Abstract
RNA interference (RNAi) is considered a highly specific approach for gene silencing and holds tremendous potential for treatment of various pathologic conditions such as cardiovascular diseases, viral infections, and cancer. Although gene silencing approaches such as RNAi are widely used in preclinical models, the clinical application of RNAi is challenging primarily because of the difficulty in achieving successful systemic delivery. Effective delivery systems are essential to enable the full therapeutic potential of RNAi. An ideal nanocarrier not only addresses the challenges of delivering naked siRNA/miRNA, including its chemically unstable features, extracellular and intracellular barriers, and innate immune stimulation, but also offers “smart” targeted delivery. Over the past decade, great efforts have been undertaken to develop RNAi delivery systems that overcome these obstacles. This review presents an update on current progress in the therapeutic application of RNAi with a focus on cancer therapy and strategies for optimizing delivery systems, such as lipid-based nanoparticles.
Keywords: RNA interference, delivery systems, nanoparticles, cancer therapy
1. Introduction
RNA interference (RNAi) is the biologic process by which RNA molecules induce sequence-specific inhibition of target gene expression or translation [1]. RNAi was demonstrated to occur in mammalian cells in 2001 [2]. Rapid development in the study of RNAi has driven it from an experimental technology to a powerful tool for therapeutic development. In 2010, the first case of systemic targeted delivery of small interfering RNA (siRNA)-nanoparticles was reported, providing a solid foundation for the clinical application of RNAi [3]. In recent years, RNAi has become more widely applied in gene silencing and drug development because of its high level of specificity, minor side effects, and ease of synthesis [4].
Noncoding RNAs (ncRNAs) are non–protein-coding transcripts that guide multiple functions, such as gene silencing, DNA imprinting, and demethylation [5]. A growing number of ncRNAs have been discovered and implicated in gene regulation and RNA processing [6]. Generally, ncRNAs are divided into small regulatory ncRNAs and long ncRNAs [7]. The small ncRNAs, which could interfere with the translation of target mRNA transcript, are the cleavage product of dsRNA, named siRNA and microRNA (miRNA). Synthetic siRNA can trigger silencing of target genes without disturbing endogenous mRNA pathways. MicroRNAs (miRNAs) are a class of natural RNAs that play a role in regulating cell differentiation, proliferation, and survival, resulting in mRNA translational inhibition or degradation [8].
The sequence-specific silencing of cognate genes by RNAi can be triggered by siRNA, short hairpin RNA (shRNA), miRNA as well as other ncRNAs such as long ncRNAs and pyknons. RNA-induced silencing complex (RISC), a multiprotein complex, is the core intermediate for the degradation and translational inhibition of target mRNA. Briefly, the process is started by breaking down long double-strand RNA (dsRNA) and complex hairpin precursors into shorter duplexes or siRNA by Dicer, an endoribonuclease [5]. The siRNAs are then loaded into RISC. Once incorporated, double-stranded siRNA is split into passenger and guide strands. The former is then degraded and released, and the RISC is activated to catalyze the guide strand to bind with target sequences. The bound mRNA is then cleaved and degraded by cellular nucleases, resulting in inhibition of target gene expression. Unlike siRNA, the use of shRNA is mostly viral vector based. The RNA sequence of shRNA has a tight hairpin structure, and it is transcribed by RNA polymerase III or modified polymerase II. In cells with lower Dicer levels, gene silencing with shRNA could be impaired. Because of the promoter-dependent expression feature, shRNAs must interact with chromosomal DNA to work [9]. Moreover, shRNAs act in the nucleus, which poses additional challenges to its use.
MiRNAs are single-stranded RNAs transcribed by RNA polymerase II and III [10]. After transcription, the primary miRNA (pri-miRNA) with a hairpin loop structure is generated. It is processed into precursor (pre)-miRNA by the Drosha-DGCR8 complex, then cleaved by Dicer. A duplex of about 22 nucleotides, which is the mature functional miRNA, is generated from the pre-miRNA. The double-stranded miRNA is then incorporated into RISC so that the guide strand is maintained and binds to the mRNA. Unlike siRNA, which displays complementary binding, miRNA binds to its target sequence imperfectly [10]. A single miRNA sequence can bind to hundreds of mRNA sequences. With a precise match, the miRNA sequences can induce endonucleolytic cleavage, which results in mRNA degradation; whereas with an imperfect match, it could regulate translation, leading to inhibition of mRNA expression.
2. Therapeutic applications of RNAi
2.1 Advantages of RNAi as a therapeutic
Therapeutic approaches such as small-molecule inhibitors and monoclonal antibodies targeting identical proteins have been successful in many cases [11,12]. However, relatively few of these drugs are available, in part because identification of specific agents to disrupt protein function has been challenging [13,14]. Small molecule inhibitors can act over multi-targets, while siRNAs have high specificity in a sequence specific manner [15].
RNAi overcomes these difficulties by targeted silencing of genes of interest. Regulatory ncRNAs have been demonstrated to either enhance or repress target gene expression, and abnormal expression of ncRNAs is a crucial component in the pathogenesis of cancer and other human disorders [10]. In most pathologic conditions, the expression or function of certain genes is impaired, and miRNAs are deregulated in many diseases, including hepatitis, cardiovascular diseases, and cancer [16,17]. Therefore, RNAi could be applied as a therapeutic to provide transcription-suppressive factors, subsets of kinases, and other signaling molecules to restore the expression of these impaired genes. In a single target cell, several copies of siRNA can regulate target gene expression. Compared with small-molecule or antibody-based drugs, the major advantage of siRNA is the restricted choice of targets determined by complementary base pairing. The RNAi-based strategy also benefits from developments in whole-genome sequencing. Comprehensive nucleotide sequence databases have been established, laying a solid foundation for the investigation of RNAi-based drugs [18,19]. Interest has also grown for therapies aimed at restoring ncRNA expression [20].
