Abstract
Molecular chaperones and protein folding factors of bacterial periplasmic space play important roles in assisting disulfide bond formation and proper protein folding. In this study, effects of disulfide bond protein (Dsb) families were investigated on assembly of 3F3 Fab, an antibody inhibitor targeting matrix metalloproteinase-14 (MMP-14). By optimizing DsbA/C co-expression, promoter for 3F3 Fab, host strains, and culture media and conditions, a high yield of 30 mg purified 3F3 Fab per liter culture was achieved. Produced 3F3 Fab exhibited binding affinity of 34 nM and inhibition potency of 970 nM. This established method of DsbA/C co-expression can be applied to produce other important disulfide bond-dependent recombinant proteins in E. coli periplasm.
Keywords: Fab, IgG, periplasm, over-expression, protein folding factor, DsbA, DsbC
INTRODUCTION
With fast growth, low cost, ease of genetic manipulation, and many molecular tools and protocols at hand, Escherichia coli is one of the most widely used microorganism species for producing recombinant proteins [1]. Particularly, as the space between its inner and outer membranes, E. coli periplasm provides beneficial properties for protein expression [2], such as multiple molecular chaperones (e.g. Skp, FkpA, SurA) [3–5], quality control mechanisms via the secretion machinery [6,7], and the oxidizing environment and folding modulators facile for disulfide bond formation. With extensive understandings of recombinant protein expression in E. coli periplasmic space, many challenging eukaryotic proteins with multiple disulfide bonds have been successfully produced [8–12].
Our lab recently isolated a panel of monoclonal antibody Fabs (e.g. 2B5, 3A2, and 3E9) which inhibited matrix metalloproteinase-14 (MMP-14) with high potency and selectivity [13]. However, one potentially valuable Fab clone 3F3 exhibiting an inhibition potency at hundreds nM range, failed to be produced at milligram level needed for full characterizations. Further analysis of 3F3 Fab production profiles demonstrated that both its light chain (VL-CL-FLAG tag) and heavy chain (VH-CH1-His tag) were expressed in E. coli periplasm and were able to be separately purified via their associated tags (Fig 1). But 3F3 Fab was not well assembled in vivo (< 10 μg per liter of culture media) in contrast to other Fab clones (typical yields of 0.5–2 mg/L). Encouraged by numerous pioneering works of multiple intramolecular and intermolecular disulfide bond formation by co-expressing folding factors involved in E. coli secretion expression systems [8–12,14], we hypothesized that similar approaches can efficiently facilitate 3F3 Fab assembly. In this study, we report the effects of Dsb (disulfide bond) protein family on 3F3 Fab production. In addition, the promoter, host strain, and culture media and conditions were also systematically optimized to yield milligrams of purified 3F3 Fab.
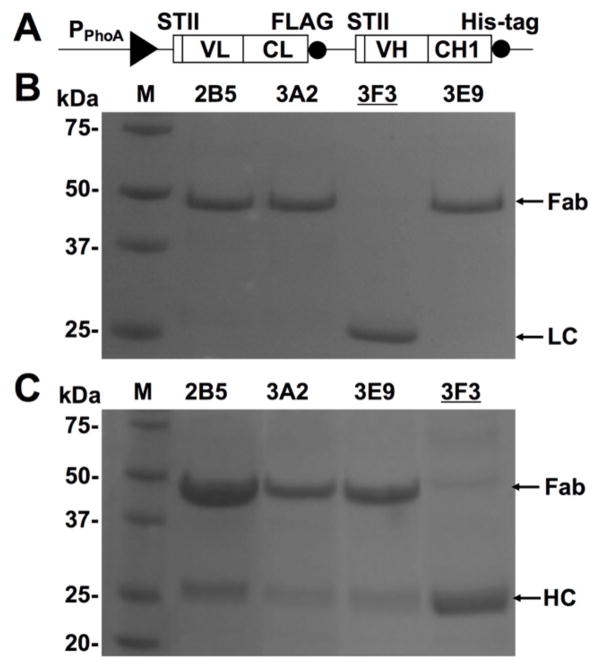
Figure 1. Expressed 3F3 heavy and light chains failed assembling to Fab.
