and ecological diversity of their eukaryotic counterparts (see above). Indeed, there are only a handful of the +RNA bacteriophages all of which belong to the family Leviviridae infecting primarily enterobacteria and some other proteobacteria (Bollback and Huelsenbeck, 2001). Likewise, only a few dsRNA bacteriophages of the family Cystoviridae that infect γ-proteobacteria of the genus Pseudomonas are currently known (Mindich, 2004) although efforts on new virus isolation might expand this range (Mantynen et al., 2015). The targeted search for extant archaeal RNA viruses so far has netted only a single +RNA virus candidate that appears to represent a novel virus family but whose host range remains to be validated (Bolduc et al., 2012). Thus, the very existence of archaeal RNA viruses remains an open question. Finally, there is no evidence of −RNA viruses infecting prokaryotes. The protoeukaryotic scenario would imply a different narrative on the origins of the RNA viruses of eukaryotes whereby the remarkable diversity of these viruses evolved within the ancient protoeukaryotic lineage due to the features of the (proto)eukaryotic cell organization, such as an intracellular membrane system, that might be conducive to RNA virus reproduction. Should that be the case, the search for bacterial or archaeal ancestry would be futile in principle. Below we discuss how the available data on the origins of different genes of RNA viruses bear on these distinct origin scenarios.
Positive-strand RNA viruses: Assembly from diverse prokaryotic progenitors and gene exchanges leading to enormous diversification
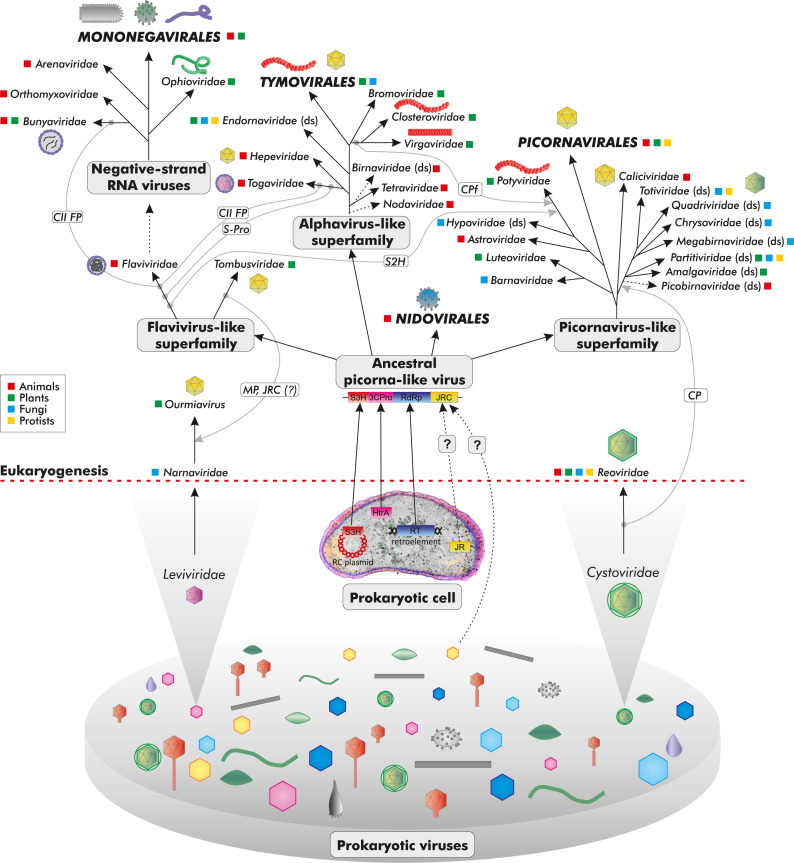
Large-scale phylogenomic analysis of the +RNA viruses of eukaryotes was initiated over two decades ago and yielded conclusions that withstood the test of time remarkably well (Goldbach and Wellink, 1988, Koonin, 1991b, Koonin and Dolja, 1993). These studies have identified three major evolutionary lineages that collectively encompass the vast majority of the +RNA viruses infecting eukaryotes: picornavirus-like, alphavirus-like and flavivirus-like superfamilies ( Fig. 2). This classification is based on a combination of evidence from the RdRp phylogeny with signature genes and gene arrangements that have been identified for the picornavirus-like and alphavirus-like superfamilies (see below). The congruence between the two lines of evidence is crucial because the high sequence divergence of the RdRp that is dictated by the overall high mutation rate of RNA viruses, despite the essentiality of the polymerase, hampers the construction of fully reliable phylogenetic trees (Zanotto et al., 1996).
Fig. 2.
Origin of the major groups of RNA viruses of eukaryotes. The depicted evolutionary reconstruction is predicated on the symbiogenetic scenario of eukaryogenesis. The host ranges of viral groups are color-coded as shown in the inset. Icons of virion structures are shown for selected groups. Ancestor-descendant relationships that are considered tentative are shown with dotted lines, and particularly weak links are additionally indicated by question marks (see text for details). Key horizontal gene transfer events are shown by gray, curved arrows. Abbreviations: CII FP, Class II fusion protein; CP, capsid protein; CPf, capsid protein of filamentous viruses; JRC, jelly roll capsid (protein); MP, movement protein; RT, reverse transcriptase; S2H, Superfamily 2 helicase; S3H, Superfamily 3 helicase.
The picornavirus-like superfamily is by far the largest, most diverse and most widely represented across the diversity of the eukaryotic hosts. In addition to a distinct RdRp lineage, the picornavirus-like superfamily is defined by the presence of a conserved array of signature genes, which encode a superfamily 3 helicase (S3H), a small genome-linked protein (VPg), a distinct chymotrypsin-like protease 3CPro and a single beta-barrel jelly-roll capsid protein (JRC), and are represented, some losses and replacements notwithstanding, in most members of this superfamily (Koonin and Dolja, 1993, Koonin et al., 2008).
The global ecology of the picornavirus-like superfamily, which spans a broad range of multicellular and unicellular eukaryotic hosts (Supplementary Table S1) points to an early origin of these viruses antedating the radiation of the eukaryotic supergroups. The core of the picornavirus-like superfamily is represented by the order Picornavirales that encompasses 5 families, several floating genera and many unclassified viruses (Le Gall et al., 2008). The viruses within this order share all the signature genes of the superfamily. Furthermore, all these viruses express their genomes via polyprotein processing (in some groups, there are two polyproteins, one encompassing the structural proteins and the other one proteins involved in replication) and package the genomic RNA into characteristic icosahedral virions with a pseudo-T=3 symmetry. Notably, Picornavirales include viruses infecting a broad range of hosts from three supergroups of eukaryotic organisms, Unikonts (vertebrates, insects), Plantae (angiosperms) and Chromalveolates (diatomes, raphidophytes, thraustrochytrids), as well as viruses from marine environments with unidentified hosts (Le Gall et al., 2008).
The family of vertebrate viruses Caliciviridae is closely related to Picornavirales, sharing a conserved S3H-VPg-3CPro-RdRp-JRC gene array and differing only in the structure of their true T=3 capsid. Strikingly, in the phylogenetic tree of the RdRp, caliciviruses confidently cluster with the members of Totiviridae, a family of dsRNA viruses that infect fungi (Unikonts) as well as Kinetoplastids, Trichomonads and Diplomonads, all of which belong to a distinct supergroup of unicellular eukaryotes, the Excavates. Because the clade that unites Caliciviradae and Totiviridae is lodged inside the picornavirus-like RdRp tree, it seems likely that this family of dsRNA viruses is a highly derived off-shoot of the picornavirus-like superfamily of +RNA viruses. The viruses in the remaining three major evolutionary lineages of picornavirus-like viruses (Fig. 2) encompass only subsets of the five picornaviral signature genes or, in the case of the family Partitiviridae, only the picornavirus-type RdRp. Each of these groups also includes viruses infecting hosts that belong to two or three eukaryotic supergroups (Koonin et al., 2008).
Thus, the evolutionary scenario best compatible with the superimposition of the phylogenetic trees of eukaryotes and picorna-like viruses involves early diversification antedating the divergence of eukaryotic supergroups. The alternative, i.e. emergence of the ancestors of each of the 6 lineages of the picornavirus-like superfamily in one of the eukaryotic supergroups followed by horizontal virus transfer (HVT) to hosts from other supergroups, appears to be decidedly less parsimonious because such a scenario would require numerous HVT events involving organisms with widely different lifestyles and ecological niches (Koonin et al., 2008). However, HVT could have played an important role in the subsequent evolution of the picorna-like viruses (Dolja and Koonin, 2011). One case in point is the phylogeny of partitiviruses in which fungal and plant viruses intermix, pointing to multiple occurrences of HVT between two widely different host taxa (Nibert et al., 2013). Another example involves the closely related plant Potiviridae and fungal Hypoviridae (Koonin et al., 1991a). The HVT between plants and fungi appears to be particularly plausible given close associations between plants and their ubiquitous fungal pathogens and symbionts.
In contrast to the picornavirus-like superfamily, the alphavirus-like and flavivirus-like superfamilies exhibit much less diversity in terms of both the numbers of included families and even more so their global ecologies (Dolja and Koonin, 2011). The alphavirus-like superfamily includes the order Tymovirales along with several other families of plant viruses and two families of animal viruses (Supplementary Table S1 and Fig. 2). All these viruses are unified by a conserved array of replication-associated genes which encode capping enzyme, superfamily 1 helicase and the RdRp (Koonin and Dolja, 1993). A recent in-depth comparative analysis of viral protein sequences has revealed a highly derived variant of the capping enzyme in the nodaviruses, an abundant family of animal +RNA viruses with small genomes (Ahola and Karlin, 2015). The RdRp of nodaviruses does not show an affinity with the alphavirus-like superfamily but rather had been tentatively included in the picorna-like superfamily on the basis of limited conservation of some sequence motifs (Koonin, 1991b, Koonin and Dolja, 1993, Koonin et al., 2008). However, there is no strong objective support for this affinity. Although nodaviruses, similar to other +RNA viruses with small genomes, lack a helicase, the presence of the predicted capping enzyme suggests their inclusion in the alphavirus-like superfamily as a deep, perhaps basal branch (Fig. 2). This affiliation is compatible with the observation that nodaviruses share a distinct variant of the JRC containing an autoprocessing domain with tetraviruses and birnaviruses that appear to share a common ancestor and are included in the alphavirus-like superfamily on the basis of the RdRp phylogeny (Wang et al., 2012a). Unlike the picorna-like viruses, the great majority of which possess JRC-based icosahedral capsids (with the exception of filamentous potyviruses and capsid-less hypoviruses), capsid architectures of alphavirus-like viruses are extremely diverse. These architectures include: (i) icosahedral virions built of either JRC or unrelated proteins; (ii) helical rod-shaped or flexible filamentous virions formed by a distinct family of four-helix bundle capsid proteins; (iii) membrane-enveloped virions. The host ranges of alpha-like viruses are limited almost exclusively to plants, where these viruses reach remarkable diversity, and animals. Only the family Endornaviridae that consists of capsid-less elements has a broader host range including “viruses” of plants and fungi, and a single “virus” of a plant-parasitic oomycete, potentially, a result of HVT from a host plant (Koonin and Dolja, 2014, Roossinck et al., 2011).
The flavivirus-like superfamily is the smallest of the three major groups of the +RNA viruses of eukaryotes and encompasses only two families that appear to be rather odd bedfellows (Fig. 2). The Flaviviridae are enveloped animal viruses that encode a specific lineage of RdRp, a superfamily 2 helicase as well as a protease and a capping enzyme that are distinct from the functionally analogous proteins of the picornavirus-like and alphavirus-like superfamilies, respectively (Koonin and Dolja, 1993). None of these genes except for RdRp is conserved in Tombusviridae, viruses with small icosahedral capsid built of JRC that infect plants (with the exception of a single marine virus that presumably infects a unicellular eukaryotic host) (Culley et al., 2006, Dolja and Koonin, 2011). Thus, the flavivirus-like superfamily is held together only by the phylogenetic affinity of the RdRPs. Although this association is consistently observed in multiple, independent phylogenetic analyses (Koonin and Dolja, 1993), the lack of additional support from signature genes makes this superfamily a tenuous group. It is not inconceivable that Flaviviridae and Tombusviridae would be best treated as separate superfamilies of +RNA viruses.
In accordance with a major, general trend of virus evolution (see also below), the histories of the three superfamilies of +RNA viruses were not completely independent but rather involved multiple gene exchanges. A striking case in point is the family Potiviridae, the largest family of plant viruses (Gibbs and Ohshima, 2010) that are confidently included in the picornavirus-like superfamily on the basis of a combination of several features including the RdRp phylogeny, the presence of two additional signature genes, namely the picornavirus-like protease and VPg, and the mode of protein expression via polyprotein processing. However, two other signature genes of the picornavirus-like superfamily, namely the S3H and the JRC, are replaced in the potyviruses, respectively, by a Superfamily 2 helicase most closely related to the homologous helicase of flaviviruses and by a four-helix bundle capsid protein related to that of filamentous plant viruses in the alphavirus-like superfamily (e.g. potexviruses) (Dolja et al., 1991, Koonin and Dolja, 1993, Koonin et al., 2008). Thus, evolution of the potyviruses involved substantial modification of the picornavirus-like scaffold (and consequently, the virion structure) through contributions from the other two superfamilies of +RNA viruses (Fig. 2). Other notable cases of intersuperfamily gene exchange include the apparent transfer of the serine protease gene between flaviviruses and togaviruses in which, strikingly, the protease was recruited for the capsid protein function (Gorbalenya et al., 1989b); spread of the genes for movement proteins between plant-infecting viruses from all three superfamilies (Mushegian and Koonin, 1993); and spread of class II fusion proteins among flaviviruses, togaviruses and bunyaviruses (Modis, 2014, Vaney and Rey, 2011).
A notable complementary trend in the evolution of +RNA viruses is the parallelism between the designs of the viral genomes in the three superfamilies. Indeed, apart from the RdRp and the CP, most of the viruses in the picorna-like and alpha-like superfamilies and the animal viruses in the flavi-like superfamily encode proteins with two types of functionality, helicases and proteases (Koonin and Dolja, 1993). The presence of these domains most likely is dictated by functional requirements such as the requirement of a helicase for the replication of (relatively) large RNA genomes. The existence of such a requirement is suggested by the clear threshold for the presence of the helicase gene which is found in all +RNA viruses with genomes larger than approximately 6 kb but not in viruses with smaller genomes (Gorbalenya and Koonin, 1989). Strikingly, however, both the helicases and the proteases in the three viral superfamilies belong to different protein families (Koonin and Dolja, 1993 and see above). Whether these analogous designs of the viral genomes evolved in parallel from a common ancestor that lacked the helicase and the protease or through displacement of the corresponding ancestral domains, is difficult to ascertain.
Elucidation of the exact evolutionary relationships among the three superfamilies of +RNA viruses of eukaryotes requires in-depth phylogenetic analyses of their RdRps which is a daunting task given the high sequence divergence of this protein outside the conserved motifs. Expansion of the collection of RdRp structures and refinement of methods for structure-based phylogeny could lead to progress. Nonetheless, the available evidence seems to support evolutionary primacy of the picornavirus-like superfamily. Most importantly, the host ranges of alphavirus-like and flavivirus-like superfamilies are limited almost exclusively to vertebrates, their arthropod parasites, and flowering plants, that is, only three groups of multicellular organisms. These narrow host ranges could point to relatively late evolutionary origins of the viruses of these superfamilies, perhaps concomitant with the emergence of the respective host groups. Furthermore, HVT, in particular via insect vectors, could have played an important role in the evolution of these viral superfamilies. In contrast, the broad host range of picorna-like viruses encompasses four eukaryotic supergroups and a great variety of both unicellular and multicellular organisms. Furthermore, multiple host-specific and metagenomic studies of marine RNA viruses (most of them demonstrated or thought to infect diverse unicellular eukaryotes) have recovered a large number of novel picorna-like viruses but only one tombus-like virus and no alpha-like viruses (Culley et al., 2006, Culley et al., 2014, Culley and Steward, 2007, Koonin et al., 2008).
The three-superfamily classification of +RNA viruses does not readily accommodate the distinct order Nidovirales which includes viruses with the largest known RNA genomes and several unique genomic features. Notably, none of these viruses encode JRC and, consistently, do not form icosahedral virions. Instead, members of the Nidovirales have enveloped virions which vary from roughly spherical to rod-shaped, depending on the organization of the helical nucleocapsids (Gorbalenya et al., 2006, Koonin and Dolja, 1993). However, certain evolutionary affinity between RdRps of picornavirus-like viruses and nidoviruses, together with the presence of distantly related proteases responsible for polyprotein processing in both of these virus groups (Gorbalenya et al., 2006, Koonin and Dolja, 1993), suggests that nidoviruses could be highly derived off-shoots of the picornavirus-like superfamily.
Thus, the extreme diversity of the picorna-like viruses, with respect to both the host range and the genome architecture, suggests that picornaviral ancestors have evolved concomitantly with or shortly after the emergence of eukaryotes, rapidly diversified and spawned the ancestors of the alphavirus-like and flavivirus-like superfamilies as well as the Nidovirales (that are known to infect only vertebrates, insects and crustaceans), perhaps later in evolution (Fig. 2).