2.2 Diseases treatable by RNAi
2.2.1 Cancer
Oncogenes, tumor suppressor genes, and other regulatory genes are all potential targets of RNAi regulation. Targeted silencing of cancer-related genes and their regulators have been the most commonly studied [21,22]. For instance, leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) is a marker for gastric cancer, and silencing of Lgr5 can inhibit cancer angiogenesis [23]. In a recent study, angiogenesis in osteosarcoma was inhibited by siRNA against hypoxia-inducible factor (HIF)-1α [24]. RNAi could also be useful for addressing the problem of drug resistance induced by VEGF inhibitors. For example, Kim et al. found that siRNA targeting VEGF in combination with bevacizumab relieved resistance to bevacizumab, and successfully extended the effects of conventional anti-VEGF treatment options [25].
Deregulation of miRNA is frequently involved in many types of cancer. Strong evidence has shown that eukaryotes contain more ncRNAs than protein-coding genes [26]. Therefore, the opportunity to identify new diagnostic and prognostic markers and therapeutics is substantially expanded. Circulating miRNA levels could represent promising biomarkers for early detection and diagnosis of carcinoma [27–29]. For example, the miR-200 family, acting as key inhibitors of epithelial-to-mesenchymal transition, could suppress cell proliferation by inhibiting self-renewal and differentiation of cancer stem cells and modulating cell division and apoptosis and thus have potential prognostic value in human malignancies including gastric, breast, and endometrial cancers [30–33]. MiRNAs could also predict sensitivity to chemotherapy and radiotherapy in a personalized manner [34,35]. Preclinical study of therapeutic miRNA mimics and molecules targeting miRNAs (anti-miRs) have shown promising results [36].
In recent years, a number of RNAi-based drugs have been successfully validated by in vivo models and introduced into clinical trials. With the development of clinically relevant delivery methods, a number of clinical trials using systemically delivered RNAi are ongoing, many of which are focused on cancer (Table 1).
Table 1.
RNAi-based clinical trials for cancer therapy
| Drug | Indications | Active ingredient |
Target | Delivery route |
Delivery system |
Phase | Current status | Trial number |
|---|---|---|---|---|---|---|---|---|
| CALAA-01 | Solid tumors | siRNAs | RRM2 | IV injection | Cyclodextrin-containing polymer | 1 | Terminated 2008–2013 |
NCT00689065 |
| ALN-VSP02 | Solid tumors | siRNAs | KSP, VEGF | IV infusion | LNP | 1 | Completed 2009–2011 |
NCT00882180 |
| Atu027 | Advanced solid tumors | siRNAs | PKN3 | IV infusion | LNP | 1 | Completed 2009–2012 |
NCT00938574 |
| TKM-080301 | Primary or secondary liver cancer | siRNAs | PLK1 | Hepatic intra-arterial injection | LNP | 1 | Completed 2011–2012 |
NCT01437007 |
| Neuroendocrine tumors | siRNAs | PLK1 | IV infusion | LNP | 1/2 | Completed 2010–2015 |
NCT01262235 | |
| Advanced hepatocellular carcinoma | siRNAs | PLK1 | IV infusion | LNP | 1/2 | Completed 2014–2016 |
NCT02191878 | |
| MRX34 | Primary liver cancer or other solid tumors or hematologic malignancies | miR-34a mimic | miR-34 | IV injection | LNP | 1 | Terminated 2013–2016 |
NCT01829971 |
| DCR-MYC | Hepatocellular carcinoma | siRNAs | MYC | IV infusion | LNP | 1/2 | Ongoing, not recruiting 2014-present |
NCT02314052 |
| Solid tumors, multiple myeloma, non-Hodgkin lymphoma, or pancreatic neuroendocrine tumors | siRNAs | MYC | IV infusion | LNP | 1 | Ongoing, not recruiting 2014-present |
NCT02110563 | |
| APN401 | Recurrent melanoma, pancreatic cancer, renal cell cancer, or other solid tumors that are metastatic or cannot be removed by surgery | siRNA-transfected PBMCs | E3 ubiquitin ligase Cbl-b | IV injection | Ex vivo transfection | 1 | Ongoing, not recruiting 2014-present |
NCT02166255 |
| Metastatic pancreatic cancer, colorectal cancer, or other solid tumors or recurrent cancer | siRNA-transfected PBMCs | E3 ubiquitin ligase Cbl-b | IV injection | Ex vivo transfection | 1 | Recruiting 2017-present |
NCT03087591 | |
| SNS01-T | Multiple myeloma | siRNAs plasmids | eIF5A | IV injection | Polyethylenimine | 1/2 | Unknown 2014-present |
NCT01435720 |
| siRNA-EphA2-DOPC | Advanced solid tumors | siRNAs | EphA2 | IV injection | LNP | 1 | Recruiting 2016-present |
NCT01591356 |
| MiHA-loaded PD-L-silenced DC Vaccination | Hematologic malignancies | siRNA-treated Dendritic cells | PD-L1/PD-L2 | IV infusion | Ex vivo transfection | 1/2 | Recruiting 2015-present |
NCT02528682 |
| siG12D-LODER | Ductal adenocarcinoma or pancreatic cancer | siRNAs | KRAS | Intratumoral administration by EUS guidance | LODER polymer | 1 | Completed 2010–2014 |
NCT01188785 |
| Pancreatic ductal adenocarcinoma or pancreatic cancer | siRNAs | KRAS | Intratumoral implantation by EUS guidance | LODER polymer | 2 | Not yet recruiting 2017-present |
NCT01676259 |
IV: intravenous, LNP: lipid nanoparticles, PBMCs: peripheral blood mononuclear cells, EUS: endoscopic ultrasound.
