Abstract
Background
The aim of this study was to discuss the possible roles and clinical value of 2 circRNAs (hsa_circ_0049224 and has_circ_0049220) in SLE patients.
Material/Methods
Reverse-transcription real-time polymerase chain reaction (RT-PCR) was conducted to detect the expressions of hsa_circ_0049224, has_circ_0049220, and DNMT1 in peripheral blood mononuclear cells (PBMCs) from 18 diagnosed SLE patients and 10 healthy controls.
Results
We found that the expressions of hsa_circ_0049224 and has_circ_0049220 in healthy control groups were both much higher than those in inactive and active SLE patients. The expression level of DNMT1 is positively correlated with the expressions of hsa_circ_0049224 and has_circ_0049220. Moreover, there was a negative correlation between the SLE Disease Activity Index (SLEDAI) and the expressions of hsa_circ_0049224 and has_circ_0049220. We also found that these 2 circRNAs are associated with some clinical characteristics of SLE.
Conclusions
Hsa_circ_0049224 and has_circ_0049220 are probable factors involved in the pathogenesis of SLE, and they have potential clinical value in SLE.
MeSH Keywords: Biological Markers, Clinical, Systemic Lupus Erythematous
Background
System lupus erythematous (SLE), characterized by production of autoantibodies directed against various autoantigens, is a complex autoimmune disease [1]. Many studies have reported that microRNAs (miRNAs), noncoding small RNAs of approximately 20 nucleotides in length, are involved with the pathological processes of SLE, which means miRNAs could be regarded as novel biomarkers and therapeutic targets for patients with SLE [2–4].
However, to the best of our knowledge, studies of circular RNAs (circRNAs) and SLE are still scarce. CircRNAs are a new class of noncoding RNAs recently shown to have huge capabilities as gene regulators in mammals, and to have diverse biological functions, which form covalently closed RNA circles and show higher degree of stability than miRNAs [5]. Some circRNAs bind with miRNAs and act as natural miRNA sponges to inhibit related miRNAs activities [6,7]. A growing number of studies have demonstrated that circRNAs participate in the occurrences and processes of many diseases, including various kinds of cancers, autoimmune diseases, and cardiovascular diseases [7–10].
In this study, we focus on hsa_circ_0049224 and has_circ_0049220, which have been demonstrated to be related with the pathological processes of rheumatoid arthritis and target DNMT1 [11]. DNMT1 is one of the key factors of DNA methylation, and DNA hypomethylation was associated with the etiology of SLE [12]. Furthermore, in our previous research, we demonstrated that gene expression of DNMT1 of SLE patients is lower than in healthy controls [13,14]. Therefore, in the present study, we discuss the roles of hsa_circ_0049224 and has_circ_0049220 in SLE and define their possible relationships with clinical parameters of SLE.
Material and Methods
Subjects
We enrolled 18 newly-diagnosed SLE patients from the Department of Dermatology of Shanghai General Hospital. All the patients were diagnosed with SLE according to the 1997 American College of Rheumatology (ACR) revised criteria. The disease activity was assessed by the SLE Disease Activity Index (SLEDAI). Active disease was defined as a SLEDAI score of 5 or more, and inactive disease was defined as a SLEDAI score of 4 or less. Ten healthy controls were enrolled from the medical staff at Shanghai General Hospital. Each subject signed informed consent before agreeing to take part in this study. This study was approved by the Shanghai General Hospital Institutional Review Board.
Isolation of peripheral blood mononuclear cells (PBMCs)
A total of 10 ml of venous peripheral blood was obtained from each subject and stored in ethylenediaminetetraacetic acid (EDTA), and PBMCs were separated from blood samples by Ficoll-Paque density centrifugation.
Extraction of RNA and reverse-transcription PCR (RT-PCR)
Total RNA was extracted from PBMCs using TRIzol according to the manufacturer’s instructions (Invitrogen). For cDNA synthesis, RNA (1 μg) was mixed with 500 ng of oligo (dT) (Promega). The RNA primer mixture was incubated at 65°C for 10 min, followed by snap freezing in an ice bath for 2 min. Samples were then incubated at 42°C for 45 min with 4 μl of 5× first-strand buffer, 2 μl of 0.1 mol dNTP, 1 μl RNasin (Takara), and distilled water to a total volume of 19 μl. The reverse transcriptase was inactivated at 70°C for 10 min and then chilled on ice.
