Abstract
We aggregated coding variant data for 81,412 type 2 diabetes cases and 370,832 controls of diverse ancestry, identifying 40 coding variant association signals (p<2.2×10−7): of these, 16 map outside known risk loci. We make two important observations. First, only five of these signals are driven by low-frequency variants: even for these, effect sizes are modest (odds ratio ≤1.29). Second, when we used large-scale genome-wide association data to fine-map the associated variants in their regional context, accounting for the global enrichment of complex trait associations in coding sequence, compelling evidence for coding variant causality was obtained for only 16 signals. At 13 others, the associated coding variants clearly represent “false leads” with potential to generate erroneous mechanistic inference. Coding variant associations offer a direct route to biological insight for complex diseases and identification of validated therapeutic targets: however, appropriate mechanistic inference requires careful specification of their causal contribution to disease predisposition.
Genome-wide association studies (GWAS) have identified thousands of association signals influencing multifactorial traits such as type 2 diabetes (T2D) and obesity1–7. Most of these associations involve common variants that map to non-coding sequence, and identification of their cognate effector transcripts has proved challenging. Identification of coding variants causally implicated in trait predisposition offers a more direct route from association signal to biological inference.
The exome occupies 1.5% of overall genome sequence, but for many common diseases, coding variants make a disproportionate contribution to trait heritability8,9. This enrichment indicates that coding variant association signals have an enhanced probability of being causal when compared to those involving an otherwise equivalent non-coding variant. This does not, however, guarantee that all coding variant associations are causal. Alleles driving common-variant (minor allele frequency [MAF] ≥5%) GWAS signals typically reside on extended risk haplotypes that, owing to linkage disequilibrium (LD), incorporate many common variants10,11. Consequently, the presence of a coding allele on the risk haplotype does not constitute sufficient evidence that it represents the causal variant at the locus, or that the gene within which it lies is mediating the association signal. Since much coding variant discovery has proceeded through exome-specific analyses (either exome-array genotyping or exome sequencing), researchers have often been poorly-placed to position coding variant associations in the context of regional genetic variation. It is unclear how often this may have led to incorrect assumptions regarding their causal role.
In our recent study of T2D predisposition12, we surveyed the exomes of 34,809 T2D cases and 57,985 controls, of predominantly European descent, and identified 13 distinct coding variant associations reaching genome-wide significance. Twelve of these associations involved common variants, but the data hinted at a substantial pool of lower-frequency coding variants of moderate impact, potentially amenable to detection in larger samples. We also reported that, whilst many of these signals fell within common variant loci previously identified by GWAS, it was far from trivial to determine, using available data, whether those coding variants were causal or ‘hitchhiking’ on risk haplotypes.
Here, we report analyses that address these two issues. First, we extend the scope of our exome-array genotyping to include data from 81,412 T2D cases and 370,832 controls of diverse ancestry, substantially increasing power to detect coding variant associations across the allele-frequency spectrum. Second, to understand the extent to which identification of coding variant associations provides a reliable guide to causal mechanisms, we undertake high-resolution fine-mapping of identified coding variant association signals in 50,160 T2D cases and 465,272 controls of European ancestry with genome-wide genotyping data.
RESULTS
Discovery study overview
First, we set out to discover coding variant association signals by aggregating T2D association summary statistics in up to 452,244 individuals (effective sample size 228,825) across five ancestry groups, performing both European-specific (EUR) and trans-ethnic (TE) meta-analyses (Supplementary Tables 1 and 2). Analysis was restricted to the 247,470 variants represented on the exome-array. Genotypes were assembled from: (a) 58,425 cases and 188,032 controls genotyped with the exome-array; (b) 14,608 cases and 174,322 controls from UK Biobank and GERA (Resource for Genetic Epidemiology on Adult Health and Aging) genotyped with GWAS arrays enriched for exome content and/or coverage of low-frequency variation across ethnic groups13,14; and (c) 8,379 cases and 8,478 controls with whole-exome sequence from GoT2D/T2D-GENES12 and SIGMA15 studies. Overall, this represented a 3-fold increase in effective sample size over our previous study of T2D predisposition within coding sequence12. To deconvolute the impact of obesity on T2D-associated variants, association analyses were conducted with and without body mass index (BMI) adjustment.
We considered p<2.2×10−7 as significant for protein truncating variants (PTVs) and moderate impact coding variants (including missense, in-frame indel and splice region variants) based on a weighted Bonferroni correction that accounts for the observed enrichment in complex trait association signals across sequence annotation16. This threshold matches those obtained through other approaches such as simple Bonferroni correction for the number of coding variants on the exome-array (Methods). Compared to our previous study12, the expanded sample size substantially increased power to detect association for common variants of modest effect (e.g. from 14.4% to 97.9% for a variant with 20% MAF and odds ratio [OR]=1.05) and lower-frequency variants with larger effects (e.g. from 11.8% to 97.5% for a variant with 1% MAF and OR=1.20) assuming homogenous allelic effects across ancestry groups (Methods).
Insights into coding variant association signals underlying T2D susceptibility
We detected significant associations at 69 coding variants under an additive genetic model (either in BMI unadjusted or adjusted analysis), mapping to 38 loci (Supplementary Fig. 1, Supplementary Table 3). We observed minimal evidence of heterogeneity in allelic OR between ancestry groups (Supplementary Table 3), and no compelling evidence for non-additive allelic effects (Supplementary Fig. 2, Supplementary Table 4). Reciprocal conditional analyses (Methods) indicated that the 69 coding variants represented 40 distinct association signals (conditional p<2.2×10−7) across the 38 loci, with two distinct signals each at HNF1A and RREB1 (Supplementary Table 5). These 40 signals included the 13 associations reported in our earlier publication12, each featuring more significant associations in this expanded meta-analysis (Supplementary Table 6). Twenty-five of the 40 signals were significant in both EUR and TE analyses. Of the other 15, three (PLCB3, C17orf58, and ZHX3) were significant in EUR, and all reached pTE<6.8×10−6 in the TE analysis: for PLCB3 and ZHX3, risk allele frequencies were substantially lower outside European descent populations. Twelve loci (Supplementary Table 3) were significant in TE alone, but for these (except PAX4 which is East Asian specific), the evidence for association was proportionate in the smaller EUR component (pEUR<8.4×10−5).
Sixteen of the 40 distinct association signals mapped outside regions previously implicated in T2D susceptibility (Methods, Table 1). These included missense variant signals in POC5 (p.His36Arg, rs2307111, pTE=1.6×10−15), PNPLA3 (p.Ile148Met, rs738409, pTE BMI-adjusted=2.8×10−11), and ZZEF1 (p.Ile2014Val, rs781831, pTE=8.3×10−11).
Table 1.
Summary of discovery and fine-mapping analyses of the 40 index coding variants associated with T2D (p<2.2×10−7).