The advantages of RNAi in cancer therapy are effective suppression of the growth of advanced-stage tumors, relatively low cost, and high specificity. RNAi can inhibit multiple genes of various pathways simultaneously, which could be conducive to reducing drug resistance. For example, Guan et al. found that inhibition of SH3GL1 using siRNA could reverse MDR by decreasing P-glycoprotein expression via the EGFR/ERK/AP-1 pathway [37]. With the development of more effective delivery systems, RNAi could also be used to develop personalized drugs for specific patients [38,39] as adjuvants to chemotherapy.
2.2.2 Viral infection
Shortly after the discovery of RNAi, synthetic siRNAs were recognized as a potential alternative to traditional antiviral therapy, which limits viral infection via direct silencing of viral genes or host-directed viral target genes regulating cellular defense function [40] [41]. Because a virus depends on the replication of a limited set of viral genes, RNAi may be ideal for treating viral infection [42]. For example, both siRNA and miRNA have demonstrated efficient inhibition of viral replication from different subtypes of HIV [43,44]. Drug-resistant mutants of HIV may be generated in response to almost all currently used anti-HIV agents; RNAi could avoid this resistance by targeting the mutated genomes [45,46]. The advanced targeted delivery of RNAi offers a practical way to protect uninfected cells and reverse drug resistance by introducing multiple silencers into infected cells [47].
Some of the earliest work using siRNA targeting respiratory syncytial virus [48] represents a well developed study against viral infection, and several preclinical and clinical trials have demonstrated its safety and tolerance [49,50]. New strategies such as second-generation siRNAs against the paramyxoviral RNA polymerase large subunit and a siRNA cocktail against influenza virus have also been applied [51]. In a recent study using dual-targeting siRNAs, which can knock down the expression of mRNA and viral genomic RNA simultaneously with its two active strands, the replication of respiratory syncytial virus was more effectively inhibited [52].
Patients with any of the three principal types of hepatitis can benefit from RNAi-based therapy. Because of the compact genome structure of the hepatitis B virus (HBV), which has overlapping regions, a single siRNA can silence multiple transcripts, resulting in direct inhibition of HBV replication [53]. RNAi can reduce viral load by knocking down the expression of pre-genomic RNA, eliminating the viral proteins. In addition, RNAi can stimulate a strong immune response, which can further amplify response to treatment [54]. To prevent the emergence of escaped mutant virus, a mixture of several HBV-siRNAs delivered by a pH-sensitive multifunctional envelope-type nanodevice was recently established, and proved more effective than a reverse transcriptase inhibitor in a mouse model [55]. With regard to hepatitis C virus, which has a much more complicated life cycle than HBV [56], miRNAs could not only inhibit viral replication but also function as biomarkers for the early detection and staging of liver disease related to hepatitis C, including liver cirrhosis and hepatocellular carcinoma [57,58]. Both serum and exosomal miRNA levels can predict the therapeutic efficacy of miRNAs against hepatitis C virus [59]. One of the most well studied miRNA targets is miR-122 [60], a highly abundant, liver-specific miRNA expressed in vertebrates that could facilitate replication of infectious virus in hepatic cells. The miR-122–specific inhibitor Miravirsen is the first miRNA-targeted antiviral drug that has undergone a phase II clinical trial, which prolonged HCV RNA reduction in a dose-dependent manner without evidence of resistance [61].
2.2.3 Cardiovascular disease
Dysregulation of genes and miRNAs in cardiomyocytes and smooth muscle cells has been demonstrated in many cardiovascular diseases [62]. Targeted delivery of miRNA therapeutics has been applied to several cardiomyopathies and related pathologies, such as hypertension, coronary disease, and atherosclerosis [63,64]. Although delivering miRNA to cardiac tissue is very challenging, interesting results have been obtained over the years, such as short peptide Arg-Glu-Asp-Val (REDV) modified PEG-trimethyl chitosan and nanofiber [65,66]. Studies have also suggested the potential of miRNAs as biomarkers in cardiovascular disease [67] for diagnosis and for predicting disease course or response to therapy [68].
2.2.4 Diabetes
A number of deregulated miRNAs have been linked to pathways associated with the metabolic process, including insulin secretion and pancreatic β-cell and adipocyte differentiation [69,70]. MiRNAs could be a key player in the molecular processes of type 2 diabetes mellitus and the associated complications. Meanwhile, since some miRNAs can direct cell differentiation toward β-cell–like cells and control islet β-cell development, it has been suggested that these miRNAs can help repair impaired islet β cells [71]. Promising results have been achieved using miRNA mimics or inhibitors to treat diabetes mellitus and its complications [72,73]. In addition, circulating microRNAs could function as biomarkers for the risk of diabetes mellitus [74,75], especially gestational diabetes [76].
2.3 RNAi in stem cell therapy
Stem cells have attracted a great deal of interest and shown potential both as a cellular therapy and as targets for RNAi. RNAi has been used to enhance the therapeutic effects of mesenchymal stem cells in several diseases, and RNAi-based functional modification is one such approach [77]. The migration or homing properties of stem cells can be regulated by miRNAs [78], and modification of miRNA could enhance the efficacy of mesenchymal stem cell–based therapy. Restrictions to the application of stem cells, such as stem-cell-related fibrosis, which is caused by the spontaneous fibroblastic differentiation of stem cells, could be well controlled by RNAi-based modification of stem cells [79,80]. RNAi-regulated gene silencing could also induce the secretion of specific cellular factors. Suppression of miR-383 can augment GDNF secretion, which could enhance the effects of human bone marrow–derived mesenchymal stem cells in treating spinal cord injury [81]. RNAi could also play a therapeutic role by targeting cancer stem cells [82]. For example, silencing stem cell factors in cancer stem cells via RNAi could inhibit the migration and epithelial-mesenchymal transition of cancer stem cells [83].