The RT-PCR reaction mixture contained 2.5 μl of qPCR mix, 0.15 μl of gene-specific forward and reverse primers, 1 μl of cDNA, and 1.2 μl of distilled water. The primers used in this study were as follows:
hsa_circ_0049224 forward: 5′-GCATTGTTGTAACGACCACCTCCG-3′,
reverse: 5′-CCGGATTGTAACTGGACCAAAGGC-3′;
hsa_circ_0049224 forward: 5′-CCAATTGTCTTGCGAACTATCGTA-3′,
reverse: 5′-GTGCCATTGTAACGACTAACGCT-3′;
DNMT1 forward: 5′-GCACCTCATTTGCCGAATACT-3′,
reverse: 5′-TCTCCTGCATCAGCCCAAATA-3′.
The relative expression levels of hsa_circ_0049224 and has_circ_0049220 were determined by RT-PCR.
Data analysis
All statistical analyses were performed with a statistical software package (SAS version 9.0; SAS Software, Cary, NC, USA). All measurement data results are displayed as mean ±SD. Data were analyzed by analysis of variance followed by the unpaired t test for multiple comparisons. P values less than or equal to 0.05 were considered significant.
Results
Clinical features of all the subjects
This study included 10 healthy subjects and 18 SLE patients. Of the SLE patients, 7 were inactive and 11 were active according to SLEDAI score. The clinical information is displayed in Table 1.
Table 1.
Clinical features of all the subjects.
| Characteristic | Healthy controls | Inactive SLE patients | Active SLE patients |
|---|---|---|---|
| No. males/females | 2/8 | 1/6 | 2/9 |
| Age (years) | 28.0±7.9 | 30.4±5.9 | 29.1±5.4 |
| Disease duration (months) | – | 19.4±2.2 | 21.4±2.7 |
| SLEDAI | – | 2.4±1.9 | 10.0±6.0 |
The expressions of hsa_circ_0049224 and has_circ_0049220 in healthy controls and patients
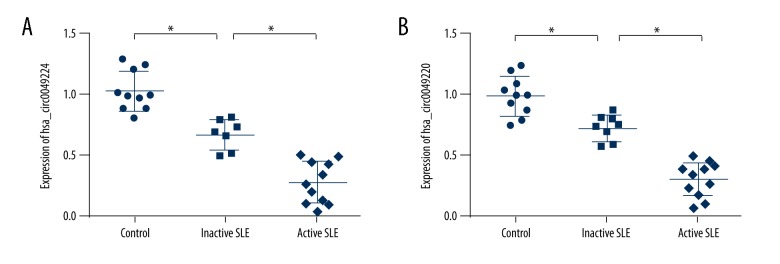
To validate the candidate hsa_circ_0049224 and has_circ_0049220, RT-PCR was conducted in an independent cohort (controls, n=10; inactive SLE patients, n=7; active SLE patients, n=11). The results are displayed in Figure 1. The levels of hsa_circ_0049224 (1.03±0.16, 0.68±0.12, 0.29±0.17; P<0.001) and has_circ_0049220 (1.00±0.16, 0.72±0.11, 0.31±0.14; P<0.001) among the 3 groups were significantly different. The expression of hsa_circ_0049224 and has_circ_0049220 in healthy controls were both much higher than in inactive and active SLE patients.
Figure 1.
Expression of hsa_circ_0049224 (A) and has_circ_0049220 (B) in healthy controls (n=10), inactive (n=7) and active SLE patients (n=11). * P<0.05.