| Discovery meta-analysis using exome-array component: 81,412 T2D cases and 370,832 controls from diverse ancestries | Fine-mapping meta-analysis using GWAS: 50,160 T2D cases and 465,272 controls from European ancestry | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Index variant | rs ID | Chr | Pos | Alleles | RAF | BMI unadjusted | BMI adjusted | RAF | OR | L95 | U95 | p-value | Group | ||||||
| R/O | OR | L95 | U95 | p-value | OR | L95 | U95 | p-value | ||||||||||||
| Previously reported T2D associated loci | ||||||||||||||||||||
| MACF1 | MACF1 p.Met1424Val | rs2296172 | 1 | 39,835,817 | G/A | 0.193 | 1.06 | 1.05 | 1.08 | 6.7×10−16 | 1.04 | 1.03 | 1.06 | 5.9×10−8 | 0.22 | 1.08 | 1.06 | 1.1 | 1.6×10−15 | 3 |
| GCKR | GCKR p.Pro446Leu | rs1260326 | 2 | 27,730,940 | C/T | 0.630 | 1.06 | 1.05 | 1.08 | 5.3×10−25 | 1.06 | 1.04 | 1.07 | 3.2×10−18 | 0.607 | 1.05 | 1.04 | 1.07 | 9.1×10−10 | 1 |
| THADA | THADA p.Cys845Tyr | rs35720761 | 2 | 43,519,977 | C/T | 0.895 | 1.08 | 1.05 | 1.1 | 4.6×10−15 | 1.07 | 1.05 | 1.10 | 8.3×10−16 | 0.881 | 1.1 | 1.07 | 1.12 | 3.4×10−12 | 2 |
| GRB14 | COBLL1 p.Asn901Asp | rs7607980 | 2 | 165,551,201 | T/C | 0.879 | 1.08 | 1.06 | 1.11 | 8.6×10−20 | 1.09 | 1.07 | 1.12 | 5.0×10−23 | 0.871 | 1.08 | 1.06 | 1.11 | 3.6×10−10 | 2 |
| PPARG | PPARG p.Pro12Ala | rs1801282 | 3 | 12,393,125 | C/G | 0.887 | 1.09 | 1.07 | 1.11 | 1.4×10−17 | 1.10 | 1.07 | 1.12 | 2.7×10−19 | 0.876 | 1.12 | 1.09 | 1.14 | 3.7×10−17 | 3 |
| IGF2BP2 | SENP2 p.Thr291Lys | rs6762208 | 3 | 185,331,165 | A/C | 0.367 | 1.03 | 1.01 | 1.04 | 1.6×10−6 | 1.03 | 1.02 | 1.05 | 3.0×10−8 | 0.339 | 1.02 | 1.01 | 1.04 | 0.01 | 2 |
| WFS1 | WFS1 p.Val333Ile | rs1801212 | 4 | 6,302,519 | A/G | 0.748 | 1.07 | 1.06 | 1.09 | 1.1×10−24 | 1.07 | 1.05 | 1.08 | 7.1×10−21 | 0.703 | 1.07 | 1.05 | 1.09 | 4.1×10−13 | 2 |
| PAM-PPIP5K2 | PAM p.Asp336Gly | rs35658696 | 5 | 102,338,811 | G/A | 0.045 | 1.13 | 1.10 | 1.17 | 1.2×10−16 | 1.13 | 1.09 | 1.17 | 7.4×10−15 | 0.051 | 1.17 | 1.13 | 1.22 | 2.5×10−17 | 1 |
| RREB1 | RREB1 p.Asp1171Asn | rs9379084 | 6 | 7,231,843 | G/A | 0.884 | 1.08 | 1.06 | 1.11 | 1.1×10−13 | 1.10 | 1.07 | 1.13 | 1.5×10−17 | 0.888 | 1.09 | 1.06 | 1.12 | 1.1×10−9 | 1 |
| RREB1 p.Ser1499Tyr | rs35742417 | 6 | 7,247,344 | C/A | 0.836 | 1.04 | 1.03 | 1.06 | 5.5×10−8 | 1.04 | 1.02 | 1.06 | 2.2×10−7 | 0.817 | 1.04 | 1.02 | 1.07 | 0.00012 | 2 | |
| MHC | TCF19 p.Met131Val | rs2073721 | 6 | 31,129,616 | G/A | 0.749 | 1.04 | 1.02 | 1.05 | 1.6×10−10 | 1.04 | 1.02 | 1.05 | 2.3×10−9 | N/A | N/A | N/A | N/A | N/A | N/A |
| PAX4 | PAX4 p.Arg190His | rs2233580 | 7 | 127,253,550 | T/C | 0.029 | 1.36 | 1.25 | 1.48 | 1.8×10−12 | 1.38 | 1.26 | 1.51 | 4.2×10−13 | 0 | N/A | N/A | N/A | N/A | N/A |
| SLC30A8 | SLC30A8 p.Arg276Trp | rs13266634 | 8 | 118,184,783 | C/T | 0.691 | 1.09 | 1.08 | 1.11 | 1.9×10−47 | 1.09 | 1.08 | 1.11 | 1.3×10−47 | 0.683 | 1.12 | 1.1 | 1.14 | 8.2×10−36 | 1 |
| GPSM1 | GPSM1 p.Ser391Leu | rs60980157 | 9 | 139,235,415 | C/T | 0.771 | 1.06 | 1.05 | 1.08 | 3.2×10−16 | 1.06 | 1.05 | 1.08 | 6.6×10−16 | 0.756 | 1.06 | 1.04 | 1.09 | 8.3×10−8 | 3 |
| KCNJ11-ABCC8 | KCNJ11 p.Lys29Glu | rs5219 | 11 | 17,409,572 | T/C | 0.364 | 1.06 | 1.05 | 1.07 | 5.7×10−22 | 1.07 | 1.05 | 1.08 | 1.5×10−22 | 0.381 | 1.07 | 1.05 | 1.09 | 8.1×10−16 | 1 |
| CENTD2 | ARAP1 p.Gln802Glu | rs56200889 | 11 | 72,408,055 | G/C | 0.733 | 1.04 | 1.02 | 1.05 | 4.8×10−8 | 1.05 | 1.03 | 1.06 | 5.2×10−10 | 0.727 | 1.05 | 1.03 | 1.07 | 2.3×10−8 | 2 |
| KLHDC5 | MRPS35 p.Gly43Arg | rs1127787 | 12 | 27,867,727 | G/A | 0.850 | 1.06 | 1.04 | 1.08 | 1.4×10−11 | 1.05 | 1.03 | 1.07 | 1.5×10−8 | 0.842 | 1.06 | 1.04 | 1.09 | 2.2×10−7 | 2 |
| HNF1A | HNF1A p.Ile75Leu | rs1169288 | 12 | 121,416,650 | C/A | 0.323 | 1.04 | 1.03 | 1.06 | 1.1×10−11 | 1.04 | 1.02 | 1.06 | 1.9×10−10 | 0.33 | 1.05 | 1.04 | 1.07 | 4.6×10−9 | 1 |
| HNF1A p.Ala146Val | rs1800574 | 12 | 121,416,864 | T/C | 0.029 | 1.11 | 1.06 | 1.15 | 6.1×10−8 | 1.10 | 1.06 | 1.15 | 1.3×10−7 | 0.03 | 1.16 | 1.1 | 1.21 | 5.0×10−9 | 1 | |
| MPHOSPH9 | SBNO1 p.Ser729Asn | rs1060105 | 12 | 123,806,219 | C/T | 0.815 | 1.04 | 1.02 | 1.06 | 5.7×10−7 | 1.04 | 1.02 | 1.06 | 1.1×10−7 | 0.787 | 1.04 | 1.02 | 1.06 | 3.6×10−5 | 2 |
| CILP2 | TM6SF2 p.Glu167Lys | rs58542926 | 19 | 19,379,549 | T/C | 0.076 | 1.07 | 1.05 | 1.10 | 4.8×10−12 | 1.09 | 1.06 | 1.11 | 3.4×10−15 | 0.076 | 1.09 | 1.05 | 1.12 | 2.0×10−7 | 1 |
| GIPR | GIPR p.Glu318Gln | rs1800437 | 19 | 46,181,392 | C/G | 0.200 | 1.03 | 1.02 | 1.05 | 7.1×10−5 | 1.06 | 1.04 | 1.07 | 6.8×10−12 | 0.213 | 1.09 | 1.06 | 1.12 | 4.6×10−9 | 1 |
| HNF4A | HNF4A p.Thr139Ile | rs1800961 | 20 | 43,042,364 | T/C | 0.032 | 1.09 | 1.05 | 1.13 | 2.6×10−8 | 1.10 | 1.06 | 1.14 | 5.0×10−8 | 0.037 | 1.17 | 1.12 | 1.22 | 1.4×10−12 | 1 |
| MTMR3-ASCC2 | ASCC2 p.Asp407His | rs28265 | 22 | 30,200,761 | C/G | 0.925 | 1.09 | 1.06 | 1.11 | 2.1×10−12 | 1.09 | 1.07 | 1.12 | 4.4×10−14 | 0.916 | 1.1 | 1.07 | 1.14 | 9.6×10−11 | 3 |
| Novel T2D associated loci | ||||||||||||||||||||
| FAM63A | FAM63A p.Tyr95Asn | rs140386498 | 1 | 150,972,959 | A/T | 0.988 | 1.21 | 1.14 | 1.28 | 7.5×10−8 | 1.19 | 1.12 | 1.26 | 6.7×10−7 | 0.986 | 1.15 | 1.06 | 1.25 | 0.00047 | 3 |
| CEP68 | CEP68 p.Gly74Ser | rs7572857 | 2 | 65,296,798 | G/A | 0.846 | 1.05 | 1.04 | 1.07 | 8.3×10−9 | 1.05 | 1.03 | 1.07 | 6.6×10−7 | 0.830 | 1.06 | 1.03 | 1.08 | 6.6×10−7 | 2 |
| KIF9 | KIF9 p.Arg638Trp | rs2276853 | 3 | 47,282,303 | A/G | 0.588 | 1.02 | 1.01 | 1.04 | 8.0×10−5 | 1.03 | 1.02 | 1.05 | 5.3×10−8 | 0.602 | 1.04 | 1.02 | 1.05 | 2.6×10−5 | 3 |
| ANKH | ANKH p.Arg187Gln | rs146886108 | 5 | 14,751,305 | C/T | 0.996 | 1.29 | 1.16 | 1.45 | 1.4×10−7 | 1.27 | 1.13 | 1.41 | 3.5×10−7 | 0.995 | 1.51 | 1.29 | 1.77 | 3.5×10−7 | 1 |
| POC5 | POC5 p.His36Arg | rs2307111 | 5 | 75,003,678 | T/C | 0.562 | 1.05 | 1.04 | 1.07 | 1.6×10−15 | 1.03 | 1.01 | 1.04 | 2.1×10−5 | 0.606 | 1.06 | 1.05 | 1.08 | 1.1×10−12 | 1 |
| LPL | LPL p.Ser474* | rs328 | 8 | 19,819,724 | C/G | 0.903 | 1.05 | 1.03 | 1.08 | 6.8×10−9 | 1.05 | 1.03 | 1.07 | 2.3×10−7 | 0.901 | 1.08 | 1.05 | 1.11 | 7.1×10−8 | 1 |
| PLCB3† | PLCB3 p.Ser778Leu | rs35169799 | 11 | 64,031,241 | T/C | 0.071 | 1.05 | 1.02 | 1.08 | 1.3×10−5 | 1.06 | 1.03 | 1.09 | 1.8×10−7 | 0.065 | 1.07 | 1.04 | 1.11 | 3.8×10−5 | 1 |
| TPCN2 | TPCN2 p.Val219Ile | rs72928978 | 11 | 68,831,364 | G/A | 0.890 | 1.05 | 1.02 | 1.07 | 5.2×10−7 | 1.05 | 1.03 | 1.07 | 1.8×10−8 | 0.847 | 1.03 | 1.00 | 1.05 | 0.042 | 2 |
| WSCD2 | WSCD2 p.Thr113Ile | rs3764002 | 12 | 108,618,630 | C/T | 0.719 | 1.03 | 1.02 | 1.05 | 3.3×10−8 | 1.03 | 1.02 | 1.05 | 1.2×10−7 | 0.736 | 1.05 | 1.03 | 1.07 | 8.1×10−7 | 1 |
| ZZEF1 | ZZEF1 p.Ile402Val | rs781831 | 17 | 3,947,644 | C/T | 0.422 | 1.04 | 1.03 | 1.05 | 8.3×10−11 | 1.03 | 1.02 | 1.05 | 1.8×10−7 | 0.407 | 1.04 | 1.02 | 1.05 | 2.1×10−5 | 2 |
| MLX | MLX p.Gln139Arg | rs665268 | 17 | 40,722,029 | G/A | 0.294 | 1.04 | 1.02 | 1.05 | 2.0×10−8 | 1.03 | 1.02 | 1.04 | 1.1×10−5 | 0.280 | 1.04 | 1.02 | 1.06 | 5.2×10−6 | 2 |
| TTLL6 | TTLL6 p.Glu712Asp | rs2032844 | 17 | 46,847,364 | C/A | 0.754 | 1.04 | 1.02 | 1.06 | 1.2×10−7 | 1.03 | 1.01 | 1.04 | 0.00098 | 0.750 | 1.04 | 1.02 | 1.06 | 9.5×10−5 | 3 |
| C17orf58† | C17orf58 p.Ile92Val | rs9891146 | 17 | 65,988,049 | T/C | 0.277 | 1.04 | 1.02 | 1.06 | 1.3×10−7 | 1.02 | 1.00 | 1.04 | 0.00058 | 0.269 | 1.05 | 1.03 | 1.07 | 1.7×10−7 | 2 |
| ZHX3† | ZHX3 p.Asn310Ser | rs17265513 | 20 | 39,832,628 | C/T | 0.211 | 1.05 | 1.03 | 1.07 | 9.2×10−8 | 1.04 | 1.02 | 1.05 | 2.9×10−6 | 0.208 | 1.02 | 1.00 | 1.04 | 0.068 | N/A |
| PNPLA3 | PNPLA3 p.Ile148Met | rs738409 | 22 | 44,324,727 | G/C | 0.239 | 1.04 | 1.03 | 1.05 | 2.1×10−10 | 1.05 | 1.03 | 1.06 | 2.8×10−11 | 0.230 | 1.05 | 1.03 | 1.07 | 5.8×10−6 | 1 |
| PIM3 | PIM3 p.Val300Ala | rs4077129 | 22 | 50,356,693 | T/C | 0.276 | 1.04 | 1.02 | 1.05 | 1.9×10−7 | 1.04 | 1.02 | 1.06 | 3.5×10−8 | 0.280 | 1.04 | 1.02 | 1.06 | 8.7×10−5 | 3 |
Chr: chromosome. Pos: Position build 37. RAF: risk allele frequency. R: risk allele. O: other allele. BMI: body mass index. OR: odds ratio. L95: lower 95% confidence interval. U95: upper 95% confidence interval. GWAS: genome wide association studies.