3. Challenges of ncRNA delivery
Despite its great promise, RNAi-based therapy is challenging because of issues related to stability and delivery of siRNA. The action site of siRNA is primarily in the cytosol; however, there are multiple barriers to its delivery that depend on administration route and targeted organs. In general, loco-regional delivery of siRNA has fewer barriers than systemic delivery. NcRNA by subcutaneous injection can access the circulation via capillaries or lymphatic drainage from interstitial space, which bypass the first-pass effect. However, the lipophilicity and size of the vectors must be carefully considered for the vectors to avoid phagocytosis. Oral administration is poor in maintaining intestinal stability and sufficient passage across the intestinal epithelium. Thus, the most common mode of administration of RNAi-based therapy is systemic delivery by intravenous or infusion injections.
3.1 Chemically unstable features of ncRNAs
The unfavorable physiochemical features of ncRNAs, such as high molecular weight, low stability, negative charge, and high structural stiffness, make it difficult to transport into the cytoplasm [84]. In its native form, ncRNAs are unstable in biologic fluids. Entering the blood, ncRNAs are easily enzymatically degraded by endogenous nucleases. The degraded ncRNAs are then filtered by the kidney and taken up by phagocytes or the reticuloendothelial system (RES). The reported half-life of unmodified siRNA in serum is only about 20 minutes [85,86]. Efforts have been made to improve the stability of siRNA in the blood by chemical or structural modification [87,88]. Various molecular positions could be chemically replaced or modified. For example, modification with hydrophilic and neutral molecules such as polyethylene glycol or non-anionic surfactants substantially prolongs the circulation time of siRNAs [89]. The half-life of the modified siRNAs could be increased by up to 20 times compared with the naked form [90,91].
3.2 Extracellular and intracellular barriers
To realize the full potential of RNAi therapy, the siRNAs and miRNAs must reach the cytoplasm of the target cells. To be delivered to the target sites, these RNA molecules must extravasate through the tight junctions of the vascular endothelium. Because of the leaky, discontinuous vessels at tumor sites, the cross-membrane transportation of siRNAs is easier in tumor tissue than in normal tissue [92]. However, tumor microvasculature also presents physiologic barriers, such as the irregular organization of vessels and heterogeneous hyperpermeability [93]. To overcome these challenges in delivering RNAi therapy across the vascular barrier, methods such as targeting of the tumor vasculature and increasing the aqueous solubility of drugs have been applied [93].
Cellular uptake is the next barrier for RNAi. The cellular membrane is made of negatively charged bilayer phospholipids consisting of functional proteins; and siRNAs, which are negatively charged, cannot diffuse passively into the cells. Chemical modification is a classic and commonly used strategy to enhance cellular uptake of oligonucleotides. Although no vehicles are provided, modifications such as lipids and cell-penetrating peptides can facilitate the siRNA delivery process. Lipidic conjugation is one of the earliest-studied methods of increasing hepatic uptake of siRNAs. The modified siRNA complex is incorporated into lipoprotein particles and internalized by hepatocytes, which promotes trans-membrane delivery of RNAi [94]. After conjugating cell-penetrating peptides with siRNAs or its delivery vectors, the complex is internalized by cells, mainly via direct penetration or endocytosis, which facilitates endosomal escape. Thus, cell-penetrating peptide conjugation increases the uptake of the siRNA complexes and enhances gene silencing [95]. Conjugating the complex, with cholesterol or aptamers in particular, substantially increases cellular uptake [96,97]. For example, dendritic cells can resist siRNA delivery, but after cholesterol modification, siRNAs could be efficiently delivered to dendritic cells to modify their function [98]. Another option is packaging siRNAs into extracellular vesicles to enter the cells by endocytosis. Conjugated with ligands such as folate or aptamers, siRNAs could be delivered by targeted endocytosis [99].
Once inside the cells, ncRNAs are trafficked intracellularly, beginning with early endosomes, which then fuse with sorting endosomes, and the contents are transferred into late endosomes. The late endosomes are then relocated to lysosomes, where the ncRNAs are degraded by various nucleases [100]. To get to the RISC complex in the cytoplasm, ncRNAs have to escape the endosome. Approaches to do so would increase either endosomolysis or rupture of the lysosome membrane. Neutrally charged ionizable lipids could be applied to induce lysosome rupture, which could release the siRNA carriers [101].
3.3 RNAi-induced innate immune stimulation and safety
The stimulation of the immune system by RNA molecules could enhance the treatment effect of RNAi against viral infection [102]. However, a high dose of siRNA is known to switch on the innate immune response and the production of cytokines [103]. RNAi could also trigger off-target effects, which is an important challenge. The induction of unanticipated phenotypes could interrupt the interpretation of therapeutic benefits. Unintended transcripts might be regulated by siRNAs, and the processing and function of miRNAs could be interrupted through saturation of the endogenous RNAi machinery by exogenous siRNAs [104]. RNAi-induced innate immune stimulation could thus undermine the safety and efficacy of RNAi, especially for clinical applications.
ARC-520, a targeted RNAi therapeutic against HBV, was among the first antiviral siRNAs to enter clinical development. ARC-520 showed promising therapeutic effects in sustaining reduced HBV antigen levels and virus titers [105]. ARC-520 injection was initially found to induce histamine release through mast cell degranulation, which is preventable. Although ARC-520 entered a phase 2 clinical trial, the U.S. Food and Drug Administration put the trial on hold because of safety concerns due to deaths in non-human primates given the highest dose. The first-in-human trial of miRNA cancer therapy, starting in 2016, investigated the therapeutic effect of MRX34, a liposomal miR-34a mimic, on multiple advanced solid tumors [106]. Unfortunately, it was closed 3 months after its start because of five reported immune-related serious adverse events, including severe cytokine release syndrome in one patient.
The activation of innate immunity might originate from the delivery vehicles or even the process of RNAi itself, and presents a challenge to the delivery of RNAi. Ideally, the delivery system should be non-immunogenic and incapable of inducing undesirable side effects. To avoid degradation before reaching the target site, the RNA molecules should be identified as host particles by the innate immune system. One newly demonstrated approach that addresses these considerations is ribose modification, which could decrease cytokine production [107].