The expressions of hsa_circ_0049224 and has_circ_0049220 were associated with expression of DNMT1
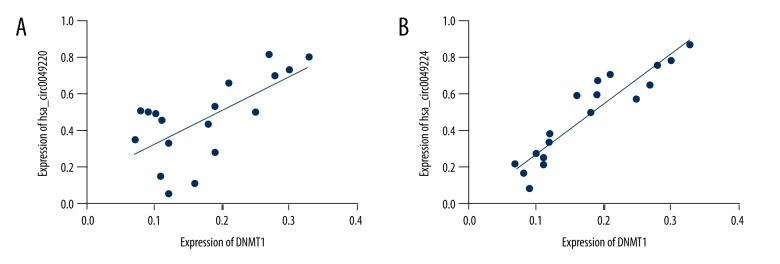
To ascertain the involvement of hsa_circ_0049224 and has_circ_0049220 in the DNA methylation of SLE, we first assessed the expression level of DNMT1 among healthy controls and inactive and active SLE patients (0.30±0.01, 0.22±0.03, 0.15±0.02; P<0.001) (Figure 2). Figure 3 shows the relationship between the expression of DNMT1 for each patient and the expression of hsa_circ_0049224 and has_circ_0049220, demonstrating that the level of DNMT1 was positively correlated with the hsa_circ_0049224 (r=0.44, P=0.002) and has_circ_0049220 expression level (r=0.86, P<0.001).
Figure 2.
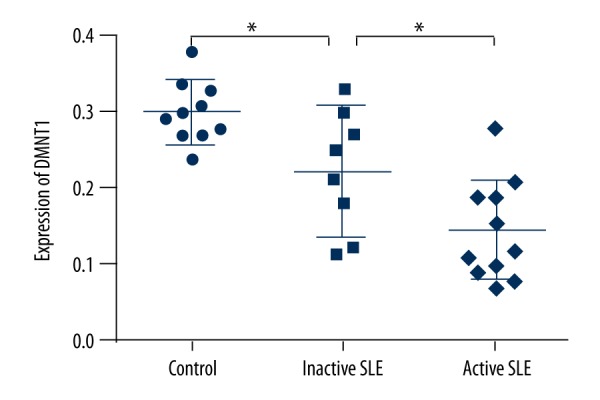
Expression levels of DNMT1 among healthy controls (n=10), inactive SLE patients (n=7), and active SLE patients (n=11). *P<0.05.
Figure 3.
Expressions of hsa_circ_0049224 (A) and has_circ_0049220 (B) were positively related with expression level of DNMT1 among SLE patients.
The expressions of hsa_circ_0049224 and has_circ_0049220 were associated with SLE clinical manifestation
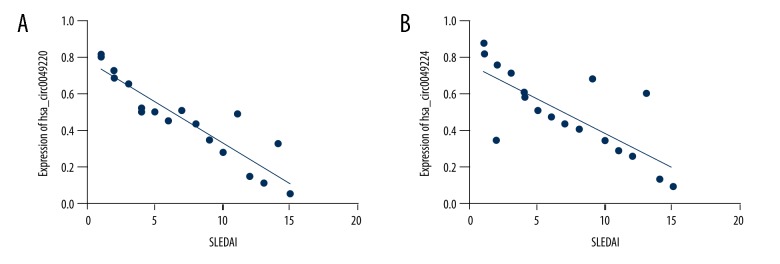
To assess the role of hsa_circ_0049224 and has_circ_0049220 in SLE, we evaluated whether hsa_circ_0049224 and has_circ_0049220 expression was associated with SLE disease activity. Figure 4 displays the relationship between the SLEDAI score and the expression of hsa_circ_0049224 and has_circ_0049220, showing that SLE disease activity was negatively related with expression of the hsa_circ_0049224 (r=−0.89, P<0.001) and has_circ_0049220 (r=−0.84, P<0.001).
Figure 4.
Expressions of hsa_circ_0049224 (A) and has_circ_0049220 (B) were negatively related with SLEDAI.
Moreover, we attempted to make a distinction between the expression of hsa_circ_0049224 and has_circ_0049220 expression in SLE patients with or without various skin features and lab biomarkers (anti-dsDNA and anti-ssDNA). Among 18 SLE patients, we found 13 had photosensitivity, 11 had a malar rash, 10 had Raynaud’s phenomenon, 8 had alopecia, 7 had discoid lupus erythematosus (DLE), 1 had subacute cutaneous LE, 12 were anti-dsDNA positive, and 10 were anti-ssDNA positive (Table 2). Because of the small number of patients, the expression of hsa_circ_0049224 and has_circ_0049220 expression in SLE patients with or without photosensitivity, malar rash, Raynaud’s phenomenon, alopecia, DLE, anti-dsDNA and anti-ssDNA were analyzed. And the results showed that the hsa_circ_0049224 level was higher in patients with photosensitivity, Raynaud’s phenomenon and anti-dsDNA positive than in patients without those symptoms. In addition, hsa_circ0049220 level was higher in patients with photosensitivity and anti-dsDNA positivity than in patients without those symptoms.