Summary statistics from European ancestry specific meta-analyses of 48,286 cases and 250,671 controls. Fine-mapping group 1: signal is driven by coding variant, group 2: signal attributable to non-coding variants, and group 3: consistent with partial role for coding variants. p-values are based on the meta-analyses of discovery stage and fine-mapping studies as appropriate.
In addition to the 69 coding variant signals, we detected significant (p<5×10−8) and novel T2D-associations for 20 non-coding variants (at 15 loci) that were also assayed on the exome-array (Supplementary Table 7). Three of these (POC5, LPL, and BPTF) overlap with novel coding signals reported here.
Contribution of low-frequency and rare coding variation to T2D susceptibility
Despite increased power and good coverage of low-frequency variants on the exome-array12, 35 of the 40 distinct coding variant association signals were common, with modest effects (allelic ORs 1.02–1.36) (Supplementary Fig. 3, Supplementary Table 3). The five signals attributable to lower-frequency variants were also of modest effect (allelic ORs 1.09–1.29) (Supplementary Fig. 3). Two of the lower-frequency variant signals were novel, and in both, the minor allele was protective against T2D: FAM63A p.Tyr95Asn (rs140386498, MAF=1.2%, OR= 0.82 [0.77–0.88], pEUR=5.8×10−8) and ANKH p.Arg187Gln (rs146886108, MAF=0.4%, OR=0.78 [0.69–0.87], pEUR=2.0×10−7). Both variants were very rare or monomorphic in non-European descent individuals.
In Fuchsberger et al.12, we highlighted a set of 100 low-frequency coding variants with allelic ORs between 1.10 and 2.66, which despite relatively large estimates for liability-scale variance explained, had not reached significance. In this expanded analysis, only five of these variants, including the two novel associations at FAM63A p.Tyr95Asn and ANKH p.Arg187Gln, attained significance. More precise effect-size estimation in the larger sample size indicates that OR estimates in the earlier study were subject to a substantial upwards bias (Supplementary Fig. 3).
To detect additional rare variant association signals, we performed gene-based analyses (burden and SKAT17) using previously-defined “strict” and “broad” masks, filtered for annotation and MAF12,18 (Methods). We identified gene-based associations with T2D susceptibility (p<2.5×10−6, Bonferroni correction for 20,000 genes) for FAM63A (10 variants, combined MAF=1.9%, pEUR=3.1×10−9) and PAM (17 variants, combined MAF=4.7%, pTE=8.2×10−9). On conditional analysis (Supplementary Table 8), the gene-based signal at FAM63A was entirely attributable to the low-frequency p.Tyr95Asn allele described earlier (conditional p=0.26EUR). The gene-based signal for PAM was also driven by a single low-frequency variant (p.Asp563Gly; conditional pTE=0.15). A second, previously-described, low-frequency variant, PAM p.Ser539Trp19, is not represented on the exome-array, and did not contribute to these analyses.
Fine-mapping of coding variant association signals with T2D susceptibility
These analyses identified 40 distinct coding variant associations with T2D, but this information is not sufficient to determine that these variants are causal for disease. To assess the role of these coding variants given regional genetic variation, we fine-mapped these association signals using a meta-analysis of 50,160 T2D cases and 465,272 controls (European-descent only; partially overlapping with the discovery samples), which we aggregated from 24 GWAS. Each component GWAS was imputed using appropriate high-density reference panels (for most, the Haplotype Reference Consortium20; Methods, Supplementary Table 9). Before fine-mapping, distinct association signals were delineated using approximate conditional analyses (Methods, Supplementary Table 5). We included 37 of the 40 identified coding variants in this fine-mapping analysis, excluding three (those at the MHC, PAX4, and ZHX3) that were, for various reasons (see Methods), not amenable to fine-mapping in the GWAS data.
For each of these 37 signals, we first constructed “functionally-unweighted” credible variant sets, which collectively account for 99% of the posterior probability of association (PPA), based exclusively on the meta-analysis summary statistics21 (Methods, Supplementary Table 10). For each signal, we calculated the proportion of PPA attributable to coding variants (missense, in-frame indel, and splice region variants; Figure 1, Supplementary Fig. 4 and 5). There were only two signals at which coding variants accounted for ≥80% of PPA: HNF4A p.Thr139Ile (rs1800961, PPA>0.999) and RREB1 p. Asp1171Asn (rs9379084, PPA=0.920). However, at other signals, including those for GCKR p.Pro446Leu and SLC30A8 p.Arg276Trp, for which robust empirical evidence has established a causal role22,23, genetic support for coding variant causation was weak. This is because coding variants were typically in high LD (r2>0.9) with large numbers of non-coding variants, such that the PPA was distributed across many sites with broadly equivalent evidence for association.
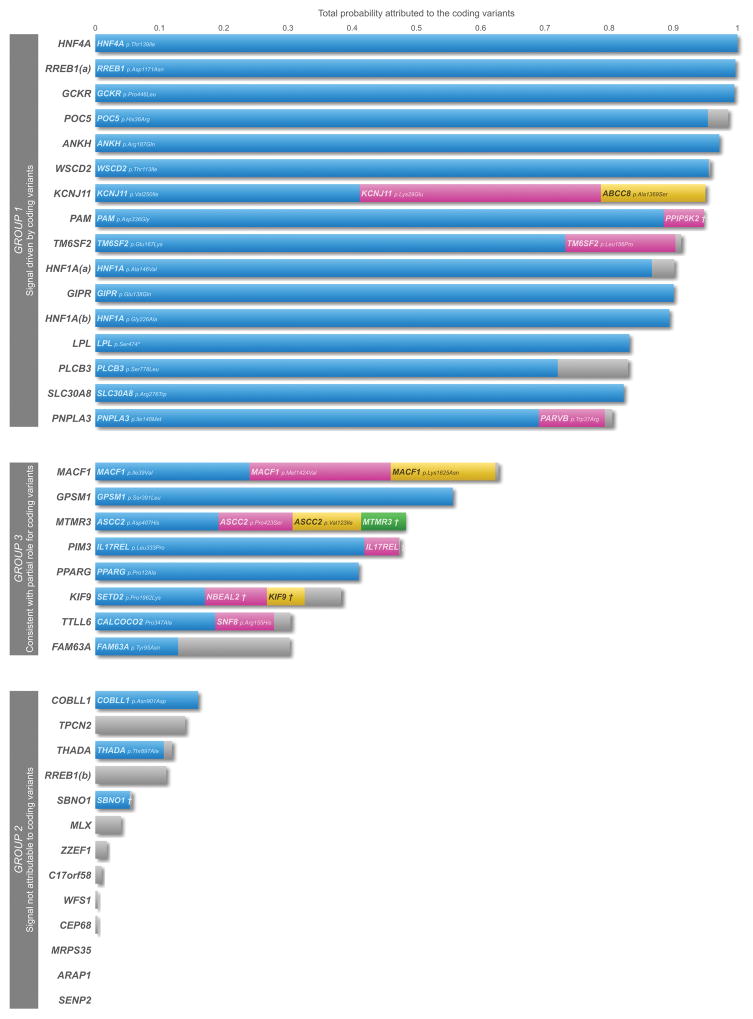
Figure 1. Posterior probabilities for coding variants across loci with annotation-informed priors.
Fine-mapping of 37 distinct association signals was performed using European ancestry GWAS meta-analysis including 50,160 T2D cases and 465,272 controls. For each signal, we constructed a credible set of variants accounting for 99% of the posterior probability of driving the association, incorporating an “annotation informed” prior model of causality which “boosts” the posterior probability of driving the association signal that is attributed to coding variants. Each bar represents a signal with the total probability attributed to the coding variants within the 99% credible set plotted on the y-axis. When the probability (bar) is split across multiple coding variants (at least 0.05 probability attributed to a variant) at a particular locus, these are indicated by blue, pink, yellow, and green colours. The combined probability of the remaining coding variants is highlighted in grey. RREB1(a): RREB1 p. Asp1171Asn; RREB1(b): RREB1 p.Ser1499Tyr; HNF1A(a): HNF1A p.Ala146Val; HNF1A(b): HNF1A p.Ile75Leu; PPIP5K2† : PPIP5K2 p.Ser1207Gly; MTMR3†: MTMR3 p.Asn960Ser; IL17REL†: IL17REL p.Gly70Arg; NBEAL2†: NBEAL2 p.Arg511Gly, KIF9†: KIF9 p.Arg638Trp.