If the picornavirus-like superfamily indeed represents the ancestral viral reservoir from which the rest of the eukaryotic +RNA viruses evolved (with some notable exceptions discussed below), then, the problem of the origin of eukaryotic +RNA viruses boils down to the origin of the ancestral picorna-like virus. This question has been addressed through a focused search for potential prokaryotic roots of picorna-like viruses (Koonin et al., 2008). In addition to validating the tight relationship between the three superfamilies of the eukaryotic positive-strand RNA viruses, in-depth sequence analysis of the RdRps of the picornavirus-like superfamily has revealed remarkably high similarity of picornavius-like RdRps to the reverse transcriptases (RTs) of the bacterial group II retroelements (self-splicing introns), in contrast to the much lower similarity to the RdRps of RNA bacteriophages (Koonin et al., 2008). Considering the wide spread of the group II retroelements in bacteria (Lambowitz and Zimmerly, 2004, Lambowitz and Zimmerly, 2011), in contrast to the scarcity of RNA bacteriophages, it appears plausible that the prokaryotic RTs were the ancestors of picornavirus-like RdRps. Search for the closest homologs of the 3CPro confidently identified bacterial and mitochondrial proteases of the HtrA family (Gorbalenya et al., 1989a, Koonin et al., 2008), suggesting direct descent of the viral protease from bacterial endosymbiont of emerging eukaryotic cell. The exact origins of the other picornaviral signature genes, S3H, JRC and VPg, proved much more difficult to trace. Nevertheless, S3H is encoded in some dsDNA bacteriophages and bacterial rolling-circle plasmids (see below) whereas the single β-barrel JRC of the picorna-like variety is present in ssDNA bacteriophages of the family Microviridae (McKenna et al., 1992, Roux et al., 2012). Additionally, the JRC-like β-barrel fold is found in various carbohydrate-binding proteins including those from bacteria (Norris et al., 1994, Wong et al., 2000), and some non-viral β-barrel proteins, such as tumor necrosis factor, are even known to form virus-like particles (Liu et al., 2002). These cellular jelly-roll proteins are considerably more compact than CPs of microviruses and thus might be more likely to have been the ancestors of JRC of RNA viruses. Consequently, bacterial origins for these genes are conceivable as well, leading to an evolutionary scenario in which the ancestral picorna-like virus was assembled from diverse building blocks derived from the proto-mitochondrial endosymbiont during eukaryogenesis (Koonin et al., 2008) (Fig. 2). Clearly, this scenario is most plausible within the framework of the symbiogenetic scenario for the origin of eukaryotes. Under the protoeukaryote scenario, the ancestral picorna-like virus could be construed as a direct descendant of the primordial RNA world that survived and thrived in the protoeukaryotic lineage (Fig. 2). In this case, the RdRp of the picorna-like viruses would be viewed as the primordial replicase, and S3H and JRC accordingly would be considered ancestral forms of the respective proteins. The ancestral picorna-like virus thus could resemble the extant nodaviruses that possess a “minimal” genome within the picornavirus-like superfamily encoding only the RdRp and the JRC. Incidentally, the only reported putative RNA virus of archaea shows a similar genome architecture although it is premature to discuss its possible role in the evolution of the viruses of eukaryotes until the archaeal host range is validated (Bolduc et al., 2012). The 3CPro, for which the bacterial origin appears undeniable, could be a later acquisition concurrent with the symbiogenesis.
Although the only known group of +RNA bacteriophages, the leviviruses, apparently have not contributed to the origin of the bulk of the eukaryotic +RNA viruses, they did give rise to two distinct, small lineages of the eukaryotic viruses (Fig. 2). Searches for the most closely related homologs of the leviviral RdRps identified the RdRps of these two narrow groups, fungal Narnaviridae and plant Ourmiavirus, as the eukaryotic descendants of the leviviruses. The narnaviruses hardly meet the narrow definition of viruses because they are neither infectious nor possess an extracellular encapsidated form (Hillman and Cai, 2013). The entire replication cycle of the narnaviruses of the genus Mitovirus takes place within fungal mitochondria. Given the origin of the mitochondria from an alphaproteobacterial endosymbiont, it appears most likely that the ancestral narnavirus evolved from an RNA bacteriophage brought along by the protomitochondrion, by losing the capsid and thus switching to the status of a mitochondrial RNA plasmid. In contrast, plant ourmiaviruses are full-fledged, infectious, encapsidated +RNA plant viruses. Because their RdRps are related to those of narnaviruses, whereas the intercellular movement and possibly capsid proteins are related to respective proteins of tombusviruses, it has been proposed that ourmiaviruses evolved via recombination between a narnavirus-like element from a plant-pathogenic fungus and a tombusvirus (Rastgou et al., 2009).
dsRNA viruses: Multiple origins from positive-strand RNA viruses
The dsRNA viruses of eukaryotes appear to be much less diverse than +RNA viruses as follows from the numbers of currently recognized families (10 versus 31, respectively; Supplementary Table S2). However, the recent accelerated pace of discovery of new, diverse dsRNA viruses might soon challenge this perception (Liu et al., 2012a, Liu et al., 2012b). Early phylogenetic analyses of the RdRps led to the conclusion that the dsRNA viruses originated on multiple occasions, mainly from different groups of +RNA viruses (Koonin, 1992, Koonin et al., 1989). The inclusion of two families of dsRNA viruses, Totiviridae and Partitiviridae, into the picornavirus-like superfamily is in full accord with this evolutionary scenario. The viruses in the family Birnaviridae share an unusual permuted RdRp, a genome-linked protein and a distinct variant of the JRC with some of the tetraviruses (the family Tetraviridae has been recently split into three distinct families, namely Alphatetraviridae, Carmotetraviridae and Permutotetraviridae; Table S1), supporting a common origin of these families of dsRNA and +RNA viruses at an early stage of the evolution of the alphavirus-like superfamily (Fig. 2) (Gorbalenya et al., 2002, Zeddam et al., 2010). Notably, the divergence of birnaviruses from tetraviruses has apparently occurred following the acquisition of the JRC protein gene by their common ancestor from a nodavirus (Wang et al., 2012a). The family of capsid-less viruses Endornaviridae that is currently classified with dsRNA viruses clearly evolved from an alphavirus-like ancestor as indicated by the conservation of a signature set of core replication genes (Koonin and Dolja, 2014).
Evolutionary scenarios based on the phylogenetic analysis of viral replication proteins often deviate from those centered on the evolution of other functional modules, in particular those of viral capsid proteins (Krupovic and Bamford, 2008, Krupovic and Bamford, 2009). Thus, for comprehensive reconstruction of virus evolution, that would reflect the intrinsic modularity of this process, it is essential to complement phylogenetic and comparative genomic analyses with the analysis of structural data (Koonin et al., 2009). The emerging picture of the evolution of dsRNA viruses is among the best illustrations of this general principle.
Structural analyses have shown that eukaryotic dsRNA viruses from the families Picobirnaviridae, Chrysoviridae, Totiviridae, Partitiviridae, Reoviridae and bacteriophages of the family Cystoviridae employ related capsid proteins to build their unique T=1 icosahedral capsids from 60 asymmetrical CP dimers (El Omari et al., 2013, Janssen et al., 2015, Luque et al., 2014, Poranen and Bamford, 2012). Based on comparisons of the virion and CP structures, it has been proposed that reoviruses are most closely related to cystoviruses whereas picobirnaviruses, partitiviruses, and totiviruses form another, distant branch of dsRNA viruses (El Omari et al., 2013); additionally, the CP of chrysoviruses has been concluded to be most closely related to that of totiviruses (Luque et al., 2014). Thus, bacterial cystoviruses appear to have contributed the structural genes to most of the dsRNA viruses infecting eukaryotes. The reoviruses, the largest family of dsRNA viruses that infect diverse eukaryotic hosts (Fig. 2 and Supplementary Table S2), appear to be direct descendants of the cystoviruses. In contrast, in the evolution of picobirnaviruses, partitiviruses, totiviruses, chrysoviruses and the related megabirnaviruses the pivotal event was recombination (or more likely, multiple, independent recombination events) with members of the picornavirus-like superfamily of +RNA viruses, resulting in chimeric genomes encoding cystovirus-derived capsid proteins and pricornavirus-like RdRps (Fig. 2).
The global ecology of the dsRNA viruses appears rather peculiar. Unlike most of the families of +RNA viruses that are confined to a relatively narrow host ranges (e.g., arthropods for Iflaviridae, vertebrates for Picornaviridae and plants for Secoviridae), extremely diverse hosts are often infected by the dsRNA viruses from the same family. As a case in point, the family Reoviridae includes viruses that infect vertebrates, arthropods, mollusks, fungi, flowering plants and a unicellular green alga. Likewise, Partitiviridae infect fungi, flowering plants and an apicomplexan unicellular eukaryote, whereas host range of Totiviridae includes fungi and several unicellular eukaryotic parasites from the Excavate supergroup (King et al., 2011). Such ecological patterns including two or three supergroups of eukaryotic hosts for each of the three largest families of the dsRNA viruses point to their ancient origins from the dsRNA bacteriophage and picornavirus-like ancestors as discussed above (Fig. 2).
The role of HVT in the evolution of the dsRNA viruses is most apparent for the family Endornaviridae where the plant and fungal virus branches in the phylogenetic trees of viral RdRps often intermingle within the same cluster (Roossinck et al., 2011). A contribution of HVT appears likely also in the evolution of reoviruses many of which, both from vertebrates and from plants, are also capable of infecting their arthropod vectors (Ng and Falk, 2006, Quito-Avila et al., 2012) that could serve as HVT intermediaries. Thus, phylogenetic, structural, and host range analyses converge in supporting the major theme in the evolution of the dsRNA viruses: ancient polyphyletic origin from dsRNA bacteriophages or distinct groups of +RNA virus ancestors, or via recombination between these distinct types of ancestors. The current spread of the dsRNA viruses, however, could have been substantially affected by more recent HVT events.
Negative-strand RNA viruses: The emerging positive-strand connection
Negative-strand RNA viruses of eukaryotes include the order Mononegavirales that consists of three related virus families with non-segmented genomes and 5 families of viruses with segmented genomes (Supplementary Table S3). For a long time, the evolutionary origin of the −RNA viruses had been veiled in mystery due to the highly derived sequences of their RdRps (Tordo et al., 1988, Xiong and Eickbush, 1990) and the lack of readily identified homologs for other proteins, with the exception of capping enzymes in Mononegavirales that also is extremely diverged from all homologs (Bujnicki and Rychlewski, 2002, Li et al., 2008). The narrow host ranges of −RNA viruses, limited to animals and plants, imply relatively recent evolutionary origin. Furthermore, it has been proposed that −RNA viruses of plants were acquired from animals via HVT (Dolja and Koonin, 2011). This scenario is compatible with the markedly higher diversity and prevalence of the animal −RNA viruses compared to the relative scarcity of these viruses in plants. The protein sequences, as well as virion and genome architectures, are highly similar between animal and plant viruses in the families Rhabdoviridae and Bunyaviridae. Furthermore, arthropod parasites of animals and plants could have readily served as HVT vehicles because both plant and animal rhabdoviruses and bunyaviruses are transmitted by and replicate in their arthropod vectors (Ammar el et al., 2009, Guu et al., 2012). The discovery of four –RNA viruses that infect soybean cyst nematodes further expands the ecological reach of these viruses within animal lineage of evolution (Bekal et al., 2011). This finding suggests a potential major route of animal-to-plant HVT of −RNA viruses given that the nematodes, many of which are plant parasites, are the most numerous animals on earth (Blaxter et al., 1998). Notably, two of these novel viruses are most closely related to bunyaviruses, and one to rhabdoviruses, the two −RNA virus families that include members infecting either animals or plants.
A major insight into the origin of −RNA viruses came from the recently solved crystal structure of the Influenza A virus RdRp that has revealed striking similarity to the structure of the flavivirus RdRps (Pflug et al., 2014). This finding strongly suggests that −RNA viruses evolved from a +RNA ancestor of the flavivirus-like superfamily but diverged from the ancestral forms beyond recognition at the sequence level due to the switch to a radically different replication cycle. Although influenza RdRp is also structurally similar to the RdRp of dsRNA bacteriophages (cystoviruses), a direct evolutionary connection seems unlikely given the significantly lower similarity than that with the flavivirus RdRp and the apparent relatively late emergence of the −RNA viruses (see above). This reasoning is further buttressed by the recent identification of a nematode-infecting flavi-like virus (Bekal et al., 2014) which suggests that nematodes could have played the role of a melting pot in which the progenitor of the −RNA viruses was conceived and that also played a key role in the spread of these viruses to new hosts. Further, in-depth phylogenetic and structural analysis of the proteins encoded by flavi-like viruses and −RNA viruses are required to develop the proposed evolutionary scenario in more detail.
Given the accumulating evidence of the origin of both dsRNA viruses and −RNA viruses from different groups of +RNA viruses, the ancestor of the picorna-like viruses appears to have been the ultimate progenitor of the great majority of eukaryotic RNA viruses. Whether this ancestral picorna-like virus was assembled from several distinct building blocks of bacterial origin during eukaryogenesis (Fig. 2) or evolved as a continuous lineage from the primordial gene pool, is an intriguing and important question. The answer critically depends on the choice of the scenario for the origin of eukaryotes that hopefully will be informed by the further advances of archaeal and bacterial genomics. Regardless of the impending solution to this key problem, a limited footprint of RNA bacteriophages on the evolution of eukaryotic RNA viruses is apparent in the origin of narnaviruses and ourmiaviruses from leviviruses, and most likely, reoviruses from cystoviruses.
Synopsis on eukaryotic RNA virome
To recapitulate the key points on the eukaryotic RNA virome, the enormous diversity of RNA viruses is a hallmark of the eukaryotic part of the virus world. We are far from a full understanding of the underlying causes of this remarkable bloom of RNA viruses but it stands to reason that the eukaryotic cytosol, with its extensive endomembrane system provides a niche that is highly conducive to RNA replication. There is sufficient evidence to derive the great majority of eukaryotic RNA viruses from a common, positive-strand ancestor that might have been assembled from several components with distinct roots in prokaryotes including a reverse transcriptase. In contrast, several isolated groups of eukaryotic RNA viruses derive directly from bacterial RNA viral ancestors. The striking diversification of RNA viruses in eukaryotes, in part, depended on switches in genome replication-expression strategies (from positive-strand to double-stranded and negative-stranded genomes) and multiple exchanges of genes between far diverged groups of viruses.
Retroelements and retroviruses: Viruses as derived forms
An extremely common and abundant class of selfish elements in eukaryotes consists of reverse-transcribing elements (or retroelements for short), including retroviruses. Similar to the case of RNA viruses, the single common denominator of these extremely diverse elements is the polymerase involved in their replication, in this case, the reverse transcriptase (RT) which defines the key feature of the reproduction cycle, namely reverse transcription of RNA into DNA (Eickbush and Jamburuthugoda, 2008, Finnegan, 2012, Kazazian, 2004, Xiong and Eickbush, 1990). Beyond this unifying step, retroelements show all conceivable reproduction strategies: some behave like mobile elements that jump around host genomes via reverse transcription and integration, and regularly degrade to become integral parts of the host genomes; others behave as DNA or RNA plasmids; yet others, the best-characterized ones, are bona fide viruses that pack in the virions either RNA or DNA, or even a DNA–RNA hybrid, and go through an essential or facultative stage of integration into the host genome during virus replication. Although all retroelements are relatively small, their genomic complexity varies greatly, from solo RT to sophisticated build-ups of viral genomes with over 10 genes, for example in the case of HIV.
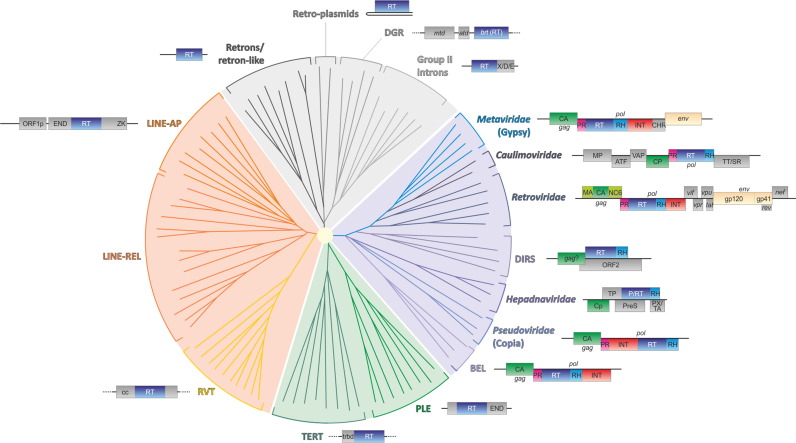
Given that the RT is the only universal gene among the retroelements, a natural approach to the reconstruction of their evolution involves using a phylogenetic tree of the RT as a framework. Phylogenetic analysis (Gladyshev and Arkhipova, 2011) divides the RTs into four major branches that include: (1) retroelements from prokaryotes including Group II self-splicing introns and retrons, (2) LINE-like elements, (3) Penelope-like elements, (4) reverse-transcribing viruses and related retrotransposons that contain Long Terminal Repeats (LTR) ( Fig. 3). Historically, all retroelements, with the exception of reverse-transcribing viruses and their relatives, are often called non-LTR retrotransposons. The 4 main branches of RTs as well as several branches within each of them (see below) are well resolved but the position of the root is not known.