Table 2.
hsa_circ_0049224 and has_circ_0049220 expression in SLE patients with different clinical characteristics.
| Group | Positive/negative | Cases | hsa_circ_0049224 expression | hsa_circ_0049224 expression |
|---|---|---|---|---|
| Photosensitivity | Positive | 13 | 0.38±0.17* | 0.39±0.11* |
| Negative | 5 | 0.67±0.18 | 0.75±0.18 | |
| Malar rash | Positive | 11 | 0.40±0.16 | 0.41±0.16 |
| Negative | 7 | 0.56±0.23 | 0.61±0.28 | |
| Raynaud’s phenomenon | Positive | 10 | 0.36±0.17* | 0.44±0.19 |
| Negative | 8 | 0.60±0.21 | 0.55±0.28 | |
| Alopecia | Positive | 8 | 0.51±0.23 | 0.45±0.21 |
| Negative | 10 | 0.43±0.22 | 0.44±0.21 | |
| DLE | Positive | 7 | 0.48±0.21 | 0.49±0.24 |
| Negative | 11 | 0.46±0.23 | 0.49±0.23 | |
| Anti-dsDNA | Positive | 12 | 0.38±0.20* | 0.41±0.22* |
| Negative | 6 | 0.64±0.15 | 0.65±0.19 | |
| Anti-ssDNA | Positive | 10 | 0.48±0.24 | 0.45±0.25 |
| Negative | 8 | 0.45±0.21 | 0.54±0.21 |
Patients with vs. patients without clinical characteristics (P<0.05).
Discussion
The pathogenesis of SLE remains poorly understood, but its various complications severely threaten human health. In this research, we used PBMCs from SLE patients and healthy controls, and identified 2 circRNAs (hsa_circ_0049224 and has_circ_0049220) that were significantly downregulated in SLE patients. Our study, for the first time, suggests that circRNAs are involved in the pathogenesis of SLE, which could contribute to our understanding of a new pathway of SLE.
Ever since the discovery of circRNAs, tremendous effort has been devoted to identifying the biological functions of circRNAs and their relevance to various kinds of diseases. CircRNAs are much more stable than linear RNAs in cells, and in some tissues their expression levels are 10 times higher, so circRNAs could be regarded as preferred biomarkers of diseases [9,15,16]. Many SLE symptoms are quite similar to those of other illnesses, which makes it vague and difficult to diagnose [17] Currently, the diagnosis of SLE often requires presence of typical clinical manifestations combined with lab indicators [18]. In the present study, we found that 2 circRNAs (hsa_circ_0049224 and has_circ_0049220) were differently expressed in the PBMCs of healthy controls vs. inactive SLE and active patients, and patients with and without some clinical characteristics of SLE had significantly different expressions of these 2 circRNAs (Table 2), which means that hsa_circ_0049224 and has_circ_0049220 may participate in the pathogenesis of SLE or even be related with the degree of disease activity. We also discovered that hsa_circ_0049224 and has_circ_0049220 expression was negatively correlated with SLEDAI and the degree of SLE severity, which means these 2 circRNAs may be involved in the progress, recrudescence, and organ injure of SLE.
DNA methylation is an epigenetic marker that may regulate gene expression. Many studies have indicated the abnormal methylation in some genes of CD4+T cells from SLE patients [3,19,20], and our results support this. For the first time, we report that the expression level of DNMT1 is positively correlated with the mRNA expression of hsa_circ_0049224 and has_circ_0049220, which means hsa_circ_0049224 and has_circ_0049220 could be used as biomarker in the evaluation of hypomethylation.
Conclusions
To the best of our knowledge, this study is the first to reveal the expression profiles of circRNAs in the peripheral blood of SLE patients and to validate the utility of hsa_circ_0049224 and hsa_circ_0049224 as diagnostic biomarkers for inactive and active SLE. However, due to the relatively small sample of subjects included, our study has limitations. Thus, further investigations with much larger sample sizes and longer follow-up periods are needed to verify this result. In our future research, we plan to focus on the function of circRNAs to determine if they have a connection with microRNA expression in SLE PBMCs or even CD4+ T cells, because circRNAs have been verified as being actual miRNA sponges in mammals in many diseases. More importantly, further studies are also needed to elucidate the regulatory mechanisms of circRNAs in the pathogenesis of SLE.