4. Nanoparticle RNAi delivery systems
To overcome barriers in the delivery process and realize the broad potential of RNAi-based therapeutics, safe and efficient delivery systems are needed to transport ncRNAs to their site of action with minimal adverse effects. These carriers can generally be divided into viral and non-viral vectors. Viral vectors are efficient delivery systems using genetically modified viruses, which offer advantages such as sustained gene silencing and ease of expression of multiple copies of RNAi molecules from one transcript [108]. However, safety concerns such as viral genome insertion into patient chromosomes, immunogenicity of viral vectors, and expensive production restrain the widespread use of viral vectors [109]. Thus, great efforts have been undertaken to introduce synthetic vehicles for the in vivo delivery of ncRNAs.
Nanoparticles are the most common choice for the delivery of RNAi. Unlike viral vectors, which deliver ncRNAs in the form of a viral genome, non-viral carriers deliver native ncRNAs [110]. To administer ncRNAs systemically and allow them to cross physiologic barriers, delivery systems must be engineered to provide serum stability, offer high structural and functional tenability, mitigate interactions with non-target cells, enhance cell entry and endosome escape, resist renal clearance, and generate low toxicity and immunogenicity. A broad diversity of materials has been explored to address the challenges of in vivo delivery, with some successful systems developed by rational design or discovered by high-throughput screens. These systems can be grouped by composition into lipid-based and polymer-based systems. The destiny of siRNA-loaded nanoparticles with desirable structural and functional properties is depicted in Fig. 1.
Fig 1.
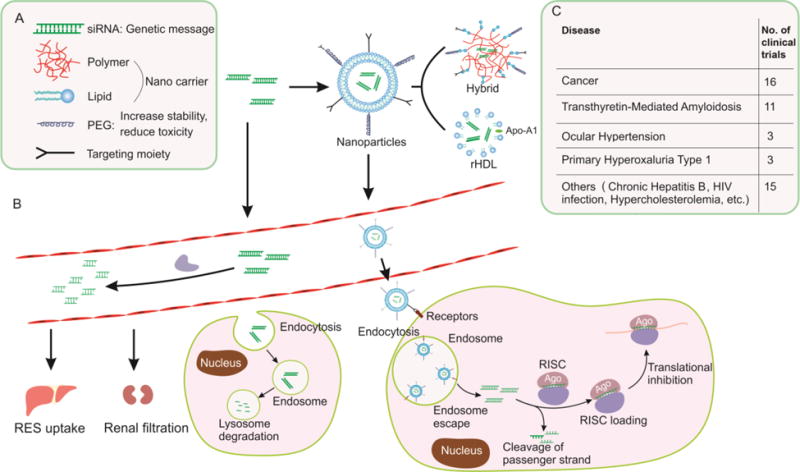
The destiny of synthetic siRNA-loaded nanoparticles with desirable structural and functional properties. (A) Components of ideally designed nanoparticles for RNAi delivery; (B) Process of RNAi; (C) Clinical application of siRNA-based drugs.
4.1 Lipid-based nanoparticle delivery systems
Lipid-based nanoparticle (LNP) systems are typically less than 100 nm in diameter and are currently the leading non-viral delivery systems employed to facilitate RNAi delivery. Since the cell membrane mainly consists of lipids and phospholipids, LNPs have a natural tendency to interact well with the cell membrane for cellular uptake [16]. Many biocompatible and biodegradable lipids and phospholipids can be applied to create LNPs. The risk of undesirable immunogenic reactions to lipids is lower than that of most of polymeric materials, which have higher molecular weights. Several classes of LNPs have been developed for RNA delivery, most notably liposomes, solid lipid nanoparticles (SLNs), and nanostructured lipid carriers (NLCs).
4.1.1 Liposomes
A liposome is structured as a spherical vesicle with an aqueous core and a bilayer lipid membrane. Unlike other lipid-based systems, which are almost exclusively used for loading lipophilic compounds, liposomes can be easily developed for the entrapment of hydrophilic and ionic molecules. As analogues of biological membranes, liposomes can be taken up by fusing with the plasma membrane; once inside the cell, they are processed via endocytosis, and the genetic material is released into the cytoplasm. Cationic liposomes can form complexes with negatively charged anionic siRNAs and polycations, forming “lipoplexes,” which offer biocompatibility and ease of production for clinical applications [111].
However, because of their positive charge, cationic liposomes can lead to dose-dependent cytotoxicity and inflammatory response, and the complexes may interact non-specifically with negatively charged serum proteins. To address these issues, neutral lipids are desired. 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) is a neutral lipid that has been demonstrated to effectively reduce cellular toxicity [112]. A successful example is DOPC nanoliposomal EphA2-targeted siRNA therapeutic (EPHARNA). EphA2 is a tyrosine kinase receptor in the ephrin family that plays an important role in multiple malignant processes [113,114]. EPHARNA effectively decreased in vivo EphA2 expression and showed 10-fold improvement in delivery compared with N-[1-(2,3-dioleoyloxyl)propyl]-N,N,N-trimethylammonium methyl sulfate (DOTAP) [115]. Preclinical research on safety in mouse models demonstrated that EPHARNA was well tolerated at all doses, and a phase 1 clinical trial of the agent is ongoing [116]. Another approach to reduce toxicity is the pH-sensitive liposome, which is usually made of neutral phosphatidylethanolamine (PE), with the addition of small pH-sensitive lipids, such as oleic acid and linoleic acid [117]. pH-sensitive liposomes can release drugs in tissues with lower pH values, such as those of the tumor environment compared with normal tissue. Besides decreasing cytotoxicity, pH-sensitive liposomes as a carrier for RNAi could also prolong gene-silencing effects and relieve resistance to chemotherapy [118]. To improve the stability of nanoparticles, the most commonly used strategy is shielding the surface with polyethylene glycol (PEG). PEGylation has been widely applied for bio-macromolecule delivery. Coating liposomes with PEG can increase the half-life of siRNA [119] and reduce its non-specific interactions with serum proteins [120].
