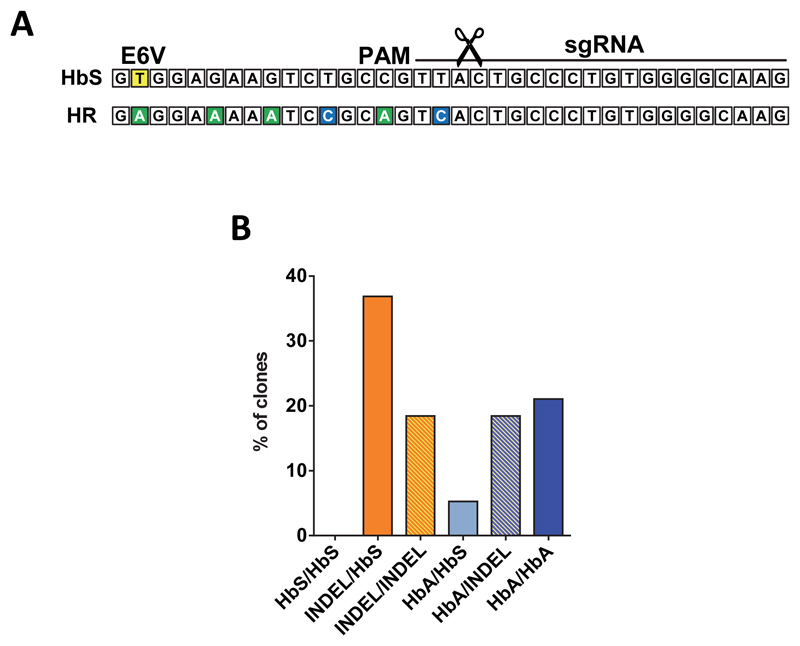
Extended Data Figure 8. Correction of the sickle cell mutation in patient-derived CD34+ HSPCs.
(A) Schematic overview of the sequence of the sickle allele aligned with the sequence of an allele that has undergone HR using the corrective SNP donor. The E6V mutation in sickle cell patients (A->T) is highlighted in yellow. The sgRNA recognition sequence, the PAM site, and the cut site (scissors) are shown. The donor carries synonymous nucleotide changes between the sickle nucleotide and the cut site to avoid premature cross-over during HR. Synonymous changes are also added to the PAM and an early nucleotide in the sgRNA target site to avoid subsequent re-cutting and potential inactivation of the corrected allele. (B) HSPCs from two different sickle cell patients were targeted with the corrective SNP donor and seeded in methylcellulose. After 14 days, In-Out PCR amplicons from a total of 38 clones were sequenced and genotypes were extracted from sequence chromatograms.