Abstract
Trophoblast stem cells (TSCs) are crucial for embryo implantation and placentation. Environmental toxicants that compromise TSC function could impact fetal viability, pregnancy, and progeny health. Understanding the effects of low, chronic EDC exposures on TSCs and pregnancy is a priority in developmental toxicology. Differences in early implantation between primates and other mammals make a nonhuman primate model ideal. We examined effects of chronic low-level exposure to atrazine, tributyltin, bisphenol A, bis(2-ethylhexyl) phthalate, and perfluorooctanoic acid on rhesus monkey TSCs in vitro by RNA sequencing. Pathway analysis of affected genes revealed negative effects on cytokine signaling related to anti-viral response, most strongly for atrazine and tributyltin, but shared with the other three EDCs. Other affected processes included metabolism, DNA repair, and cell migration. Low-level chronic exposure of primate TSCs to EDCs may thus compromise trophoblast development in vivo, inhibit responses to infection, and negatively affect embryo implantation and pregnancy.
Key words/phrases: atrazine, perfluorooctanoic acid, phthalate, obesogen, tributyltin, bisphenol A, transcriptome, implantation, gene-environment interaction, developmental origins of disease
Introduction
Endocrine disrupting chemicals (EDCs) are widespread in the environment and detectable in serum, cord blood, placenta and other tissues, demonstrating chronic low-level exposure outside of occupational exposures in at least some populations [1]. Animal model studies have revealed adverse effects of EDCs on fetal development [2], and on embryo implantation, placental cells, and pregnancy [3]. Additional studies revealed adverse effects on trophoblast cell viability, lipid metabolism, in vitro invasiveness, and steroid biogenesis, along with disruptions in the expression of select genes examined [4-14]. EDCs can also negatively affect the immune system, which is also crucial for embryo implantation and defense of fetus and placenta against infection. Atrazine, for example, negatively impacts immune systems in a range of organisms, including effects in developing fish and both age-and sex-dependent effects in mammals [15, 16]. These adverse effects on placenta and trophoblast are of concern due to the crucial role of the placenta in mammalian reproduction, and the impact of placenta function on progeny health.
Investigating the effects of EDCs on early implantation and placenta formation presents challenges because most animal models are phylogenetically distant from humans with significant differences in anatomical structure [17, 18]. While Old World nonhuman primates, such as rhesus monkeys, share many reproductive features with humans, in vivo studies with this species can be prohibitively expensive. The function of trophoblast cells in the rhesus placenta is very similar to humans, including the nature of the interhemal barrier during trophoblast invasion [19]. Implantation and early placenta function are particularly difficult to study in vivo due to relative inaccessibility of embryos and early implantation sites for observation. Nonhuman primate cytotrophoblast cells recovered from term placentas have been used for decades for in vitro studies to elucidate early placenta function [20], but that model has limitations. More recently, trophoblast stem cells (TSCs) generated directly from rhesus monkey embryos have been characterized and utilized to understand better trophoblast function pathways [20, 21].
Developing an in vitro placenta model to test the effects of EDCs on early pregnancy faces many challenges. Trophoblast cells display unique patterns of gene expression and specialized modes of epigenetic gene regulation, including genome imprinting and X chromosome dosage compensation [22-25], and a global pattern of DNA hypomethylation relative to embryonic stem cells (ESCs) and somatic tissues [26]. These unique properties of trophoblast cells limit the ability of data from somatic lineage tissues and cells to be extrapolated to understand potential trophoblast sensitivities and responses to environmental insults, such as EDC exposure. Additionally, in vitro studies employing either high concentrations and/or short durations of EDC exposure do not accurately model actual environmental exposure and do not capture potential long-term adaptive responses in target cells. Similarly, studies employing transformed cell lines and rodent models do not precisely model human trophoblast function and placentation. Moreover, studies that examine effects of EDCs on limited sets of marker genes provide limited mechanistic understanding.
To understand better the potential effects of chronic low-level exposure to EDCs on embryo implantation and potential impact on pregnancy, we applied low, environmentally relevant doses of five EDCs [atrazine (ATR), tributyltin (TBT), bisphenol A (BPA), bis(2-ethylhexyl) phthalate (DEHP), and perfluorooctanoic acid (PFOA)] to a rhesus monkey trophoblast stem cell (TSC) line in vitro for four weeks. These EDCs were selected because of their prevalence in the environment and numerous reports of effects on developing systems (reviewed, [1]). Atrazine is widely used as a pesticide, particularly in agricultural areas growing corn, detectable in some water supplies [27], is associated with CNS, endocrine, obesity, insulin resistance, cancer, mitochondrial dysfunction, immune system, and other effects, and may work through multiple mechanism, including epigenetic changes [28]. Perinatal and lactational ATR exposure also affects progeny immune function [16, 29]. Tributyltin use has been discontinued, but remains present in the environment, and has obesogenic activities, in part attributable to transcriptional signaling and changes in DNA methylation effects, and rts diverse effects on reproduction mediated by interference with endocrine signaling (e.g., follicle stimulating hormone, testosterone, aldosterone, estradiol), as well as carcinogenesis and respiratory system effects [33]. BPA is also widely used in household products and prevalent in the environment, has been extensively studied for its estrogenic effects, but impacts other endocrine pathways, and has non-estrogenic effects and effects on DNA methylation [34, 35]. PFOA and other perfluoroalkyl substances are widespread in the environment around the world, impact estrogen and thyroid signaling, have immunotoxic effects, and are associated with negative effects on fetal growth and other health effects [36]. The four-week treatment period was selected to encompass four passages during treatment, in order to allow time for any DNA replication-dependent epigenetic changes that would generate stable changes in gene expression and phenotype, and thereby reveal potential effects of low-level, constant, environmentally relevant exposures on the expression of genes contributing to trophoblast functions, without the complications of acute toxicity. Because the intent was to assess effects of environmentally relevant exposures without overt toxicity, a single comparatively low concentration was selected for each compound. We then determined the global effects of these treatments on cellular phenotype by RNA deep sequencing (RNAseq) followed by Ingenuity Pathway Analysis to identify major affected biological pathways, processes and functions, and pertinent upstream regulators associated with those effects. The combination of RNAseq and pathway analysis thus provided a global assessment of altered functional states of TSCs following low level chronic EDC exposure.
To our knowledge, this is the first systematic and in-depth analysis of the effects of chronic low-level EDC exposure on trophoblast gene expression in an animal model closely related to the human. The major outcomes of the analysis and the potential relevance of these results to human reproductive health and risk assessment are discussed.
Materials and Methods
Cells and cell culture
TSCs (line 119-T) were isolated previously from rhesus monkey blastocysts and characterized using a panel of antibodies to detect TSC biomarkers [21]. On the basis of X-and Y-linked gene expression in this study (absence of expression of two Y-linked genes that are expressed in male blastocysts, RPS4Y1, RPS4Y2, and biallelic expression of genes that escape X chromosome inactivation), we determined that the 119-T TSCs were derived from a female embryo. Additionally, to assess more thoroughly the transcriptional state of our cells, RNAseq data acquired in this study were used to compare marker gene expression profiles to those seen in rhesus monkey ORMES6 ESCs [37], and human embryo trophectoderm and epiblast lineages [38], and trophoblast cells [39, 40]. This included examination of 14 genes expressed more highly in human blastocyst trophectoderm (TE) cells compared to human epiblast, 15 additional genes reported as TE markers, and 13 genes expressed more highly in human epiblast as compared to TE cells [38].
TSCs were maintained in DMEM/F-12 (Invitrogen) supplemented with 15% fetal bovine serum (FBS; Hyclone), 1% minimum essential medium (MEM) nonessential amino acids (Invitrogen), 1 mM glutamine (Sigma), 0.1 mM β -mercaptoethanol, and 1× penicillin/streptomycin sulfate (Invitrogen) on plates coated with human placental collagen (Sigma) [21]. Medium was changed daily. The cells were grown to confluence and passaged weekly by rinsing wells with PBS, incubating with 1.0 mL trypsin-EDTA, and transferring 20% of cell suspension to each new well. Treatments (see below) began when cells first reached confluence and continued for 4 weeks, including during passage. Untreated and vehicle-treated control cultures were also processed for analysis. Cells in the different groups grew at the same rate. There was no apparent cell death and cultures in all groups reached confluence as expected prior to passage. Because there were no obvious effects on cell survival and because the levels of toxins applied were low and below toxic levels reported elsewhere, no detailed measurements of cell survival were made. Expression of alpha fetoprotein, CDX2, cytokeratin 7, vimentin and hCG were consistent between start and end of study (data not shown).
After 4 weeks of treatment with the toxicant or control media, cells were detached with 0.25% trypsin-EDTA (Gibco). They were then centrifuged at 3000 × g for 3 min, supernatant removed, and the cell pellet resuspended in 100 μL Picopure lysis buffer (Applied Biosystems). Lysates were stored at -80°C until shipment.
The study design provides a relatively continuous low-dose toxicant exposure, which is likely a more realistic model of implantation, as well as the fetal and placental compartments during pregnancy. In the case of BPA, for example, urinary clearance occurs through liver glycosylation, and thus the level of unconjugated BPA exposure is dependent on the route of exposure. Non-oral exposure results in higher levels of unconjugated BPA than oral doses that are passed through the liver after absorption in the gastro-intestinal track [41-43]. Human exposure to BPA through dermal, buccal and inhalation routes can lead to essentially continuous levels of serum BPA. The levels of toxicants that may be present in the fetal and placental compartments are even more complex. Very few studies have been performed that compare maternal and fetal levels of toxicants over time. A recent study of BPA exposure in rhesus monkeys showed that BPA quickly entered the fetal compartment and that rapid maternal glycosylation of BPA did not prevent fetal exposure [44]. In fact, that study supported the findings in other species that BPA metabolites may be trapped in the fetal compartment and that the placenta may have the ability to de-conjugate and prolong fetal exposure [45, 46]. Thus, the continuous level of exposure in this in vitro TSC model is likely most relevant to real world human exposure.