4.1.2 Solid lipid nanoparticles
SLNs, with a size range of 50–1000 nm, are composed of a solid lipid core surrounded by a layer of surfactants in an aqueous dispersion, with multiple potential combinations of lipids and surfactants [121]. The lipid components are in a solid state at both body and ambient temperatures. SLNs are among the most effective carriers for hydrophilic and hydrophobic drugs owing to their inclusion of cationic lipids, which provide a positive surface potential that favors binding to nucleic acids; and SLNs can be used for gene delivery [122]. In RNAi delivery, siRNA can be incorporated into SLNs and provide sustained release of the material [123]. In a study, paclitaxel and human MCL1-specific siRNA (siMCL1) were loaded into SLNs. Compared with paclitaxel or siMCL1 alone, this co-delivery system exerted greater in vitro anti-cancer effects in the human epithelial carcinoma cell line KB [124]. In other studies, a siRNA-PEG/SLN system containing siRNA against c-Met (an oncogene overexpressed in a variety of carcinomas) was demonstrated to cross the blood-brain barrier to the tumor site after intravenous administration. By silencing of c-Met, tumor growth was inhibited in a dose-dependent manner [125] Xue et al. evaluated the in vivo biodistribution of SLNs in a prostate cancer model [126] and found extravascular delivery.
The solid, lipophilic core of SLNs presents an inherent difficulty in fully incorporating RNA molecules, which are hydrophilic and polyanionic. However, this structure can be coated with a RNA molecule or small-molecule drug on the surface, incorporating the lipophilic drugs in the core. Other advantages of SLNs include ease of preparation, excellent physical stability, and low toxicity [127].
4.1.3 Nanostructured lipid carriers
NLCs are next-generation lipid nanoparticles that combine advantages of different nanocarriers. They are modified SLNs with a solid lipid core in which the lipidic phase can consist of either solid or liquid forms at an ambient temperature [127]. Compared with SLNs, NLCs have more loading capacity and less water in the dispersion, making them more stable for storage. No difference between SLNs and NLCs in biotoxicity has been reported.
Studies have demonstrated that NLCs can be used as a novel delivery tool for the genetic therapy of cancer [128,129]. NLCs could also be used in co-delivery strategies. For instance, Taratula and colleagues set up a multifunctional NLC system that contained an anticancer drug (doxorubicin or paclitaxel), two siRNAs against cellular resistance (one targeted to MRP1 and the other targeted to BCL2), and a modified analogue of luteinizing-hormone-releasing hormone (LHRH) for targeting lung cancer cells. The system successfully enhanced the antitumor activity of the anticancer drug [130]. NLCs could also be modified to achieve targeted delivery and sustained effects. For example, by manipulation of the degradation process of NLCs, intracellular siRNA kinetics could be tailored to sustain the effects of the siRNAs [131]. This design extended the sustained knockdown period of siRNAs to 9 days, which would be better for clinical use than a shorter duration of effect.
Various strategies have been undertaken to overcome the drawbacks of the current lipid-based systems. Modification and engineering of the lipid structure could enhance transfection with reduced toxicity. A phospholipid molecule typically consists of a cationic head, a hydrophobic hydrocarbon backbone, and a linker region; selectively altering the structure of one or more of these components could be an effective approach. For example, the linker of DOTAP consists of biodegradable ester bonds, so its toxic effects are relatively short term and moderate [132]. The incorporation of fusogenic lipids, such as dioleoylphosphatidyl-ethanolamine, by increasing the interactions between the liposomal and endosomal membranes, could promote endosomal release of the siRNA[133].
In terms of clinical development, stabilized nucleic acid lipid particles (SNALPs) are among the most studied nanocarriers for the targeted delivery of RNAi [134]. The SNALP system consists of siRNAs surrounded by a lipid bilayer containing a mixture of cationic and fusogenic lipids coated with diffusible PEG. SNALPs, with their high bioavailability, can accumulate at the sites of leaky vasculature. Once accumulated, SNALPs can be internalized easily by cancer cells and the siRNA can be delivered.
4.2 Polymer-based delivery
Polymer-based nanoparticles are another well-studied system for the delivery of RNAi. Negatively charged nucleic acids, such as siRNAs, easily form complexes with positively charged polymers, forming polyplexes [135]. Commonly used carriers for this purpose are cationic polymers such as natural DNA-binding proteins, synthetic polypeptides, poly-ethylenimine, and carbohydrate-based polymers such as chitosan. Chitosan is a biocompatible, biodegradable polysaccharide with low immunogenic properties. However, naked chitosan is incompatible with biologic fluids, which cause particle degradation and reduce the working efficiency. Therefore, structure modification is necessary. PEGylated chitosan polyplexes have distinct mechanisms of endocytosis and macrophage phagocytosis, and show reduced non-specific interaction with red blood cells [136]. Polypeptide-modified chitosan is believed to have higher working efficiency and better endosomal escape [137]. Antibody-conjugated chitosan nanoparticles can deliver siRNA to specific cells. In a recent study, transferrin antibody and bradykinin B2 antibody were chemically conjugated with chitosan polyplexes, which delivered siRNA across the blood-brain barrier in a targeted manner, and increased its gene-silencing efficiency in astrocytes [138].