These functionally-unweighted sets are based on genetic fine-mapping data alone, and do not account for the disproportionate representation of coding variants amongst GWAS associations for complex traits8,9. To accommodate this information, we extended the fine-mapping analyses by incorporating an “annotation-informed prior” model of causality. We derived priors from estimates of the enrichment of association signals by sequence annotation from analyses conducted by deCODE across 96 quantitative and 123 binary phenotypes16 (Methods). This model “boosts” the prior, and hence the posterior probabilities (we use ‘aiPPA’ to denote annotation-informed PPAs) of coding variants. It also takes account (in a tissue-non-specific manner) of the GWAS enrichment of variants within enhancer elements (as assayed through DNase I hypersensitivity) when compared to non-coding variants mapping elsewhere. The annotation-informed model generated smaller 99% credible sets across most signals, corresponding to fine-mapping at higher resolution (Supplementary Table 10). As expected, the contribution of coding variants was increased under the annotation-informed model. At these 37 association signals, we distinguished three broad patterns of causal relationships between coding variants and T2D risk.
Group 1: T2D association signal is driven by coding variants
At 16 of the 37 distinct signals, coding variation accounted for >80% of the aiPPA (Fig. 1, Table 2, Supplementary Table 10). This was attributable to a single coding variant at 12 signals and multiple coding variants at four. Reassuringly, group 1 signals confirmed coding variant causation for several loci (GCKR, PAM, SLC30A8, KCNJ11-ABCC8) at which functional studies have established strong mechanistic links to T2D pathogenesis (Table 2). T2D association signals at the 12 remaining signals (Fig. 1, Supplementary Table 10) had not previously been shown to be driven by coding variation, but our fine-mapping analyses pointed to causal coding variants with high aiPPA values: these included HNF4A, RREB1 (p. Asp1171Asn), ANKH, WSCD2, POC5, TM6SF2, HNF1A (p.Ala146Val; p.Ile75Leu), GIPR, LPL, PLCB3, and PNPLA3 (Table 2). At several of these, independent evidence corroborates the causal role of the genes harbouring the associated coding variants. For example, rare coding mutations at HNF1A and HNF4A are causal for monogenic, early-onset forms of diabetes24; and at TM6SF2 and PNPLA3, the associated coding variants are implicated in the development of non-alcoholic fatty liver disease (NAFLD)25,26.
Table 2.
Posterior probabilities for coding variants within 99% credible set across loci with annotation-informed and functionally-unweighted prior based on fine-mapping analysis performed using 50,160 T2D cases and 465,272 controls of European ancestry.
| Locus | Variant | rs ID | Chr | Position | Posterior probability | Cumulative posterior probability attributed to coding variants | ||
|---|---|---|---|---|---|---|---|---|
| PPA | aiPPA | PPA | aiPPA | |||||
| MACF1 | MACF1 p.Ile39Val | rs16826069 | 1 | 39,797,055 | 0.012 | 0.240 | 0.032 | 0.628 |
| MACF1 p.Met1424Val | rs2296172 | 1 | 39,835,817 | 0.011 | 0.224 | |||
| MACF1 p.Lys1625Asn | rs41270807 | 1 | 39,801,815 | 0.008 | 0.163 | |||
| FAM63A | FAM63A p.Tyr95Asn | rs140386498 | 1 | 150,972,959 | 0.005 | 0.129 | 0.012 | 0.303 |
| GCKR | GCKR p. Pro 446Leu | rs1260326 | 2 | 27,730,940 | 0.773 | 0.995 | 0.773 | 0.995 |
| THADA | THADA p.Cys845Tyr | rs35720761 | 2 | 43,519,977 | <0.001 | 0.011 | 0.003 | 0.120 |
| THADA p.Thr897Ala | rs7578597 | 2 | 43,732,823 | 0.003 | 0.107 | |||
| CEP68 | CEP68 p.Gly74Ser | rs7572857 | 2 | 65,296,798 | <0.001 | 0.004 | <0.001 | 0.004 |
| GRB14 | COBLL1 p.Asn901Asp | rs7607980 | 2 | 165,551,201 | 0.006 | 0.160 | 0.006 | 0.160 |
| PPARG | PPARG p.Pro12Ala | rs1801282 | 3 | 12,393,125 | 0.023 | 0.410 | 0.024 | 0.410 |
| KIF9 | SETD2 p.Pro1962Lys | rs4082155 | 3 | 47,125,385 | 0.008 | 0.171 | 0.018 | 0.384 |
| NBEAL2 p.Arg511Gly | rs11720139 | 3 | 47,036,756 | 0.005 | 0.097 | |||
| KIF9 p.Arg638Trp | rs2276853 | 3 | 47,282,303 | 0.003 | 0.059 | |||
| IGF2BP2 | SENP2 p.Thr291Lys | rs6762208 | 3 | 185,331,165 | <0.001 | <0.001 | <0.001 | <0.001 |
| WFS1 | WFS1 p.Val333Ile | rs1801212 | 4 | 6,302,519 | <0.001 | 0.001 | <0.001 | 0.004 |
| ANKH | ANKH p.Arg187Gln | rs146886108 | 5 | 14,751,305 | 0.459 | 0.972 | 0.447 | 0.972 |
| POC5 | POC5 p.His36Arg | rs2307111 | 5 | 75,003,678 | 0.697 | 0.954 | 0.702 | 0.986 |
| PAM-PPIP5K2 | PAM p.Asp336Gly | rs35658696 | 5 | 102,338,811 | 0.288 | 0.885 | 0.309 | 0.947 |
| PPIP5K2 p.Ser1207Gly | rs36046591 | 5 | 102,537,285 | 0.020 | 0.063 | |||
| RREB1 p.Asp1171Asn | RREB1 p.Asp1171Asn | rs9379084 | 6 | 7,231,843 | 0.920 | 0.997 | 0.920 | 0.997 |
| RREB1 p.Ser1499Tyr | RREB1 p.Ser1499Tyr | rs35742417 | 6 | 7,247,344 | <0.001 | 0.013 | 0.005 | 0.111 |
| LPL | LPL p.Ser474* | rs328 | 8 | 19,819,724 | 0.023 | 0.832 | 0.023 | 0.832 |
| SLC30A8 | SLC30A8 p.Arg276Trp | rs13266634 | 8 | 118,184,783 | 0.295 | 0.823 | 0.295 | 0.823 |
| GPSM1 | GPSM1 p.Ser391Leu | rs60980157 | 9 | 139,235,415 | 0.031 | 0.557 | 0.031 | 0.557 |
| KCNJ11-ABCC8 | KCNJ11 p.Val250Ile | rs5215 | 11 | 17,408,630 | 0.208 | 0.412 | 0.481 | 0.951 |
| KCNJ11 p.Lys29Glu | rs5219 | 11 | 17,409,572 | 0.190 | 0.376 | |||
| ABCC8 p.Ala1369Ser | rs757110 | 11 | 17,418,477 | 0.083 | 0.163 | |||
| PLCB3 | PLCB3 p.Ser778Leu | rs35169799 | 11 | 64,031,241 | 0.113 | 0.720 | 0.130 | 0.830 |
| TPCN2 | TPCN2 p.Val219Ile | rs72928978 | 11 | 68,831,364 | <0.001 | 0.004 | 0.006 | 0.140 |
| CENTD2 | ARAP1 p.Gln802Glu | rs56200889 | 11 | 72,408,055 | <0.001 | <0.001 | <0.001 | <0.001 |
| KLHDC5 | MRPS35 p.Gly43Arg | rs1127787 | 12 | 27,867,727 | <0.001 | <0.001 | <0.001 | <0.001 |
| WSCD2 | WSCD2 p.Thr113Ile | rs3764002 | 12 | 108,618,630 | 0.281 | 0.955 | 0.282 | 0.958 |
| HNF1A p.Ile75Leu | HNF1A_Gly226Ala | rs56348580 | 12 | 121,432,117 | 0.358 | 0.894 | 0.358 | 0.894 |
| HNF1A p.Ile75Leu | rs1169288 | 12 | 121,416,650 | <0.001 | <0.001 | |||
| HNF1A p.Ala146Val | HNF1A p.Ala146Val | rs1800574 | 12 | 121,416,864 | 0.269 | 0.867 | 0.280 | 0.902 |
| MPHOSPH9 | SBNO1 p.Ser729Asn | rs1060105 | 12 | 123,806,219 | 0.002 | 0.054 | 0.002 | 0.057 |
| ZZEF1 | ZZEF1 p.Ile402Val | rs781831 | 17 | 3,947,644 | <0.001 | 0.001 | <0.001 | 0.018 |
| MLX | MLX p.Gln139Arg | rs665268 | 17 | 40,722,029 | 0.002 | 0.038 | 0.002 | 0.039 |
| TTLL6 | TTLL6 p.Glu712Asp | rs2032844 | 17 | 46,847,364 | <0.001 | <0.001 | 0.016 | 0.305 |
| CALCOCO2 p.Pro347Ala | rs10278 | 17 | 46,939,658 | 0.0100 | 0.187 | |||
| SNF8 p.Arg155His | rs57901004 | 17 | 47,011,897 | 0.005 | 0.092 | |||
| C17orf58 | C17orf58 p.Ile92Val | rs9891146 | 17 | 65,988,049 | <0.001 | 0.009 | <0.001 | 0.009 |
| CILP2 | TM6SF2 p.Glu167Lys | rs58542926 | 19 | 19,379,549 | 0.211 | 0.732 | 0.263 | 0.913 |
| TM6SF2 p.Leu156Pro | rs187429064 | 19 | 19,380,513 | 0.049 | 0.172 | |||
| GIPR | GIPR p.Glu318Gln | rs1800437 | 19 | 46,181,392 | 0.169 | 0.901 | 0.169 | 0.901 |
| ZHX3 | ZHX3 p.Asn310Ser | rs17265513 | 20 | 39,832,628 | <0.001 | 0.003 | 0.003 | 0.110 |
| HNF4A | HNF4A p.Thr139Ile | rs1800961 | 20 | 43,042,364 | 1.000 | 1.000 | 1.00 | 1.000 |
| MTMR3-ASCC2 | ASCC2 p.Asp407His | rs28265 | 22 | 30,200,761 | 0.011 | 0.192 | 0.028 | 0.481 |
| ASCC2 p.Pro423Ser | rs36571 | 22 | 30,200,713 | 0.007 | 0.116 | |||
| ASCC2 p.Val123Ile | rs11549795 | 22 | 30,221,120 | 0.006 | 0.107 | |||
| MTMR3 p.Asn960Ser | rs41278853 | 22 | 30,416,527 | 0.004 | 0.065 | |||
| PNPLA3 | PNPLA3 p.Ile148Met | rs738409 | 22 | 44,324,727 | 0.112 | 0.691 | 0.130 | 0.806 |
| PARVB p.Trp37Arg | rs1007863 | 22 | 44,395,451 | 0.017 | 0.103 | |||
| PIM3 | IL17REL p.Leu333Pro | rs5771069 | 22 | 50,435,480 | 0.041 | 0.419 | 0.047 | 0.475 |
| IL17REL p.Gly70Arg | rs9617090 | 22 | 50,439,194 | 0.005 | 0.054 | |||
| PIM3 p.Val300Ala | rs4077129 | 22 | 50,356,693 | <0.001 | 0.002 | |||
Chr: chromosome. Pos: Position build 37. PPA: functionally-unweighted prior; aiPPA: annotation informed prior. Index coding variants are highlighted in bold.