Chemicals for testing and treatment groups
The chemicals were purchased from Sigma (DEHP: Bis(2-ethylhexyl) phthalate – 47994; PFOA: Perfluorooctanoic acid – 171468; ATR: Atrazine – 45330; TBT: Tributyltin chloride – 442869; BPA: Bisphenol A – 133027). Treatment groups included a vehicle control (methanol) and each of five EDCs at environmentally relevant doses [10 nM BPA, 5 μM DEHP, 30 μM ATR, 100 nM PFOA, and 25 nM TBT], as described in our previous studies of rhesus monkey embryonic stem cells [37]. The vehicle treated cultures, receiving equal amounts of vehicle as toxicant treated cultures, were controls for possible effects of vehicle and other characteristics of the culture system. EDCs for testing were made as 1000× stocks in methanol and were added to aliquots of DMEM/F12 medium up to 48 hours before use and stored at 4°C until use. The above concentrations are generally within the ranges reported for human serum and drinking water, and in studies of responses of mammalian oocytes, preimplantation stage embryos, or pluripotent cells to the chemicals in vitro [47-52]. The ATR concentration is higher than the maximum concentration reported in one study for drinking water [53] and human serum level (up to 245 nM) [54], but affects placenta cell gene expression in vitro [55] and is much lower than the doses (200-300 mg/kg) typically applied in rodent studies to test for reproductive effects. The EDCs selected for study affect rhesus monkey gonads, embryo or fetal development, and progeny phenotype, as well as human reproductive tissues or stem cells. For example, 10-15 nM BPA affects fetal lung and mammary gland development [56, 57], DEHP at 25 μM affects monkey Sertoli cell development [58], TBT (100 nM) modifies human embryonal carcinoma cells [59, 60], and ATR (200 mg/kg) broadly affects vertebrate gonadogenesis [61]. It should also be noted that the concentrations of EDCs applied were chosen to avoid overt toxicity. For example, ATR shows only limited toxicity to human trophoblast cells (30%) at 1 mM concentration [62]. For each treatment, an assessment of toxicity and cell death is also achieved in the course of the transcriptome analysis.
Preparation and sequencing of libraries for RNAseq
Six replicate cultures of untreated control, vehicle treated control, and EDC treated cells were treated in parallel during a single treatment period. Each replicate culture was maintained in a separate well, even after passage and was never mixed or pooled with cells from any other well throughout the experiment. The replicate cultures were processed for RNA extraction and RNAseq analysis. RNA was isolated following the PicoPure™ RNA Extraction kit manufacturer protocol, with DNAse digestion to remove any contaminating DNA. To produce libraries for sequencing, 100 ng of each RNA sample were processed first using a mixture of random and oligo(dT) primers and reverse transcription to generate double stranded cDNA using the Ovation Universal RNA-Seq System (NuGen, San Carlos, CA). This was followed by cDNA fragmentation to an average of 300 bp using a Covaris-2 sonicator, and then a brief S1 nuclease digestion as described [63]. After purification, the cDNA was processed further through the Ovation Universal RNA-Seq System (NuGen) for end repair, barcoding, InDA-C mediated ribosomal RNA depletion, and final library production with the addition of unique nucleotide barcodes to each library. Barcoded libraries were pooled and sequenced with an Illumina HiSeq 4000 to generate 50 nt single end reads. The total numbers of PF (passed-filter) reads ranged from 17.2 M to 55.0 M, the fraction of Q30 bases from 96.6% to 97.0% and average Q from 39.3 to 39.4 (Table S1). Sequencing data will be available in Gene Expression Omnibus (GSE103033) and at our Primate Embryo Gene Expression Resource (www.preger.org).
RNAseq data analysis and Ingenuity Pathway analysis
Reads were aligned to the rhesus monkey genome (MacaM v7, [64]) using HISAT2 [65]. Reads aligned to ribosomal RNA (rRNA) or rRNA-like genes were removed, as were the “ExAmp” duplicates – caused by the sequencing technology – which were defined as one read in a pair of identical reads found within the distance of 2500 units on the same tile of a sequencing lane. After analysis of clusters using Multidimensional Scaling (Figure S1), libraries considered to be outliers in their respective treatment groups (one Control, one DEHP, three PFOA and one TBT) were removed from further analyses. A total of 3.2M to 14.2M reads per library were successfully aligned to unique non-rRNA gene transcript sequences (Table S1). Cuffdiff [66] was used for quantification and differential expression analyses between the vehicle and five EDC treatment groups. Differentially expressed genes (DEGs) were defined as those with q-value (false discovery rate) ≤ 0.05.
QIAGEN Ingenuity Pathway Analysis® (IPA) was used to analyze the biological relevance of DEGs. Analysis tools applied from IPA included Canonical Pathway (CP), Disease and Functions (DF), and Upstream Regulator (UR). For CP analysis, IPA calculates overlap p-values, taking into account the number of DEGs and the number of molecules in the knowledge database associated with that pathway, and the number of DEGs and the number of molecules in the knowledge database. For DF analysis, overlap p-values are based on the number of DEGs associated with increase or decrease of a given DF. For UR analysis, results are based on the number of DEGs regulated by a given UR. In addition to overlap p-values, z-scores are calculated for CPs, DFs, and URs. The z-score reflects activation (z>0) or inhibition (z<0) of CPs and URs, or increase (z>0) or decrease (z<0) of DFs, and is based on the number of associated DEGs for which the direction of regulation (up- or down-) is consistent with activation/increase or with inhibition/decrease. Because P(|z|>1.96) ∼ 0.05 for normal N(0,1) distribution, we consider CPs, URs and DFs with z>1.96 to be significantly activated or increased, and those with z < -1.96 to be significantly inhibited or decreased.
Results
Overview of TSC gene expression characteristics and their responses to treatments
No changes were noted in growth rate or morphology characteristics of TSCs during treatment, which demonstrates that the doses selected were low enough to avoid acute toxicity. Comparisons of expression of human embryo trophectoderm and epiblast marker genes between our rhesus monkey TSC line and rhesus monkey ORMES6 in (Table S2) revealed that the 119-T TSCs closely resemble the human TE cells. Of the 29 TE marker genes, 22 displayed higher expression values in control TSCs as compared to control ESCs (2 qualitative differences). CLDN10, FHL1, HIP1, HMGCS1, TIPIN, ELF5, and EOMES were expressed at lesser levels in TSCs than ESCs among the TE markers. CGB mRNA was low but detectable in both cell types, as was ESR2. Excluding the above 9 markers, the median ratio of TSC:ESC mRNA expression was 10.1, with a maximum of >2000-fold higher expression in TSCs. All of the 13 markers of human epiblast were expressed at lower levels in TSCs. None of the marker genes was significantly affected by vehicle control treatment. We noted a high level of expression of fibronectin 1 (FN1) mRNA (a possible survival factor in trophoblast) across all samples, and this was also unaffected by EDC treatment (data not shown).
Because in female embryo-derived cells X-chromosome inactivation can be disrupted by a loss of DNA methylation, and the EDCs might affect DNA methylation, we examined the DEG lists from all five EDCs for possible over-representation of genes on the X chromosome. We used chi-square test to compare the fractions of X-chromosome genes in the sets of DEGs to the fraction of X-chromosome genes in the set of expressed genes (with FPKM ≥ 1). While for most EDCs (all except PFOA), the fraction of X-chromosome genes in the set of DEGs was slightly increased compared to the set of expressed genes (average of 4.4% vs. 3.5%), none of these differences were statistically significant at α = 0.05; the lowest p-values were obtained for TBT (p = 0.056) and ATR (p = 0.085).
The effects of low-level chronic exposure to the five EDCs on TSC gene expression profile and cell functions were determined (Supplemental Tables S3-S26). The number of genes with significantly (q ≤ 0.05) affected expression differed widely with the EDC applied (Figure 1). The numbers of affected genes with statistical significance (q ≤ 0.05) were modest for BPA, DEHP, and PFOA (n = 115, 112, & 32 total, and n= 6, 50, & 14 at FC ≤ 1.5, respectively). Observed effects were much greater with ATR and TBT. ATR treatment significantly altered the expression of 1491 genes (431 > 1.5-fold). TBT treatment significantly altered the expression of 1961 genes (623 > 1.5-fold). A lack of toxicity was confirmed by IPA results for all five treatments, showing either no change or a decrease in cell death-related pathways (Table 1, excerpted from Tables S4, S8, S12, S16, and S20). With the exception of a result of reduced cell viability results for two IPA DF annotations for DEHP treatment, IPA results generally yielded no significant negative effects indicating increased cell death. For both ATR and TBT, which had the largest overall effects on cells, two or more IPA results indicated reduced cell death, and one IPA result for ATR indicated increased viability. Details of the effects for each treatment are provided in the sections below.
Figure 1.
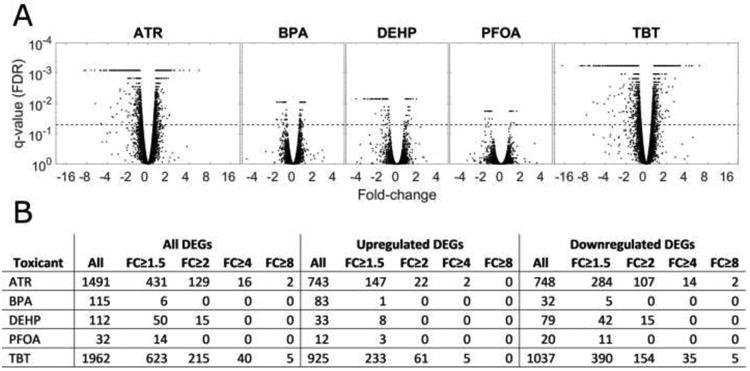
Summary of differential gene expression in toxicant treated samples: A) Volcano plots showing q-values (false discovery rate; genes above the dashed lines are considered differentially expressed) and fold-change (positive for upregulated, negative for downregulated genes); B) Number of differentially expressed genes (DEGs) for five toxicant treatments, broken down by direction (upregulated and downregulated) and magnitude of fold change (FC).
Table 1. Effects of treatments on IPA® Diseases & Functions related to cell death and survival.
| IPA Diseases & Functions | PFOA | BPA | z-score 1 DEHP | ATR | TBT |
|---|---|---|---|---|---|
| Necrosis | 1.885 | -1.109 | |||
| necrosis of liver | 1.067 | ||||
| necrosis epithelial cells | 1.054 | ||||
| necrosis prostate cancer cells | -1.555 | ||||
| Apoptosis | 1.434 | 1.037 | -1.723 | ||
| apoptosis tumor cells | 1.386 | ||||
| apoptosis leukocytes | -0.851 | ||||
| apoptosis breast cancer | 1.925 | ||||
| apoptosis prostate cancer cells | -1.660 | ||||
| cell death of immune cells | -1.237 | -2.139 | -2.449 | ||
| cell death of lymphocytes | -1.640 | -1.924 | |||
| cell death of macrophages | -1.853 | ||||
| cell death of epithelial cells | 1.150 | ||||
| cell death of osteosarcoma cells | -3.162 | ||||
| cell death of cancer cells | -3.049 | ||||
| cell death tumor cells | -3.049 | ||||
| cell death connective tissue | -1.201 | ||||
| cell death fibroblasts | 1.066 | ||||
| cell death of blood cells | -2.238 | -2.236 | |||
| cell death | -0.992 | ||||
| cell death cervical cancer | -0.847 | ||||
| cell survival | -2.981 | ||||
| cell viability cancer/carcinoma | -1.964 | 2.066 | |||
| cell viability | 2.952 | ||||
| cell proliferation | 2.025 |
Z-scores indicating significant increase (z > 1.96) or decrease (z < - 1.96) of disease/functions are shown in bold.