In summary, hsa_circ_0049224 and has_circ_0049220 are probable factors involved in the pathogenesis of SLE, and they have potential clinical value in SLE.
Footnotes
Conflicts of interest
None.
Source of support: National Natural Science Foundation of China (Grant No. 81573031)
References
- 1.Ferrera F, Filaci G, Rizzi M, et al. New therapies in SLE. Recent Pat Inflamm Allergy Drug Discov. 2008;2(1):11–23. doi: 10.2174/187221308783399243. [DOI] [PubMed] [Google Scholar]
- 2.Smith S, Fernando T, Wu PW, et al. MicroRNA-302d targets IRF9 to regulate the IFN-induced gene expression in SLE. J Autoimmun. 2017;79:105–11. doi: 10.1016/j.jaut.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Zhao M, Liu S, Luo S, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J Autoimmun. 2014;54:127–36. doi: 10.1016/j.jaut.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Lashine YA, Salah S, Aboelenein HR, Abdelaziz AI. Correcting the expression of miRNA-155 represses PP2Ac and enhances the release of IL-2 in PBMCs of juvenile SLE patients. Lupus. 2015;24(3):240–47. doi: 10.1177/0961203314552117. [DOI] [PubMed] [Google Scholar]
- 5.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–88. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription. 2015;6(4):61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–61. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Gonzalez-Cano L, Menzl I, Tisserand J, et al. Parkinson’s disease-associated mutant LRRK2-mediated inhibition of miRNA activity is antagonized by TRIM32. Mol Neurobiol. 2017 doi: 10.1007/s12035-017-0570-y. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–56. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. 2017;487(4):769–75. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 11.Song XQZL, Zhao SQ, Wang XQ, et al. Peripheral blood circRNA expression profile analysis in patients with rheumatoid arthritis. Chin J Rheumatol. 2016;20(8):541–46. [Google Scholar]
- 12.Nawrocki MJ, Majewski D, Puszczewicz M, Jagodzinski PP. Decreased mRNA expression levels of DNA methyltransferases type 1 and 3A in systemic lupus erythematosus. Rheumatol Int. 2017;37(5):775–83. doi: 10.1007/s00296-017-3711-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z, Li X, Qin H, et al. Ultraviolet B enhances DNA hypomethylation of CD4+ T cells in systemic lupus erythematosus via inhibiting DNMT1 catalytic activity. J Dermatol Sci. 2013;71(3):167–73. doi: 10.1016/j.jdermsci.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z, Mei X, Ying Z, et al. Ultraviolet B inhibition of DNMT1 activity via AhR activation dependent SIRT1 suppression in CD4+ T cells from systemic lupus erythematosus patients. J Dermatol Sci. 2017;86(3):230–37. doi: 10.1016/j.jdermsci.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Cui X, Niu W, Kong L, et al. hsa_circRNA_103636: Potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark Med. 2016;10(9):943–52. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 16.Gruner H, Cortes-Lopez M, Cooper DA, et al. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-Hishmeh M, Sattar A, Zarlasht F, et al. Systemic lupus erythematosus presenting as refractory thrombotic thrombocytopenic purpura: A diagnostic and management challenge. A case report and concise review of the literature. Am J Case Rep. 2016;17:782–87. doi: 10.12659/AJCR.898955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesa A, Fernandez M, Wu W, et al. Can SLE classification rules be effectively applied to diagnose unclear SLE cases? Lupus. 2016 doi: 10.1177/0961203316655212. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JM, Ratliff ML, Dozmorov MG, et al. Expression and methylation data from SLE patient and healthy control blood samples subdivided with respect to ARID3a levels. Data Brief. 2016;9:213–19. doi: 10.1016/j.dib.2016.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang J, Zhu XH, Qin HH, et al. A correlation study on the effects of DNMT1 on methylation levels in CD4(+) T cells of SLE patients. Int J Clin Exp Med. 2015;8(10):19701–8. [PMC free article] [PubMed] [Google Scholar]