(A) Construct of 3F3 Fab expression cassette with PPhoA promotor and STII leaders. FLAG tag and His-tag are at the C-termini of light and heavy chains. (B) Purification results of four Fabs 2B5, 3A2, 3F3, and 3E9 [13] from their BL21 periplasmic fractions using anti-FLAG resin, resulting in capture of Fabs and light chain (LC) fragments (VL-CL). 3F3 LC MW = 27 kDa. (C) Purification results of four Fabs from their BL21 periplasmic fractions using Ni-NTA resin, resulting in capture of Fabs and heavy chain (HC) fragments (VH-CH1). 3F3 HC MW = 30 kDa. Further quantification showed assembled 3F3 Fab (MW = 57 kDa) had a yield <10 μg per litter of culture.
MATERIALS AND METHODS
Construction of 3F3 Fab Expression Plasmids
The fragment encoding VL-CL-VH-CH1 genes of antibody clone 3F3 [13] were amplified by PCR, digested with NsiI and SalI, and cloned into the same sites on a Fab expression vector containing a phoA promoter and STII leader peptide sequences (Fig 1A, [15]). On this plasmid, a polyhistidine tag and a FLAG tag were placed at the C-termini of heavy and light chains. To construct 3F3 Fab expression vector with a Lac promoter and pelB leader peptides, its VH and VL fragments were amplified by PCR and cloned into NcoI/NotI and NheI/HindIII sites on pMAZ360-IgG [16] respectively, and a 10×His tag with a stop codon was introduced at downstream of CH1 domain to obtain pLac-Fab3F3.
Construction of DsbA/C Plasmids
The plasmids encoding both DsbA and DsbC, either under one promoter (1P) or two separate promotors (2P), were constructed by overlap extension PCR cloning [17]. Briefly, DsbC gene cassette with its promoter region was PCR amplified using pBAD-DsbC [18] as the template, and the 1.6 kb product was gel purified and mixed with pBAD-DsbA [18] for overlap extension PCR. The product was digested with DpnI and transformed to obtain pBAD-DsbAC2P (under two separate promoters) (Fig 2A). Similarly, DsbC gene cassette without its promoter region was amplified and overlap extended with pBAD-DsbA to construct pBAD-DsbAC1P (under one promoter). All resulting plasmids were confirmed by DNA sequencing.
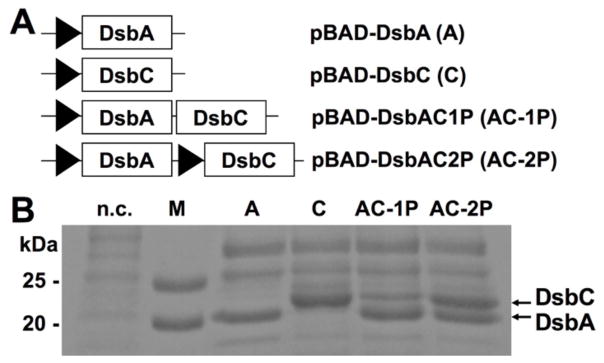
Figure 2. Folding factor DsbA/C construct designs and expression results.
(A) Four constructs were applied in this paper: expression of either DsbA or DsbC under an arabinose promoter PBAD; bicistronic co-expression of DsbA and DsbC under one PBAD promoter (AC-1P); monocistronic co-expression of DsbA and DsbC under two separate PBAD promoters (AC-2P). (B) SDS-PAGE analysis of periplasmic fractions. Cultures were induced with 0.2% arabinose. DsbA MW = 23 kDa; DsbC MW = 25 kDa; n.c. = negative control; M = protein marker.