Effects of PFOA
PFOA treatment yielded the smallest number of significant differences in gene expression (32 genes, highest fold-change 1.74) (Figure 1, Table S3). The small number of PFOA treated libraries included in the analysis may have limited detection of some gene expression effects. The IPA results for affected diseases and biological functions (DFs) revealed significant decreases in several biological functions including cell movement, epithelial tissue growth, and vasculogenesis (Table S4). No significant z-scores were returned for CP analysis (Table S5), but pathways with significant overlap of PFOA affected DEGs were seen related to cysteine metabolism, and signaling through interleukins 6, 10, and 17A, Tolllike receptor, TGF-β, PDGF, PPAR, MAPK, Endothelin 1, TNRF2, and tight junctions. Upstream regulators (UR) analysis of the observed effects indicated significant inhibition of actions of several cytokines including IFNγ and IFNα (Table S6). UR analysis also revealed potential inhibition (z-score -1.407) related to FOS signaling (a regulator of trophoblast function, [67]), but note that FOS mRNA expression itself is increased by PFOA treatment.
Effects of BPA
Cultures treated with BPA displayed significant effects on gene expression of 115 genes, with highest fold-change of 1.77 (Figure 1, Table S7) and more than twice as many genes being upregulated than downregulated. The IPA DF analysis revealed significant decreases for cancer, cell death, cell proliferation, cell viability, and protein translation (Table S8). A significant z-score was obtained for EIF2 signaling, and other significantly affected (p ≤ 0.05) CPs included regulation of EIF4 and p7056k signaling, MTOR signaling, oxidative phosphorylation, mitochondrial dysfunction, and nucleotide excision pathway (Table S9). UR analysis indicated significant activation of gene regulation by MYCN, MYC, MAPK1, MTOR, GATA1, and CEBPA, and inhibition of gene regulation by RICTOR, IFNγ IFNL1, and CD28, and lesser effects related to other regulators, including XBP1, PPARGC1A, NUPR1, NKX2-3, TGFB1, FOS, HRAS, and others (Table S10).
Effects of DEHP
DEHP treatment yielded significant effects on 112 genes, with over 70% of them being downregulated (Figure 1, Table S11). Unlike for PFOA and BPA treatment, 13% of genes were affected by more than 2-fold, and the highest change was 3.98-fold. Genes with the largest fold-changes included genes related to trophoblast development and implantation (e.g., FOSB, EGR1, WNT7A, HAND1, INHBA, keratins) and immunomodulation (e.g., CEACAM6, and interferon-regulated genes). The IPA DF analysis revealed a significant decrease of cell growth and proliferation, cell invasion, endothelial development, and inflammatory response (Table S12). Canonical Pathway analysis revealed no significantly activated or inhibited pathways, but significant overlap (p ≤ 0.05) for Toll-receptor signaling, and TNF and cytokine signaling (Table S13). The CP effects were also evident in the UR analysis, which revealed significant inhibition for gene regulation by TNF, TGFB1, PDGF, interferons and cytokines, and LPS (Table S14).
Effects of ATR
Our analysis revealed effects on ten times as many genes with ATR treatment than with BPA, PFOA, or DEHP, with effects ranging to as high as 8.62-fold (Figure 1, Table S15). The most prominent effects included reduced expression of interferon-regulated and anti-viral genes (e.g., IFI44, IFI27, IFI44L, IFI35, IFITM1, OASL, APOBEC3G), TNF and cytokine signaling related genes (e.g. TNFSF18), immunomodulation genes (e.g., CEACAM6), genes related to trophoblast function (e.g., HAND1, MAMUF, INHBA) and other signaling pathway genes. Among top DF analysis results from IPA were decrease of metabolic disease, cell death (including cell death of immune cells) and cell movement, and increase of viral replication, cell division, cell viability, and protein synthesis (Table 2, S16). The z-scores from the IPA CP analysis revealed significant activation for EIF2 signaling (protein synthesis), embryonic stem cell pluripotency, spingosine-1-phosphate signaling, and mitotic role of PLK1, and significant inhibition of DNA damage checkpoint and interferon signaling (Table 3, S17). High z-scores below the 1.96 significance threshold were also seen for STAT3 signaling, VDR/RxR activation, and other key signaling pathways. The upstream regulator analysis revealed strong inhibition of responses driven by interferon signaling, LPS signaling, TNF signaling, anti-viral functions, and immune function (Table 4, S18). These effects encompassed strong inhibition of signaling through multiple pro-inflammatory and anti-inflammatory mediators, such as IFNG, LPS, IFNA, IFNB, TNF, multiple IRFs, multiple interleukins, STAT1, and OSM, as well as inhibition of signaling through immunomodulatory TLR3. Many of the implicated upstream regulators related to cytokine signaling and inflammatory response were themselves partially repressed at the mRNA level by ATR, including IRF7, TNF, IFNL1*, IL1B*, IFNB1, STAT1*, IRF1, TLR3*, TMEM173*, DDX58*, and STAT2 (* denotes statistically significant reduction in expression). Regulators of other functions also displayed repression, such as MYD88 (cytokine and TOLL receptor signaling), and PTGER4 (possible role in implantation, and itself significantly downregulated) (Table S18). There was also activation of genes regulated by TRIM24 (itself significantly increased in expression), a mediator of estrogen signaling. Overall, these results indicate a sweeping repression of cytokine signaling mechanisms in ATR treated trophoblast cells, with additional compromise in other trophoblast-related functions and marker genes.
Table 2. Top 20 Increased and Decreased IPA® Diseases and Functions in ATR and TBT Treated Samples.
| Toxicant: ATR | Toxicant: TBT | ||||||
|---|---|---|---|---|---|---|---|
| Diseases/Functions | Predicted Activation State and z-score 1 | # DE Genes 2 | Diseases/Functions | Predicted Activation State and z-score 1 | # DE Genes 2 | ||
| replication of virus | Increased | 4.003 | 120 | replication of Herpesviridae | Increased | 3.714 | 23 |
| replication of RNA virus | Increased | 3.516 | 107 | replication of virus | Increased | 3.094 | 131 |
| Viral Infection | Increased | 3.142 | 279 | disorder of basal ganglia | Increased | 2.878 | 155 |
| replication of Flaviviridae | Increased | 2.825 | 25 | migration of connective tissue cells | Decreased | -2.760 | 39 |
| glucose metabolism disorder | Decreased | -2.783 | 159 | glucose metabolism disorder | Decreased | -2.579 | 207 |
| replication of Hepatitis C virus | Increased | 2.562 | 20 | cell movement of connective tissue cells | Decreased | -2.563 | 51 |
| replication of Herpesviridae | Increased | 2.511 | 20 | cell death of immune cells | Decreased | -2.449 | 144 |
| S phase of fibroblasts | Increased | 2.387 | 13 | replication of RNA virus | Increased | 2.398 | 120 |
| squamous-cell carcinoma | Increased | 2.345 | 119 | contact growth inhibition | Decreased | -2.395 | 39 |
| re-entry into S phase | Increased | 2.254 | 12 | progressive motor neuropathy | Decreased | -2.353 | 105 |
| cell death of blood cells | Decreased | -2.238 | 120 | squamous-cell carcinoma | Increased | 2.331 | 141 |
| interphase of fibroblasts | Increased | 2.232 | 19 | cell death of blood cells | Decreased | -2.236 | 150 |
| interphase of connective tissue cells | Increased | 2.232 | 20 | formation of muscle | Decreased | -2.161 | 55 |
| colony formation | Increased | 2.221 | 97 | cell death of lymphoid organ | Decreased | -2.160 | 37 |
| survival of organism | Increased | 2.218 | 143 | synthesis of reactive oxygen species | Decreased | -2.096 | 97 |
| diabetes mellitus | Decreased | -2.195 | 133 | cell movement of fibroblasts | Decreased | -2.095 | 43 |
| cell death of immune cells | Decreased | -2.139 | 113 | metabolism of reactive oxygen species | Decreased | -2.047 | 98 |
| expression of prot. | Increased | 2.114 | 38 | G1 phase of tumor cell lines | Decreased | -2.046 | 53 |
| Dysplasia | Decreased | -2.089 | 55 | cell proliferation of breast cell lines | Decreased | -2.003 | 36 |
| cell viability of carcinoma cell lines | Increased | 2.066 | 33 | formation of actin filaments | Decreased | -1.986 | 57 |
Activation state is predicted Increased for z > 1.96 and Decreased for z < -1.96.
Number of genes affected by toxicant treatment that are involved in function/disease.
Table 3. Top 10 Activated and Inhibited IPA® Canonical Pathways in ATR and TBT Treated Samples.
| Toxicant: ATR | Toxicant: TBT | ||||||
|---|---|---|---|---|---|---|---|
| Ingenuity Canonical Pathways | Predicted Activation State and z-score 1 | # DE Genes2 | Ingenuity Canonical Pathways | Predicted Activation State and z-score 1 | # DE Genes2 | ||
| EIF2 Signaling | Activated | 4.041 | 38 | Interferon Signaling | Inhibited | -2.309 | 12 |
| Mouse Embryonic Stem Cell Pluripotency | Activated | 2.673 | 14 | Cell Cycle: G2/M DNA Damage Checkpoint Regulation | Inhibited | -2.121 | 10 |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | Inhibited | -2.449 | 9 | Mouse Embryonic Stem Cell Pluripotency | 1.789 | 20 | |
| Role of BRCA1 in DNA Damage Response | Inhibited | -2.236 | 12 | Death Receptor Signaling | -1.698 | 17 | |
| Sphingosine-1-phosphate Signaling | Activated | 2.183 | 17 | VDR/RXR Activation | -1.633 | 14 | |
| Mitotic Roles of Polo-Like Kinase | Activated | 2.121 | 9 | Activation of IRF by Cytosolic Pattern Recognition Receptors | -1.604 | 14 | |
| Interferon Signaling | Inhibited | -2.111 | 11 | Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes | 1.604 | 14 | |
| STAT3 Pathway | 1.897 | 10 | IL-8 Signaling | 1.567 | 36 | ||
| VDR/RXR Activation | -1.890 | 11 | α-Adrenergic Signaling | 1.508 | 13 | ||
| IGF-1 Signaling | 1.732 | 16 | SAPK/JNK Signaling | 1.500 | 16 | ||
Activation state is predicted Activated if z-score > 1.96 and Inhibited for z-score < -1.96.
umber of genes affected by toxicant treatment that are involved in function/disease.