The use of priors to capture the enrichment of coding variants seems a reasonable model, genome-wide. However, at any given locus, strong priors (especially for PTVs) might elevate to apparent causality, variants that would have been excluded from a causal role on the basis of genetic fine-mapping alone. Comparison of the annotation-informed and functionally-unweighted credible sets for group 1 signals indicated that this scenario was unlikely. For 11 of the 16 (GCKR, PAM, KCNJ11-ABCC8, HNF4A, RREB1 [p.Asp1171Asn], ANKH, POC5, TM6SF2, HNF1A [p.Ala146Val], PLCB3, PNPLA3), the coding variant had the highest PPA in the fine-mapping analysis (Table 2) even under the functionally-unweighted model. At SLC30A8, WSCD2, and GIPR, the coding variants had similar PPAs to the lead non-coding SNPs under the functionally-unweighted prior (Table 2). At these 14 signals therefore, coding variants have either greater or equivalent PPA to the best flanking non-coding SNPs under the functionally-unweighted model, but receive a boost in PPA after incorporating the annotation weights.
The situation is less clear at LPL. Here, fine-mapping resolution is poor under the functionally-unweighted prior, and the coding variant sits on an extended haplotype in strong LD with non-coding variants, some with higher PPA, such as rs74855321 (PPA=0.048) (compared to LPL p.Ser474* [rs328, PPA=0.023]). However, LPL p.Ser474* is annotated as a PTV, and benefits from a substantially-increased prior that boosts its annotation-informed ranking (Table 2). Ultimately, decisions regarding the causal role of any such variant must rest on the amalgamation of evidence from diverse sources including detailed functional evaluation of the coding variants, and of other variants with which they are in LD.
Group 2: T2D association signals are not attributable to coding variants
At 13 of the 37 distinct signals, coding variation accounted for <20% of the PPA, even after applying the annotation-informed prior model. These signals are likely to be driven by local non-coding variation and mediated through regulatory mechanisms. Five of these signals (TPCN2, MLX, ZZEF1, C17orf58, and CEP68) represent novel T2D-association signals identified in the exome-focused analysis. Given the exome-array discoveries, it would have been natural to consider the named genes at these, and other loci in this group, as candidates for mediation of their respective association signals. However, the fine-mapping analyses indicate that these coding variants do not provide useful mechanistic inference given low aiPPA (Fig. 1, Table 2).
The coding variant association at the CENTD2 (ARAP1) locus is a case-in-point. The association with the p.Gln802Glu variant in ARAP1 (rs56200889, pTE=4.8×10−8 but aiPPA<0.001) is seen in the fine-mapping analysis to be secondary to a substantially stronger non-coding association signal involving a cluster of variants including rs11603334 (pTE=9.5×10−18, aiPPA=0.0692) and rs1552224 (pTE=2.5×10−17, aiPPA=0.0941). The identity of the effector transcript at this locus has been the subject of detailed investigation, and some early studies used islet expression data to promote ARAP127. However, a more recent study integrating human islet genomics and murine gene knockout data establishes STARD10 as the gene mediating the GWAS signal, consistent with the reassignment of the ARAP1 coding variant association as irrelevant to causal inference28.
Whilst, at these loci, the coding variant associations represent “false leads”, this does not necessarily exclude the genes concerned from a causal role. At WFS1 for example, coding variants too rare to be visible to the array-based analyses we performed, and statistically independent of the common p.Val333Ile variant we detected, cause an early-onset form of diabetes that renders WFS1 the strongest local candidate for T2D predisposition.
Group 3: Fine-mapping data consistent with partial role for coding variants
At eight of the 37 distinct signals, the aiPPA attributable to coding variation lay between 20% and 80%. At these signals, the evidence is consistent with “partial” contributions from coding variants, although the precise inference is likely to be locus-specific, dependent on subtle variations in LD, imputation accuracy, and the extent to which global priors accurately represent the functional impact of the specific variants concerned.
This group includes PPARG for which independent evidence corroborates the causal role of this specific effector transcript with respect to T2D-risk. PPARG encodes the target of antidiabetic thiazolidinedione drugs and harbours very rare coding variants causal for lipodystrophy and insulin resistance, conditions highly-relevant to T2D. The common variant association signal at this locus has generally been attributed to the p.Pro12Ala coding variant (rs1801282) although empirical evidence that this variant influences PPARG function is scant29–31. In the functionally-unweighted analysis, p.Pro12Ala had an unimpressive PPA (0.0238); after including annotation-informed priors, the same variant emerged with the highest aiPPA (0.410), although the 99% credible set included 19 non-coding variants, spanning 67kb (Supplementary Table 10). These credible set variants included rs4684847 (aiPPA=0.0089), at which the T2D-associated allele has been reported to impact PPARG2 expression and insulin sensitivity by altering binding of the homeobox transcription factor PRRX132. These data are consistent with a model whereby regulatory variants contribute to altered PPARG activity in combination with, or potentially to the exclusion of, p.Pro12Ala. Future improvements in functional annotation for regulatory variants (gathered from relevant tissues and cell types) should provide increasingly granular priors that allow fine-tuned assignment of causality at loci such as this.
Functional impact of coding alleles
In other contexts, the functional impact of coding alleles is correlated with: (i) variant-specific features, including measures of conservation and predicted impact on protein structure; and (ii) gene-specific features such as extreme selective constraints as quantified by the intolerance to functional variation33. To determine whether similar measures could capture information pertinent to T2D causation, we compared coding variants falling into the different fine-mapping groups for a variety of measures including MAF, Combined Annotation Dependent Depletion (CADD) score34, and loss-of-function (LoF)-intolerance metric, pLI33 (Methods, Fig. 2). Variants from group 1 had significantly higher CADD-scores than those in group 2 (Kolmogorov-Smirnov p=0.0031). Except for the variants at KCNJ11-ABCC8 and GCKR, all group 1 coding variants considered likely to be driving T2D association signals had CADD-score ≥20. On this basis, we predict that the East-Asian specific coding variant at PAX4, for which the fine-mapping data were not informative, is also likely causal for T2D.
Figure 2. Plot of measures of variant-specific and gene-specific features of distinct coding signals to access the functional impact of coding alleles.
Each point represents a coding variant with the minor allele frequency plotted on the x-axis and the Combined Annotation Dependent Depletion score (CADD-score) plotted on the y-axis. Size of each point varies with the measure of intolerance of the gene to loss of function variants (pLI) and the colour represents the fine-mapping group each variant is assigned to. Group 1: signal is driven by coding variant. Group 2: signal attributable to non-coding variants. Group 3: consistent with partial role for coding variants. Group 4: Unclassified category; includes PAX4, ZHX3, and signal at TCF19 within the MHC region where we did not perform fine-mapping. Inset: plot shows the distribution of CADD-score between different groups. The plot is a combination of violin plots and box plots; width of each violin indicates frequency at the corresponding CADD-score and box plots show the median and the 25% and 75% quantiles. P value indicates significance from two-sample Kolmogorov-Smirnov test.
T2D loci and physiological classification
The development of T2D involves dysfunction of multiple mechanisms. Systematic analysis of the physiological effects of known T2D-risk alleles has improved understanding of the mechanisms through which they exert their primary impact on disease risk35. We obtained association summary statistics for diverse metabolic traits (and other outcomes) for 94 T2D-associated index variants. These 94 were restricted to sites represented on the exome-array and included the 40 coding signals plus 54 distinct non-coding signals (12 novel and 42 previously-reported non-coding GWAS lead SNPs). We applied clustering techniques (Methods) to generate multi-trait association patterns, allocating 71 of the 94 loci to one of three main physiological categories (Supplementary Figs. 6, Supplementary Table 11). The first category, comprising nine T2D-risk loci with strong BMI and dyslipidemia associations, included three of the novel coding signals: PNPLA3, POC5 and BPTF. The T2D associations at both POC5 and BPTF were substantially attenuated (>2-fold decrease in −log10p) after adjusting for BMI (Table 1, Supplementary Table 3, Supplementary Fig. 7), indicating that their impact on T2D-risk is likely mediated by a primary effect on adiposity. PNPLA3 and POC5 are established NAFLD25 and BMI6 loci, respectively. The second category featured 39 loci at which multi-trait profiles indicated a primary effect on insulin secretion. This set included four of the novel coding variant signals (ANKH, ZZEF1, TTLL6, ZHX3). The third category encompassed 23 loci with primary effects on insulin action, including signals at the KIF9, PLCB3, CEP68, TPCN2, FAM63A, and PIM3 loci. For most variants in this category, the T2D-risk allele was associated with lower BMI, and T2D association signals were more pronounced after adjustment for BMI. At a subset of these loci, including KIF9 and PLCB3, T2D-risk alleles were associated with higher waist-hip ratio and lower body fat percentage, indicating that the mechanism of action likely reflects limitations in storage capacity of peripheral adipose tissue36.
DISCUSSION
The present study adds to mounting evidence constraining the contribution of lower-frequency variants to T2D-risk. Although the exome-array interrogates only a subset of the universe of coding variants, it captures the majority of low-frequency coding variants in European populations. The substantial increase in sample size in the present study over our previous effort12 (effective sample sizes of 228,825 and 82,758, respectively), provides more robust evaluation of the effect size distribution in this low-frequency variant range, and indicates that previous analyses are likely, if anything, to have overestimated the contribution of low-frequency variants to T2D-risk.