Fig. 3.
Evolution of retroelements and reverse-transcribing viruses. Genomic organizations of selected representatives of the major groups of retroelements overlay the phylogenetic tree of the reverse transcriptases. The topology of the tree is from (Gladyshev and Arkhipova, 2011). Abbreviations: DGR, diversity-generating retroelements; X/D/E, maturase, DNA binding, and endonuclease domains, respectively, of the intron-encoded protein; mtd, major tropism determinant; atd, accessory tropism determinant; brt, bacteriophage reverse transcriptase; LINE, long interspersed nucleotide elements; END, endonuclease; ZK, zinc knuckle; gag, group-specific antigen; env, envelope; pol, polymerase; PR, aspartate protease; RT, reverse transcriptase; RH, RNase H; INT, integrase; CHR, chromodomain; MA, matrix protein; CA/Cp, capsid protein; NC, nucleocapsid; 6, 6-kDa protein; vif, vpr, vpu, tat, rev, and nef, regulatory proteins encoded by spliced mRNAs; gp120 and gp41, the 120- (surface) and 41-kDa (transmembrane) glycoproteins; ATF, aphid transmission factor; VAP, virion-associated protein; TT/SR, translation trans-activator/suppressor of RNA interference; TP, terminal protein; P, polymerase; PreS, pre-surface protein (envelope); PX/TA, protein X/transcription activator; trbd, telomerase RNA-binding domain; cc, coiled-coil.
The archaeal and bacterial retroelements that comprise one of the 4 major clades in the RT tree (Fig. 3) include 3 well-characterized groups of bacterial retroelements (represented also in some archaea): (i) Group II introns, (ii) retrons and (iii) diversity-generating retroelements (DGR) (Robart and Zimmerly, 2005; Simon and Zimmerly, 2008; Toro and Nisa-Martínez, 2014). The fourth group in this clade of RTs includes the so-called retroplasmids that replicate in fungal mitochondria, and given the endosymbiotic origin of the mitochondria, are likely to be of bacterial origin (Griffiths, 1995). In addition, analysis of bacterial and archaeal genomes revealed many RTs of unclear provenance that are likely to constitute or derive from uncharacterized retroelements (Simon and Zimmerly, 2008).
The Group II self-splicing introns are by far the most common retroelements in archaea and bacteria representing over 70% of the RTs detected by a survey of bacterial and archaeal genomes, and are the only group of prokaryotic retroelements with demonstrated independent horizontal mobility (Lambowitz and Zimmerly, 2004, Lambowitz and Zimmerly, 2011, Simon and Zimmerly, 2008). In addition to bacteria and some archaea, Group II introns are commonly present in mitochondrial genomes of fungi, plants and some protists. The large protein encoded in Group II introns, in addition to the RT, encompasses an endonuclease domain that is involved in transposition. This endonuclease domain belongs to the HNH family which is one of the nucleases frequently encoded also in Group I introns (Stoddard, 2005). Thus, from the evolutionary standpoint, Group II introns are likely to have evolved from self-splicing, endonuclease-encoding introns (similar in architecture to Group I introns but with a distinct ribozyme structure) that acquired an RT gene resulting in a more autonomous reproduction strategy.
Retrons are retroelements that consist of a solo RT gene and are vertically inherited in bacteria suggestive of some ‘normal’ function(s) in bacterial cells; to date, however, there is no indication of the nature of such a presumptive function of the retrons (Lampson et al., 2005). The RT of the retrons makes multiple copies of a branched RNA-DNA hybrid but accumulation of these unusual molecules does not result in any discernible phenotype in the bacteria.
The DGRs are unusual retroelements that are present in some bacteriophage and bacterial genomes and have been shown to employ the RT to modify specific target genes and accordingly their protein products in a specific fashion resulting in changes in phage receptor specificity, helping the phage to evade bacterial resistance (Medhekar and Miller, 2007).
Bacterial retroelements, primarily Group II introns, have reached substantial diversity, with several distinct groups revealed by phylogenetic analysis, and invaded most of the bacterial divisions (Simon et al., 2008). In contrast, in archaea, the spread of these elements is restricted to a few groups of mesophiles, such as Methanosarcina, that appear to have acquired numerous bacterial genes via HGT. The same route has been proposed for the retroelements (Rest and Mindell, 2003).
In a stark contrast to the prokaryotic retroelements that are rather sparsely represented among bacteria, are rare in archaea and do not reach high copy numbers, diverse eukaryotic genomes are replete with retroelements of different varieties. By conservative estimates, retroelement-derived sequences account for over 50% of mammalian genomes (mostly non-LTR elements) and over 75% of some plant genomes, e.g. maize (Defraia and Slotkin, 2014, Lee and Kim, 2014, Solyom and Kazazian, 2012). Although usually not reaching such extravagant excesses, retroelements are abundant also in genomes of diverse unicellular eukaryotes (Bhattacharya et al., 2002, Lorenzi et al., 2008). The eukaryotic retroelements show limited diversity of the RT sequences compared to the prokaryotic retroelements which is in sharp contrast with the enormous diversity of genome organizations and reproduction strategies. We discuss these elements in accord with their branching in the phylogenetic trees of the RTs (Fig. 3).
Penelope-like retroelements (PLE) are simple retrotransposons that typically encode a single large protein that in the originally discovered group of PLE is a fusion of the RT with a GIY-YIG endonuclease (Fig. 3) (Evgen’ev, 2013, Lyozin et al., 2001). This complete form of PLE so far has been identified only in animals. However, a shorter PLE variants that lack the endonuclease are integrated in subtelomeric regions of chromosomes in a broad variety of eukaryotes (Gladyshev and Arkhipova, 2011). In the phylogenetic tree of the RT, the PLE confidently cluster with the telomerase RT (TERT), a pan-eukaryotic enzyme that is essential for the replication of the ends of linear chromosomes (Chan and Blackburn, 2004). This relationship implies that the PLE-like branch of retroelements antedates the LECA although the complete, endonuclease-encoding PLE apparently evolved later. The recruitment of the PLE-related RT for the telomerase function clearly was an early, pivotal event during the evolution of the eukaryotic cell. Remarkably, several groups of eukaryotes, in particular insects, have lost the TERT gene and instead use a distinct variety of non-LTR retrotransposons as telomeric repeats (Pardue and DeBaryshe, 2011). Thus, it seems that retroelements provide for the replication of chromosome ends in all eukaryotes thanks to their intrinsic ability to generate sequence repeats.
The GIY-YIG endonuclease domains are widely represented in Group I introns and are also present in the repair endonuclease UvrC that is strongly conserved among bacteria (Aravind et al., 1999). These endonuclease domains are small and highly diverged, so establishing evolutionary relationships is difficult. Nevertheless, it is interesting to note that the Penelope endonuclease domain shows the strongest similarity to GIY-YIG endonucleases from Group I introns of some large DNA viruses such as phycodnaviruses (Van Etten, 2003). Thus, the complete forms of PLE found in animals might have evolved by fusing a viral intron-encoded endonuclease domain to the ancestral RT.
The LINE elements (Long Interspersed Nuclear Elements) comprise another group of simple retroelements that appear to be both the most common retroelements in eukaryotes, being represented in the genomes of diverse organisms of all major eukaryotic groups, and the most abundant among the extant retroelements as they reach extremely high copy numbers in animal genomes (de Koning et al., 2011, Kazazian, 2004). Most of these LINE elements are inactivated and decaying but a small fraction remains active and spawns new copies. In addition, the active LINE RT mediate the retrotransposition of SINEs (such as the Alu elements that are extremely abundant in primate genomes), small elements that lack any protein-coding genes but still follow the retrotransposon life style and propagate to extremely high numbers in animal genomes (de Koning et al., 2011).
A typical, complete vertebrate LINE consists of two genes one of which encodes the RT and endonuclease domains whereas the second one encodes an RNA-binding domain that is required for transposition. The RTs of the LINEs form two distinct branches in the phylogenetic tree (Fig. 3), and the respective elements also encode distinct endonucleases. The ‘classic’ LINEs including all elements found in mammals encode an apurinic/apyrimidinic (AP) endonuclease that also possesses RNase H activity and is essential for transposition. In contrast, a subset of LINEs from diverse eukaryotes encode a bona fide RNase H (Fig. 3). Although some phylogenetic analyses suggest that RNase H is a late acquisition in the history of non-LTR retroelements (Malik, 2005), it does not appear possible to rule out that this is the ancestral architecture among the LINEs. Another branch of LINEs encode a RLE (Restriction-like Endonuclease) domain that, similar to the AP endonuclease, introduces a nick into the target and thus initiates transposition (Mandal et al., 2004, Yang et al., 1999). Furthermore, comparative analysis of the LINEs in plants has shown that, in addition to the AP endonuclease, a group of these elements acquired a distinct RNase H domain, surprisingly, of apparent archaeal origin (Smyshlyaev et al., 2013).
In the phylogenetic tree of the RT (Fig. 3), the LINEs cluster (albeit with limited statistical support) with a recently discovered distinct group of RT (denoted RVT) that contain no identifiable domains other than the RT proper, are not currently known to behave as mobile elements, are present in a single copy in the genomes of diverse eukaryotes, and hence are likely to fulfill some still uncharacterized function(s) in eukaryotic cells. Members of the RVT group have been identified also in several bacterial genomes suggesting the possibility of horizontal gene transfer the direction of which remains uncertain (Gladyshev and Arkhipova, 2011).
Among the RT-elements, bona fide viruses, with genomes encased in virus particles, and typical infection cycles including an extracellular phase, are a minority (Supplementary Table S4). Importantly, capsid-less retroelements are found in all major divisions of cellular organisms, and by inference, are ancestral to this entire class of genetic elements. By contrast, reverse-transcribing viruses are derived forms that apparently evolved at an early stage in the evolution of eukaryotes (see below).
The reproduction strategy of the retroviruses (family Retroviridae) partly resembles that of RNA viruses, combining aspects analogous to both positive-strand RNA viruses and negative-strand RNA viruses. The retroviruses are effectively RNA viruses that have evolved the capacity to convert to DNA, integrate into the host genome and then exploit the host replication and transcription machinery. In addition to the typical infectious retroviruses, vertebrate genomes carry numerous endogenous retroviruses that are largely transmitted vertically and are often inactivated by mutation but, until that happens, have the potential to get activated and yield infectious virus (Stoye, 2012, Weiss, 2013).
The two other families of reverse-transcribing viruses, Hepadnaviridae infecting animals and Caulimoviridae infecting plants (collectively often denoted pararetroviruses), have ventured further into the DNA world: these viruses package the DNA form of the genome (or sometimes a DNA–RNA, in the case of hepadnaviruses) into the virions but retain the reverse transcription stage in the reproduction cycle (Nassal, 2008, Rothnie et al., 1994, Seeger and Hu, 1997). In contrast to the retroviruses, for viruses of these families, integration into the host genome is not an essential stage of the reproduction cycle although apparent spurious integration is common among caulimoviruses (Harper et al., 2002, Staginnus and Richert-Poggeler, 2006). The remaining two families of reverse-transcribing viruses, Metaviridae and Pseudoviridae, include RT-encoding elements that are traditionally not even considered viruses but rather retrotransposons because they normally do not infect new cells, although it has been suggested that Gypsy elements of Drosophila are infectious (Kim et al., 1994, Song et al., 1994). In any case, these elements, e.g. Gypsy/Ty3-like elements (Metaviridae) in animals or Copia/Ty1-like elements in fungi (Pseudoviridae), encode virion proteins and form particles, and thus meet the definition of a virus.
Among all retroelements, the reverse-transcribing viruses possess the most complex genomes (Fig. 3). All retroviruses share 3 major genes that are traditionally denoted pol, gag and env, and in many cases, also additional, variable genes. The retrovirus RT is a domain of the Pol polyprotein. In the viral branch of retroelements, the strictly conserved module consists of the RT together with the RNase H (RH) domain that is essential for the removal of the RNA strand during the synthesis of the DNA provirus. Two other domains, integrase and aspartic protease, are found only in a subset of pol polyproteins. However, superposition of the domain architectures of the pol polyproteins over the phylogenetic tree of the RTs strongly suggests that the common ancestor of the reverse-transcribing viruses encoded the complex form of Pol, most likely one with the PR-RT-RH-INT arrangement that is shared between retroviruses and metaviruses (Fig. 3). The phylogenies of the RT, RH and INT domains of reverse-transcribing viruses appear to be concordant and cluster metaviruses with retroviruses to the exclusion of pseudoviruses (Malik and Eickbush, 1999), in agreement with the RT phylogeny in Fig. 3 and the above evolutionary scenario. Under this scenario, caulimoviruses have lost the integrase domain whereas hepadnaviruses have lost both the integrase and the protease but acquired the terminal protein domain that is involved in the initiation of DNA synthesis.
A more complete phylogenetic analysis of the RNase H that involved also the RH from non-LTR retroelements of the LINE branch as well as bacterial and eukaryotic RNH I indicated, first, that the non-LTR retroelements in eukaryotes were older than the LTR elements, and second, quite unexpectedly, that in retroviruses, the ancestral RH apparently was secondarily replaced with the eukaryotic homolog (Malik and Eickbush, 2001). The ultimate origin of the RH in retroelements is not easy to decipher because, for this short domain, the topology of the deep branches in the tree is unreliable. However, a “smoking gun” has been detected that links the RH in retroelements with eukaryotic homologs, namely a distinct DNA–RNA hybrid and dsRNA-binding domain that is shared by eukaryotic RNH I and a subset of the retroelement RH (Majorek et al., 2014, Smyshlyaev et al., 2013). The presence of this derived shared character indicates that the retroelements have acquired a eukaryotic RNH I at an early stage of their evolution.
The INT domain of the LTR retroelements belongs to the DDE family of transposases (named after the distinct catalytic triad) that mediate transposition of numerous DNA transposons in eukaryotes and prokaryotes (Nesmelova and Hackett, 2010). Therefore, it has been proposed that the founder of the LTR retrotransposon branch emerged as a result of recombination between a non-LTR retrotransposon and a DNA transposon (Capy and Maisonhaute, 2002, Malik and Eickbush, 2001). Notably, the Gypsy/Ty3 retrotransposons have acquired a chromodomain (a widespread domain involved in chromatin remodeling in eukaryotes) that is fused to the integrase of these elements and modulates the specificity of integration (Novikova et al., 2010).
The aspartic protease of the LTR retroelements is homologous to the pan-eukaryotic protein DDI1, an essential, ubiquitin-dependent regulator of the cell cycle whereas DDI1 itself appears to have been derived from a distinct group of bacterial aspartyl proteases (Krylov and Koonin, 2001, Sirkis et al., 2006). Thus, strikingly, the ancestral Pol polyprotein of the LTR retroelements seems to have evolved through assembly from 4 distinct components only one of which, the RT, derives from a pre-existing retroelement.
Apart from the Pol polyprotein, the relationships between genes in different groups of reverse-transcribing viruses are convoluted. The capsid protein domain of the Gag polyprotein is conserved between retroviruses and the Ty3/Gypsy metaviruses. The conserved region of the nucleocapsid (NC) protein consists of a distinct C2HC Zn-knuckle that at least in retroviruses is involved in RNA and DNA binding (Darlix et al., 2014). The retroviral capsid (CA) protein contains a conserved C-terminal α-helical domain known as SCAN that mediates protein dimerization (Ivanov et al., 2005). Phylogenetic analysis of the conserved portion of Gag suggests that the 3 classes of retroviruses evolved from 3 distinct lineages of metaviruses as suggested by the so-called ”three kings” hypothesis (Llorens et al., 2008). However, it is unclear whether the Gag-like protein of Copia/Ty1 (pseudoviruses) is homologous as well, and neither is the ultimate origin of this protein outside of the retroelements. Although homologs of the Gag proteins in animals have been discovered and shown to be important in development, the respective genes apparently have been transferred from retroviruses to the host genomes (Kaneko-Ishino and Ishino, 2012).
Strikingly, in the evolution of retroviruses, the env genes have been apparently acquired by LTR retrotransposons on at least three independent occasions from different groups of RNA and DNA viruses: gypsy/metaviruses have acquired their env-like gene from insect baculoviruses (dsDNA viruses); the envelope genes of the Cer retroelements in the Caenorhabditis elegans genome appear to derive from a phlebovirus (−RNA virus) source; and the Tas retroviral envelope (Ascaris lumricoides) might have been obtained from herpesviruses (dsDNA viruses) (Malik et al., 2000). The origin of the env genes of the vertebrate retroviruses that appear not to be homologous to any of the above env genes remains obscure. Interestingly, however, in vertebrate retroviruses, such as HIV, the gp41 domain of env is a class I fusion protein which is also found in many −RNA viruses, including orthomyxoviruses, paramyxoviruses, coronaviruses, filoviruses and arenaviruses (Kielian and Rey, 2006, White et al., 2008). Thus, despite the lack of a readily traceable ancestral relationship, it is thus conceivable that vertebrate retroviruses assembled their env proteins from preexisting protein domains of other eukaryotic viruses.