Table 4. Top 20 Activated and Inhibited IPA® Upstream Regulators in ATR and TBT Treated Samples.
| Toxicant: ATR | Toxicant: TBT | ||||||
|---|---|---|---|---|---|---|---|
| Upstream Regulator | Predicted Activation State and z-score 1 | # DE Genes 2 | Upstream Regulator | Predicted Activation State and z-score 1 | # DE Genes 2 | ||
| IFNG | Inhibited | -8.278 | 197 | IFNG | Inhibited | -7.802 | 249 |
| lipopolysaccharide | Inhibited | -7.115 | 248 | lipopolysaccharide | Inhibited | -6.957 | 305 |
| IRF7 | Inhibited | -7.095 | 57 | ↓IRF7 | Inhibited | -6.851 | 62 |
| IFNA2 | Inhibited | -6.932 | 65 | IFNA2 | Inhibited | -6.676 | 78 |
| Interferon α | Inhibited | -6.793 | 79 | Interferon α | Inhibited | -6.605 | 86 |
| poly rI:rC-RNA | Inhibited | -6.782 | 87 | poly rI:rC-RNA | Inhibited | -6.308 | 104 |
| TNF | Inhibited | -6.624 | 255 | ↓IFNL1 | Inhibited | -6.241 | 40 |
| ↓IFNL1 | Inhibited | -6.114 | 38 | IFN Beta | Inhibited | -5.779 | 44 |
| ↓IL1B | Inhibited | -5.961 | 123 | IFNB1 | Inhibited | -5.591 | 72 |
| IFNB1 | Inhibited | -5.785 | 55 | ↓TNF | Inhibited | -5.468 | 323 |
| ↓STAT1 | Inhibited | -5.330 | 71 | Ifnar | Inhibited | -5.393 | 35 |
| IFN Beta | Inhibited | -5.330 | 37 | NKX2-3 | Activated | 5.314 | 82 |
| NKX2-3 | Activated | 5.197 | 69 | IRF3 | Inhibited | -5.269 | 55 |
| ↓IL1RN | Activated | 5.155 | 42 | IRF1 | Inhibited | -4.985 | 51 |
| IRF1 | Inhibited | -5.083 | 44 | EIF2AK2 | Inhibited | -4.933 | 33 |
| TLR9 | Inhibited | -5.019 | 38 | TLR7 | Inhibited | -4.928 | 29 |
| ↑TRIM24 | Activated | 5.000 | 37 | ↓STAT1 | Inhibited | -4.917 | 94 |
| IRF3 | Inhibited | -4.936 | 53 | TLR9 | Inhibited | -4.885 | 44 |
| MAPK1 | Activated | 4.845 | 91 | TRIM24 | Activated | 4.864 | 43 |
| Ifnar | Inhibited | -4.746 | 30 | CHUK | Inhibited | -4.839 | 53 |
Activation state is predicted Activated if z-score > 1.96 and Inhibited for z-score < -1.96.
Number of genes affected by toxicant treatment that are involved in functions/disease.
Effects of TBT
The largest number of significantly affected genes was achieved with TBT treatment, with effects ranging as high as 17-fold (CDH13) (Table S19). Many of the genes, DFs, CPs, and URs affected by ATR were also affected by TBT (Tables 2-4, S20-S22). As with ATR, many of the implicated upstream regulators related to cytokine signaling and inflammatory response were themselves repressed at the mRNA level by TBT, including IRF7* IFNL1* IFNB1, TNF* STAT1* TLR3* IL1RN, IL1B, TMEM173* STAT2* DDX58*. Other regulators also significantly reduced in expression included NFATC2 (cell invasion), as well as MYD88 and PTGER4. Subtle differences between ATR and TBT were evident in the IPA DF analysis, such as a stronger decrease of muscle formation, free radical scavenging and G1 phase categories. The CP analysis for TBT treatment yielded a positive z-score for embryonic stem cell pluripotency genes, below the level of significance but indicating a possible trend toward activation of pluripotency-related genes. Notable z-scores, although below the 1.96 significance threshold, also indicated potential decreases in death receptor signaling, VDR/RxR signaling, and IRF signaling, and increases in IL-8 and α-adrenergic signaling, and SAPK/JNK signaling. As with ATR treatment, TBT treatment yielded an overall repression of cytokine signaling mechanisms (see UR analysis), changes of gene expression consistent with increase of viral infection, and a decrease in other trophoblast-related functions and marker genes.
Overlap in EDC effects
The foregoing summaries of effects of the five EDCs on trophoblast cells indicate substantial overlap in effects. A summary of these overlaps is provided (Table 5, Tables S23, S24). A total of 892 genes were affected similarly (same direction) by ATR and TBT, accounting for 62 % of the 1491 genes affected by ATR (53% of upregulated and 66% of downregulated genes). Additionally, there were 27 genes affected oppositely between ATR and TBT. Three of the 32 genes affected by PFOA were increased by all five EDCs, six genes were decreased by all five EDCs, and two showed mixed effects. The greatest percentage overlaps in effects (>80%) were observed between ATR and DEHP, and between ATR and PFOA. A small number of genes were affected by multiple treatments but displayed differences in effects (increase or decrease) between the different treatments, particularly FOS, FOSB, MME, CAV1, and ATP6V0A4.
Table 5. Top 12 differentially expressed genes shared across treatment groups (from Table S24).
| Fold-change | Num. Tox. w/DE | |||||||
|---|---|---|---|---|---|---|---|---|
| ATR | BPA | DEHP | PFOA | TBT | GeomMean | |||
| IFI27 | interferon alpha inducible protein 27 | -8.18 | -1.34 | -1.54 | -1.55 | -9.43 | -3.01 | 5 |
|
| ||||||||
| DTX3L | deltex E3 ubiquitin ligase 3L | -4.39 | -1.57 | -2.07 | -1.74 | -4.75 | -2.60 | 5 |
|
| ||||||||
| KRT17 | keratin 17 | -2.87 | -1.48 | -1.99 | -1.50 | -5.97 | -2.38 | 5 |
|
| ||||||||
| CEACAM6 | carcinoembryonic antigen related cell adhesion molecule 6 | -3.19 | -1.67 | -3.07 | -1.72 | -1.73 | -2.17 | 5 |
|
| ||||||||
| INHBA | inhibin beta A subunit | -3.13 | -1.28 | -1.62 | -1.45 | -3.25 | -1.98 | 5 |
|
| ||||||||
| FOSB | FosB proto-oncogene, AP-1 transcription factor subunit | 1.69 | 1.45 | -3.98 | 1.50 | 1.95 | 1.95 | 5 |
|
| ||||||||
| MME | membrane metalloendopeptidase | -2.23 | -1.39 | -1.61 | -1.60 | 2.82 | -1.86 | 5 |
|
| ||||||||
| IL1RL1 | interleukin 1 receptor like 1 | -2.67 | -1.28 | -1.38 | -1.57 | -1.69 | -1.66 | 5 |
|
| ||||||||
| HMCN1 | hemicentin 1 | 2.45 | 1.30 | 1.93 | 1.43 | 1.29 | 1.63 | 5 |
|
| ||||||||
| ADIRF | adipogenesis regulatory factor | 2.02 | 1.25 | 1.33 | 1.35 | 2.15 | 1.58 | 5 |
|
| ||||||||
| FOXO4 | forkhead box O4 | 1.91 | 1.26 | 1.59 | 1.43 | 1.72 | 1.57 | 5 |
|
| ||||||||
| IFI44L | interferon induced protein 44 like | -6.07 | -1.27 | -1.36 | -5.33 | -2.73 | 4 | |
Substantial overlap was also evident in the IPA analysis. As indicated above, ATR and TBT displayed very similar effects in DF, CP and UR results. There are also features shared across all five or a majority of the five EDCs tested. This becomes most apparent in comparing the results of the UR analysis (Tables 6 and S25), as different subsets of downstream mediators may be affected by different treatments but impact a common process controlled by specific upstream regulators. IFNγ emerges as the top UR for all five treatments, with poly-rI:poly-rC RNA (activator of antiviral response), TGFB, NFKB, and FOS also showing inhibition for all five EDCs, albeit with some z-scores below the +/- 1.96 threshold. UR analysis further revealed some level of inhibition of downstream functions mediated by TNF, LPS, IFNL1, IL1B, STAT1, OSM, NFKB, IL1, and LPS in four of the five treatments, and TLR3 emerged for three of the EDCs. Suppression of PRL and TRIM24 response genes was also seen in four of the five treatments, and MYD88 regulated genes were affected in three of the treatments. Overall, sweeping inhibition in cytokine signaling, repression of pro-inflammatory and anti-inflammatory pathways, and suppression of anti-viral defense gene expression were the major effects of EDC treatment of trophoblast cells in vitro. This was accompanied by increased expression of genes associated with viral replication and infection in TBT and ATR treatments. Comparing the expression of genes regulated by specific upstream factors across the five treatments further emphasizes conservation of effects across the EDC treatments. Figure 2 shows the effects of five treatments on genes downstream of predicted upstream regulators, representing significant downregulation (q < 0.05) of a gene with green, significant (q < 0.05) upregulation with red, genes with reduced expression (blue) or increased expression (orange) expression for which the change was not statistically significant (q > 0.05), and overall activation/inhibition states of upstream regulators (+ and – annotations below heatmaps) Scanning left to right across the heat maps highlights that many of the UR effects are shared across two, three or more of the treatments. There is a substantial overlap of genes affected by ATR and TBT, and for most of these genes the direction of change is the same for TBT and ATR treatments. Furthermore, many of the genes that are significantly affected by ATR and/or TBT treatment show modulations in the same direction with the other treatment groups, some with large fold-changes. Specific z-scores for the upstream regulators in Figure 2 are provided in Table 6, which shows top ranked results for UR analysis ranked by sum of z-score. Z-scores exhibit consistent sign (positive or negative) for majority of the upstream regulators, further highlighting that these effects are shared across treatments. These results also highlight the shared aspect of strong downregulation of genes related to cytokine signaling, also seen in Table S25. Those effects on cytokine signaling may be functionally linked to effects on responsiveness to infection. Indeed, the effects evident in shared DFs (Table S26) are also consistent with increased viral replication (or susceptibility thereto), as well as decreased expression of genes associated with cell migration, cell death, inflammatory response, and immune function. Examining individual genes with common effects across treatments (Tables 5, S23 and S24) also highlights the categories of cytokine signaling and cell adhesion, among others. Of further note is that the expression of mRNAs encoding several of the upstream regulator genes identified in the UR analysis are themselves moderately elevated in cells treated with ATR, TBT or both, including TNF, IFNL1, IRF7, ILIB, STAT1, MYD88, and IL1RN (Tables 6, S15, S19).