To address the limitations of polymeric and lipid-based nanoparticles, lipid–polymer hybrid nanoparticles (LPNs) have been developed. The unique structure of LPNs, which comprise polymer cores and lipid shells, exhibits complementary characteristics of both of the materials. In one study focusing on the application of LPNs in cancer treatment, lipid/rPAA-Chol polymer hybrid nanoparticles were modified by PEG and T7 peptide; the hybrid greatly inhibited tumor growth without any activation of immune responses [139]. In a later study, siRNAs and chemotherapeutic drug were co-delivered via LPNs, which enhanced the antitumor effect in pancreatic tumor model [140]. The preparation of LPNs consists of a conventional two-step method or an updated one-step method. In the updated method, instead of preparing polymeric nanoparticles and lipid vesicles separately, a single step such as nanoprecipitation after simply mixing polymer and lipid solutions is all that is needed [141]. In a recent study, to address the poor working efficiency of PEI-800, that is less toxic than the ordinarily used PEI-25K, PEI-800 and pH-sensitive lipid based LPNs were synthesized by microfluidic approaches. By this optimization, the delivery ability of PEI-800 was substantially improved, as shown by the stable and efficient knocking down of the target gene [142].
Other kinds of hybrid carriers besides LPNs have been investigated. For example, hybrid nanocomplexes composed of chitosan, protamine, lecithin, and thiamine pyrophosphate were prepared for the delivery of siRNAs against survivin. The hybrid system had suitable physicochemical properties, superior cellular uptake, and highly efficient gene silencing [143]. Another hybrid carrier was developed for the delivery of siRNAs against neurite growth-promoting factor 2 (NEGF2). With modifications using 2-chloroethylamine hydrochloride and N,N-dimethyl-2-chloroethylamine hydrochloride, gene silencing and inhibition of proliferation were greatly enhanced [144].
4.3 Other delivery systems
4.3.1 Exosomes
Exosomes are natural bio-carriers that have been investigated for the delivery of nucleotides and chemical drugs. Exosomes are phospholipid bilayer vesicles with a diameter of 40–120 nm that originate from endosomes. Exosomes are the smallest type of extracellular vesicle; when multi-vesicular bodies fuse with the plasma membrane, exosomes are secreted out of the cell. They contain a series of cell-specific transmembrane proteins that could guide exosomes to the target cells [145–147]. When exosomes reach the target, exosomal content is released into the cytoplasm, which results in changes to the intracellular compartment of the recipient cell. Accumulating evidence suggests that exosomes are a superior choice for delivering genetic materials between cells in a natural pathway [148,149]. An ideal natural nanocarrier, exosomes are suitable for hosting soluble drugs, and have high capacity for overcoming biologic barriers, few off-target effects, and low immunogenicity [150,151].
Exosomes can be loaded with therapeutic miRNAs and siRNAs using various methods. In brief, by means of a certain treatment of donor cells, synthetic miRNAs or other exogenous RNAs can be introduced into exosomes [152,153]. A wise choice of donor cell is crucial for achieving an efficient delivery. To date, some have used cell types such as HeLa, HEK-293, and dendritic cells. Because of their small size, exosomes can extravasate selectively into specific lesion sites and can even cross the blood-brain barrier, which is promising for the treatment of neurologic diseases [151]. To improve treatment and reduce side effects, exosomes can be genetically engineered to display ligands/homing peptides on their surface [154,155]. For example, by placing the GE11 peptide on the surface of exosomes loaded with let-7a miRNA, which can bind specifically with EGFR, let-7a miRNA can be delivered in a targeted manner to breast cancer tissues [156].
4.3.2 High-density lipoprotein
High-density lipoprotein (HDL), the smallest plasma lipoprotein, is well known as “good” cholesterol because of its cardio-protective properties. Mechanistically, HDL functions as an endogenous vehicle for the transportation and metabolism of cholesterol, phospholipids, and triglycerides. HDL can also transport endogenous proteins, hormones, and miRNA to various organs, indicating that it could serve as a delivery vehicle for nucleic acids. In recent years, HDL has drawn attention as a potential tumor-targeting carrier for RNAi-based therapeutics [157]. Scavenger receptor B type I (SCARB1) is a major receptor of HDL; furthermore, because of the biogenesis requirements of tumor growth, many tumor cells overexpress SCARB1. This receptor thus offers a natural, tumor-specific, HDL-dependent delivery method for RNAi [158]. It has been reported that HDL can selectively deliver siRNAs to various types of cancer cells, including breast, ovarian, and liver cancers [159–161]. In a study comparing tumor accumulation, synthetic HDL was demonstrated to penetrate much deeper into tumor spheroids than liposomes and PEG-liposomes [162]. In addition to having the basic properties of endogenous HDL, synthetic HDL may enable tailored and unique functions. In addition, synthetic HDL could be produced at a large scale, which would facilitate clinical applications.
5. Targeted delivery
Although improvements in biodistribution of nanoparticles could be attained using techniques such as PEGylation, most nanoparticle therapeutics will inevitably be concentrated in the reticuloendothelial organs such as the liver and spleen. However, the therapeutic window of nanoparticles could be increased by targeted delivery to specific tissues, cells, or microenvironments. Targeted delivery can reduce off-target effects and increase the bioavailability of the therapeutic agent at the target site. This targeting ability is usually conferred by adding a ligand to the ncRNA itself or to the nanoparticle. The molecules most commonly applied as these ligands are described below.
5.1 Aptamer
Aptamers are single-stranded short DNAs or RNAs that can specifically bind to various small molecules with high affinity. They are nucleotide analogues of antibodies, which bind with targets in a similar way, in antigen-antibody interactions [163]. Moreover, aptamers are neither immunogenic nor toxic. Thus, they have been widely used for diagnosis and drug delivery. In terms of RNAi delivery, aptamers can facilitate targeted delivery by non-covalent conjugation, forming an aptamer-RNA complex or combination with nanoparticles [164,165]. A lot of work has explored the potential of aptamer-RNA in cancer treatment [166,167]. In a recent study, RNA nanoparticles were constructed using anti–prostate-specific membrane antigen RNA aptamer as a targeting ligand and anti-miR-17 or anti-miR-21 as therapeutic modules. After being administered systemically, the complex bonded selectively with the tumor tissue and repressed tumor growth [168]. In another study to target metastatic breast cancer cells, aptamer-labeled hybrid nanoparticles were used to deliver siRNA. In vitro studies demonstrated that this aptamer-functionalized complex significantly increased the knockdown efficacy of the siRNA [97].