Caulimoviruses and especially hepadnaviruses are highly derived forms that apparently have lost and/or displaced several genes of the ancestral reverse-transcribing virus, with the exception of RT and RH, and also PR in the case of caulimoviruses (Fig. 3). In addition, the capsid proteins of caulimoviruses share the C2HC Zn-knuckle with the NCs of retroviruses and metaviruses (Covey, 1986). Thus, at least one domain of the ancestral nucleocapsid protein of reverse-transcribing viruses survives in caulimoviruses. In contrast, the core protein of hepadnaviruses shows no significant sequence similarity to capsid proteins of retroviruses or caulimoviruses, and might be a displacement of uncertain provenance. However, based on similar dimerization principles and sequence conservation patterns, it has been suggested that the capsid protein of hepadnaviruses and the C-terminal domain of retroviral CA actually are distant homologs (Steven et al., 2005).
The origins of the family-specific genes of reverse-transcribing viruses remain uncertain, with the notable exception of the movement protein (MP) of caulimoviruses. The MP is conserved in a great variety of plant viruses including positive-strand RNA viruses, negative strand RNA viruses and ssDNA viruses. Clearly, the MP gene horizontally spread among diverse viruses driven by selection for the ability to cross plasmodesmata and hence cause systemic infection in plants (Koonin et al., 1991b, Melcher, 2000, Mushegian and Elena, 2015, Mushegian and Koonin, 1993). A much better known, textbook case of viral genes with a clear provenance are the oncogenes of numerous animal retroviruses (e.g. such thoroughly characterized oncogenes as v-src, v-ras or v-abl) which are mutated versions of host genes involved in cell cycle control that cause cell transformation when expressed from an integrated DNA copy of the viral genome (Maeda et al., 2008).
Most likely, retroelements have been an integral part of biological systems since the stage of the primordial replicators when they gave rise to the first DNA genomes (Koonin, 2009). Indeed, under the RNA World scenario, the transition to DNA genomes would necessarily require reverse transcription, with the implication that some varieties of retroelements already existed at that stage of evolution. However, in prokaryotes, retroelements maintain a low profile and never attain complex genomic architectures. In eukaryotes, the fortunes of retroelements have turned around: they proliferated dramatically, have become a defining factor of genome evolution and spawned several families of reverse-transcribing viruses. The wide spread of each of the major groups of retroelements across the diversity of eukaryotes indicates that the principal events in the evolution of retroelements occurred before the radiation of the eukaryotic supergroups. The PLE appear to be the best candidates for the role of the founder eukaryotic retroelements that gave rise to other simple, widespread non-LTR elements, such as the LINEs, as well as fully ‘domesticated’ RTs such as TERT and RVT that are conserved throughout the eukaryotic domain. A much more complex series of events led to the emergence of the LTR retroelements (in particular, reverse-transcribing viruses) including highly derived forms such as caulimoviruses and hepadnaviruses.
The parsimonious version of the scenario for the origin of the eukaryotic retroelements depends on the scenario for the origin of eukaryotes. The symbiogenetic scenario would root the entire diversity of the eukaryotic retroelements in prokaryotic ones, most likely, Group II introns. This origin of the eukaryotic retroelements appears compatible with the ancestral relationship between Group II introns and the eukaryotic spliceosomal introns (that have lost both protein-coding genes and the self-splicing capacity) as well as the snoRNAs, the catalytic components of the spliceosome (Chalamcharla et al., 2010, Dai et al., 2008, Lambowitz and Zimmerly, 2011, Robart et al., 2014, Toor et al., 2008). Remarkably, the essential, highly conserved (yet functionally poorly characterized) pan-eukaryotic protein subunit of the spliceosome, Prp8, also is an inactivated RT derivative that most likely evolved from the Group II intron RT (Dlakic and Mushegian, 2011). Thus, under the symbiogenetic scenario, prokaryotic retroelements provide intermediates between the primordial genetic pool and the diversity of the eukaryotic retroelements. In contrast, the protoeukaryote scenario implies that both prokaryotic and eukaryotic retroelements are direct descendants of primordial genetic entities that adopted distinct routes of evolution in prokaryotes and eukaryotes.
The sequence variability of the prokaryotic RTs is extremely high, with only the essential motifs of the RT domain conserved throughout, by far exceeding the variation among the eukaryotic retroelements (Simon and Zimmerly, 2008). This greater sequence diversity of the RTs in prokaryotes, despite their relatively low abundance, seems to be compatible with the origin of all eukaryotic retroelements from a distinct branch of prokaryotic retroelements, such as Group II introns. Furthermore, given the apparent origin of the eukaryotic splicing from Group II introns, the symbiogenetic scenario seems to offer a simpler evolutionary narrative than the protoeukaryotic scenario. Regardless, the remarkable diversification of the retroelements in eukaryotes could have been triggered by the (typically) weaker purifying selection compared to prokaryotes which allowed for the massive proliferation of integrated retroelements and provided the playground for their further evolution (Lynch, 2007, Lynch and Conery, 2003).
Synopsis on eukaryotic retroelements
To summarize, the retroelements enjoyed no less success in eukaryotes than RNA viruses with which they could share the ultimate common origin from prokaryotic Group II elements (self-splicing introns). However, bona fide reverse-transcribing viruses are derived forms and show limited diversity. Notably, although all these viruses share a common origin, they seem to have acquired the envelope proteins from different sources and on independent occasions. Retroelements including retro-transcribing viruses evolve in a much closer integration with the eukaryotic hosts than RNA viruses and sequences from these elements have been extensively recruited by eukaryotes for a variety of cellular functions at all stages of evolution.
Origins of ssDNA viruses of eukaryotes: Multiple crosses between plasmids and RNA viruses
Viruses with ssDNA genomes are increasingly appreciated as a rapidly expanding, highly diverse class of economically, medically and ecologically important pathogens. They infect hosts from all three domains of cellular life and are present in all conceivable environments, from near-surface atmosphere (Whon et al., 2012) to soil (Kim et al., 2008), from freshwater and marine habitats (Labonte and Suttle, 2013, Rosario et al., 2009, Roux et al., 2012, Zawar-Reza et al., 2014) to the most extreme settings, such as terrestrial hot springs (Mochizuki et al., 2012). Bacterial and archaeal ssDNA viruses are grouped into four families, whereas the eukaryotic ssDNA viruses are classified into 6 families, Anelloviridae, Bidnaviridae, Circoviridae, Geminiviridae, Nanoviridae and Parvoviridae, and one unassigned genus (Bacilladnavirus) (Supplementary Table S5). Anelloviruses appear to be restricted to various mammals (Okamoto, 2009); circoviruses are known to infect different avian species and pigs (Delwart and Li, 2012); nanoviruses and geminiviruses infect plants (Grigoras et al., 2014, Hanley-Bowdoin et al., 2013); parvoviruses replicate in vertebrates and arthropods (Cotmore et al., 2014); bidnaviruses are restricted to insects (Hu et al., 2013); bacilladnaviruses replicate in marine algae (Nagasaki et al., 2005), whereas members of the proposed genus “Gemycircularvirus” infect fungi (Jiang et al., 2013). Thus, ssDNA viruses prey on a wide range of eukaryotic hosts; however, numerous metagenomic and paleovirological studies suggest that the host range of eukaryotic ssDNA viruses might be even considerably broader (Labonte and Suttle, 2013, Rosario et al., 2012).
All eukaryotic ssDNA viruses, except for the members of the family Bidnaviridae (see below), replicate their genomes using a rolling-circle (or rolling-hairpin) mechanism which involves nicking of the viral genome by a virus-encoded rolling-circle replication initiation endonuclease, RC-Rep. The same replication mechanism is also used by most prokaryotic ssDNA viruses, many plasmids and some transposons (Chandler et al., 2013, Krupovic, 2013, Krupovic and Forterre, 2015, Rosario et al., 2012). Perhaps unexpectedly, the RC-Reps of eukaryotic ssDNA viruses bear only limited similarity to the RC-Reps of bacterial and archaeal ssDNA viruses. The RC-Reps of eukaryotic ssDNA viruses show a distinct two-domain organization (Koonin and Ilyina, 1993) ( Fig. 4): the N-terminal endonuclease domain is followed by the S3H domain which is required for genome replication as well as other processes, such as viral genome encapsidation (King et al., 2001). By contrast, none of the known prokaryotic ssDNA viruses encodes a S3H domain, whereas the endonuclease domains are not significantly similar to those of eukaryotic viruses, except for the short regions encompassing the three diagnostic sequence motifs that are common to all endonucleases of the HUH superfamily (Chandler et al., 2013, Ilyina and Koonin, 1992, Koonin and Ilyina, 1993) and the overall shared structural fold (Fig. 4). Thus, it appears extremely unlikely that ssDNA viruses of eukaryotes are direct descendants of their prokaryotic counterparts; the distantly related endonuclease domains involved in the mechanistically similar replication initiation processes probably were acquired independently and from different sources.
Fig. 4.
The conserved RC-Rep proteins of ssDNA viruses and their homologs: key motifs, domain architectures and structures. (A) The catalytic motifs of the nicking endonuclease and superfamily 3 helicase (S3H) domains. Note the absence of the S3H domain in the prokaryotic ssDNA viruses and the inactivation of the endonuclease domain in the dsDNA-containing papillomaviruses and polyomaviruses. (B) Homologous structures of the endonuclease domains. The structures are colored according to the secondary structure elements: α-helixes, blue; β-strands, green. Abbreviations and PDB accession numbers: PCV2, porcine circovirus type 2 (2HW0); TYLCV, tomato yellow leaf curl virus (1L2M); FBNYV, faba bean necrotic yellows virus (2HWT); AAV5, adeno-associated virus type 5 (1M55); SV40, simian virus 40 (1TBD); BPV, bovine papilloma virus (1F08).
In contrast, the eukaryotic ssDNA viruses share the endonuclease-helicase domain architecture with the RC-Reps of various bacterial plasmids (Fig. 4). Furthermore, RC-Reps from different families of eukaryotic ssDNA viruses are typically more similar to homologs form different groups of bacterial plasmids than they are to each other, suggesting a close evolutionary relationship between bacterial plasmids and eukaryotic ssDNA viruses (Koonin and Ilyina, 1992). In particular, RC-Reps of geminiviruses and fungal gemycircularviruses cluster in phylogenetic trees with the homologous proteins encoded by plasmids of phytoplasmas (parasitic wall-less bacteria replicating in plant and insect cells) rather than the RC-Reps of other plant or animal ssDNA viruses, such as nanoviruses and circoviruses (Krupovic et al., 2009, Liu et al., 2011). Accordingly, it has been hypothesized that geminiviruses have evolved from bacterial replicons (Koonin and Ilyina, 1992), and specifically, from phytoplasmal plasmids (Krupovic et al., 2009). In contrast, RC-Reps of circoviruses show closer similarity to proteins from a different group of bacterial plasmids, represented by the plasmid p4M of Bifidobacterium pseudocatenulatum (Gibbs et al., 2006, Krupovic et al., 2009). Furthermore, phylogenetic analysis of the RC-Rep encoded by an uncultivated Gastropod-associated circular ssDNA virus (GaCSV), isolated from the mollusk Amphibola crenata, showed that the viral protein is nested within the clade containing RC-Reps of bacterial origin (Dayaram et al., 2013). A striking, independent finding that is compatible with an evolutionary relationship between bacterial RC replicons and eukaryotic ssDNA viruses is that genomes of certain plant geminiviruses retain functional bacterial promoters and can replicate in different bacterial cells in an RC-Rep-dependent manner (Rigden et al., 1996, Selth et al., 2002, Wang et al., 2013, Wu et al., 2007). Although it is usually difficult to pinpoint the exact origin of viral RC-Reps, the above examples strongly suggest that RC-Reps of eukaryotic ssDNA viruses are polyphyletic and their roots are in different groups of bacterial plasmids.
The key step in the transformation of a plasmid into a virus is the acquisition of the genetic determinants allowing genome encapsidation and inter-cellular transfer. Indeed, some cryptic bacterial RC plasmids encode a single protein, the RC-Rep, and thus the only qualitative difference between such plasmids and the simplest eukaryotic ssDNA viruses, such as circoviruses, is the presence of a capsid protein (CP) gene in the latter (Krupovic and Bamford, 2010). All eukaryotic ssDNA viruses, for which structural information is available or the fold of the CP could be inferred using in silico analyses, possess structurally similar CPs with the jelly-roll fold (Krupovic, 2012, Krupovic, 2013). As discussed above, the jelly-roll fold is the most common fold in the CPs of icosahedral +RNA viruses and is also found in CPs of some dsRNA viruses ( Fig. 5) (Koonin et al., 2008, Krupovic, 2013, Rossmann and Johnson, 1989). Strikingly, CPs of some ssDNA viruses are more similar to the CPs of +RNA viruses than they are to the CPs of other ssDNA viruses, mirroring the relationships between the viral and plasmid RC-Reps. For example, the CP of geminiviruses is most closely related to the CP from satellite tobacco necrosis virus (STNV; Fig. 5) (Bottcher et al., 2004, Krupovic et al., 2009, Zhang et al., 2001). Thus, the genomes of eukaryotic ssDNA viruses appear to be chimeras composed of RC-Rep genes inherited from bacterial plasmids and CP genes derived from different groups of +RNA viruses ( Fig. 6). The exact circumstances under which bacterial plasmids crossed paths with eukaryotic +RNA viruses and gave rise to ssDNA viruses remain obscure. It is clear, however, that each such event would involve recombination between two unrelated RNA and DNA replicons. Recent findings discussed below indicate that such RNA-DNA recombination occasionally does take place and indeed is likely to play an important role in the emergence of new virus types.
Fig. 5.
Homologous single jelly-roll structures of the capsid proteins of RNA and DNA viruses of eukaryotes. The structures are colored according to the secondary structure elements: α-helixes, blue; β-strands, green. Depicted structures and their PDB accession numbers: Tymoviridae, turnip yellow mosaic virus (1AUY); Picornaviridae, rhinovirus 16 (1ND2); Satellite virus, satellite tobacco necrosis virus (2BUK); Birnaviridae, infectious bursal disease virus (1WCD); Circoviridae, porcine circovirus type 2 (3R0R); Parvoviridae, adeno-associated virus type 2 (1LP3); Papillomaviridae, human papillomavirus 16 (1DZL); Polyomaviridae, simian virus 40 (1SVA).
Fig. 6.
Evolution of ssDNA viruses of eukaryotes: polyphyletic origin from different plasmids and multiple cases of recombination with ssRNA viruses. Abbreviations: JRC, jelly roll capsid protein; pPolB, protein-primed DNA polymerase of family B; RC-Rep, rolling circle replication protein. Different colors of JRC and RC-Rep denote distinct variants of the respective genes.
Metagenomic exploration of viral diversity in the Boiling Springs Lake (BSL) at Lassen Park, California, has led to the discovery of a novel ssDNA viral genome (Diemer and Stedman, 2012). This virus, named BSL RDHV (RNA-DNA hybrid virus), encodes an RC-Rep closely related to those of circoviruses and a CP which, unexpectedly, is not related to circoviral CPs but instead has a domain organization specific to CPs of icosahedral +RNA viruses of the family Tombusviridae (Diemer and Stedman, 2012). Subsequent discovery of many additional BSL RDHV-like genomes enabled a more detailed analysis of this peculiar virus group, dubbed chimeric viruses (CHIV) (Roux et al., 2013). It has been shown that in the history of the CHIV group, there was a single event of CP gene acquisition from an RNA virus, followed by a recurrent replacement of the RC-Rep genes as well as gene fragments in CHIVs with distant counterparts from diverse ssDNA viruses representing three families, Circoviridae, Nanoviridae and Geminiviridae (Krupovic et al., 2015, Roux et al., 2013). Thus, recombination between contemporary RNA and DNA viruses appear to be relatively common, and a similar event or, more likely, several independent events involving different groups of bacterial RC plasmids and RNA viruses, gave rise to the ancestors of eukaryotic ssDNA viruses (Krupovic, 2013, Stedman, 2013) (Fig. 6).
Once in existence, eukaryotic ssDNA viruses have undergone substantial diversification, giving rise to several new groups of viruses and other mobile genetic elements. One of the most striking examples of such diversification is presented by members of the family Bidnaviridae. Bidnaviruses do not encode RC-Reps and accordingly do not replicate by the rolling-circle mechanism; instead, these viruses encode protein-primed family B DNA polymerases (Hu et al., 2013). Recent reconstruction of the evolutionary history of these insect viruses has shown that in all likelihood, they evolved from an insect-infecting parvovirus ancestor (Krupovic and Koonin, 2014). The key event in the evolution of bidnaviruses involved replacement of the typical parvovirus-like RC-Rep gene with a family B DNA polymerase gene acquired from large, virus-like DNA transposons of the Polinton/Maverick superfamily (see below), followed by acquisition of additional genes from insect baculoviruses that have dsDNA genomes and reoviruses that contain segmented dsRNA genomes (Krupovic and Koonin, 2014). Evolution of bidnaviruses from genes of four widely different groups of viruses is a striking example emphasizing the central role of recombination and genomic plasticity in virus evolution.