Table 6. Top shared Upstream Regulators related to DEGs for five EDC treatments ranked by sum of z-score (from Table S25).
| z-score2 | |||||||
|---|---|---|---|---|---|---|---|
| Regulator1 | ATR | BPA | DEHP | PFOA | TBT | SUM | Num. p≤.05 |
| IFNG | -8.28 | -2.02 | -3.32 | -2.58 | -7.80 | -24.00 | 5 |
| poly rI:rC-RNA | -6.78 | -1.14 | -3.15 | -1.23 | -6.31 | -18.60 | 5 |
| TNF (T) | -6.62 | -4.10 | -1.81 | -5.47 | -18.00 | 4 | |
| lipopolysaccharide | -7.12 | -2.35 | -1.18 | -6.96 | -17.60 | 4 | |
| Interferon alpha | -6.79 | -1.00 | -2.20 | -6.60 | -16.60 | 4 | |
| IFNL1 (A,T) | -6.11 | -2.00 | -6.24 | -14.35 | 4 | ||
| NKX2-3 | 5.20 | 1.89 | 1.91 | 5.31 | 14.31 | 5 | |
| IRF7 (T) | -7.09 | -6.85 | -13.95 | 2 | |||
| IL1B (A) | -5.96 | -2.29 | -1.00 | -4.39 | -13.65 | 4 | |
| IFNA2 | -6.93 | -6.68 | -13.61 | 4 | |||
| MAPK1 | 4.84 | 2.24 | -0.46 | 1.92 | 4.57 | 13.12 | 5 |
| tributyrin | -4.21 | -1.95 | -2.78 | -1.95 | -2.15 | -13.03 | 5 |
| STAT1 (A,T) | -5.33 | -1.50 | -0.85 | -4.92 | -12.60 | 4 | |
| PRL | -4.44 | -0.64 | -2.04 | -0.50 | -4.21 | -11.83 | 5 |
| TGFB1 | -2.97 | -1.85 | -3.52 | -1.24 | -2.04 | -11.63 | 4 |
| IFNB1 | -5.78 | -5.59 | -11.38 | 4 | |||
| MYD88 (T) | -4.48 | -3.07 | -3.82 | -11.37 | 4 | ||
| TLR7 | -4.45 | -1.98 | -4.93 | -11.35 | 4 | ||
| mycophenolic acid | -3.71 | -1.96 | -2.25 | -1.22 | -2.21 | -11.35 | 5 |
| OSM | -4.32 | -2.03 | -0.94 | -3.95 | -11.24 | 4 | |
| IFN Beta | -5.33 | -5.78 | -11.11 | 3 | |||
| TLR9 | -5.02 | -1.13 | -4.88 | -11.03 | 4 | ||
| IL1RN (A) | 5.15 | 1.27 | 4.56 | 10.99 | 5 | ||
| EIF2AK2 | -4.07 | -1.98 | -4.93 | -10.98 | 4 | ||
| IKBKB | -3.50 | -2.57 | -0.96 | -3.84 | -10.87 | 4 | |
| IFN | -4.57 | -2.00 | -4.25 | -10.82 | 4 | ||
(A) and (T) indicate the UR itself is affected in ATR and TBT treated cultures
Activation state is predicted Activated if z-score > 1.96 and Inhibited for z-score < -1.96.
Figure 2.
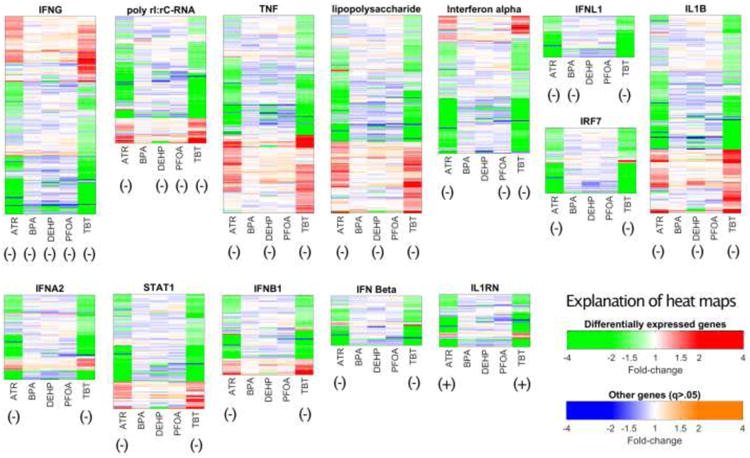
Heat maps summarizing differential expression of genes downstream of the indicated (above heat maps) upstream regulators. Each heat map illustrates the degree to which effects on individual gene expression (increase or decrease) are shared by two or more treatments, for downstream genes affected by at least one toxicant. Green and red denote genes that are significantly (q < 0.05) downregulated (green) or upregulated (red). Blue and orange indicate genes with reduced (blue) or increased (orange) expression that was not statistically significant (q > 0.05). Overall predictions of activation or inhibition of upstream regulator actions are indicated by annotations below each map, with (+) indicating activation (z > 1.96) and (-) indicating inhibition (z < - 1.96). Details of effects are provided in Tables 6 and S25.
Expression of IFI27, DTX3L, CEACAM6, INHBA, KRT17, and IL1RL1 mRNAs was significantly reduced with all five EDCs (Table S24). In addition to these, expression of SCEL, MME, OASL, GPRC58, C1orf116, MAP18, PPP1R1C, ZFP36L2, PARP9, EDN1, AQPEP, F3, and CXCL2 was reduced in four of the five treatments. Expression of HMCN1, C10orf116, and FOXO4 was significantly increased in all five treatments and additionally expression of SMARCA1, CD36, FOSB, and FRAS1 was elevated in four of the five treatments. Some of these genes are related to cytokine signaling, but some are not. This further demonstrates the sensitivity of TS cells to chronic low-level exposure to a variety of EDCs, with effects on a range of trophoblast functions.
Several other overlapping IPA results related to apparent disruption in metabolic processes, quantity of cells, and cell invasion. Both ATR and TBT treated cells displayed significant decrease (z < -1.96) for glucose metabolism disorder in the DF analysis; ATR treatment also yielded significant decrease for diabetes mellitus, and TBT treatment yielded a significant decrease for carbohydrate metabolism (Table S26). Synthesis of reactive oxygen species was significantly decreased for ATR and TBT with non-significant decrease (-1.96 < z < 0) for DEHP and PFOA treatment. The DF category for invasion of cells tended to be reduced in all but BPA treatments with strongest z-scores for DEHP and PFOA. There were additional overlaps in effects with z-scores below the level of significance but with significant over-representation of affected genes (p ≤ 0.05), indicating that the different treatments likely shared other effects on the cells but with greater variability in outcomes. Comparing the expression of genes associated with specific DFs or CPs across the five treatments further emphasizes conservation of effects across the EDC treatments. As with the UR analysis, there is a substantial overlap of genes affected by ATR and TBT, and many of these genes show modulations in the same direction with the other treatment groups, some significantly altered, and others with large fold-changes, but with q values > 0.05.
Discussion
The results presented here demonstrate for the first time that long-term exposure of nonhuman primate trophoblast stem cells to comparatively low concentrations of five EDCs leads to significant disruption in the expression of genes related to cytokine signaling and antiviral mechanisms. Both pro-inflammatory and anti-inflammatory pathways are affected, including genes regulated by multiple interferons, interleukins, tumor necrosis factor, LPS stimulation, viral infection, Oncostatin M, and STAT1, and NFKB signaling. This indicates suppression of response to viral infection. This effect was seen to varying degrees with all five EDCs tested. This discovery is significant because it highlights a previously unrecognized risk to human pregnancy of chronic exposures to low, environmentally relevant concentrations of multiple EDCs. Adverse effects in women exposed to these EDCs could include greater susceptibility of the embryo and placenta to maternal viral infection. Additionally, disruptions in cytokine signaling could inhibit embryo hatching, attachment, invasion, and implantation into the uterus, and increase the risk pregnancy loss due to insufficient vasculogenesis or maternal tolerance of the conceptus.
Fetal membranes produce a wide array of pro- and anti-inflammatory cytokines, and this is regulated by a number of physical and physiological factors or stimuli, and by pathological factors such as viral infection [68]. Viral infection can lead to cellular apoptosis and following induction of pro-inflammatory cytokines as part of a natural defense mechanism in the placenta against infection [68]. However, this can have negative consequences, such as preterm birth.
Although high circulating levels of cytokines, infections, and uterine inflammation are associated with pregnancy loss [69-71] and failure in implantation [72, 73], other studies point to crucial roles for local cytokine signaling between the uterine endothelium and the embryo (trophoblast cells) in facilitating embryo attachment and implantation, and local remodeling of vascular supply [57, 74-83]. These local effects at the site of implantation have been most thoroughly studied in rodents but have also been seen in humans, indicating that although the details of placentation differ markedly between humans and rodents, this crucial role for local cytokine signaling in the maternal-embryo dialog is conserved. Additionally, autocrine cytokine signaling participates in blastocyst hatching [84]. The importance of cytokine signaling in embryo implantation and pregnancy across mammalian species coupled with the strong negative effects of low-level chronic exposure to EDCs on cytokine signaling shown here raise new concerns about the effects of environmental EDCs on human reproduction as well as propagation and breeding in other species.
Other effects of EDCs on reproduction have been reported. PFOS (perfluorooctanesulfonic acid) and PFOA inhibit prolactin signaling [5, 85], inhibit aromatase activity [4], and affect lipid metabolism [4] in placental cells. Organotin compounds induce progesterone synthesis in Jar cells [6]. TBT mediates intrauterine growth and post-natal growth restriction in rats [86], and inhibits aromatase activity in the placenta. Phthalates inhibit trophoblast invasion via PPARγ inhibition [7], disrupt placentation [8], and modify expression of lipid metabolism genes and fatty acid placental transport [9, 87]. BPA reduces invasiveness of trophoblast [10] and alters trophoblast cell proliferation even at concentrations that do not produce overt toxicity [11]. Some of these effects were also detected in our RNAseq data. Our data substantially extend what is known about effects of these EDCs on trophoblast stem cells, but using long-term low-level dosing and whole transcriptome analysis in a nonhuman primate cell that closely resembles human trophoblast, to discover the long-term effects on trophoblast stem cell properties.