3F3 Fab Expression
Constructed Fab expression plasmids were transformed into E. coli Jude-I or BL21 and cultivated overnight in 2×YT or TB supplemented with 100 μg/ml ampicillin at 30 °C or 37 °C. For pLac-Fab3F3, cells were induced with 0.1 mM IPTG. To co-express Dsb proteins, pBAD-DsbA/-DsbC/-DsbAC1P/-DsbAC2P were transformed into Jude-I or BL21 harboring pLac-Fab3F3. The co-transformed hosts were grown in 2×YT or TB media supplemented with 100 μg/ml ampicillin and 35 μg/ml chloramphenicol at 37 °C. When cell density reached OD600 0.6–0.8, arabinose was added to a final concentration of 0.2% (w/v) and cells were further cultured for 30 minutes before 0.1 mM IPTG was added for Fab induction at either 30 or 37°C overnight. To further improve Fab production in simulated fed-batch and high-glucose conditions [19], BL21 harboring both pLac-Fab3F3 and pBAD-DsbA was cultivated in EnBase media following manufacturer’s instructions (BioSilta).
Periplasmic Fraction Preparation and Western Blotting
A modified cold osmotic shock protocol was applied to release expressed antibody fragments from the periplasmic space [20]. After cells were pelleted by centrifugation at 16,000 ×g for 1 min, for every two OD600 cells, the pellet was resuspended in 350 μl periplasmic buffer (200 mM Tris-HCl, pH 7.5, 20% sucrose, 30 U/μl lysozyme), and incubated at RT for 5 min. 350 μl ice-cold DDW was then added for osmotic shock and incubation on ice for 10 min. The resulting suspensions were centrifuged at 16,000 ×g for 2 min and clarified supernatants were subjected to Western blotting. Either anti-His-HRP (Abcam) or anti-Fab-HRP (Sigma-Aldrich) was used as the second antibody. Signals were developed with chemiluminescent substrate (Thermo Scientific) and analyzed with an imager (Bio-Rad).
3F3 Fab Purification
From periplasmic fractions, 3F3 Fab was purified by affinity chromatography using Ni-NTA resin (Qiagen), and dialyzed against 50 mM HEPES (pH 6.8), 150 mM NaCl to remove residual imidazole (a weak inhibitor of MMPs). The homogeneities of purified antibody fragments were verified by SDS-PAGE (non-reducing), and 3F3 Fab concentration was measured with NanoDrop 2000 (Thermo Scientific).
Fab ELISA
Maxisorp 96-well immunoplates (Nunc) were coated with 5 μg/ml streptavidin in TBS buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl2, 100 μM ZnCl2) overnight at 4 °C and washed twice with TBST (TBS with 0.1% Tween-20). The plates were blocked with 0.5% gelatin in TBS for 2 hr at RT followed by washing twice with TBST. Biotinylated-cdMMP-14 [21] in TBS was incubated at RT for 15 min followed by washing twice with TBST. Purified Fab was serially diluted into cdMMP-14 coated wells and incubated at RT for 1 hr. Bound 3F3 Fab was detected using anti-Fab-HRP. TMB (3,3′,5,5′-tetramethylbenzidine) solution was added to develop signals. The reaction was stopped by acidification using 1 M sulfuric acid. The absorbance was measured at 450 nm. The half-maximal effective concentration (EC50) was calculated from a four-parametric logistic curve-fitting analysis.
MMP Inhibition Assay
The enzymatic activities of 1 nM cdMMP-14 in the presence of 4 nM - 3μM of 3F3 Fab were measured at 37 °C by monitoring the hydrolysis of 1 μM fluorogenic peptide Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 (Bachem) at λex = 328 nm and λem = 393 nm in TBS [20,22]. Fluorescence was recorded continuously for 30 min and the initial reaction rates and inhibition constants were calculated by fitting the data to the equation (1), where Vi is initial velocity in the presence of the inhibitor, Vo is the initial velocity in the absence of inhibitor, and [I] is the inhibitor concentration.
| (1) |
RESULTS
Over-expression of DsbA/C in Periplasm
To construct plasmids encoding both folding factors, DsbC gene cassettes with or without its arabinose-inducible promoter PBAD were cloned into pBAD-DsbA [18] by overlap extension PCR [17]. The resulting plasmids encoded DsbAC either bicistronically (pBAD-DsbAC1P) or monocistronically (pBAD-DsbAC2P) (Fig 2A). These two plasmids together with pBAD-DsbA/-DsbC were transformed to E. coli strain Jude-I, and cells were induced with 0.2% arabinose. Expression profile analysis by SDS-PAGE confirmed that DsbA and DsbC were over-expressed in the periplasmic space with their expected MWs of 23 kDa and 25 kDa (Fig 2B). In contrast, no DsbA/C associated bands were detected in the control cells without transformation. For AC-2P construct, both Dsb proteins were produced at comparable levels. While for AC-1P, DsbA was expressed twice as much as DsbC, likely because DsbC gene was placed in the downstream of the bicistronic structure. Similar results were observed when BL21 was applied as the expression host (data not shown).