Many groups of prokaryotic and eukaryotic ssDNA viruses have the ability to integrate into the genomes of their cellular hosts. In bacterial and archaeal viruses, this process is mediated by dedicated integrases or transposases. By contrast, integration of eukaryotic ssDNA virus genomes primarily depends on the endonuclease activity of their RC-Reps (Krupovic and Forterre, 2015, Liu et al., 2011). Whereas most groups of eukaryotic ssDNA viruses integrate only sporadically, some have evolved towards more aggressive proliferation within host genomes, akin to transposable elements. For example, a group of parvovirus-like transposons, encoding both CP and RC-Rep proteins, has been discovered in the genome of acorn worm, Saccoglossus kowalevskii, where these putative transposons are present in over 50 copies per genome (Liu et al., 2011). Some ssDNA viruses have apparently abandoned the virus-like propagation in favor of the transposon-like life style: elements encoding parvoviral RC-Reps (but lacking the CP genes) and flanked by typical terminal inverted repeat sequences have been identified in the genomes of Hydra magnipapillata and Schmidtea mediterranea in over 400 and 100 copies per genome, respectively (Liu et al., 2011).
Yet another distinct evolutionary trajectory leads from ssDNA viruses to small dsDNA viruses of the families Papillomaviridae and Polyomaviridae. From their ssDNA virus ancestors, members of both these families inherited genes for capsid and replication proteins (Fig. 4, Fig. 5), albeit both underwent major modifications (see below in the section on the origin of eukaryotic dsDNA viruses).
Synopsis on ssDNA virus origins
Taken together, the results of comparative genomic analysis clearly indicate that eukaryotic ssDNA viruses evolved on several independent occasions from bacterial plasmids via acquisition of CP genes from pre-existing +RNA viruses (Fig. 6). This scenario is neutral with respect to the two eukaryogenesis scenarios outlined above because it predicts de novo origin of ssDNA viruses postdating the emergence of eukaryotes. Considering that plasmid-carrying bacteria often establish mutualistic and parasitic interactions with diverse modern eukaryotes or simply serve as a food source for the latter (in the case of grazing protists), different groups of ssDNA viruses probably emerged at different time points during eukaryal evolution. Some groups, such as parvoviruses, could have arisen before the radiation of major eukaryotic kingdoms, whereas other lineages, such as bidnaviruses, have a more recent history. Mixing-and-matching of different functional modules from widely different plasmid and virus groups representing both RNA and DNA virospheres is an ongoing process which continues to generate new groups of ssDNA viruses (Krupovic, 2013, Stedman, 2013). The extent of gene shuffling is such that it can completely obliterate the ancestral evolutionary signal, as in the case of CHIVs, where original genes for both CP and RC-Rep have been replaced in some of the viruses. Furthermore, during the course of evolution, ssDNA viruses have taken different evolutionary paths which allowed them to explore diverse replication mechanisms, including switch to dsDNA genomes, expand the host range and occasionally step away from the bona fide viral propagation and switch to transposon-like life-styles, reversibly or otherwise.
Origins and primary diversification of eukaryotic dsDNA viruses: The bacteriophage and transposable element connections
Compared to RNA viruses and retroelements, dsDNA viruses and mobile elements are somewhat less diverse and less abundant in eukaryotes but nevertheless have been identified in all major eukaryotic groups. All in all, there are 18 formally recognized families of dsDNA viruses and many unclassified viruses that infect a broad spectrum of unicellular and multicellular hosts and span almost the entire range of viral genome sizes, from about 4 kb to almost 2.5 Mb (Supplementary Table S6).
By far the largest and most common group of DNA viruses in eukaryotes (Supplementary Table S6) consists of 7 families of large and giant viruses including mimiviruses and pandoraviruses, with genomes in the megabase range. All these viruses that infect diverse eukaryotes including animals and a variety of protists are thought to share a common ancestry as indicated by the conservation of a substantial number of genes encoding essential proteins involved in viral genome replication and virion formation. Although only 5 genes are strictly conserved in all viruses of this group, maximum likelihood evolutionary reconstructions led to the inference of an ancestral gene set consisting of approximately 50 genes (Iyer et al., 2001, Iyer et al., 2006, Koonin and Yutin, 2010). This major group of eukaryotic viruses has become known as the Nucleo-Cytoplasmic Large DNA Viruses (NCLDV)(Iyer et al., 2001) or more recently, the proposed order “Megavirales” (Colson et al., 2013).
The viruses of the family Mimiviridae are hosts to a distinct class of satellite viruses, the virophages, that reproduce within the viral “factories” inside protist cells infected by the giant virus and depend on the latter for their replication (Claverie and Abergel, 2009, Desnues et al., 2012, Krupovic and Cvirkaite-Krupovic, 2011, La Scola et al., 2008). Recently, an evolutionary connection between the virophages and large eukaryotic dsDNA transposons of the Polinton/Maverick group (hereinafter Polintons) has been identified (Fischer and Suttle, 2011, Yutin et al., 2013). The polintons are common in diverse unicellular protists and animals (Krupovic and Koonin, 2015), indicative of their ancient origin, perhaps concomitant with the origin of eukaryotes. Recently, it has been shown that the majority of the Polintons encode two proteins homologous to the version of the JRC that is typical of the capsids of icosahedral dsDNA viruses that infect bacteria, eukaryotes and some archaea (double beta-barrel) (Krupovic et al., 2014). All key structural elements of the capsid proteins are preserved in the polinton-encoded homologs suggesting that these proteins are indeed functional. The Polintons also encode two proteins that are essential for morphogenesis in members of the “Megavirales”, namely an FtsK-like ATPase and a Ulp1-like protease. The presence of these genes, together with those for capsid proteins, leaves little doubt that, under some still unknown conditions, the polintons actually produce virions that might possess the ability to infect new hosts (Krupovic et al., 2014). Thus, the Polintons, perhaps to be renamed Polintoviruses (the term we use hereinafter), combine central features of viruses and transposons, and seem to represent the second major group of eukaryotic dsDNA viruses, after the “Megavirales”, that infect numerous hosts across the entire eukaryotic diversity (Krupovic and Koonin, 2015).
Polintoviruses share blocks of homologous genes with diverse viruses, transposons and plasmids (Krupovic and Koonin, 2015). In particular, bacteriophages of the family Tectiviridae, Polintons and the Mavirus virophage all share 4 genes encoding two capsid proteins, DNA-packaging ATPase and protein-primed DNA polymerase (pPolB). The Polintoviruses share two additional genes with the Mavirus, namely those for the capsid maturation protease and the RVE integrase, whereas the rest of the virophages also encode the capsid proteins, ATPase and protease, but lack pPolB and the integrase (Yutin et al., 2013). Adenoviruses join this network of related viruses through pPolB, the two capsid proteins and the protease, whereas the much larger “Megavirales” connect through the capsid proteins, the ATPase and the protease. Thus, the morphogenetic module is the common denominator that links all these diverse families of viruses. The yeast linear cytoplasmic plasmids (Klassen and Meinhardt, 2007) provide additional connections between Polintons and the incomparably more complex members of the “Megavirales”: these plasmids lack the morphogenetic module but encode pPolB along with four key proteins required for cytoplasmic transcription that are conserved in most of the “Megavirales”.
The multiple connections between the Polintoviruses and various other groups of viruses and plasmids have prompted a unifying scenario under which Polintoviruses were the first group of eukaryotic dsDNA viruses that, on different occasions, gave rise to several groups of eukaryotic viruses, transposons and plasmids ( Fig. 7) (Krupovic and Koonin, 2015). The Polintoviruses most likely evolved from bacteriophages of the family Tectiviridae that entered the protoeukaryotic cell along with the α-proteobacterial endosymbiont, the ancestor of the mitochondria (Fig. 7). This scenario is compatible with the presence of linear plasmids that encode pPolB in fungal mitochondria (Handa, 2008). In phylogenetic trees, these pPolBs form a deep branch that is distinct from the rest of the eukaryotic plasmids and viruses, suggestive of early divergence of the descendants of the ancestral tectivirus into mitochondrial and cytoplasmic or nuclear lineages of mobile elements (Krupovic and Koonin, 2015).
Fig. 7.
Evolution of large dsDNA viruses of eukaryotes from two distinct groups of bacteriophages. The dotted line with a question mark shows a tenuous evolutionary relationship. The host ranges of the eukaryotic virus groups are color-coded as shown in the inset. The hatched yellow square for the virophages indicates that these viruses parasitize on the giant viruses of the family Mimivirdae which themselves infect amoeba and other protists. For each family of large eukaryotic viruses, a simplified schematic depiction of the virion structure is included.
The key event in the evolution of the Polintoviruses from the ancestral tectivirus apparently was the acquisition of the RVE family integrase and the Ulp1-like cysteine protease, conceivably via a single recombination event with a eukaryotic Ginger 1-like transposon (Bao et al., 2010, Krupovic and Koonin, 2015) (Fig. 7). The capture of the integrase was pivotal in the evolution of the Polintoviruses, endowing them with the ability to combine two alternative lifestyles, those typical of transposable elements and bona fide viruses. This “bet-hedging” strategy, that is also characteristic of Mu-like bacteriophages and eukaryotic Ty1-copia retrotransposons (pseudoviruses) and Ty3-gypsy retrotransposons (metaviruses) (Koonin and Dolja, 2014, Krupovic et al., 2011a, Sandmeyer and Menees, 1996) (and see above), would provide the flexibility of parasite-host relationships that conceivably underlies the diversification and successful spread of Polintoviruses in diverse eukaryotes.
Some Polintoviruses apparently abandoned the virus lifestyle after losing the genes involved in virion formation and became pure transposons (it seems prudent to reserve the term Polintons for these elements) (Krupovic and Koonin, 2015). Adenoviruses followed the opposite course of evolution, having lost the integrase gene and thereby committing to the strict viral lifestyle. Polintons also contributed the pPolB gene to the evolution of a distinct family of ssDNA viruses, the Bidnaviridae, which emerged via extensive gene shuffling between four groups of selfish elements (Krupovic and Koonin, 2014) (and see above).
The “Megavirales”, the largest, most diverse group of eukaryotic dsDNA viruses, apparently inherited from the Polintoviruses the virion morphogenesis module including the major and minor capsid proteins, genome packaging ATPase and maturation protease (Krupovic and Koonin, 2015). Among the numerous double-JRC proteins, the predicted major capsid protein of the Polintoviruses is most similar to the capsid proteins of phycodnaviruses (Krupovic et al., 2014), suggesting a direct evolutionary link between Polintoviruses and the “Megavirales”. Although the packaging ATPases and the maturation proteases are highly diverged, the topologies of the respective phylogenetic trees are compatible with the Polintovirus—“Megavirales” link (Yutin et al., 2013).
Polintons reside in the nucleus of the host cell, and most likely, their predicted viral forms, the Polintoviruses, also reproduce in the nucleus and thus rely on the host enzymatic machinery for transcription. A key event in the evolution of the “Megavirales” was the escape from the nucleus, most likely concomitant with the acquisition of the RNA polymerase and the capping apparatus from the host. The escaped element that would replicate in the cytoplasm using the ancestral Polinton pPolB spawned two groups of mobile elements, namely cytoplasmic plasmids (surviving in fungi) and the “Megavirales” that share with these plasmids the distinct three-domain capping enzyme, two RNA polymerase subunits and the D11-like helicase (Krupovic and Koonin, 2015). The cytoplasmic plasmids retain pPolB but have lost the morphogenesis module and are thus restricted to the intracellular lifestyle. By contrast, evolution of the “Megavirales” took the route of increasing complexity and autonomy from host functions. The major events in the evolution of “Megavirales” from the putative cytoplasmic Polintovirus-like ancestor include the displacement of pPolB with a RNA/DNA-primed PolB and acquisition of the D5-like helicase-primase (Krupovic and Koonin, 2015). It seems likely that pPolB that initiates DNA replication at genome termini cannot efficiently replicate genomes above a certain threshold (probably, about 45 kb, as in adenoviruses). Replication of larger genomes would become efficient upon the recruitment of a dedicated primase-helicase. Some Polintons encode divergent D5-like primases-helicases that typically cluster in phylogenetic trees with the primases-helicases of the “Megavirales” (Yutin et al., 2013). Several additional genes that belong to the inferred ancestral gene set of the “Megavirales” are also shared with various Polintons (Yutin et al., 2013). Thus, Polintoviruses could have donated a substantial fraction of the ancestral genes of the “Megavirales”. A notable exception is the PolB gene that replaced the ancestral pPolB and most likely was acquired from the eukaryotic host (Yutin and Koonin, 2012). The acquisition of this form of PolB, jointly with the primase-helicase, provided the opportunity for almost unlimited genome expansion in the “Megavirales”, yielding the giant viruses.
A radically different scenario of the origin of the giant viruses among the “Megavirales”, such as the mimiviruses and pandoraviruses, has been proposed on the strength of their microbe-like size and genomic complexity, and most important, the presence of genes encoding some components of the translation system, such as several aminoacyl-tRNA synthetases, that are universally present in cellular life forms (Koonin, 2003). The initial and subsequent phylogenetic analysis of these universal genes has suggested that the giant viruses did not fall into any of three domains of cellular life (bacteria, archaea and eukaryote) and prompted the hypothesis that these viruses evolved by reductive evolution from a hypothetical (conceivably, extinct) cellular domain (Colson et al., 2012, Colson et al., 2011, Nasir et al., 2012, Raoult et al., 2004). However, independent phylogenetic studies that employed representative sets of cellular life forms from the three domains and more advanced phylogenetic methods have effectively refuted the fourth domain hypothesis by showing that nearly all universal genes of the giant viruses were nested within the eukaryotic domain of the respective phylogenetic trees (Williams et al., 2011, Yutin et al., 2014). Moreover, in different groups of giant viruses, these genes were affiliated with different eukaryotes, suggestive of independent acquisition. Consistent with this conclusion, reconstruction of the evolution of the gene repertoire of the “Megavirales” indicates that the giant viruses most likely evolved from smaller viruses in this group via the acquisition of numerous genes from different sources and gene duplication (Filee, 2013, Yutin and Koonin, 2013, Yutin et al., 2014). Thus, notwithstanding their complexity that is unprecedented in the virus world, the giant viruses share a common history with the rest of the “Megavirales” and thus ultimately appear to have evolved from Polintoviruses.
The virophages retain many ancestral features of the Polintoviruses, in particular the complete morphogenesis module. Unlike the ancestors of the “Megavirales”, these smaller viruses have not acquired the molecular machinery required for the reproduction in the cytoplasm of the host cells and instead evolved to parasitize on their giant relatives by exploiting their transcription apparatus and other functions (Claverie and Abergel, 2009, Desnues et al., 2012, Fischer and Suttle, 2011, Krupovic and Cvirkaite-Krupovic, 2011).
Ten recognized families of eukaryotic dsDNA viruses do not show clear evolutionary relationship to the Polintovirus-centered assemblage of the eukaryotic dsDNA viruses (Supplementary Table S6) (Koonin et al., 2015). All these viruses have narrow host ranges compared to the “Megavirales”, mostly infecting members of a particular animal phylum such as chordates or arthropods. The evolution of these viruses so far has not been reconstructed in a comprehensive manner as it had been the case with the “Megavirales”. Nevertheless, some general trends have become apparent. Five families of large eukaryotic dsDNA viruses, namely Baculoviridae, Hytrosaviridae, Nimaviridae, Nudiviridae, and Polydnaviridae, so far have been isolated exclusively from arthropods. Although these viruses, particularly the latter three families, mostly encode highly diverged (presumably, fast-evolving) protein sequences and are currently represented by only a few genomes each, phylogenomic analysis suggests that they comprise a monophyletic group, with several signature genes that are not found in other viruses (Jehle et al., 2013, Wang et al., 2012b, Wang and Jehle, 2009). Polydnaviruses represent a unique group of viruses that are only vertically transmitted, with the virus genomes permanently integrated in the genomes of the insect hosts. Nevertheless, even in this unusual case, phylogenetic analysis of the retained viral genes indicates that polydnaviruses are highly derived descendants of nudiviruses (Herniou et al., 2013, Theze et al., 2011). Preliminary phylogenetic analysis of several essential genes that are shared by all these arthoropod viruses and the “Megavirales”, such as PolB, RNAP subunits, helicase-primase and thiol oxidoreductase, has suggested that this group of viruses might be a highly derived offshoot of the “Megavirales” (Wang et al., 2012b) (Fig. 7). However, this remains but a tentative clue until a comprehensive study on the evolution of these unusual viruses is performed.
The highly diversified order Herpesvirales is of special interest from the standpoint of virus evolution because of a distinct connection with tailed viruses of the order Caudovirales which includes three families, namely Siphoviridae, Podoviridae and Myoviridae. Caudovirales are nearly ubiquitous in Bacteria (Ackermann and Prangishvili, 2012) and are also present in diverse orders of Archaea, including the deeply branching archaeal phylum Thaumarchaeota (Krupovic et al., 2011b). The putative bacterial or archaeal virus ancestors of the herpesviruses are unrelated to the tectiviruses, the likely ancestors of the Polintovirus-related majority of eukaryotic dsDNA viruses (Fig. 7). Herpesviruses share with the Caudovirales homologous major capsid proteins of the HK97 fold that is unrelated to the double jelly-roll fold present in the capsid proteins of numerous groups of icosahedral viruses (including the Polintovirus-centered assemblage), terminases (packaging ATPases-nucleases), and capsid maturation proteases as well as several other proteins (Pietila et al., 2013, Selvarajan Sigamani et al., 2013, Baker et al., 2005, Krupovic and Bamford, 2011, Krupovic et al., 2010, Rixon and Schmid, 2014). Thus, tailed prokaryotic viruses and herpesviruses share a complex and unique virion assembly and maturation program which is not found in other dsDNA viruses.