In addition to the pronounced effects on cytokine signaling and antiviral pathways, our analysis revealed other less pronounced effects, such as diminished DNA damage checkpoint signaling, a marginally increased expression of ESC pluripotency-related genes, disruptions in metabolic processes, and disruptions of cell movement and invasion. These effects could also compromise embryo viability and embryo implantation.
The effects of the five EDCs shown here when applied to a rhesus monkey TSC line are substantially greater than when the same treatments were applied to a rhesus monkey ESC line [37]. This suggests that the TSCs are much more sensitive to EDCs than ESCs. This may reflect the generally lower state of DNA methylation in trophoblast lineage (placental) cells [26]. Because some of the effects of these EDCs include epigenetic effects at the level of DNA methylation [88, 89], TSCs may be more susceptible to gene expression changes. Effects on DNA methylation of the X chromosome could be especially noticeable in TSCs, given plasticity of X chromosome inactivation in these cells [90]. Additionally, trophoblast-specific variation and change in DNA methylation [22] and X-chromosome inactivation [91] may be more susceptible to disruption. The elevated sensitivity of primate TSCs to EDC effects suggests that strategies for screening of compounds for reproductive toxicity may be improved by including long-term exposures of primate TSCs. Studies of EDC toxicity to TSCs may be essential to detect potential toxic effects on conception and pregnancy.
The broader impact of chronic low-level EDC exposure on human reproduction via effects on TSCs remains to be assessed. Our results indicate that environmentally relevant levels of EDC exposures may contribute to diminished pregnancy rates by inhibiting embryo implantation and interfering with other processes. The magnitude of effects may depend on a range of maternal, genetic, and other environmental factors, particularly factors related to pro-inflammatory and anti-inflammatory signaling, reproductive tract infection, and control of maternal immune function in response to the invading embryo. Further functional studies to assess changes in protein expression and specific cellular activities (e.g., anti-viral responsiveness, cytokine signaling), and epigenetic changes would be useful future areas of study. Additionally, studies of low level exposures to combinations of EDCs should be pursued, as many such chemicals co-exist in the environment. The extensive data set presented here provides a broad foundation to warrant many future studies targeting specific endpoints to better understand the impact of low-level chronic EDC exposure on trophoblast function and pregnancy. Greater awareness of maternal EDC exposures, monitoring maternal serum EDC levels, or strategies to modulate embryonic cytokine signaling activity in vitro could provide useful components of enhanced strategies for increasing rates of embryo implantation, particularly in assisted reproduction programs.
Supplementary Material
Highlights.
Effects of chronic exposure to low concentrations of five endocrine disrupting chemicals (EDCs) were examined in rhesus monkey trophoblast stem cells
RNA sequencing revealed largest numbers of affected genes following tributyltin and atrazine treatment, with substantial overlap in effects between these two toxicants
The most prominent effect for all five compounds was the suppression of pathways related to cytokine signaling and anti-viral response
Other effects observed predominantly with tributyltin or atrazine included diminished DNA damage repair and cell movement functions, increased cell viability and proliferation functions, and disruption of metabolic processes.
These results from an animal model closely related to humans indicate that chronic low-level exposure to EDCs could impair trophoblast stem cell function and diminish human pregnancy outcomes by compromising trophoblast invasiveness, embryo implantation and placenta defense against viral pathogens.
Acknowledgments
The authors thank Dana Dagget for her technical assistance, and Dr. Almudena Veiga-Lopez for her helpful comments on the manuscript. This work was supported by grants from the Office of Research Infrastructure Programs Division of Comparative Medicine Grants R24 OD-012221 (to K.E.L.), and OD011107/RR00169 (California National Primate Research Center) and OD010967/RR025880 (to C.A.V.), by MSU AgBioResearch, and Michigan State University.
Abbreviations
- ATR
Atrazine
- BPA
bisphenol A
- CP
canonical pathway
- DEG
differentially expressed gene
- DEHP
di-(2-ethylhexyl) phthalate
- DF
disease and function
- EDC
endocrine disrupting chemical
- ESC
embryonic stem cell
- TSC
trophoblast stem cell
- IPA
QIAGEN Ingenuity Pathway Analysis®
- PFOA
perfluorooctanoic acid
- RNAseq
RNA sequencing
- TBT
tributyltin
- UR
upstream regulator
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Latham KE, Sapienza C, Engel N. The epigenetic lorax: gene-environment interactions in human health. Epigenomics. 2012;4(4):383–402. doi: 10.2217/epi.12.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallozzi M, Bordi G, Garo C, Caserta D. The effect of maternal exposure to endocrine disrupting chemicals on fetal and neonatal development: A review on the major concerns. Birth Defects Res C Embryo Today. 2016;108(3):224–242. doi: 10.1002/bdrc.21137. [DOI] [PubMed] [Google Scholar]
- 3.Krieg SA, Shahine LK, Lathi RB. Environmental exposure to endocrine-disrupting chemicals and miscarriage. Fertil Steril. 2016;106(4):941–7. doi: 10.1016/j.fertnstert.2016.06.043. [DOI] [PubMed] [Google Scholar]
- 4.Gorrochategui E, Perez-Albaladejo E, Casas J, Lacorte S, Porte C. Perfluorinated chemicals: differential toxicity, inhibition of aromatase activity and alteration of cellular lipids in human placental cells. Toxicology and applied pharmacology. 2014;277(2):124–30. doi: 10.1016/j.taap.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Suh CH, Cho NK, Lee CK, Lee CH, Kim DH, Kim JH, Son BC, Lee JT. Perfluorooctanoic acid-induced inhibition of placental prolactin-family hormone and fetal growth retardation in mice. Molecular and cellular endocrinology. 2011;337(1-2):7–15. doi: 10.1016/j.mce.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Hiromori Y, Yui H, Nishikawa J, Nagase H, Nakanishi T. Organotin compounds cause structure-dependent induction of progesterone in human choriocarcinoma Jar cells. The Journal of steroid biochemistry and molecular biology. 2016;155(Pt B):190–8. doi: 10.1016/j.jsbmb.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Gao F, Hu W, Li Y, Shen H, Hu J. Mono-2-ethylhexyl phthalate inhibits human extravillous trophoblast invasion via the PPARgamma pathway. Toxicology and applied pharmacology. 2017;327:23–29. doi: 10.1016/j.taap.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Zong T, Lai L, Hu J, Guo M, Li M, Zhang L, Zhong C, Yang B, Wu L, Zhang D, Tang M, Kuang H. Maternal exposure to di-(2-ethylhexyl) phthalate disrupts placental growth and development in pregnant mice. Journal of hazardous materials. 2015;297:25–33. doi: 10.1016/j.jhazmat.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Agrawal S, Cook TJ, Knipp GT. Maternal di-(2-ethylhexyl)-phthalate exposure influences essential fatty acid homeostasis in rat placenta. Placenta. 2008;29(11):962–9. doi: 10.1016/j.placenta.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZY, Lu J, Zhang YZ, Zhang M, Liu T, Qu XL. Effect of Bisphenol A on invasion ability of human trophoblastic cell line BeWo. Int J Clin Exp Pathol. 2015;8(11):14355–64. [PMC free article] [PubMed] [Google Scholar]
- 11.Spagnoletti A, Paulesu L, Mannelli C, Ermini L, Romagnoli R, Cintorino M, Ietta F. Lowconcentrations of Bisphenol A and para-Nonylphenol affect extravillous pathway of human trophoblast cells. Molecular and cellular endocrinology. 2015;412:56–64. doi: 10.1016/j.mce.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 12.Morice L, Benaitreau D, Dieudonne MN, Morvan C, Serazin V, de Mazancourt P, Pecquery R, Dos Santos E. Antiproliferative and proapoptotic effects of bisphenol A on human trophoblastic JEG-3 cells. Reprod Toxicol. 2011;32(1):69–76. doi: 10.1016/j.reprotox.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Rajakumar C, Guan H, Langlois D, Cernea M, Yang K. Bisphenol A disrupts gene expression in human placental trophoblast cells. Reprod Toxicol. 2015;53:39–44. doi: 10.1016/j.reprotox.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Benachour N, Aris A. Toxic effects of low doses of Bisphenol-A on human placental cells, Toxicology and applied pharmacology. 2009;241(3):322–8. doi: 10.1016/j.taap.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Chen R, Liu W, Fu Z. Effect of endocrine disrupting chemicals on the transcription of genes related to the innate immune system in the early developmental stage of zebrafish (Danio rerio) Fish Shellfish Immunol. 2010;28(5-6):854–61. doi: 10.1016/j.fsi.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Rowe AM, Brundage KM, Barnett JB. Developmental immunotoxicity of atrazine in rodents. Basic & clinical pharmacology & toxicology. 2008;102(2):139–45. doi: 10.1111/j.1742-7843.2007.00175.x. [DOI] [PubMed] [Google Scholar]
- 17.Grigsby PL. Animal Models to Study Placental Development and Function throughout Normal and Dysfunctional Human Pregnancy. Seminars in reproductive medicine. 2016;34(1):11–6. doi: 10.1055/s-0035-1570031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter AM. Animal models of human placentation--a review. Placenta. 2007;28 Suppl A:S41–7. doi: 10.1016/j.placenta.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Martin CB, Jr, Ramsey EM, Donner MW. The fetal placental circulation in rhesus monkeys demonstrated by radioangiography. Am J Obstet Gynecol. 1966;95(7):943–7. doi: 10.1016/0002-9378(66)90542-4. [DOI] [PubMed] [Google Scholar]
- 20.Douglas GC, VandeVoort CA, Kumar P, Chang TC, Golos TG. Trophoblast stem cells: models for investigating trophectoderm differentiation and placental development. Endocrine reviews. 2009;30(3):228–40. doi: 10.1210/er.2009-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandevoort CA, Thirkill TL, Douglas GC. Blastocyst-derived trophoblast stem cells from the rhesus monkey. Stem Cells Dev. 2007;16(5):779–88. doi: 10.1089/scd.2007.0020. [DOI] [PubMed] [Google Scholar]