DsbA/C Improved 3F3 Fab Assembly
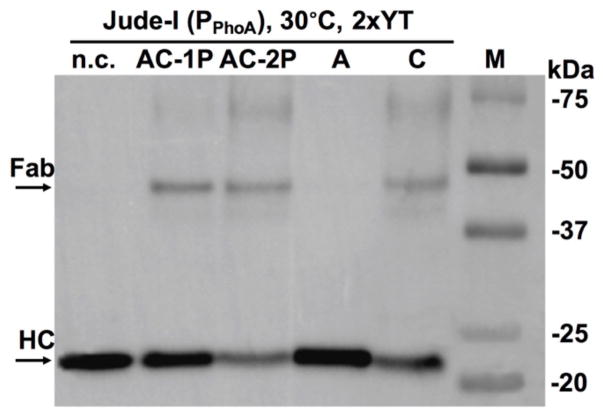
To test the hypothesis that co-expression of DsbA/C can facilitate 3F3 Fab assembly, and to identify the most efficient Dsb construct for 3F3 Fab production, each plasmid of pBAD-DsbA/-DsbC/-DsbAC1P(one promoter)/-DsbAC2P(two separate promoters) was transformed to Jude-I cells harboring 3F3 Fab gene which was at downstream of alkaline phosphatase (phoA) promoter and STII signal peptides (Fig 1A). Arabinose was added to induce DsbA/C, and PPhoA was auto-induced following depletion of phosphate from culture media 2×YT. Periplasmic fractions were prepared and subjected for Western blotting at non-reducing conditions using anti-His-HRP, which recognized the polyhistidine tag at C-terminal of heavy chain (HC). Results indicated that in addition to large amounts of unassembled HC, trace amounts of 3F3 Fab formed (with an apparent MW ~50 kDa) when co-expressed with DsbC, AC-1P or AC-2P (Fig 3). However, without Dsb folding factor (n.c. in Fig 3) or co-expressing DsbA alone showed no assembled 3F3 Fab. Overall, AC-1P promoted the highest Fab expression when PPhoA was used.
Figure 3. Effects of disulfide bond enzymes DsbA/C on 3F3 Fab assembly.
Western blotting results of overnight cultures at 30°C in 2×YT. Jude-I was the host. 3F3 Fab was expressed under a PPhoA promotor. No DsbA/C co-expression served as the negative control (n.c.). Bands were detected with anti-His-HRP.
Promoter, Host and Culture Temperature Optimizations
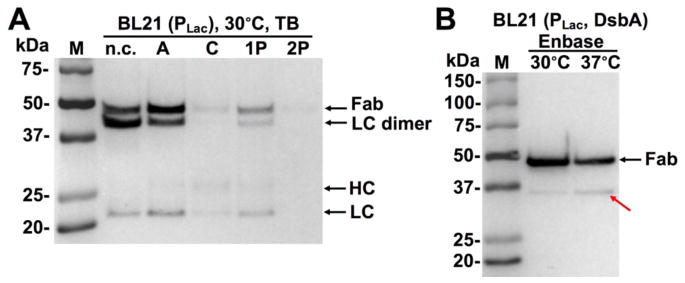
To improve Fab production, 3F3 LC and HC genes were cloned into a periplasmic expression vector carrying a strong inducible PLac promotor and pelB leader peptides [23]. This pLac-Fab3F3 (Fig 4A) was co-transformed with each of Dsb plasmids A/C/1P/2P into Jude-I cells. In 2×YT, 0.1 mM IPTG and 0.2% arabinose were used for Fab and DsbA/C inductions. After culture at 30 °C, periplasmic fractions were analyzed by Western blotting (non-reducing) using anti-Fab-HRP, which bound to both assembled 3F3 Fab and unassembled HC/LC fragments. Results indicated that significant amounts of 3F3 Fab were produced (Fig 4B). In addition, unassembled LC monomer was detected, suggesting that more LC was produced than HC. Associated with this unbalanced expression, LC dimer (with a MW less than that of Fab) was also present. Notably, when co-expressed with DsbA alone, much less LC monomer and no LC dimer were detected (arrow in Fig 4B), suggesting DsbA was optimal for 3F3 Fab production under PLac.