The apparent bacteriophage origin of the herpesvirus morphogenesis module that consists of a capsid protein, an ATPase and a protease is a striking parallel with the similar evolutionary route of the Polintovirus ancestor but the actual proteins involved are unrelated (or in the case of the ATPase, distantly related). This evolutionary parallelism clearly reflects a general trend in the origins of the largest, most complex viruses of eukaryotes. Somewhat ironically, bacteriophages of the order Caudovirales, which are the most common viruses on earth, gave rise to a single (even if diverse) group of eukaryotic dsDNA viruses, whereas the bulk of eukaryotic dsDNA viruses seem to originate from the narrowly spread tectiviruses. Conceivably, the key event behind the success of the Polintoviruses that defined the wide spread of their descendants was the acquisition of the transposase (see above). Furthermore, the fact that herpesviruses seem to be limited to animal hosts might indicate that this group of viruses emerged relatively late in the course of eukaryotic evolution, with the ancestor bacteriophage coming not from the proto-mitochondrion but from a distinct (perhaps transient) bacterial symbiont of early animals. Paradoxically, however, the proto-mitochondrial symbiont apparently did contain a provirus derived from a tailed bacteriophage and this provirus had a significant effect on the evolution of mitochondria: in modern mitochondria, ancestral bacterial genes for RNA polymerase, DNA polymerase and DNA primase have been all replaced with the counterparts from the resident prophage early in eukaryogenesis (Filee and Forterre, 2005, Shutt and Gray, 2006).
Finally, the two families of dsDNA viruses with small, circular genomes, Papillomaviridae and Polyomaviridae, appear to have evolved via a route that is completely distinct from the origins of all larger dsDNA viruses of eukaryotes. The capsids of papillomaviruses and polyomaviruses are constructed from JRC proteins homologous to those of eukaryotic ssDNA viruses (Fig. 5). Furthermore, the single multidomain replicative protein of these viruses, known as the large T antigen in polyomaviruses and the E1 protein in papillomaviruses, is homologous to the replication proteins of ssDNA viruses, such as circoviruses, nanoviruses, parvoviruses and geminiviruses (Fig. 4 and see above). This large protein has a typical domain architecture consisting of a S3H and a rolling circle replication initiation endonuclease that, however, is inactivated in papillomaviruses and polyomaviruses (Fig. 4). This inactivation of the key enzyme of RCR is concomitant with the switch from rolling circle to the “theta-like” replication mode and from ssDNA to dsDNA genome (Ilyina and Koonin, 1992, Iyer et al., 2005). Thus, the small dsDNA viruses of eukaryotes apparently are derivatives of ssDNA viruses which themselves evolved via recombination of bacterial rolling circle-replicating plasmids and ssRNA viruses (see above).
Synopsis of dsDNA virus evolution
Overall, the emerging picture of the origin of dsDNA viruses of eukaryotes reveals three readily identifiable bacterial roots (Fig. 7; see also Fig. 6). Two of these lines of descent come from distinct groups of bacteriophages and gave rise to the majority of large eukaryotic viruses, whereas the third one comes from plasmids and yielded the two families of small dsDNA viruses that actually are derivatives of ssDNA viruses. There is no evidence of a direct contribution of viruses infecting archaea to the emergence of eukaryotic virome, despite the remarkable diversity and abundance of archaeal dsDNA viruses (Prangishvili, 2013, Prangishvili et al., 2006a, Prangishvili et al., 2006b) (a caveat to be addressed in future studies is that most of the current knowledge on archaeal viruses comes from hyperthermophilic Crenarchaeota not from mesophilic members of the TACK superphylum which seem to be the likely ancestors of eukaryotes). Given this demonstrable bacterial ancestry, the reconstruction of the evolution of eukaryotic dsDNA viruses seems to be best compatible with the symbiogenetic scenario of eukaryogenesis. Acquisition of DNA polymerases and primases from the eukaryotic hosts opened the route of genome expansion to the evolving dsDNA viruses, resulting in acquisition of numerous genes from the hosts and exaptation (recruitment) of the acquired genes for virus-host interaction.
Conclusions
The recent dramatic expansion of the collection of viral genome sequences, combined with the concerted efforts in evolutionary genomics, translates into a new level of understanding of the origins of the major groups of eukaryotic viruses and the key events in their evolution. We now can delineate both the major general trends in the evolution of eukaryotic viruses and specific scenarios for different virus classes. One of the most striking trends is the distinct composition of the eukaryotic virome compared to the viromes of archaea and bacteria, namely, the high prevalence and enormous diversity of RNA viruses. It might be tempting to directly derive the eukaryotic RNA virome from the hypothetical primordial RNA world but the plausibility of this link depends on the adopted scenario for the origin of eukaryotes. The primordial origin of eukaryotic RNA viruses appears to be compatible with the protoeukaryotic but not with the symbiogenetic scenario. If, under the latter scenario, the host of the mitochondrial endosymbiont was a typical archaeon, the existence of a diverse RNA virome in such an organism appears exceedingly unlikely. Instead, a more circuitous path to the eukaryotic RNA virome would have to be postulated, with traceable contributions from bacterial retroelements as well as bona fide bacterial genes. This type of chimeric origin is a pervasive theme in the evolution of all classes of eukaryotic viruses that is particularly apparent in the emerging histories of dsRNA viruses, ssDNA viruses and dsDNA viruses. Strikingly, in each of these cases, the morphogenetic and replication-expression modules appear to be of different evolutionary provenances, and recombination between these distinct modules gave rise to a novel type of viruses. At least in some cases, the recombination of modules and spread of individual genes, such as the movement protein gene in plants, seems to have a clear adaptive value by opening up a major new niche for viruses with different particular replication-expression strategies and virion structures.
Another major trend in the evolution of the viruses of eukaryotes is the pervasive evolutionary connection between bona fide viruses and non-viral mobile genetic elements, such as transposons and plasmids. These non-viral elements appear to have made major contributions to the evolution of all classes of eukaryotic viruses as well as the hosts. Furthermore, elements with a dual life style, such as metaviruses and pseudoviruses as well as polintoviruses (polintons), appear to have played central roles in the evolution of the retroviruses and large dsDNA viruses of eukaryotes, respectively. Perhaps, the most remarkable aspect of the evolution of the viruses of eukaryotes is that it seems to be tractable, at least in its central features.
Acknowledgments
The authors thank David Karlin and Tero Ahola for the kind permission to cite the results of their work before publication. EVK is supported by the intramural funds of the US Department of Health and Human Services (National Library of Medicine).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2015.02.039.
Contributor Information
Eugene V. Koonin, Email: koonin@ncbi.nlm.nih.gov.
Valerian V. Dolja, Email: doljav@science.oregonstate.edu.
Mart Krupovic, Email: krupovic@pasteur.fr.
Appendix A. Supplementary materials
Supplementary material
References
- Ackermann H.W., Prangishvili D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 2012;157(10):1843–1849. doi: 10.1007/s00705-012-1383-y. [DOI] [PubMed] [Google Scholar]
- Adriaenssens E.M., Edwards R., Nash J.H., Mahadevan P., Seto D., Ackermann H.W., Lavigne R., Kropinski A.M. Integration of genomic and proteomic analyses in the classification of the Siphoviridae family. Virology. 2014 doi: 10.1016/j.virol.2014.10.016. [DOI] [PubMed] [Google Scholar]
- Agol V.I. Towards the system of viruses. Biosystems. 1974;6(2):113–132. doi: 10.1016/0303-2647(74)90003-3. [DOI] [PubMed] [Google Scholar]
- Ahola T., Karlin D.G. Sequence analysis reveals a conserved extension in the methyltransferase guanylyltransferase of the alphavirus supergroup, and a homologous domain in the nodavirus supergroup. Biol. Direct. 2015 doi: 10.1186/s13062-015-0050-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar el D., Tsai C.W., Whitfield A.E., Redinbaugh M.G., Hogenhout S.A. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 2009;54:447–468. doi: 10.1146/annurev.ento.54.110807.090454. [DOI] [PubMed] [Google Scholar]
- Aravind L., Walker D.R., Koonin E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27(5):1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Jiang W., Rixon F.J., Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79(23):14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35(3):235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Kapitonov V.V., Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob. DNA. 2010;1(1):3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal S., Domier L.L., Gonfa B., McCoppin N.K., Lambert K.N., Bhalerao K. A novel flavivirus in the soybean cyst nematode. J. Gen. Virol. 2014;95(Pt 6):1272–1280. doi: 10.1099/vir.0.060889-0. [DOI] [PubMed] [Google Scholar]
- Bekal S., Domier L.L., Niblack T.L., Lambert K.N. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J. Gen. Virol. 2011;92(Pt 8):1870–1879. doi: 10.1099/vir.0.030585-0. [DOI] [PubMed] [Google Scholar]
- Belov G.A. Modulation of lipid synthesis and trafficking pathways by picornaviruses. Curr. Opin. Virol. 2014;9C:19–23. doi: 10.1016/j.coviro.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Bernhardt H.S. The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others)(a) Biol. Direct. 2012;7:23. doi: 10.1186/1745-6150-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Bakre A., Bhattacharya A. Mobile genetic elements in protozoan parasites. J. Genet. 2002;81(2):73–86. doi: 10.1007/BF02715903. [DOI] [PubMed] [Google Scholar]
- Blaxter M.L., De Ley P., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Vida J.T., Thomas W.K. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392(6671):71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Bolduc B., Shaughnessy D.P., Wolf Y.I., Koonin E.V., Roberto F.F., Young M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J. Virol. 2012;86(10):5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback J.P., Huelsenbeck J.P. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae) J. Mol. Evol. 2001;52(2):117–128. doi: 10.1007/s002390010140. [DOI] [PubMed] [Google Scholar]
- Bottcher B., Unseld S., Ceulemans H., Russell R.B., Jeske H. Geminate structures of African cassava mosaic virus. J. Virol. 2004;78(13):6758–6765. doi: 10.1128/JVI.78.13.6758-6765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.R., Doolittle W.F. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 1997;61(4):456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki J.M., Rychlewski L. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. 2002;15(2):101–108. doi: 10.1093/protein/15.2.101. [DOI] [PubMed] [Google Scholar]
- Capy P., Maisonhaute C. Acquisition/loss of modules: the construction set of transposable elements. Russ. J. Genet. 2002;38:594–601. [PubMed] [Google Scholar]
- Chalamcharla V.R., Curcio M.J., Belfort M. Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry. Genes Dev. 2010;24(8):827–836. doi: 10.1101/gad.1905010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.R., Blackburn E.H. Telomeres and telomerase. Philos. Trans. R. Soc. London, B: Biol. Sci. 2004;359(1441):109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., de la Cruz F., Dyda F., Hickman A.B., Moncalian G., Ton-Hoang B. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat. Rev. Microbiol. 2013;11(8):525–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.H., Rossmann M.G. RNA-dependent RNA polymerases from Flaviviridae. Curr. Opin. Struct. Biol. 2009;19(6):746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Claverie J.M., Abergel C. Mimivirus and its virophage. Annu. Rev. Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- Claverie J.M., Ogata H., Audic S., Abergel C., Suhre K., Fournier P.E. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117(1):133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Colson P., de Lamballerie X., Fournous G., Raoult D. Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology. 2012;55(5):321–332. doi: 10.1159/000336562. [DOI] [PubMed] [Google Scholar]
- Colson, P., De Lamballerie, X., Yutin, N., Asgari, S., Bigot, Y., Bideshi, D.K., Cheng, X.W., Federici, B.A., Van Etten, J.L., Koonin, E.V., La Scola, B., Raoult, D. (2013). “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. [DOI] [PMC free article] [PubMed]
- Colson P., Gimenez G., Boyer M., Fournous G., Raoult D. The giant Cafeteria roenbergensis virus that infects a widespread marine phagocytic protist is a new member of the fourth domain of Life. PLoS One. 2011;6(4):e18935. doi: 10.1371/journal.pone.0018935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-McKenna M., Chiorini J.A., Mukha D.V., Pintel D.J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., Gatherer D., Davison A.J. The family Parvoviridae. Arch. Virol. 2014;159(5):1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey S.N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14(2):623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A.I., Lang A.S., Suttle C.A. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312(5781):1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Culley A.I., Mueller J.A., Belcaid M., Wood-Charlson E.M., Poisson G., Steward G.F. The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. MBio. 2014;5(3):e01210–e01214. doi: 10.1128/mBio.01210-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A.I., Steward G.F. New genera of RNA viruses in subtropical seawater, inferred from polymerase gene sequences. Appl. Environ. Microbiol. 2007;73(18):5937–5944. doi: 10.1128/AEM.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Chai D., Gu S.Q., Gabel J., Noskov S.Y., Blocker F.J., Lambowitz A.M., Zimmerly S. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Mol. Cell. 2008;30(4):472–485. doi: 10.1016/j.molcel.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J.L., de Rocquigny H., Mauffret O., Mely Y. Retrospective on the all-in-one retroviral nucleocapsid protein. Virus Res. 2014;193:2–15. doi: 10.1016/j.virusres.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaram A., Goldstien S., Zawar-Reza P., Gomez C., Harding J.S., Varsani A. Novel ssDNA virus recovered from estuarine Mollusc (Amphibola crenata) whose replication associated protein (Rep) shares similarities with Rep-like sequences of bacterial origin. J. Gen. Virol. 2013;94(Pt 5):1104–1110. doi: 10.1099/vir.0.050088-0. [DOI] [PubMed] [Google Scholar]
- de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defraia C., Slotkin R.K. Analysis of retrotransposon activity in plants. Methods Mol. Biol. 2014;1112:195–210. doi: 10.1007/978-1-62703-773-0_13. [DOI] [PubMed] [Google Scholar]
- Delwart E., Li L. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 2012;164(1–2):114–121. doi: 10.1016/j.virusres.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Desnues C., Boyer M., Raoult D. Sputnik, a virophage infecting the viral domain of life. Adv. Virus Res. 2012;82:63–89. doi: 10.1016/B978-0-12-394621-8.00013-3. [DOI] [PubMed] [Google Scholar]
- Diemer G.S., Stedman K.M. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct. 2012;7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener T.O. Circular RNAs: relics of precellular evolution? Proc. Natl. Acad. Sci. U.S.A. 1989;86(23):9370–9374. doi: 10.1073/pnas.86.23.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M., Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17(5):799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V.V., Boyko V.P., Agranovsky A.A., Koonin E.V. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology. 1991;184(1):79–86. doi: 10.1016/0042-6822(91)90823-t. [DOI] [PubMed] [Google Scholar]
- Dolja V.V., Koonin E.V. Common origins and host-dependent diversity of plant and animal viromes. Curr. Opin. Virol. 2011;1(5):322–331. doi: 10.1016/j.coviro.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R.A., Rohwer F. Viral metagenomics. Nat. Rev. Microbiol. 2005;3(6):504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Eickbush T.H., Jamburuthugoda V.K. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134(1–2):221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K., Sutton G., Ravantti J.J., Zhang H., Walter T.S., Grimes J.M., Bamford D.H., Stuart D.I., Mancini E.J. Plate tectonics of virus shell assembly and reorganization in phage phi8, a distant relative of mammalian reoviruses. Structure. 2013;21(8):1384–1395. doi: 10.1016/j.str.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley T.M., Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440(7084):623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Esser C., Ahmadinejad N., Wiegand C., Rotte C., Sebastiani F., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Bryant D., Steel M.A., Lockhart P.J., Penny D., Martin W. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 2004;21(9):1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Evgen’ev M.B. What happens when Penelope comes? An unusual retroelement invades a host species genome exploring different strategies. Mob. Genet. Elem. 2013;3(2):e24542. doi: 10.4161/mge.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Orta C., Arias A., Escarmis C., Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006;16(1):27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Filee J. Route of NCLDV evolution: the genomic accordion. Curr. Opin. Virol. 2013;3(5):595–599. doi: 10.1016/j.coviro.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Filee J., Forterre P. Viral proteins functioning in organelles: a cryptic origin? Trends Microbiol. 2005;13(11):510–513. doi: 10.1016/j.tim.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Finnegan D.J. Retrotransposons. Curr. Biol. 2012;22(11):R432–R437. doi: 10.1016/j.cub.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Fischer M.G., Suttle C.A. A virophage at the origin of large DNA transposons. Science. 2011;332(6026):231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- Fuhrman J.A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399(6736):541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- Gibbs A., Ohshima K. Potyviruses and the digital revolution. Annu. Rev. Phytopathol. 2010;48:205–223. doi: 10.1146/annurev-phyto-073009-114404. [DOI] [PubMed] [Google Scholar]
- Gibbs M.J., Smeianov V.V., Steele J.L., Upcroft P., Efimov B.A. Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol. Biol. Evol. 2006;23(6):1097–1100. doi: 10.1093/molbev/msj122. [DOI] [PubMed] [Google Scholar]
- Gilbert W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- Gladyshev E.A., Arkhipova I.R. A widespread class of reverse transcriptase-related cellular genes. Proc. Natl. Acad. Sci. U.S.A. 2011;108(51):20311–20316. doi: 10.1073/pnas.1100266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R., Wellink J. Evolution of plus-strand RNA viruses. Intervirology. 1988;29(5):260–267. doi: 10.1159/000150054. [DOI] [PubMed] [Google Scholar]