- 22.Hamada H, Okae H, Toh H, Chiba H, Hiura H, Shirane K, Sato T, Suyama M, Yaegashi N, Sasaki H, Arima T. Allele-Specific Methylome and Transcriptome Analysis Reveals Widespread Imprinting in the Human Placenta. Am J Hum Genet. 2016;99(5):1045–1058. doi: 10.1016/j.ajhg.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Delgado M, Court F, Vidal E, Medrano J, Monteagudo-Sanchez A, Martin-Trujillo A, Tayama C, Iglesias-Platas I, Kondova I, Bontrop R, Poo-Llanillo ME, Marques-Bonet T, Nakabayashi K, Simon C, Monk D. Human Oocyte-Derived Methylation Differences Persist in the Placenta Revealing Widespread Transient Imprinting. PLoS Genet. 2016;12(11):e1006427. doi: 10.1371/journal.pgen.1006427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo J, He H, Liu H, Liu Q, Zhang L, Liu B, Sugimoto K, Wu Q. Aquaporin-1, a New Maternally Expressed Gene, Regulates Placental Development in the Mouse. Biology of reproduction. 2016;95(2):40. doi: 10.1095/biolreprod.116.138636. [DOI] [PubMed] [Google Scholar]
- 25.Monk D. Genomic imprinting in the human placenta. Am J Obstet Gynecol. 2015;213(4 Suppl):S152–62. doi: 10.1016/j.ajog.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee A, Macaulay EC, Rodger EJ, Stockwell PA, Parry MF, Roberts HE, Slatter TL, Hung NA, Devenish CJ, Morison IM. Placental Hypomethylation Is More Pronounced in Genomic Loci Devoid of Retroelements. G3 (Bethesda) 2016;6(7):1911–21. doi: 10.1534/g3.116.030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wirbisky SE, Freeman JL. Atrazine Exposure and Reproductive Dysfunction through the Hypothalamus-Pituitary-Gonadal (HPG) Axis. Toxics. 2015;3(4):414–450. doi: 10.3390/toxics3040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBirney M, King SE, Pappalardo M, Houser E, Unkefer M, Nilsson E, Sadler-Riggleman I, Beck D, Winchester P, Skinner MK. Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS One. 2017;12(9):e0184306. doi: 10.1371/journal.pone.0184306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rooney AA, Matulka RA, Luebke RW. Developmental atrazine exposure suppresses immune function in male, but not female Sprague-Dawley rats. Toxicological sciences : an official journal of the Society of Toxicology. 2003;76(2):366–75. doi: 10.1093/toxsci/kfg250. [DOI] [PubMed] [Google Scholar]
- 30.Kirchner S, Kieu T, Chow C, Casey S, Blumberg B. Prenatal exposure to the environmental obesogen tributyltin predisposes multipotent stem cells to become adipocytes. Mol Endocrinol. 2010;24(3):526–39. doi: 10.1210/me.2009-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Ycaza J, Blumberg B. The environmental obesogen tributyltin chloride acts via peroxisome proliferator activated receptor gamma to induce adipogenesis in murine 3T3-L1 preadipocytes. The Journal of steroid biochemistry and molecular biology. 2011;127:9–15. doi: 10.1016/j.jsbmb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corton JC, Cunningham ML, Hummer BT, Lau C, Meek B, Peters JM, Popp JA, Rhomberg L, Seed J, Klaunig JE. Mode of action framework analysis for receptor-mediated toxicity: The peroxisome proliferator-activated receptor alpha (PPARalpha) as a case study. Critical reviews in toxicology. 2014;44(1):1–49. doi: 10.3109/10408444.2013.835784. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Arguelles DB, Papadopoulos V. Prenatal phthalate exposure: epigenetic changes leading to lifelong impact on steroid formation. Andrology. 2016;4(4):573–84. doi: 10.1111/andr.12175. [DOI] [PubMed] [Google Scholar]
- 34.MacKay H, Abizaid A. A plurality of molecular targets: The receptor ecosystem for bisphenol-A (BPA) Hormones and behavior. 2017 doi: 10.1016/j.yhbeh.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Mileva G, Baker SL, Konkle AT, Bielajew C. Bisphenol-A: epigenetic reprogramming and effects on reproduction and behavior. International journal of environmental research and public health. 2014;11(7):7537–61. doi: 10.3390/ijerph110707537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bach CC, Bech BH, Brix N, Nohr EA, Bonde JP, Henriksen TB. Perfluoroalkyl and polyfluoroalkyl substances and human fetal growth: a systematic review. Critical reviews in toxicology. 2015;45(1):53–67. doi: 10.3109/10408444.2014.952400. [DOI] [PubMed] [Google Scholar]
- 37.Midic U, Vincent KA, VandeVoort CA, Latham KE. Effects of long-term endocrine disrupting compound exposure on Macaca mulatta embryonic stem cells. Reprod Toxicol. 2016;65:382–393. doi: 10.1016/j.reprotox.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blakeley P, Fogarty NM, del Valle I, Wamaitha SE, Hu TX, Elder K, Snell P, Christie L, Robson P, Niakan KK. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142(18):3151–65. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omigbodun A, Coukos G, Ziolkiewicz P, Wang CL, Coutifaris C. Macrophage-colony stimulating factor (M-CSF) regulates the expression of fibronectin and its alpha5 integrin receptor in human trophoblasts. Endocrinology. 1998;139(4):2190–3. doi: 10.1210/endo.139.4.6031. [DOI] [PubMed] [Google Scholar]
- 40.Pijnenborg R, Luyten C, Vercruysse L, Keith JC, Jr, Van Assche FA. Cytotoxic effects of tumour necrosis factor (TNF)-alpha and interferon-gamma on cultured human trophoblast are modulated by fibronectin. Molecular human reproduction. 2000;6(7):635–41. doi: 10.1093/molehr/6.7.635. [DOI] [PubMed] [Google Scholar]
- 41.Stahlhut RW, Welshons WV, Swan SH. Bisphenol A data in NHANES suggest longer than expected half-life, substantial nonfood exposure, or both. Environ Health Perspect. 2009;117(5):784–9. doi: 10.1289/ehp.0800376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hormann AM, Vom Saal FS, Nagel SC, Stahlhut RW, Moyer CL, Ellersieck MR, Welshons WV, Toutain PL, Taylor JA. Holding thermal receipt paper and eating food after using hand sanitizer results in high serum bioactive and urine total levels of bisphenol A (BPA) PLoS One. 2014;9(10):e110509. doi: 10.1371/journal.pone.0110509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vom Saal FS, Vande Voort CA, Taylor JA, Welshons WV, Toutain PL, Hunt PA. Bisphenol A (BPA) pharmacokinetics with daily oral bolus or continuous exposure via silastic capsules in pregnant rhesus monkeys: Relevance for human exposures. Reprod Toxicol. 2014;45:105–16. doi: 10.1016/j.reprotox.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vande Voort CA, Gerona RR, Vom Saal FS, Tarantal AF, Hunt PA, Hillenweck A, Zalko D. Maternal and Fetal Pharmacokinetics of Oral Radiolabeled and Authentic Bisphenol A in the Rhesus Monkey. PLoS One. 2016;11(12):e0165410. doi: 10.1371/journal.pone.0165410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zalko D, Soto AM, Dolo L, Dorio C, Rathahao E, Debrauwer L, Faure R, Cravedi JP. Biotransformations of bisphenol A in a mammalian model: answers and new questions raised by low-dose metabolic fate studies in pregnant CD1 mice. Environ Health Perspect. 2003;111(3):309–19. doi: 10.1289/ehp.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbel T, Gayrard V, Viguie C, Puel S, Lacroix MZ, Toutain PL, Picard-Hagen N. Bisphenol A disposition in the sheep maternal-placental-fetal unit: mechanisms determining fetal internal exposure. Biology of reproduction. 2013;89(1):11. doi: 10.1095/biolreprod.112.106369. [DOI] [PubMed] [Google Scholar]
- 47.Huang XF, Li Y, Gu YH, Liu M, Xu Y, Yuan Y, Sun F, Zhang HQ, Shi HJ. The effects of Di-(2-ethylhexyl)-phthalate exposure on fertilization and embryonic development in vitro and testicular genomic mutation in vivo. PLoS One. 2012;7(11):e50465. doi: 10.1371/journal.pone.0050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graves JE, Richardson ME, Bernard RS, Camper ND, Bridges WC. Atrazine effects on in vitro maturation and in vitro fertilization in the bovine oocyte. Journal of environmental science and health Part B, Pesticides, food contaminants, and agricultural wastes. 2002;37(2):103–12. doi: 10.1081/PFC-120002982. [DOI] [PubMed] [Google Scholar]
- 49.Casas E, Bonilla E, Ducolomb Y, Betancourt M. Differential effects of herbicides atrazine and fenoxaprop-ethyl, and insecticides diazinon and malathion, on viability and maturation of porcine oocytes in vitro. Toxicology in vitro : an international journal published in association with BIBRA. 2010;24(1):224–30. doi: 10.1016/j.tiv.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Mlynarcikova A, Nagyova E, Fickova M, Scsukova S. Effects of selected endocrine disruptors on meiotic maturation, cumulus expansion, synthesis of hyaluronan and progesterone by porcine oocyte-cumulus complexes. Toxicology in vitro : an international journal published in association with BIBRA. 2009;23(3):371–7. doi: 10.1016/j.tiv.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 51.Ferris J, Mahboubi K, MacLusky N, King WA, Favetta LA. BPA exposure during in vitro oocyte maturation results in dose-dependent alterations to embryo development rates, apoptosis rate, sex ratio and gene expression. Reprod Toxicol. 2016;59:128–38. doi: 10.1016/j.reprotox.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Yamada S, Asanagi M, Hirata N, Itagaki H, Sekino Y, Kanda Y. Tributyltin induces mitochondrial fission through Mfn1 degradation in human induced pluripotent stem cells. Toxicology in vitro : an international journal published in association with BIBRA. 2016;34:257–263. doi: 10.1016/j.tiv.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Fan A, Alexeef GV Office of Environmental Health Hazard Assessment. California Environmental Protection Agency. Public Health Goal for Atrazine In Drinking Water. 1999:1–44. [Google Scholar]
- 54.Scutaru B, Giersch T, Cozmei C, Hock B. Immunoenzymatic determination of atrazine in rat tissue samples. Toxicology. 1998;127(1-3):11–6. doi: 10.1016/s0300-483x(98)00007-9. [DOI] [PubMed] [Google Scholar]