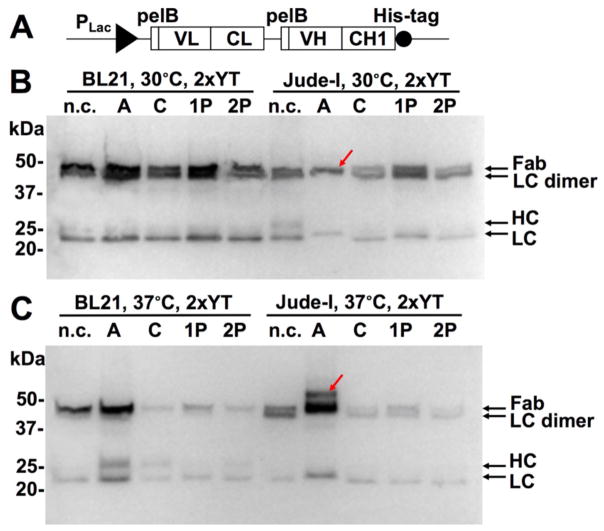
Figure 4. Promoter, host and culture temperature optimizations.
(A) 3F3 Fab expression construct with PLac and pelB leaders. 0.2% arabinose was used for DsbA/C expression. 0.1 mM IPTG was used for Fab induction. Western blotting results using host cells Jude-I or BL21 at (B) 30 °C or (C) 37°C overnight in 2×YT. Bands were detected with anti-Fab-HRP. The red arrows in (B) and (C) indicate the assembled 3F3 Fab and 3F3 Fab with leader peptide respectively.
The same set of Fab3F3-DsbA/C co-expression plasmids were also transformed into BL21, a commonly used B strain with deficiency in lon and ompT proteases. Transformed cells were cultured in 2×YT at 30 °C with 0.2% arabinose and 0.1 mM IPTG. Comparing to same amounts of Jude-I cells, BL21 produced several folds more 3F3 Fab for all cases (Fig 4B), suggesting that BL21 was superior for Fab expression.
Although low culture temperatures (e.g. 30 °C) were often used for proper protein folding, to further understand the effects of DsbA/C, we repeated the above experiments (Fig 4B) at culture temperature of 37 °C. Under this challenge, all constructs except DsbA exhibited inferior 3F3 Fab production as expected (Fig 4C). Interestingly, when cultured at 37 °C, BL21 co-transformed with pLac-Fab3F3 and pBAD-DsbA produced fully assembled 3F3 Fab as the single major band, with significantly decreased amounts of unassembled LC/HC fragments and only a trace amount of LC dimer. Presumably, the appropriate folding factor (DsbA in this case) facilitated efficiently assembling of produced 3F3 LC and HL, while other constructs failed on Fab assembly resulting in degradation of LC and HC fragments. Culturing the same plasmid combination (pLac-Fab3F3 and pBAD-DsbA) in Jude-I at 37 °C exhibited similar results with an even higher Fab yield. However, there was an additional band of a larger MW than that of Fab (arrow in Fig 4C), likely associated with Fab molecules carrying unprocessed leader peptides due to the high secretion demand. Collectively, results of Fig 4 suggested that among all tested conditions, the combination of PLac, BL21 and DsbA was the most beneficial for 3F3 Fab production.