The present study is less informative regarding rare variants. These are sparsely captured on the exome-array. In addition, the combination of greater regional diversity in rare allele distribution and the enormous sample sizes required to detect rare variant associations (likely to require meta-analysis of data from diverse populations) acts against their identification. Our complementary genome and exome sequence analyses have thus far failed to register strong evidence for a substantial rare variant component to T2D-risk12. It is therefore highly unlikely that rare variants missed in our analyses are causal for any of the common or low-frequency variant associations we have detected and fine-mapped. On the other hand, it is probable that rare coding alleles, with associations that are distinct from the common variant signals we have examined and detected only through sequence based analyses, will provide additional clues to the most likely effector transcripts at some of these signals (WFS1 provides one such example).
Once a coding variant association is detected, it is natural to assume a causal connection between that variant, the gene in which it sits, and the phenotype of interest. Whilst such assignments may be robust for many rare protein-truncating alleles, we demonstrate that this implicit assumption is often inaccurate, particularly for associations attributable to common, missense variants. A third of the coding variant associations we detected were, when assessed in the context of regional LD, highly unlikely to be causal. At these loci, the genes within which they reside are consequently deprived of their implied connection to disease risk, and attention redirected towards nearby non-coding variants and their impact on regional gene expression. As a group, coding variants we assign as causal are predicted to have a more deleterious impact on gene function than those that we exonerate, but, as in other settings, coding annotation methods lack both sensitivity and specificity. It is worth emphasising that empirical evidence that the associated coding allele is “functional” (i.e. can be shown to influence cognate gene function in some experimental assay) provides limited reassurance that the coding variant is responsible for the T2D association, unless that specific perturbation of gene function can itself be plausibly linked to the disease phenotype.
Our fine-mapping analyses make use of the observation that coding variants are globally enriched across GWAS signals8,9,16 with greater prior probability of causality assigned to those with more severe impact on biological function. We assigned diminished priors to non-coding variants, with lowest support for those mapping outside of DNase I hypersensitive sites. The extent to which our findings corroborate previous assignments of causality (often substantiated by detailed, disease-appropriate functional assessment and other orthogonal evidence) suggests that even these sparse annotations provide valuable information to guide target validation. Nevertheless, there are inevitable limits to the extrapolation of these ‘broad-brush’ genome-wide enrichments to individual loci: improvements in functional annotation for both coding and regulatory variants, particularly when gathered from trait-relevant tissues and cell types, should provide more granular, trait-specific priors to fine-tune assignment of causality within associated regions. These will motivate target validation efforts that benefit from synthesis of both coding and regulatory mechanisms of gene perturbation. It also needs to be acknowledged that, without whole genome sequencing data on sample sizes comparable to those we have examined here, imperfections arising from the imputation may confound fine-mapping precision at some loci, and that robust inference will inevitably depend on integration of diverse sources of genetic, genomic and functional data.
The term “smoking gun” has often been used to describe the potential of functional coding variants to provide causal inference with respect to pathogenetic mechanisms37. This study provides a timely reminder that, even when a suspect with a smoking gun is found at the scene of a crime, it should not be assumed that they fired the fatal bullet.
ONLINE METHODS
Ethics statement
All human research was approved by the relevant institutional review boards, and conducted according to the Declaration of Helsinki. All participants provided written informed consent.
Derivation of significance thresholds
We considered five categories of annotation16 of variants on the exome array in order of decreasing effect on biological function: (1) PTVs (stop-gain and stop-loss, frameshift indel, donor and acceptor splice-site, and initiator codon variants, n1=8,388); (2) moderate-impact variants (missense, in-frame indel, and splice region variants, n2=216,114); (3) low-impact variants (synonymous, 3′ and 5′ UTR, and upstream and downstream variants, n3=8,829); (4) other variants mapping to DNase I hypersensitive sites (DHS) in any of 217 cell types8 (DHS, n4=3,561); and (5) other variants not mapping to DHS (n5=10,578). To account for the greater prior probability of causality for variants with greater effect on biological function, we determined a weighted Bonferroni-corrected significance threshold on the basis of reported enrichment16, denoted wi, in each annotation category, i: w1=165; w2=33; w3=3; w4=1.5; w5=0.5. For coding variants (annotation categories 1 and 2):
We note that this threshold is similar to a simple Bonferroni correction for the total number of coding variants on the array, which would yield:
For non-coding variants (annotation categories 3, 4 and 5) the weighted Bonferroni-corrected significance threshold is:
DISCOVERY: Exome-array study-level analyses
Within each study, genotype calling and quality control were undertaken according to protocols developed by the UK Exome Chip Consortium or the CHARGE central calling effort38 (Supplementary Table 1). Within each study, variants were then excluded for the following reasons: (i) not mapping to autosomes or X chromosome; (ii) multi-allelic and/or insertion-deletion; (iii) monomorphic; (iv) call rate <99%; or (v) exact p<10−4 for deviation from Hardy-Weinberg equilibrium (autosomes only).
We tested association of T2D with each variant in a linear mixed model, implemented in RareMetalWorker17, using a genetic relationship matrix (GRM) to account for population structure and relatedness. For participants from family-based studies, known relationships were incorporated directly in the GRM. For founders and participants from population-based studies, the GRM was constructed from pair-wise identity by descent (IBD) estimates based on LD pruned (r2<0.05) autosomal variants with MAF≥1% (across cases and controls combined), after exclusion of those in high LD and complex regions39,40, and those mapping to established T2D loci. We considered additive, dominant, and recessive models for the effect of the minor allele, adjusted for age and sex (where appropriate) and additional study-specific covariates (Supplementary Table 2). Analyses were also performed with and without adjustment for BMI (where available Supplementary Table 2).
For single-variant association analyses, variants with minor allele count ≤10 in cases and controls combined were excluded. Association summary statistics for each analysis were corrected for residual inflation by means of genomic control41, calculated after excluding variants mapping to established T2D susceptibility loci. For gene-based analyses, we made no variant exclusions on the basis of minor allele count.
DISCOVERY: Exome-sequence analyses
We used summary statistics of T2D association from analyses conducted on 8,321 T2D cases and 8,421 controls across different ancestries, all genotyped using exome sequencing. Details of samples included, sequencing, and quality control are described elsewhere12,15 (http://www.type2diabetesgenetics.org/). Samples were subdivided into 15 sub-groups according to ancestry and study of origin. Each sub-group was analysed independently, with sub-group specific principal components and genetic relatedness matrices. Association tests were performed with both a linear mixed model, as implemented in EMMAX42, using covariates for sequencing batch, and the Firth test, using covariates for principal components and sequencing batch. Related samples were excluded from the Firth analysis but maintained in the linear mixed model analysis. Variants were then filtered from each sub-group analysis, according to call rate, differential case-control missing-ness, or deviation from Hardy-Weinberg equilibrium (as computed separately for each sub-group). Association statistics were then combined via a fixed-effects inverse-variance weighted meta-analysis, at both the level of ancestry as well as across all samples. P-values were taken from the linear mixed model analysis, while effect sizes estimates were taken from the Firth analysis. Analyses were performed with and without adjustment for BMI. From exome sequence summary statistics, we extracted variants passing quality control and present on the exome array.
DISCOVERY: GWAS analyses
The UK Biobank is a large detailed prospective study of more than 500,000 participants aged 40–69 years when recruited in 2006–201013. Prevalent T2D status was defined using self-reported medical history and medication in UK Biobank participants43. Participants were genotyped with the UK Biobank Axiom Array or UK BiLEVE Axiom Array, and quality control and population structure analyses were performed centrally at UK Biobank. We defined a subset of “white European” ancestry samples (n=120,286) as those who both self-identified as white British and were confirmed as ancestrally “Caucasian” from the first two axes of genetic variation from principal components analysis. Imputation was also performed centrally at UK Biobank for the autosomes only, up to a merged reference panel from the 1000 Genomes Project (multi-ethnic, phase 3, October 2014 release)44 and the UK10K Project9. We used SNPTESTv2.545 to test for association of T2D with each SNP in a logistic regression framework under an additive model, and after adjustment for age, sex, six axes of genetic variation, and genotyping array as covariates. Analyses were performed with and without adjustment for BMI, after removing related individuals.
GERA is a large multi-ethnic population-based cohort, created for investigating the genetic and environmental basis of age-related diseases [dbGaP phs000674.p1]. T2D status is based on ICD-9 codes in linked electronic medical health records, with all other participants defined as controls. Participants have previously been genotyped using one of four custom arrays, which have been designed to maximise coverage of common and low-frequency variants in non-Hispanic white, East Asian, African American, and Latino ethnicities46,47. Methods for quality control have been described previously14. Each of the four genotyping arrays were imputed separately, up to the 1000 Genomes Project reference panel (autosomes, phase 3, October 2014 release; X chromosome, phase 1, March 2012 release) using IMPUTEv2.348,49. We used SNPTESTv2.545 to test for association of T2D with each SNP in a logistic regression framework under an additive model, and after adjustment for sex and nine axes of genetic variation from principal components analysis as covariates. BMI was not available for adjustment in GERA.
For UK Biobank and GERA, we extracted variants passing standard imputation quality control thresholds (IMPUTE info≥0.4)50 and present on the exome array. Association summary statistics under an additive model were corrected for residual inflation by means of genomic control41, calculated after excluding variants mapping to established T2D susceptibility loci: GERA (λ=1.097 for BMI unadjusted analysis) and UK Biobank (λ=1.043 for BMI unadjusted analysis, λ=1.056 for BMI adjusted analysis).
DISCOVERY: Single-variant meta-analysis
We aggregated association summary statistics under an additive model across studies, with and without adjustment for BMI, using METAL51: (i) effective sample size weighting of Z-scores to obtain p-values; and (ii) inverse variance weighting of log-odds ratios. For exome-array studies, allelic effect sizes and standard errors obtained from the RareMetalWorker linear mixed model were converted to the log-odds scale prior to meta-analysis to correct for case-control imbalance52.