- Goodier J.L., Kazazian H.H., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135(1):23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Donchenko A.P., Blinov V.M., Koonin E.V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989;243(2):103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Donchenko A.P., Koonin E.V., Blinov V.M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989;17(21):8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Pringle F.M., Zeddam J.L., Luke B.T., Cameron C.E., Kalmakoff J., Hanzlik T.N., Gordon K.H., Ward V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324(1):47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L. Picornavirus-host interactions to construct viral secretory membranes. Prog. Mol. Biol. Transl. Sci. 2015;129:189–212. doi: 10.1016/bs.pmbts.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Griffiths A.J. Natural plasmids of filamentous fungi. Microbiol. Rev. 1995;59(4):673–685. doi: 10.1128/mr.59.4.673-685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoras I., Ginzo A.I., Martin D.P., Varsani A., Romero J., Mammadov A., Huseynova I.M., Aliyev J.A., Kheyr-Pour A., Huss H., Ziebell H., Timchenko T., Vetten H.J., Gronenborn B. Genome diversity and evidence of recombination and reassortment in nanoviruses from Europe. J. Gen. Virol. 2014;95(Pt 5):1178–1191. doi: 10.1099/vir.0.063115-0. [DOI] [PubMed] [Google Scholar]
- Guu T.S., Zheng W., Tao Y.J. Bunyavirus: structure and replication. Adv. Exp. Med. Biol. 2012;726:245–266. doi: 10.1007/978-1-4614-0980-9_11. [DOI] [PubMed] [Google Scholar]
- Guy L., Saw J.H., Ettema T.J. The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 2014;6(10):a016022. doi: 10.1101/cshperspect.a016022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H. Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions? Mitochondrion. 2008;8(1):15–25. doi: 10.1016/j.mito.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Bejarano E.R., Robertson D., Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013;11(11):777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- Harper G., Hull R., Lockhart B., Olszewski N. Viral sequences integrated into plant genomes. Annu. Rev. Phytopathol. 2002;40:119–136. doi: 10.1146/annurev.phyto.40.120301.105642. [DOI] [PubMed] [Google Scholar]
- Hendrix R.W. Bacteriophage genomics. Curr. Opin. Microbiol. 2003;6(5):506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Herniou E.A., Huguet E., Theze J., Bezier A., Periquet G., Drezen J.M. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos. Trans. R. Soc. London, B: Biol. Sci. 2013;368(1626):20130051. doi: 10.1098/rstb.2013.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman B.I., Cai G. The family narnaviridae: simplest of RNA viruses. Adv. Virus Res. 2013;86:149–176. doi: 10.1016/B978-0-12-394315-6.00006-4. [DOI] [PubMed] [Google Scholar]
- Hjort K., Goldberg A.V., Tsaousis A.D., Hirt R.P., Embley T.M. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos. Trans. R. Soc. London, B: Biol. Sci. 2010;365(1541):713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. What does virus evolution tell us about virus origins? J. Virol. 2011;85(11):5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.Y., Li G.H., Li G.T., Yao Q., Chen K.P. Bombyx mori bidensovirus: the type species of the new genus Bidensovirus in the new family Bidnaviridae. Chin. Sci. Bull. 2013;58:4528–4532. [Google Scholar]
- Husnik F., Nikoh N., Koga R., Ross L., Duncan R.P., Fujie M., Tanaka M., Satoh N., Bachtrog D., Wilson A.C., von Dohlen C.D., Fukatsu T., McCutcheon J.P. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153(7):1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- Ilyina T.V., Koonin E.V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20(13):3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D., Stone J.R., Maki J.L., Collins T., Wagner G. Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain. Mol. Cell. 2005;17(1):137–143. doi: 10.1016/j.molcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Iyer L.M., Aravind L., Koonin E.V. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75(23):11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Balaji S., Koonin E.V., Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117(1):156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Iyer L.M., Koonin E.V., Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Koonin E.V., Leipe D.D., Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33(12):3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen M.E., Takagi Y., Parent K.N., Cardone G., Nibert M.L., Baker T.S. Three- dimensional structure of a protozoal double-stranded RNA virus that infects the enteric pathogen Giardia lamblia. J. Virol. 2015;89(2):1182–1194. doi: 10.1128/JVI.02745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle J.A., Abd-Alla A.M., Wang Y. Phylogeny and evolution of Hytrosaviridae. J. Invertebr. Pathol. 2013;112(Suppl) doi: 10.1016/j.jip.2012.07.015. S62-7. [DOI] [PubMed] [Google Scholar]
- Jiang D., Fu Y., Guoqing L., Ghabrial S.A. Viruses of the plant pathogenic fungus Sclerotinia sclerotiorum. Adv. Virus Res. 2013;86:215–248. doi: 10.1016/B978-0-12-394315-6.00008-8. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Ishino T., Ishino F. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front. Microbiol. 2012;3:262. doi: 10.3389/fmicb.2012.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H.H., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303(5664):1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kidmose R.T., Vasiliev N.N., Chetverin A.B., Andersen G.R., Knudsen C.R. Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. U.S.A. 2010;107(24):10884–10889. doi: 10.1073/pnas.1003015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4(1):67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A., Terzian C., Santamaria P., Pelisson A., Purd’homme N., Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 1994;91(4):1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Chang H.W., Nam Y.D., Roh S.W., Kim M.S., Sung Y., Jeon C.O., Oh H.M., Bae J.W. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 2008;74(19):5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.M.Q., Lefkowitz E., Adams M.J., Carstens B., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam. Elsevier; 2011. [Google Scholar]
- King J.A., Dubielzig R., Grimm D., Kleinschmidt J.A. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20(12):3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R., Meinhardt F. Linear protein-primed replicating plasmids in eukaryotic microbes. Microbiol. Monogr. 2007;7:188–216. [Google Scholar]
- Koonin E.V. Genome replication/expression strategies of positive-strand RNA viruses: a simple version of a combinatorial classification and prediction of new strategies. Virus Genes. 1991;5(3):273–281. doi: 10.1007/BF00568977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991;72(Pt 9):2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. Evolution of double-stranded RNA viruses: a case for polyphyletic origin from different groups of positive-stranded RNA viruses. Semin. Virol. 1992;3:327–339. [Google Scholar]
- Koonin E.V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003;1(2):127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol. Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. On the origin of cells and viruses: primordial virus world scenario. Ann. N.Y. Acad. Sci. 2009;1178:47–64. doi: 10.1111/j.1749-6632.2009.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Choi G.H., Nuss D.L., Shapira R., Carrington J.C. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. U.S.A. 1991;88(23):10647–10651. doi: 10.1073/pnas.88.23.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28(5):375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013;3(5):546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 2014;78(2):278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Gorbalenya A.E., Chumakov K.M. Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett. 1989;252(1–2):42–46. doi: 10.1016/0014-5793(89)80886-5. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Ilyina T.V. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J. Gen. Virol. 1992;73(Pt 10):2763–2766. doi: 10.1099/0022-1317-73-10-2763. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Ilyina T.V. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30(1–3):241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Krupovic M., Yutin N. Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses. Ann. N.Y. Acad. Sci. 2015 doi: 10.1111/nyas.12728. in press, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Mushegian A.R., Ryabov E.V., Dolja V.V. Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J. Gen. Virol. 1991;72(Pt 12):2895–2903. doi: 10.1099/0022-1317-72-12-2895. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Senkevich T.G., Dolja V.V. The ancient Virus World and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Wolf Y.I., Nagasaki K., Dolja V.V. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 2008;6(12):925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Wolf Y.I., Nagasaki K., Dolja V.V. The complexity of the virus world. Nat. Rev. Microbiol. 2009;7(3):250. [Google Scholar]
- Koonin E.V., Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53(5):284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Yutin N. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb. Perspect. Biol. 2014;6(4):a016188. doi: 10.1101/cshperspect.a016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen D.M., Mushegian A.R., Dolja V.V., Koonin E.V. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18(1):11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen D.M., Waller A.S., Yamada T., Bork P., Mushegian A.R., Koonin E.V. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J. Bacteriol. 2013;195(5):941–950. doi: 10.1128/JB.01801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M. Recombination between RNA viruses and plasmids might have played a central role in the origin and evolution of small DNA viruses. Bioessays. 2012;34(10):867–870. doi: 10.1002/bies.201200083. [DOI] [PubMed] [Google Scholar]
- Krupovic M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr. Opin. Virol. 2013;3(5):578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Bamford D.H. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 2008;6(12):941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Bamford D.H. Does the evolution of viral polymerases reflect the origin and evolution of viruses? Nat. Rev. Microbiol. 2009;7(3):250. doi: 10.1038/nrmicro2030-c1. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Bamford D.H. Order to the viral universe. J. Virol. 2010;84(24):12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Bamford D.H. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr. Opin. Virol. 2011;1(2):118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Bamford D.H., Koonin E.V. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol. Direct. 2014;9(1):6. doi: 10.1186/1745-6150-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Cvirkaite-Krupovic V. Virophages or satellite viruses? Nat. Rev. Microbiol. 2011;9(11):762–763. doi: 10.1038/nrmicro2676. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Forterre P. Single-stranded DNA viruses employ a variety of mechanisms for integration into the host genomes. Ann. N.Y. Acad. Sci. 2015 doi: 10.1111/nyas.12675. in press, [DOI] [PubMed] [Google Scholar]
- Krupovic M., Forterre P., Bamford D.H. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 2010;397(1):144–160. doi: 10.1016/j.jmb.2010.01.037. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Koonin E.V. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci. Rep. 2014;4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Koonin E.V. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat. Rev. Microbiol. 2015;13(2):105–115. doi: 10.1038/nrmicro3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Prangishvili D., Hendrix R.W., Bamford D.H. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 2011;75(4):610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Ravantti J.J., Bamford D.H. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol. Biol. 2009;9:112. doi: 10.1186/1471-2148-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Spang A., Gribaldo S., Forterre P., Schleper C. A thaumarchaeal provirus testifies for an ancient association of tailed viruses with archaea. Biochem. Soc. Trans. 2011;39(1):82–88. doi: 10.1042/BST0390082. [DOI] [PubMed] [Google Scholar]
- Krupovic M., Zhi N., Li J., Hu G., Koonin E.V., Wong S., Shevchenko S., Zhao K., Young N.S. Multiple layers of chimerism in a single-stranded DNA virus discovered by deep sequencing. Genome Biol. Evol. 2015 doi: 10.1093/gbe/evv034. in press, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D.M., Koonin E.V. A novel family of predicted retroviral-like aspartyl proteases with a possible key role in eukaryotic cell cycle control. Curr. Biol. 2001;11(15):R584–R587. doi: 10.1016/s0960-9822(01)00357-8. [DOI] [PubMed] [Google Scholar]
- Kurland C.G., Collins L.J., Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312(5776):1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- La Scola B., Desnues C., Pagnier I., Robert C., Barrassi L., Fournous G., Merchat M., Suzan-Monti M., Forterre P., Koonin E., Raoult D. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455(7209):100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- Labonte J.M., Suttle C.A. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 2013;7(11):2169–2177. doi: 10.1038/ismej.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A.M., Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 2011;3(8):a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson B.C., Inouye M., Inouye S. Retrons, msDNA, and the bacterial genome. Cytogenet. Genome Res. 2005;110(1–4):491–499. doi: 10.1159/000084982. [DOI] [PubMed] [Google Scholar]
- Lane N., Martin W. The energetics of genome complexity. Nature. 2010;467(7318):929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- Lane N., Martin W.F. The origin of membrane bioenergetics. Cell. 2012;151(7):1406–1416. doi: 10.1016/j.cell.2012.11.050. [DOI] [PubMed] [Google Scholar]
- Le Gall O., Christian P., Fauquet C.M., King A.M., Knowles N.J., Nakashima N., Stanway G., Gorbalenya A.E. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch. Viro. 2008 doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- Lee S.I., Kim N.S. Transposable elements and genome size variations in plants. Genomics Inform. 2014;12(3):87–97. doi: 10.5808/GI.2014.12.3.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M., Bartoli J., Shmakova L., Jeudy S., Labadie K., Adrait A., Lescot M., Poirot O., Bertaux L., Bruley C., Coute Y., Rivkina E., Abergel C., Claverie J.M. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U.S.A. 2014;111(11):4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rahmeh A., Morelli M., Whelan S.P. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 2008;82(2):775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fu Y., Li B., Yu X., Xie J., Cheng J., Ghabrial S.A., Li G., Yi X., Jiang D. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol. Biol. 2011;11:276. doi: 10.1186/1471-2148-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fu Y., Xie J., Cheng J., Ghabrial S.A., Li G., Peng Y., Yi X., Jiang D. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol. Biol. 2012;12:91. doi: 10.1186/1471-2148-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fu Y., Xie J., Cheng J., Ghabrial S.A., Li G., Yi X., Jiang D. Discovery of novel dsRNA viral sequences by in silico cloning and implications for viral diversity, host range and evolution. PLoS One. 2012;7(7):e42147. doi: 10.1371/journal.pone.0042147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu L., Opalka N., Kappler J., Shu H.B., Zhang G. Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands. Cell. 2002;108(3):383–394. doi: 10.1016/s0092-8674(02)00631-1. [DOI] [PubMed] [Google Scholar]
- Llorens C., Fares M.A., Moya A. Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol. Biol. 2008;8:276. doi: 10.1186/1471-2148-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi H., Thiagarajan M., Haas B., Wortman J., Hall N., Caler E. Genome wide survey, discovery and evolution of repetitive elements in three Entamoeba species. BMC Genomics. 2008;9:595. doi: 10.1186/1471-2164-9-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque D., Gomez-Blanco J., Garriga D., Brilot A.F., Gonzalez J.M., Havens W.M., Carrascosa J.L., Trus B.L., Verdaguer N., Ghabrial S.A., Caston J.R. Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage. Proc. Natl. Acad. Sci. U.S.A. 2014;111(21):7641–7646. doi: 10.1073/pnas.1404330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl. Acad. Sci. U.S.A. 2007;104(Suppl. 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. The origins of genome complexity. Science. 2003;302(5649):1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Lyozin G.T., Makarova K.S., Velikodvorskaja V.V., Zelentsova H.S., Khechumian R.R., Kidwell M.G., Koonin E.V., Evgen’ev M.B. The structure and evolution of Penelope in the virilis species group of Drosophila: an ancient lineage of retroelements. J. Mol. Evol. 2001;52(5):445–456. doi: 10.1007/s002390010174. [DOI] [PubMed] [Google Scholar]
- Maeda N., Fan H., Yoshikai Y. Oncogenesis by retroviruses: old and new paradigms. Rev. Med. Virol. 2008;18(6):387–405. doi: 10.1002/rmv.592. [DOI] [PubMed] [Google Scholar]
- Majorek K.A., Dunin-Horkawicz S., Steczkiewicz K., Muszewska A., Nowotny M., Ginalski K., Bujnicki J.M. The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014;42(7):4160–4179. doi: 10.1093/nar/gkt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S. Ribonuclease H evolution in retrotransposable elements. Cytogenet. Genome Res. 2005;110(1–4):392–401. doi: 10.1159/000084971. [DOI] [PubMed] [Google Scholar]
- Malik H.S., Eickbush T.H. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 1999;73(6):5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S., Eickbush T.H. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11(7):1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- Malik H.S., Henikoff S., Eickbush T.H. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10(9):1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- Mandal P.K., Bagchi A., Bhattacharya A., Bhattacharya S. An Entamoeba histolytica LINE/SINE pair inserts at common target sites cleaved by the restriction enzyme-like LINE-encoded endonuclease. Eukaryot. Cell. 2004;3(1):170–179. doi: 10.1128/EC.3.1.170-179.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantynen S., Laanto E., Kohvakka A., Poranen M.M., Bamford J.K., Ravantti J.J. New enveloped dsRNA phage from freshwater habitat. J. Gen. Virol. 2015 doi: 10.1099/vir.0.000063. [DOI] [PubMed] [Google Scholar]
- Martin W., Dagan T., Koonin E.V., Dipippo J.L., Gogarten J.P., Lake J.A. The evolution of eukaryotes. Science. 2007;316(5824):542–543. doi: 10.1126/science.316.5824.542c. author reply 542-3. [DOI] [PubMed] [Google Scholar]