30 mg/L 3F3 Fab Yielded in Rich Culture Media
We next tested the best combination (promoter, host and Dsb protein) identified with rich media TB and EnBase [19]. As results shown in Fig 5A, when BL21 (pLac-Fab3F3) was cultured in TB at 30 °C, constructs C/1P/2P reduced 3F3 Fab production levels while DsbA delivered a marginal improvement, comparing to the same cells cultured in 2×YT at 30 °C (Fig 4B). However, when cultured at the simulated fed-batch and high-glucose conditions using EnBase media, fully assembled 3F3 Fab bands represented > 90% the total signal intensities in Western blotting, and unassembled LC/HC or LC dimer were not detected (Fig 5B). Furthermore, the expression level at 30 °C was two-fold higher than that at 37 °C. In addition, there was a band with an apparent MW of ~35 kDa in the 37 °C culture sample (arrow in Fig 5B), likely due to a low degree of cleavage at the long CDR-H3 region, observed phenomena for certain protease inhibitory antibodies [24, 25]. But the level of truncated Fab was reduced when cultured at 30 °C. Significantly, with OD600/mL reaching 15.7 at the time of harvest, 30 mg purified 3F3 Fab per liter of culture was achieved (inset of Fig 6), which represented a four orders of magnitude improvement compared to the initial result (< 10μg, Fig 1C).
Figure 5. High yields of 3F3 Fab using (A) rich media TB and (B) fed-batch media EnBase.
Co-expression was induced with 0.2% arabinose and 0.1 mM IPTG. Western blotting bands were detected with anti-Fab-HRP. The red arrow in (B) indicates truncated Fab.
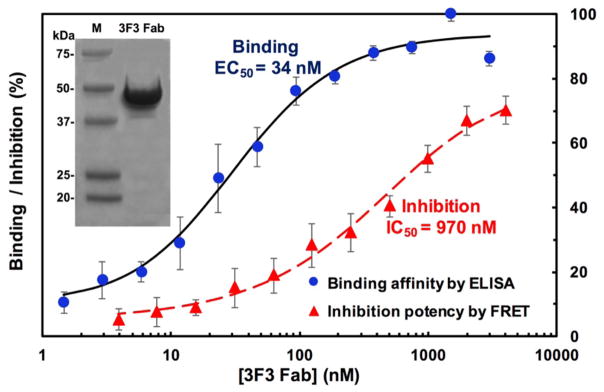
Figure 6. Binding affinity and inhibition potency of 3F3 Fab.
Binding EC50 and inhibition IC50 were determined using ELISA and FRET assays respectively. Purified 3F3 Fab sample after concentration (inset) was used for measurements.
Characterization of Produced 3F3 Fab
From the optimized expression, purified 3F3 Fab was subjected for biochemical characterizations. Its binding affinity was measured by direct ELISA, in which serially diluted 3F3 Fab samples were incubated in 96-well plates sequentially coated with streptavidin and biotinylated cdMMP-14 [21]. Captured 3F3 Fab was then detected with anti-Fab-HRP and signals were developed with chromogenic substrates. As shown in Fig 6, a sigmoidal dose-response curve was observed allowing the calculation of an affinity EC50 value as 34 nM. Next, inhibition function of 3F3 Fab on cdMMP-14 was studied using a fluorogenic peptide substrate. When 3F3 Fab concentrations increased from 4 nM to 3 μM, decreased activity of 1 nM cdMMP-14 displayed a dose-response curve. At the highest concentration tested (3 μM) 3F3 Fab inhibited ~70% activity of cdMMP-14, and the 3F3 Fab concentration gave 50% inhibition (IC50) was determined as 970 nM (Fig 6).
DISCUSSION
In periplasm of E. coli, the formation of protein disulfide bonds is catalyzed by Dsb family folding factors [18,26]. DsbA serves as an oxidase in vivo and is maintained in its oxidized state by membrane-bound DsbB. DsbC catalyzes disulfide reduction and exhibits isomerization activity. It breaks non-native disulfide bonds and acts as a proofreader for the formation of native disulfide bonds. DsbC is kept reduced by another membrane modulator DsbD. Although DsbA and DsbC catalyze the redox reactions in opposite directions, synergy between them has been reported to facilitate protein folding by reducing mismatched disulfide pairs and forming the proper ones [8,18,27].