The European-specific meta-analyses aggregated association summary statistics from a total of 48,286 cases and 250,671 controls from: (i) 33 exome-array studies of European ancestry; (ii) exome-array sequence from individuals of European ancestry; and (iii) GWAS from UK Biobank. Note that non-coding variants represented on the exome array were not available in exome sequence. The European-specific meta-analyses were corrected for residual inflation by means of genomic control41, calculated after excluding variants mapping to established T2D susceptibility loci: λ=1.091 for BMI unadjusted analysis and λ=1.080 for BMI adjusted analysis.
The trans-ethnic meta-analyses aggregated association summary statistics from a total of 81,412 cases and 370,832 controls across all studies (51 exome array studies, exome sequence, and GWAS from UK Biobank and GERA), irrespective of ancestry. Note that non-coding variants represented on the exome array were not available in exome sequence. The trans-ethnic meta-analyses were corrected for residual inflation by means of genomic control41, calculated after excluding variants mapping to established T2D susceptibility loci: λ=1.073 for BMI unadjusted analysis and λ=1.068 for BMI adjusted analysis. Heterogeneity in allelic effect sizes between exome-array studies contributing to the trans-ethnic meta-analysis was assessed by Cochran’s Q statistic53.
DISCOVERY: Detection of distinct association signals
Conditional analyses were undertaken to detect association signals by inclusion of index variants and/or tags for previously reported non-coding GWAS lead SNPs as covariates in the regression model at the study level. Within each exome-array study, approximate conditional analyses were undertaken under a linear mixed model using RareMetal17, which uses score statistics and the variance-covariance matrix from the RareMetalWorker single-variant analysis to estimate the correlation in effect size estimates between variants due to LD. Study-level allelic effect sizes and standard errors obtained from the approximate conditional analyses were converted to the log-odds scale to correct for case-control imbalance52. Within each GWAS, exact conditional analyses were performed under a logistic regression model using SNPTESTv2.545. GWAS variants passing standard imputation quality control thresholds (IMPUTE info≥0.4)50 and present on the exome array were extracted for meta-analysis.
Association summary statistics were aggregated across studies, with and without adjustment for BMI, using METAL51: (i) effective sample size weighting of Z-scores to obtain p-values; and (ii) inverse variance weighting of log-odds ratios.
We defined novel loci as mapping >500kb from a previously reported lead GWAS SNP. We performed conditional analyses where a novel signal mapped close to a known GWAS locus, and the lead GWAS SNP at that locus is present (or tagged) on the exome array (Supplementary Table 5).
DISCOVERY: Non-additive association models
For exome-array studies only, we aggregated association summary statistics under recessive and dominant models across studies, with and without adjustment for BMI, using METAL51: (i) effective sample size weighting of Z-scores to obtain p-values; and (ii) inverse variance weighting of log-odds ratios. Allelic effect sizes and standard errors obtained from the RareMetalWorker linear mixed model were converted to the log-odds scale prior to meta-analysis to correct for case-control imbalance52. The European-specific meta-analyses aggregated association summary statistics from a total of 41,066 cases and 136,024 controls from 33 exome-array studies of European ancestry. The European-specific meta-analyses were corrected for residual inflation by means of genomic control41, calculated after excluding variants mapping to established T2D susceptibility loci: λ=1.076 and λ=1.083 for BMI unadjusted analysis, under recessive and dominant models respectively, and λ=1.081 and λ=1.062 for BMI adjusted analysis, under recessive and dominant models respectively. The trans-ethnic meta-analyses aggregated association summary statistics from a total of 58,425 cases and 188,032 controls across all exome-array studies, irrespective of ancestry. The trans-ethnic meta-analyses were corrected for residual inflation by means of genomic control41, calculated after excluding variants mapping to established T2D susceptibility loci: λ=1.041 and λ=1.071 for BMI unadjusted analysis, under recessive and dominant models respectively, and λ=1.031 and λ=1.063 for BMI adjusted analysis, under recessive and dominant models respectively.
DISCOVERY: Gene-based meta-analyses
For exome-array studies only, we aggregated association summary statistics under an additive model across studies, with and without adjustment for BMI, using RareMetal17. This approach uses score statistics and the variance-covariance matrix from the RareMetalWorker single-variant analysis to estimate the correlation in effect size estimates between variants due to LD. We performed gene-based analyses using a burden test (assuming all variants have same direction of effect on T2D susceptibility) and SKAT (allowing variants to have different directions of effect on T2D susceptibility). We used two previously defined filters for annotation and MAF18 to define group files: (i) strict filter, including 44,666 variants; and (ii) broad filter, including all variants from the strict filter, and 97,187 additional variants.
We assessed the contribution of each variant to gene-based signals by performing approximate conditional analyses. We repeated RareMetal analyses for the gene, excluding each variant in turn from the group file, and compared the strength of the association signal.
Fine-mapping of coding variant association signals with T2D susceptibility
We defined a locus as mapping 500kb up- and down-stream of each index coding variant (Supplementary Table 5), excluding the MHC. Our fine-mapping analyses aggregated association summary statistics from 24 GWAS incorporating 50,160 T2D cases and 465,272 controls of European ancestry from the DIAGRAM Consortium (Supplementary Table 9). Each GWAS was imputed using miniMAC12 or IMPUTEv248,49 up to high-density reference panels: (i) 22 GWAS were imputed up to the Haplotype Reference Consortium20; (ii) the UK Biobank GWAS was imputed to a merged reference panel from the 1000 Genomes Project (multi-ethnic, phase 3, October 2014 release)44 and the UK10K Project9; and (iii) the deCODE GWAS was imputed up to the deCODE Icelandic population-specific reference panel based on whole-genome sequence data19. Association with T2D susceptibility was tested for each remaining variant using logistic regression, adjusting for age, sex, and study-specific covariates, under an additive genetic model. Analyses were performed with and without adjustment for BMI. For each study, variants with minor allele count<5 (in cases and controls combined) or those with imputation quality r2-hat<0.3 (miniMAC) or proper-info<0.4 (IMPUTE2) were removed. Association summary statistics for each analysis were corrected for residual inflation by means of genomic control41, calculated after excluding variants mapping to established T2D susceptibility loci.
We aggregated association summary statistics across studies, with and without adjustment for BMI, in a fixed-effects inverse variance weighted meta-analysis, using METAL51. The BMI unadjusted meta-analysis was corrected for residual inflation by means of genomic control (λ=1.012)41, calculated after excluding variants mapping to established T2D susceptibility loci. No adjustment was required for BMI adjusted meta-analysis (λ=0.994). From the meta-analysis, variants were extracted that were present on the HRC panel and reported in at least 50% of total effective sample size.
We included 37 of the 40 identified coding variants in fine-mapping analyses, excluding three that were not amenable to fine-mapping in the GWAS data sets: (i) the locus in the major histocompatibility complex because of the extended and complex structure of LD across the region, which complicates fine-mapping efforts; (ii) the East Asian specific PAX4 p.Arg190His (rs2233580) signal, since the variant was not present in European ancestry GWAS; and (iii) ZHX3 p.Asn310Ser (rs4077129) because the variant was only weakly associated with T2D in the GWAS data sets used for fine-mapping.
To delineate distinct association signals in four regions, we undertook approximate conditional analyses, implemented in GCTA54, to adjust for the index coding variants and non-coding lead GWAS SNPs: (i) RREB1 p. Asp1171Asn (rs9379084), p.Ser1499Tyr (rs35742417), and rs9505118; (ii) HNF1A p.Ile75Leu (rs1169288) and p.Ala146Val (rs1800574); (iii) GIPR p.Glu318Gln (rs1800437) and rs8108269; and (iv) HNF4A p.Thr139Ile (rs1800961) and rs4812831. We made use of summary statistics from the fixed-effects meta-analyses (BMI unadjusted for RREB1, HNF1A, and HNF4A, and BMI adjusted for GIPR as this signal was only seen in BMI adjusted analysis) and genotype data from 5,000 random individuals of European ancestry from the UK Biobank, as reference for LD between genetic variants across the region.
For each association signal, we first calculated an approximate Bayes’ factor55 in favour of association on the basis of allelic effect sizes and standard errors from the meta-analysis. Specifically, for the jth variant,
where βj and Vj denote the estimated allelic effect (log-OR) and corresponding variance from the meta-analysis. The parameter ω denotes the prior variance in allelic effects, taken here to be 0.0455.
We then calculated the posterior probability that the jth variant drives the association signal, given by
In this expression, ρj denotes the prior probability that the jth variant drives the association signal, and the summation in the denominator is over all variants across the locus. We considered two prior models: (i) functionally unweighted, for which ρj = 1 for all variants; and (ii) annotation informed, for which ρj is determined by the functional severity of the variant. For the annotation informed prior, we considered five categories of variation16, such that: (i) ρj = 165 for PTVs; (ii) ρj = 33 for moderate-impact variants; (iii) ρj = 3 for low-impact variants; (iv) ρj = 1.5 for other variants mapping to DHS; and (v) ρj = 0.5 for all other variants.
For each locus, the 99% credible set21 under each prior was then constructed by: (i) ranking all variants according to their posterior probability of driving the association signal; and (ii) including ranked variants until their cumulative posterior probability of driving the association attained or exceeded 0.99.
Functional impact of coding alleles
We used CADD34 to obtain scaled Combined Annotation Dependent Depletion score (CADD-score) for each of the 40 significantly associated coding variants. The CADD method objectively integrates a range of different annotation metrics into a single measure (CADD-score), providing an estimate of deleteriousness for all known variants and an overall rank for this metric across the genome. We obtained the estimates of the intolerance of a gene to harbouring loss-of-function variants (pLI) from the ExAC data set33. We used the Kolmogorov-Smirnov test to determine whether fine-mapping groups 1 and 2 have the same statistical distribution for each of these parameters.
T2D loci and physiological classification
To explore the different patterns of association between T2D and other anthropometric/metabolic/endocrine traits and diseases, we performed hierarchical clustering analysis. We obtained association summary statistics for a range of metabolic traits and other outcomes for 94 coding and non-coding variants that were significantly associated with T2D through collaboration or by querying publically available GWAS meta-analysis datasets. The z-score (allelic effect/SE) was aligned to the T2D-risk allele. We obtained the distance matrix amongst z-score of the loci/traits using the Euclidean measure and performed clustering using the complete agglomeration method. Clustering was visualised by constructing a dendogram and heatmap.