- Martin W., Koonin E.V. Introns and the origin of nucleus-cytosol compartmentation. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- McKenna R., Xia D., Willingmann P., Ilag L.L., Krishnaswamy S., Rossmann M.G., Olson N.H., Baker T.S., Incardona N.L. Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications. Nature. 1992;355(6356):137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhekar B., Miller J.F. Diversity-generating retroelements. Curr. Opin. Microbiol. 2007;10(4):388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher U. The ‘30K’ superfamily of viral movement proteins. J. Gen. Virol. 2000;81(Pt 1):257–266. doi: 10.1099/0022-1317-81-1-257. [DOI] [PubMed] [Google Scholar]
- Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 2004;101(1):83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Mochizuki T., Krupovic M., Pehau-Arnaudet G., Sako Y., Forterre P., Prangishvili D. Archaeal virus with exceptional virion architecture and the largest single-stranded DNA genome. Proc. Natl. Acad. Sci. U.S.A. 2012;109(33):13386–13391. doi: 10.1073/pnas.1203668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y. Relating structure to evolution in class II viral membrane fusion proteins. Curr. Opin. Virol. 2014;5:34–41. doi: 10.1016/j.coviro.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monttinen H.A., Ravantti J.J., Stuart D.I., Poranen M.M. Automated structural comparisons clarify the phylogeny of the right-hand-shaped polymerases. Mol. Biol. Evol. 2014;31(10):2741–2752. doi: 10.1093/molbev/msu219. [DOI] [PubMed] [Google Scholar]
- Mushegian A.R., Elena S.F. Evolution of plant virus movement proteins from the 30 K superfamily and of their homologs integrated in plant genomes. Virology. 2015;476C:304–315. doi: 10.1016/j.virol.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Mushegian A.R., Koonin E.V. Cell-to-cell movement of plant viruses. Insights from amino acid sequence comparisons of movement proteins and from analogies with cellular transport systems. Arch. Virol. 1993;133(3–4):239–257. doi: 10.1007/BF01313766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K., Tomaru Y., Takao Y., Nishida K., Shirai Y., Suzuki H., Nagumo T. Previously unknown virus infects marine diatom. Appl. Environ. Microbiol. 2005;71(7):3528–3535. doi: 10.1128/AEM.71.7.3528-3535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012;10(2):137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir A., Kim K.M., Caetano-Anolles G. Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol. Biol. 2012;12:156. doi: 10.1186/1471-2148-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134(1–2):235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Nesmelova I.V., Hackett P.B. DDE transposases: structural similarity and diversity. Adv. Drug Delivery Rev. 2010;62(12):1187–1195. doi: 10.1016/j.addr.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu M., Kim H.J., Benner S.A. The “strong” RNA world hypothesis: fifty years old. Astrobiology. 2013;13(4):391–403. doi: 10.1089/ast.2012.0868. [DOI] [PubMed] [Google Scholar]
- Ng J.C., Falk B.W. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 2006;44:183–212. doi: 10.1146/annurev.phyto.44.070505.143325. [DOI] [PubMed] [Google Scholar]
- Nibert M.L., Tang J., Xie J., Collier A.M., Ghabrial S.A., Baker T.S., Tao Y.J. 3D structures of fungal partitiviruses. Adv. Virus Res. 2013;86:59–85. doi: 10.1016/B978-0-12-394315-6.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris G.E., Stillman T.J., Anderson B.F., Baker E.N. The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum. Structure. 1994;2(11):1049–1059. doi: 10.1016/s0969-2126(94)00108-1. [DOI] [PubMed] [Google Scholar]
- Novikova O., Smyshlyaev G., Blinov A. Evolutionary genomics revealed interkingdom distribution of Tcn1-like chromodomain-containing Gypsy LTR retrotransposons among fungi and plants. BMC Genomics. 2010;11:231. doi: 10.1186/1471-2164-11-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H. TT viruses in animals. Curr. Top. Microbiol. Immunol. 2009;331:35–52. doi: 10.1007/978-3-540-70972-5_3. [DOI] [PubMed] [Google Scholar]
- Pardue M.L., DeBaryshe P.G. Retrotransposons that maintain chromosome ends. Proc. Natl. Acad. Sci. U.S.A. 2011;108(51):20317–20324. doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflug A., Guilligay D., Reich S., Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516(7531):355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- Philippe N., Legendre M., Doutre G., Coute Y., Poirot O., Lescot M., Arslan D., Seltzer V., Bertaux L., Bruley C., Garin J., Claverie J.M., Abergel C. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341(6143):281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- Pietila M.K., Laurinmaki P., Russell D.A., Ko C.C., Jacobs-Sera D., Hendrix R.W., Bamford D.H., Butcher S.J. Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc. Natl. Acad. Sci. U.S.A. 2013;110(26):10604–10609. doi: 10.1073/pnas.1303047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A., Jeffares D., Penny D. Early evolution: prokaryotes, the new kids on the block. Bioessays. 1999;21(10):880–889. doi: 10.1002/(SICI)1521-1878(199910)21:10<880::AID-BIES11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Poole A., Penny D. Eukaryote evolution: engulfed by speculation. Nature. 2007;447(7147):913. doi: 10.1038/447913a. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Bamford D.H. Assembly of large icosahedral double-stranded RNA viruses. Adv. Exp. Med. Biol. 2012;726:379–402. doi: 10.1007/978-1-4614-0980-9_17. [DOI] [PubMed] [Google Scholar]
- Prangishvili D. The wonderful world of archaeal viruses. Annu. Rev. Microbiol. 2013;67:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- Prangishvili D., Forterre P., Garrett R.A. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 2006;4(11):837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- Prangishvili D., Garrett R.A., Koonin E.V. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 2006;–(1):52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Quito-Avila D.F., Lightle D., Lee J., Martin R.R. Transmission biology of Raspberry latent virus, the first aphid-borne reovirus. Phytopathology. 2012;102(5):547–553. doi: 10.1094/PHYTO-12-11-0331. [DOI] [PubMed] [Google Scholar]
- Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306(5700):1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Raoult D., Forterre P. Redefining viruses: lessons from Mimivirus. Nat. Rev. Microbiol. 2008;6(4):315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- Rastgou M., Habibi M.K., Izadpanah K., Masenga V., Milne R.G., Wolf Y.I., Koonin E.V., Turina M. Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 2009;90(Pt 10):2525–2535. doi: 10.1099/vir.0.013086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest J.S., Mindell D.P. Retroids in archaea: phylogeny and lateral origins. Mol. Biol. Evol. 2003;20(7):1134–1142. doi: 10.1093/molbev/msg135. [DOI] [PubMed] [Google Scholar]
- Rigden J.E., Dry I.B., Krake L.R., Rezaian M.A. Plant virus DNA replication processes in Agrobacterium: insight into the origins of geminiviruses? Proc. Natl. Acad. Sci. U.S.A. 1996;93(19):10280–10284. doi: 10.1073/pnas.93.19.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon F.J., Schmid M.F. Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr. Opin. Virol. 2014;5:105–110. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Robart A.R., Chan R.T., Peters J.K., Rajashankar K.R., Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514(7521):193–197. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart A.R., Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet. Genome Res. 2005;110(1–4):589–597. doi: 10.1159/000084992. [DOI] [PubMed] [Google Scholar]
- Robertson M.P., Joyce G.F. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 2012;4:5. doi: 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F. Global phage diversity. Cell. 2003;113(2):141. doi: 10.1016/s0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- Rohwer F., Thurber R.V. Viruses manipulate the marine environment. Nature. 2009;459(7244):207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- Roossinck M.J., Sabanadzovic S., Okada R., Valverde R.A. The remarkable evolutionary history of endornaviruses. J. Gen. Virol. 2011;92(Pt 11):2674–2678. doi: 10.1099/vir.0.034702-0. [DOI] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 2009;90(Pt 10):2418–2424. doi: 10.1099/vir.0.012955-0. [DOI] [PubMed] [Google Scholar]
- Rosario K., Duffy S., Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch. Virol. 2012;157(10):1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Johnson J.E. Icosahedral RNA virus structure. Annu. Rev. Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- Rothnie H.M., Chapdelaine Y., Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv. Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- Roux S., Enault F., Bronner G., Vaulot D., Forterre P., Krupovic M. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat. Commun. 2013;4:2700. doi: 10.1038/ncomms3700. [DOI] [PubMed] [Google Scholar]
- Roux S., Krupovic M., Poulet A., Debroas D., Enault F. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One. 2012;7(7):e40418. doi: 10.1371/journal.pone.0040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado P.S., Koivunen M.R., Makeyev E.V., Bamford D.H., Stuart D.I., Grimes J.M. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4(12):e434. doi: 10.1371/journal.pbio.0040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S.B., Menees T.M. Morphogenesis at the retrotransposon-retrovirus interface: gypsy and copia families in yeast and Drosophila. Curr. Top. Microbiol. Immunol. 1996;214:261–296. doi: 10.1007/978-3-642-80145-7_9. [DOI] [PubMed] [Google Scholar]
- Seeger C., Hu J. Why are hepadnaviruses DNA and not RNA viruses? Trends Microbiol. 1997;5(11):447–450. doi: 10.1016/s0966-842x(97)01141-4. [DOI] [PubMed] [Google Scholar]
- Selth L.A., Randles J.W., Rezaian M.A. Agrobacterium tumefaciens supports DNA replication of diverse geminivirus types. FEBS Lett. 2002;516(1–3):179–182. doi: 10.1016/s0014-5793(02)02539-5. [DOI] [PubMed] [Google Scholar]
- Selvarajan Sigamani S., Zhao H., Kamau Y.N., Baines J.D., Tang L. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J. Virol. 2013;87(12):7140–7148. doi: 10.1128/JVI.00311-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt T.E., Gray M.W. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22(2):90–95. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Simon D.M., Clarke N.A., McNeil B.A., Johnson I., Pantuso D., Dai L., Chai D., Zimmerly S. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA. 2008;14(9):1704–1713. doi: 10.1261/rna.1056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D.M., Zimmerly S. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008;36(22):7219–7229. doi: 10.1093/nar/gkn867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirkis R., Gerst J.E., Fass D. Ddi1, a eukaryotic protein with the retroviral protease fold. J. Mol. Biol. 2006;364(3):376–387. doi: 10.1016/j.jmb.2006.08.086. [DOI] [PubMed] [Google Scholar]
- Smyshlyaev G., Voigt F., Blinov A., Barabas O., Novikova O. Acquisition of an Archaea-like ribonuclease H domain by plant L1 retrotransposons supports modular evolution. Proc. Natl. Acad. Sci. U.S.A. 2013;110(50):20140–20145. doi: 10.1073/pnas.1310958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyom S., Kazazian H.H., Jr. Mobile elements in the human genome: implications for disease. Genome Med. 2012;4(2):12. doi: 10.1186/gm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.U., Gerasimova T., Kurkulos M., Boeke J.D., Corces V.G. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8(17):2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- Staginnus C., Richert-Poggeler K.R. Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends Plant Sci. 2006;11(10):485–491. doi: 10.1016/j.tplants.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Stedman K. Mechanisms for RNA capture by ssDNA viruses: grand theft RNA. J. Mol. Evol. 2013;76(6):359–364. doi: 10.1007/s00239-013-9569-9. [DOI] [PubMed] [Google Scholar]
- Steven A.C., Conway J.F., Cheng N., Watts N.R., Belnap D.M., Harris A., Stahl S.J., Wingfield P.T. Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv. Virus Res. 2005;64:125–164. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- Stoddard B.L. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38(1):49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- Stoye J.P. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10(6):395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- Suttle C.A. Viruses in the sea. Nature. 2005;437(7057):356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Suttle C.A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5(10):801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Szathmary E., Demeter L. Group selection of early replicators and the origin of life. J. Theor. Biol. 1987;128(4):463–486. doi: 10.1016/s0022-5193(87)80191-1. [DOI] [PubMed] [Google Scholar]
- Takeuchi N., Hogeweg P. The role of complex formation and deleterious mutations for the stability of RNA-like replicator systems. J. Mol. Evol. 2007;65(6):668–686. doi: 10.1007/s00239-007-9044-6. [DOI] [PubMed] [Google Scholar]
- Takeuchi N., Hogeweg P. Evolutionary dynamics of RNA-like replicator systems: a bioinformatic approach to the origin of life. Phys. Life Rev. 2012;9(3):219–263. doi: 10.1016/j.plrev.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N., Hogeweg P., Koonin E.V. On the origin of DNA genomes: evolution of the division of labor between template and catalyst in model replicator systems. PLoS Comput. Biol. 2011;7(3):e1002024. doi: 10.1371/journal.pcbi.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theze J., Bezier A., Periquet G., Drezen J.M., Herniou E.A. Paleozoic origin of insect large dsDNA viruses. Proc. Natl. Acad. Sci. U.S.A. 2011;108(38):15931–15935. doi: 10.1073/pnas.1105580108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N., Keating K.S., Taylor S.D., Pyle A.M. Crystal structure of a self-spliced group II intron. Science. 2008;320(5872):77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordo N., Poch O., Ermine A., Keith G., Rougeon F. Completion of the rabies virus genome sequence determination: highly conserved domains among the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology. 1988;165(2):565–576. doi: 10.1016/0042-6822(88)90600-9. [DOI] [PubMed] [Google Scholar]
- Toro N., Nisa-Martínez R. Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0114083. e114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J. Eukaryot. Microbiol. 2009;56(3):221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- van der Giezen M., Tovar J. Degenerate mitochondria. EMBO Rep. 2005;6(6):525–530. doi: 10.1038/sj.embor.7400440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J.L. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- Vaney M.C., Rey F.A. Class II enveloped viruses. Cell. Microbiol. 2011;13(10):1451–1459. doi: 10.1111/j.1462-5822.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- von Dohlen C.D., Kohler S., Alsop S.T., McManus W.R. Mealybug beta-proteobacterial endosymbionts contain gamma-proteobacterial symbionts. Nature. 2001;412(6845):433–436. doi: 10.1038/35086563. [DOI] [PubMed] [Google Scholar]
- Wang Q., Han Y., Qiu Y., Zhang S., Tang F., Wang Y., Zhang J., Hu Y., Zhou X. Identification and characterization of RNA duplex unwinding and ATPase activities of an alphatetravirus superfamily 1 helicase. Virology. 2012;433(2):440–448. doi: 10.1016/j.virol.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.C., Hsu Y.H., Lin N.S., Wu C.Y., Lai Y.C., Hu C.C. A novel prokaryotic promoter identified in the genome of some monopartite begomoviruses. PLoS One. 2013;8(7):e70037. doi: 10.1371/journal.pone.0070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bininda-Emonds O.R.P., Jehle J.A. Nudivirus genomics and phylogeny. In: Garcia M., editor. Molecular Structure, Diversity, Gene Expression Mechanisms and Host-Virus Interactions. InTech; Rijeka: 2012. [Google Scholar]
- Wang Y., Jehle J.A. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J. Invertebr. Pathol. 2009;101(3):187–193. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Weiss R.A. On the concept and elucidation of endogenous retroviruses. Philos. Trans. R. Soc. London, B: Biol. Sci. 2013;368(1626):20120494. doi: 10.1098/rstb.2012.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whon T.W., Kim M.S., Roh S.W., Shin N.R., Lee H.W., Bae J.W. Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J. Virol. 2012;86(15):8221–8231. doi: 10.1128/JVI.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.A., Embley T.M., Heinz E. Informational gene phylogenies do not support a fourth domain of life for nucleocytoplasmic large DNA viruses. PLoS One. 2011;6(6):e21080. doi: 10.1371/journal.pone.0021080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T.Y., Preston L.A., Schiller N.L. ALGINATE LYASE: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000;54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- Wu C.Y., Yang S.H., Lai Y.C., Lin N.S., Hsu Y.H., Hu C.C. Unit-length, single-stranded circular DNAs of both polarity of begomoviruses are generated in Escherichia coli harboring phage M13-cloned begomovirus genome with single copy of replication origin. Virus Res. 2007;125(1):14–28. doi: 10.1016/j.virusres.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Malik H.S., Eickbush T.H. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. U.S.A. 1999;96(14):7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Koonin E.V. Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes. Virol. J. 2012;–(1):161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Koonin E.V. Pandoraviruses are highly derived phycodnaviruses. Biol. Direct. 2013;8:25. doi: 10.1186/1745-6150-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Makarova K.S., Mekhedov S.L., Wolf Y.I., Koonin E.V. The deep archaeal roots of eukaryotes. Mol. Biol. Evol. 2008;25(8):1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Raoult D., Koonin E.V. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol. J. 2013;10:158. doi: 10.1186/1743-422X-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Wolf M.Y., Wolf Y.I., Koonin E.V. The origins of phagocytosis and eukaryogenesis. Biol. Direct. 2009;4:9. doi: 10.1186/1745-6150-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Wolf Y.I., Koonin E.V. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology. 2014;466–467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotto P.M., Gibbs M.J., Gould E.A., Holmes E.C. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 1996;70(9):6083–6096. doi: 10.1128/jvi.70.9.6083-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawar-Reza P., Arguello-Astorga G.R., Kraberger S., Julian L., Stainton D., Broady P.A., Varsani A. Diverse small circular single-stranded DNA viruses identified in a freshwater pond on the McMurdo Ice Shelf (Antarctica) Infect. Genet. Evol. 2014;26:132–138. doi: 10.1016/j.meegid.2014.05.018. [DOI] [PubMed] [Google Scholar]
- Zeddam J.L., Gordon K.H., Lauber C., Alves C.A., Luke B.T., Hanzlik T.N., Ward V.K., Gorbalenya A.E. Euprosterna elaeasa virus genome sequence and evolution of the Tetraviridae family: emergence of bipartite genomes and conservation of the VPg signal with the dsRNA Birnaviridae family. Virology. 2010;397(1):145–154. doi: 10.1016/j.virol.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Zhang W., Olson N.H., Baker T.S., Faulkner L., Agbandje-McKenna M., Boulton M.I., Davies J.W., McKenna R. Structure of the Maize streak virus geminate particle. Virology. 2001;279(2):471–477. doi: 10.1006/viro.2000.0739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material