In this study, the effects of Dsb family on 3F3 Fab production, especially its heavy chain and light chain assembly, were investigated. Particularly, 3F3 Fab was co-expressed with either DsbA/C alone or with both DsbA and DsbC proteins, and the expression levels of Fab as well as unassembled heavy and light chains were analyzed. Results indicated that when phoA promotor was used for Fab expression, DsbC is beneficial for Fab assembly (Fig 3); while under Lac promotor, DsbA significantly improved Fab production and decreased LC dimer amounts (Fig 4). Presumably, the fast expression rate of Fab driven by Lac promoter required more DsbA to facilitate the formation of disulfide bonds; while at a low expression rate under phoA promoter, resolving the non-native disulfide bridges by DsbC was apparently more important. All these results demonstrated that the optimal folding factor(s) for disulfide protein formation was sensitive to various factors, therefore, needs to be studied case-by-case.
To promote Fab formation, this study applied a sequential induction protocol, in which DsbA/C (under PBAD, Fig 2A) were induced first, and cells were cultured for 30 min before Fab induction (under PLac, Fig 4A). Presumably, this approach allowed Fab to be synthesized with the presence of DsbA/C. Using this method, we optimized the promoter (Fig 3), host (Fig 4), culture temperature (Figs 4&5), and media (Fig 5), to achieve the yield of 30 mg purified 3F3 Fab per liter of culture, a four orders of magnitude improvement compared to initial expression result (10 μg/L, Fig 1).
We further demonstrated that co-expressing DsbA and DsbC promoted assembly of aglycosylated IgG in E. coli periplasm (Supplementary Fig 1). Due to toxicity of heterologous proteins to the expression host, it is not uncommon that over-expression often results in unhealthy growth and low cell density. Our results demonstrate that by co-expressing folding factor DsbA/C, the cell culture OD600 dramatically increased (Supplementary Fig 1A). We further showed that construct DsbAC-2P with EnBase media led the production of 2 mg aglycosylated full IgG per liter culture (Supplementary Fig 1B).
In summary, antibody fragment assembly in E. coli periplasm was significantly improved by disulfide bond proteins DsbA and DsbC, and this approach can be applied for production of other important proteins containing disulfide bonds.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation the Faculty Early Career Development (CAREER) Program 1453645, and National Institutes of Health Grant R01GM115672. C.R. received UCR Chancellor’s award for Excellence in Undergraduate Research.
Abbreviations
- CDR
complementarity determining region
- DsbA/C
disulfide bond A and C
- ELISA
enzyme linked immunosorbent assay
- Fab
fragment of antigen binding
- FRET
fluorescence resonance energy transfer assay
- IgG
immunoglobulin G
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MMP
matrix metalloprotease
- PCR
polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- VH
variable heavy
- VL
variable light
Footnotes
Author contributions: C.R., D.H.N. and X.G. designed research; C.R., D.H.N. and E.K. performed research; C.R., D.H.N. and X.G. analyzed data; and C.R., D.H.N. and X.G. wrote the paper.