- 55.Thibeault AA, Deroy K, Vaillancourt C, Sanderson JT. A unique co-culture model for fundamental and applied studies of human fetoplacental steroidogenesis and interference by environmental chemicals. Environ Health Perspect. 2014;122(4):371–7. doi: 10.1289/ehp.1307518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Winkle LS, Murphy SR, Boetticher MV, VandeVoort CA. Fetal exposure of rhesus macaques to bisphenol a alters cellular development of the conducting airway by changing epithelial secretory product expression. Environ Health Perspect. 2013;121(8):912–8. doi: 10.1289/ehp.1206064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guzeloglu-Kayisli O, Kayisli UA, Taylor HS. The role of growth factors and cytokines during implantation: endocrine and paracrine interactions. Seminars in reproductive medicine. 2009;27(1):62–79. doi: 10.1055/s-0028-1108011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez-Sosa JR, Bondareva A, Tang L, Avelar GF, Coyle KM, Modelski M, Alpaugh W, Conley A, Wynne-Edwards K, Franca LR, Meyers S, Dobrinski I. hthalate esters affect maturation and function of primate testis tissue ectopically grafted in mice. Molecular and cellular endocrinology. 2014;398(1-2):89–100. doi: 10.1016/j.mce.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamada S, Kotake Y, Nakano M, Sekino Y, Kanda Y. Tributyltin induces mitochondrial fission through NAD-IDH dependent mitofusin degradation in human embryonic carcinoma cells. Metallomics : integrated biometal science. 2015;7(8):1240–6. doi: 10.1039/c5mt00033e. [DOI] [PubMed] [Google Scholar]
- 60.Yamada S, Kotake Y, Demizu Y, Kurihara M, Sekino Y, Kanda Y. NAD-dependent isocitrate dehydrogenase as a novel target of tributyltin in human embryonic carcinoma cells. Scientific reports. 2014;4:5952. doi: 10.1038/srep05952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayes TB, Anderson LL, Beasley VR, de Solla SR, Iguchi T, Ingraham H, Kestemont P, Kniewald J, Kniewald Z, Langlois VS, Luque EH, McCoy KA, Munoz-de-Toro M, Oka T, Oliveira CA, Orton F, Ruby S, Suzawa M, Tavera-Mendoza LE, Trudeau VL, Victor-Costa AB, Willingham E. Demasculinization and feminization of male gonads by atrazine: consistent effects across vertebrate classes. The Journal of steroid biochemistry and molecular biology. 2011;127(1-2):64–73. doi: 10.1016/j.jsbmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bechi N, Sorda G, Spagnoletti A, Bhattacharjee J, Vieira Ferro EA, de Freitas Barbosa B, Frosini M, Valoti M, Sgaragli G, Paulesu L, Ietta F. Toxicity assessment on trophoblast cells for some environment polluting chemicals and 17beta-estradiol. Toxicology in vitro : an international journal published in association with BIBRA. 2013;27(3):995–1000. doi: 10.1016/j.tiv.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 63.Head SR, Komori HK, Hart GT, Shimashita J, Schaffer L, Salomon DR, Ordoukhanian PT. Method for improved Illumina sequencing library preparation using NuGEN Ovation RNA-Seq system. Biotechniques. 2011;50(3):177–181. doi: 10.2144/000113613. [DOI] [PubMed] [Google Scholar]
- 64.Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, Tharp GK, Marcais G, Roberts M, Ferguson B, Fox HS, Treangen T, Salzberg SL, Yorke JA, Norgren RB., Jr A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biology direct. 2014;9(1):20. doi: 10.1186/1745-6150-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Renaud SJ, Kubota K, Rumi MA, Soares MJ. The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem. 2014;289(8):5025–39. doi: 10.1074/jbc.M113.523746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uchide N, Ohyama K, Bessho T, Toyoda H. Induction of pro-inflammatory cytokine gene expression and apoptosis in human chorion cells of fetal membranes by influenza virus infection: possible implications for maintenance and interruption of pregnancy during infection. Med Sci Monit. 2005;11(1):RA7–16. [PubMed] [Google Scholar]
- 69.Fasouliotis SJ, Spandorfer SD, Witkin SS, Schattman G, Liu HC, Roberts JE, Rosenwaks Z. Maternal serum levels of interferon-gamma and interleukin-2 soluble receptor-alpha predict the outcome of early IVF pregnancies. Hum Reprod. 2004;19(6):1357–63. doi: 10.1093/humrep/deh169. [DOI] [PubMed] [Google Scholar]
- 70.Giakoumelou S, Wheelhouse N, Cuschieri K, Entrican G, Howie SE, Horne AW. The role of infection in miscarriage. Hum Reprod Update. 2016;22(1):116–33. doi: 10.1093/humupd/dmv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwak-Kim J, Bao S, Lee SK, Kim JW, Gilman-Sachs A. Immunological modes of pregnancy loss: inflammation, immune effectors, and stress. Am J Reprod Immunol. 2014;72(2):129–40. doi: 10.1111/aji.12234. [DOI] [PubMed] [Google Scholar]
- 72.Montazeri M, Sanchez-Lopez JA, Caballero I, Maslehat Lay N, Elliott S, Lopez-Martin S, Yanez-Mo M, Fazeli A. Activation of Toll-like receptor 3 reduces actin polymerization and adhesion molecule expression in endometrial cells, a potential mechanism for viral-induced implantation failure. Hum Reprod. 2015;30(4):893–905. doi: 10.1093/humrep/deu359. [DOI] [PubMed] [Google Scholar]
- 73.Akopians AL, Pisarska MD, Wang ET. The Role of Inflammatory Pathways in Implantation Failure: Chronic Endometritis and Hydrosalpinges. Seminars in reproductive medicine. 2015;33(4):298–304. doi: 10.1055/s-0035-1554916. [DOI] [PubMed] [Google Scholar]
- 74.Pathare ADS, Zaveri K, Hinduja I. Downregulation of genes related to immune and inflammatory response in IVF implantation failure cases under controlled ovarian stimulation. Am J Reprod Immunol. 2017;78(1) doi: 10.1111/aji.12679. [DOI] [PubMed] [Google Scholar]
- 75.Murphy SP, Tayade C, Ashkar AA, Hatta K, Zhang J, Croy BA. Interferon gamma in successful pregnancies. Biology of reproduction. 2009;80(5):848–59. doi: 10.1095/biolreprod.108.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bazer FW, Spencer TE, Johnson GA. Interferons and uterine receptivity. Seminars in reproductive medicine. 2009;27(1):90–102. doi: 10.1055/s-0028-1108013. [DOI] [PubMed] [Google Scholar]
- 77.Kapiteijn K, Koolwijk P, van der Weiden RM, van Nieuw Amerongen G, Plaisier M, van Hinsbergh VW, Helmerhorst FM. Human embryo-conditioned medium stimulates in vitro endometrial angiogenesis. Fertil Steril. 2006;85(1):1232–9. doi: 10.1016/j.fertnstert.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 78.Pereira de Sousa FL, Chaiwangyen W, Morales-Prieto DM, Ospina-Prieto S, Weber M, Photini SM, Sass N, Daher S, Schleussner E, Markert UR. Involvement of STAT1 in proliferation and invasiveness of trophoblastic cells. Reproductive biology. 2017 doi: 10.1016/j.repbio.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 79.Baston-Buest DM, Altergot-Ahmad O, Pour SJ, Krussel JS, Markert UR, Fehm TN, Bielfeld AP. Syndecan-1 Acts as an Important Regulator of CXCL1 Expression and Cellular Interaction of Human Endometrial Stromal and Trophoblast Cells. Mediators Inflamm. 2017;2017:8379256. doi: 10.1155/2017/8379256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Furmento VA, Marino J, Blank VC, Cayrol MF, Cremaschi GA, Aguilar RC, Roguin LP. Granulocyte colony-stimulating factor (G-CSF) upregulates beta1 integrin and increases migration of human trophoblast Swan 71 cells via PI3K and MAPK activation. Exp Cell Res. 2016;342(2):125–34. doi: 10.1016/j.yexcr.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma S, Godbole G, Modi D. Decidual Control of Trophoblast Invasion. Am J Reprod Immunol. 2016;75(3):341–50. doi: 10.1111/aji.12466. [DOI] [PubMed] [Google Scholar]
- 82.Dimitriadis E, Menkhorst E, Salamonsen LA, Paiva P. Review: LIF and IL11 in trophoblast-endometrial interactions during the establishment of pregnancy. Placenta. 2010;31 Suppl:S99–104. doi: 10.1016/j.placenta.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 83.Dimitriadis E, Nie G, Hannan NJ, Paiva P, Salamonsen LA. Local regulation of implantation at the human fetal-maternal interface. Int J Dev Biol. 2010;54(2-3):313–22. doi: 10.1387/ijdb.082772ed. [DOI] [PubMed] [Google Scholar]
- 84.Seshagiri PB, Vani V, Madhulika P. Cytokines and Blastocyst Hatching. Am J Reprod Immunol. 2016;75(3):208–17. doi: 10.1111/aji.12464. [DOI] [PubMed] [Google Scholar]
- 85.Lee CK, Kang SG, Lee JT, Lee SW, Kim JH, Kim DH, Son BC, Kim KH, Suh CH, Kim SY, Park YB. Effects of perfluorooctane sulfuric acid on placental PRL-family hormone production and fetal growth retardation in mice. Molecular and cellular endocrinology. 2015;401:165–72. doi: 10.1016/j.mce.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 86.Asakawa H, Tsunoda M, Kaido T, Hosokawa M, Sugaya C, Inoue Y, Kudo Y, Satoh T, Katagiri H, Akita H, Saji M, Wakasa M, Negishi T, Tashiro T, Aizawa Y. Enhanced inhibitory effects of TBT chloride on the development of F1 rats. Archives of environmental contamination and toxicology. 2010;58(4):1065–73. doi: 10.1007/s00244-009-9421-9. [DOI] [PubMed] [Google Scholar]
- 87.Xu Y, Knipp GT, Cook TJ. Effects of di-(2-ethylhexyl)-phthalate and its metabolites on the lipid profiling in rat HRP-1 trophoblast cells. Archives of toxicology. 2006;80(5):293–8. doi: 10.1007/s00204-005-0047-z. [DOI] [PubMed] [Google Scholar]
- 88.van den Dungen MW, Murk AJ, Kok DE, Steegenga WT. Persistent organic pollutants alter DNA methylation during human adipocyte differentiation. Toxicology in vitro : an international journal published in association with BIBRA. 2017;40:79–87. doi: 10.1016/j.tiv.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 89.Bastos Sales L, Kamstra JH, Cenijn PH, van Rijt LS, Hamers T, Legler J. Effects of endocrine disrupting chemicals on in vitro global DNA methylation and adipocyte differentiation. Toxicology in vitro : an international journal published in association with BIBRA. 2013;27(6):1634–43. doi: 10.1016/j.tiv.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 90.Dubois A, Deuve JL, Navarro P, Merzouk S, Pichard S, Commere PH, Louise A, Arnaud D, Avner P, Morey C. Spontaneous reactivation of clusters of X-linked genes is associated with the plasticity of X-inactivation in mouse trophoblast stem cells. Stem Cells. 2014;32(2):377–90. doi: 10.1002/stem.1557. [DOI] [PubMed] [Google Scholar]
- 91.Moreira de Mello JC, de Araujo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One. 2010;5(6):e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