Supplementary Material
Acknowledgments
A full list of acknowledgments appears in the Supplementary Information. Part of this work was conducted using the UK Biobank Resource under Application Number 9161.
DISCLOSURES
Jose C Florez has received consulting honoraria from Merck and from Boehringer-Ingelheim. Daniel I Chasman received funding for exome chip genotyping in the WGHS from Amgen. Oscar H Franco works in ErasmusAGE, a center for aging research across the life course funded by Nestlé Nutrition (Nestec Ltd.), Metagenics Inc., and AXA. Nestlé Nutrition (Nestec Ltd.), Metagenics Inc., and AXA had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review or approval of the manuscript. Erik Ingelsson is an advisor and consultant for Precision Wellness, Inc., and advisor for Cellink for work unrelated to the present project. Bruce M Psaty serves on the DSMB for a clinical trial funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. Inês Barroso and spouse own stock in GlaxoSmithKline and Incyte Corporation. Timothy Frayling has consulted for Boeringer IngelHeim and Sanofi on the genetics of diabetes. Danish Saleheen has received support from Pfizer, Regeneron, Genentech and Eli Lilly. Mark I McCarthy has served on advisory panels for NovoNordisk and Pfizer, and received honoraria from NovoNordisk, Pfizer, Sanofi-Aventis and Eli Lilly.
AUTHOR CONTRIBUTIONS
Project co-ordination. A.Mahajan, A.P.M., J.I.R., M.I.M.
Core analyses and writing. A.Mahajan, J.W., S.M.W, W.Zhao, N.R.R., A.Y.C., W.G., H.K., R.A.S., I.Barroso, T.M.F., M.O.G., J.B.M., M.Boehnke, D.S., A.P.M., J.I.R., M.I.M.
Statistical Analysis in individual studies. A.Mahajan, J.W., S.M.W., W.Zhao, N.R.R., A.Y.C., W.G., H.K., D.T., N.W.R., X.G., Y.Lu, M.Li, R.A.J., Y.Hu, S.Huo, K.K.L., W.Zhang, J.P.C., B.P., J.Flannick, N.G., V.V.T., J.Kravic, Y.J.K., D.V.R., H.Y., M.M.-N., K.M., R.L.-G., T.V.V., J.Marten, J.Li, A.V.S., P.An, S.L., S.G., G.M., A.Demirkan, J.F.T., V.Steinthorsdottir, M.W., C.Lecoeur, M.Preuss, L.F.B., P.Almgren, J.B.-J., J.A.B., M.Canouil, K.-U.E., H.G.d.H., Y.Hai, S.Han, S.J., F.Kronenberg, K.L., L.A.L., J.-J.L., H.L., C.-T.L., J.Liu, R.M., K.R., S.S., P.S., T.M.T., G.T., A.Tin, A.R.W., P.Y., J.Y., L.Y., R.Y., J.C.C., D.I.C., C.v.D., J.Dupuis, P.W.F., A.Köttgen, D.M.-K., N.Soranzo, R.A.S., A.P.M.
Genotyping. A.Mahajan, N.R.R., A.Y.C., Y.Lu, Y.Hu, S.Huo, B.P., N.G., R.L.-G., P.An, G.M., E.A., N.A., C.B., N.P.B., Y.-D.I.C., Y.S.C., M.L.G., H.G.d.H., S.Hackinger, S.J., B.-J.K., P.K., J.Kriebel, F.Kronenberg, H.L., S.S.R., K.D.T., E.B., E.P.B., P.D., J.C.F., S.R.H., C.Langenberg, M.A.P., F.R., A.G.U., J.C.C., D.I.C., P.W.F., B.-G.H., C.H., E.I., S.L.K., J.S.K., Y.Liu, R.J.F.L., N.Soranzo, N.J.W., R.A.S., T.M.F., A.P.M., J.I.R., M.I.M.
Cross-trait lookups in unplublished data. S.M.W., A.Y.C., Y.Lu, M.Li, M.G., H.M.H., A.E.J., D.J.L., E.M., G.M.P., H.R.W., S.K., C.J.W.
Phenotyping. Y.Lu, Y.Hu, S.Huo, P.An, S.L., A.Demirkan, S.Afaq, S.Afzal, L.B.B., A.G.B., I.Brandslund, C.C., S.V.E., G.G., V.Giedraitis, A.T.-H., M.-F.H., B.I., M.E.J., T.J., A.Käräjämäki, S.S.K., H.A.K., P.K., F.Kronenberg, B.L., H.L., K.-H.L., A.L., J.Liu, M.Loh, V.M., R.M.-C., G.N., M.N., S.F.N., I.N., P.A.P., W.R., L.R., O.R., S.S., E.S., K.S.S., A.S., B.T., A.Tönjes, A.V., D.R.W., H.B., E.P.B., A.Dehghan, J.C.F., S.R.H., C.Langenberg, A.D.Morris, R.d.M., M.A.P., A.R., P.M.R., F.R.R., V.Salomaa, W.H.-H.S., R.V., J.C.C., J.Dupuis, O.H.F., H.G., B.-G.H., T.H., A.T.H., C.H., S.L.K., J.S.K., A.Köttgen, L.L., Y.Liu, R.J.F.L., C.N.A.P., J.S.P., O.P., B.M.P., M.B.S., N.J.W., T.M.F., M.O.G.
Individual study design and principal investigators. N.G., P.An, B.-J.K., P.Amouyel, H.B., E.B., E.P.B., R.C., F.S.C., G.D., A.Dehghan, P.D., M.M.F., J.Ferrières, J.C.F., P.Frossard, V.Gudnason, T.B.H., S.R.H., J.M.M.H., M.I., F.Kee, J.Kuusisto, C.Langenberg, L.J.L., C.M.L., S.M., T.M., O.M., K.L.M., M.M., A.D.Morris, A.D.Murray, R.d.M., M.O.-M., K.R.O., M.Perola, A.P., M.A.P., P.M.R., F.R., F.R.R., A.H.R., V.Salomaa, W.H.-H.S., R.S., B.H.S., K.Strauch, A.G.U., R.V., M.Blüher, A.S.B., J.C.C., D.I.C., J.Danesh, C.v.D., O.H.F., P.W.F., P.Froguel, H.G., L.G., T.H., A.T.H., C.H., E.I., S.L.K., F.Karpe, J.S.K., A.Köttgen, K.K., M.Laakso, X.L., L.L., Y.Liu, R.J.F.L., J.Marchini, A.Metspalu, D.M.-K., B.G.N., C.N.A.P., J.S.P., O.P., B.M.P., R.R., N.Sattar, M.B.S., N.Soranzo, T.D.S., K.Stefansson, M.S., U.T., T.T., J.T., N.J.W., J.G.W., E.Z., I.Barroso, T.M.F., J.B.M., M.Boehnke, D.S., A.P.M., J.I.R., M.I.M.
DATA AVAILABILITY STATEMENT
Summary level data of the exome-array component of this project can be downloaded from the DIAGRAM consortium website http://diagram-consortium.org/ and Accelerating Medicines Partnership T2D portal http://www.type2diabetesgenetics.org/.
MATERIALS & CORRESPONDENCE
Correspondence and requests for materials should be addressed to mark.mccarthy@drl.ox.ac.uk and anubha@well.ox.ac.uk. Reprints and permissions information is available at www.nature.com/reprints.
Footnotes
URLs
Type 2 Diabetes Knowledge Portal: http://www.type2diabetesgenetics.org/
References
- 1.Kooner JS, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–9. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho YS, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2011;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng MC, et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014;10:e1004517. doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gusev A, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter K, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–25. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horikoshi M, et al. Transancestral fine-mapping of four type 2 diabetes susceptibility loci highlights potential causal regulatory mechanisms. Hum Mol Genet. 2016;25:2070–2081. doi: 10.1093/hmg/ddw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–7. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook JP, Morris AP. Multi-ethnic genome-wide association study identifies novel locus for type 2 diabetes susceptibility. Eur J Hum Genet. 2016;24:1175–80. doi: 10.1038/ejhg.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrada K, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311:2305–14. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sveinbjornsson G, et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat Genet. 2016;48:314–7. doi: 10.1038/ng.3507. [DOI] [PubMed] [Google Scholar]
- 17.Liu DJ, et al. Meta-analysis of gene-level tests for rare variant association. Nat Genet. 2014;46:200–4. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinthorsdottir V, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–8. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flannick J, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–63. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beer NL, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081–8. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4:200–13. doi: 10.1038/ncpendmet0778. [DOI] [PubMed] [Google Scholar]
- 25.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozlitina J, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–6. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulzer JR, et al. A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am J Hum Genet. 2014;94:186–97. doi: 10.1016/j.ajhg.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrat GR, et al. Decreased STARD10 expression is associated with defective insulin secretion in humans and mice. Am J Hum Genet. 2017;100:238–256. doi: 10.1016/j.ajhg.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deeb SS, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–7. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 30.Majithia AR, et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci U S A. 2014;111:13127–32. doi: 10.1073/pnas.1410428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majithia AR, et al. Prospective functional classification of all possible missense variants in PPARG. Nat Genet. 2016;48:1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claussnitzer M, et al. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156:343–58. doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimas AS, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63:2158–71. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotta LA, et al. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49:17–26. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altshuler D, Daly M. Guilt beyond a reasonable doubt. Nat Genet. 2007;39:813–5. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 38.Grove ML, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–5. doi: 10.1016/j.ajhg.2008.06.005. author reply 135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weale ME. Quality control for genome-wide association studies. Methods Mol Biol. 2010;628:341–72. doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 41.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 42.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eastwood SV, et al. Algorithms for the Capture and Adjudication of Prevalent and Incident Diabetes in UK Biobank. PLoS One. 2016;11:e0162388. doi: 10.1371/journal.pone.0162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann TJ, et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann TJ, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–30. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler TW, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook JP, Mahajan A, Morris AP. Guidance for the utility of linear models in meta-analysis of genetic association studies of binary phenotypes. Eur J Hum Genet. 2017;25:240–245. doi: 10.1038/ejhg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. s1–3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–27. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary level data of the exome-array component of this project can be downloaded from the DIAGRAM consortium website http://diagram-consortium.org/ and Accelerating Medicines Partnership T2D portal http://www.type2diabetesgenetics.org/.