References
- 1.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nature Biotechnology. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 2.Georgiou G, Segatori L. Preparative expression of secreted proteins in bacteria: Status report and future prospects. Current Opinion in Biotechnology. 2005;16:538–545. doi: 10.1016/j.copbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes and Development. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlapschy M, Skerra A. Periplasmic chaperones used to enhance functional secretion of proteins in E. coli. Methods in Molecular Biology. 2011;705:211–224. doi: 10.1007/978-1-61737-967-3_12. [DOI] [PubMed] [Google Scholar]
- 5.Levy R, Weiss R, Chen G, Iverson BL, Georgiou G. Production of correctly folded Fab antibody fragment in the cytoplasm of Escherichia coli trxB gor mutants via the coexpression of molecular chaperones. Protein Expression and Purification. 2001;23:338–347. doi: 10.1006/prep.2001.1520. [DOI] [PubMed] [Google Scholar]
- 6.Miot M, Betton JM. Protein quality control in the bacterial periplasm. Microbial Cell Factories. 2004;3:4. doi: 10.1186/1475-2859-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duguay AR, Silhavy TJ. Quality control in the bacterial periplasm. Biochimica et Biophysica Acta. 2004;1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu J, Swartz JR, Georgiou G. Expression of active human tissue-type plasminogen activator in Escherichia coli. Applied and Environmental Microbiology. 1998;64:4891–4896. doi: 10.1128/aem.64.12.4891-4896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makino T, Skretas G, Kang TH, Georgiou G. Comprehensive engineering of Escherichia coli for enhanced expression of IgG antibodies. Metabolic Engineering. 2011;13:241–251. doi: 10.1016/j.ymben.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons LC, Reilly D, Klimowski L, Shantha Raju T, Meng G, Sims P, Hong K, Shields RL, Damico LA, Rancatore P, Yansura DG. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. Journal of Immunological Methods. 2002;263:133–147. doi: 10.1016/s0022-1759(02)00036-4. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer JV, Plückthun A. Improving expression of scFv fragments by co-expression of periplasmic chaperones. Antibody Engineering. 2010;2:345–361. [Google Scholar]
- 13.Nam DH, Rodriguez C, Remacle AG, Strongin AY, Ge X. Active-site MMP-selective antibody inhibitors discovered from convex paratope synthetic libraries. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14970–14975. doi: 10.1073/pnas.1609375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reilly DE, Yansura DG. Production of Monoclonal Antibodies in E. coli. In: Shire SJ, Gombotz W, Bechtold-Peters K, Andya J, editors. Current Trends in Monoclonal Antibody Development and Manufacturing. Springer; New York, NY: 2010. pp. 295–308. [Google Scholar]
- 15.Fellouse F, Sidhu S. Making Antibodies in Bacteria. In: Howard GC, Kaser MR, editors. Making and Using Antibodies A Practical Handbook. 2. Bocca Raton: Taylor & Francis/CRC Press; 2006. pp. 157–180. [Google Scholar]
- 16.Mazor Y, Van Blarcom T, Mabry R, Iverson BL, Georgiou G. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nature Biotechnology. 2007;25:563–565. doi: 10.1038/nbt1296. [DOI] [PubMed] [Google Scholar]
- 17.Bryksin AV, Matsumura I. Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. BioTechniques. 2010;48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segatori L, Paukstelis PJ, Gilbert HF, Georgiou G. Engineered DsbC chimeras catalyze both protein oxidation and disulfide-bond isomerization in Escherichia coli: Reconciling two competing pathways. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10018–10023. doi: 10.1073/pnas.0403003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause M, Ukkonen K, Haataja T, Ruottinen M, Glumoff T, Neubauer A, Neubauer P, Vasala A. A novel fed-batch based cultivation method provides high cell-density and improves yield of soluble recombinant proteins in shaken cultures. Microbial Cell Factories. 2010;9:11. doi: 10.1186/1475-2859-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam DH, Ge X. Development of a periplasmic FRET screening method for protease inhibitory antibodies. Biotechnology and Bioengineering. 2013;110:2856–2864. doi: 10.1002/bit.24964. [DOI] [PubMed] [Google Scholar]
- 21.Nam DH, Ge X. Direct production of functional matrix metalloproteinase-14 without refolding or activation and its application for in vitro inhibition assays. Biotechnology and Bioengineering. 2016;113:717–723. doi: 10.1002/bit.25840. [DOI] [PubMed] [Google Scholar]
- 22.Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Letters. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 23.Harvey BR, Georgiou G, Hayhurst A, Jeong KJ, Iverson BL, Rogers GK. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9193–9198. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider EL, Lee MS, Baharuddin A, Goetz DH, Farady CJ, Ward M, Wang CI, Craik CS. A reverse binding motif that contributes to specific protease inhibition by antibodies. Journal of Molecular Biology. 2012;415(4):699–715. doi: 10.1016/j.jmb.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez T, Nam DH, Kaihara E, Mustafa Z, Ge X. Identification of highly selective MMP-14 inhibitory Fabs by deep sequencing. Biotechnology and Bioengineering. 2017 doi: 10.1002/bit.26248. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joly JC, Swartz JR. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry. 1997;36:10067–10072. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 27.de Marco A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microbial Cell Factories. 2009;8:26. doi: 10.1186/1475-2859-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.