5.2 Peptide and antibody
Molecules such as peptide ligands and antibodies can selectively bind with their receptors. Conjugated with siRNA-loaded nanoparticles, both peptide ligands and antibodies can direct targeted delivery of RNAi [169]. Studies have investigated the potential of peptides in siRNA delivery, and most have focused on cyclic Arg-Gly-Asp (RGD) peptides. The receptor of RGD, alpha V/β integrin, is ubiquitously expressed in tumor endothelial cells and thus offers a potential target. In one study, siRNAs were selectively delivered to the tumor site using an RGD-modified liposomal-siRNA system, resulting in a substantial delay in tumor growth [167]. Rengaswamy and colleagues developed integrin receptor–targeted liposome-protamine-siRNA nanoparticles using RGD peptide [170]. Loaded with siRNA against PAX3-FOXO1, the specific fusion transcript for alveolar rhabdomyosarcoma, the complex efficiently inhibited tumor cell proliferation, suggesting that this system is a promising approach to the management of residual disease. Owing to their superior stability and specificity, monoclonal antibodies and antibody parts have attracted much attention as a powerful tool for targeted delivery of RNAi. By linking specific antibodies to nanocarriers, this tumor-specific recognition process could greatly enhance the effect of siRNAs [171,172].
5.3 Folate
The folate receptor is another popular option for cancer-specific RNAi delivery. Folic acid is needed for rapid cell growth, and the folate receptor is overexpressed in many types of cancer cells. Thus, folate can be used in molecular imaging for cancer detection and can function as a tumor-specific ligand for tumor-targeted delivery. Once the folate-conjugated nanoparticle binds with a folic acid receptor, the complex is transferred through receptor-mediated endocytosis. In an ovarian cancer study, folate- conjugated nanoparticles selectively delivered siRNA into the cancer cells and achieved a drastic and sustained knockdown of toll-like receptor 4, which helped re-sensitize the cancer cells to paclitaxel [173]. Some of these promising conjugated delivery systems are already being tested in clinical trials [174].
6. Monitoring delivery
To optimize the efficacy of gene silencing and guide the treatment plan, one must track the delivery of RNAi in a non-invasive manner. Generally, monitoring is achieved by RNAi imaging, a process that visualizes the in vivo behavior of RNAi therapeutics in a quantitative and qualitative manner. To date, a variety of imaging modalities have been used alone or in combination, including bioluminescence imaging, magnetic resonance imaging, positron emission tomography, single-photon emission computed tomography, and targeted ultrasonography [175,176]. The RNAi imaging system includes a nanoparticle whose core has imaging properties as a carrier and reporter, a ligand for targeting, and gene-specific siRNA. One of the most widely studied optical techniques in this setting is fluorescence. By chemically conjugating an exogenous contrast agent to delivery vehicles, fluorescence imaging represents a powerful monitoring strategy [177,178].
Magnetic particles are another popular choice for siRNA monitoring. In vivo studies have shown that a magnetic core is capable of probing the site and size of cancer in vivo on magnetic resonance imaging. Magnetic particles have also been shown to effectively monitor treatment effects such as retardation of tumor growth [179,180]. The imaging of RNAi can monitor the target cells as well, which helps ensure gene-silencing efficacy indirectly. A synthesized siRNA conjugated with magnetic nanoparticles and labeled with a near-infrared fluorescent agent Cy5.5 was introduced to islet transplantation for the treatment of type 1 diabetes [181]. At the time of gene silencing, the islets were labeled by Cy5.5. Magnetic resonance imaging and NIGF imaging allowed monitoring of the survival of the transplanted islets.
7. RNAi delivery addressing MDR in cancer therapy
Despite advances in early detection and chemotherapy, successful cancer management remains a formidable challenge. One reason for this difficulty is the frequent development of MDR after long-term chemotherapy [182,183]. Overexpression of MDR1 is a widely studied reason for MDR [184]. Nevertheless, a variety of MDR1 inhibitors failed to provide clinical benefit [185,186]. RNAi-based genetic therapy, with its ability to selectively transport siRNA/miRNA to the cancer tissue in vivo, could offer opportunities to avoid MDR arising from chemotherapy through the strategy of co-delivery of siRNA and chemotherapy.
This co-delivery incorporates chemotherapy and a MDR modulator into the same nanoparticle to simultaneously deliver two payloads to the same cancer cell population. This strategy could maximize therapeutic effects with a minimal chemotherapy dose; compared with monotherapy, co-delivery leads to a better therapeutic response and increased survival rate in mouse models [187,188]. RNAi can silence chemotherapy export genes to increase drug accumulation in chemotherapy-resistant cells [189]. To achieve highly efficient co-delivery, some have been focused on targeting or using stimuli-responsive delivery systems [190,191]. Recently, a tumor-targeted nano-vehicle peptide-conjugated PSPG (PSPGP) was synthesized for co-delivery of paclitaxel and TR3 siRNA [192]. This system enhanced endosomal escape and intracellular degradation, which increased sensitivity to paclitaxel. Overexpression of the erythroblastosis virus E26 oncogene homolog 1 (ETS1) gene is believed to correlate with tumor progression and poor chemotherapy response [193,194]; supramolecular nano-assemblies that co-deliver siETS1 and doxorubicin could be another promising approach to reverse MDR [195]. The complex also increases drug residence time at the tumor site, which could further enhance inhibition of tumor growth. The basic mechanisms of chemo-resistance in most cancers with MDR include pump and non-pump resistance. Pump resistance, induced by membrane proteins such as P-glycoprotein (MDR1/ABCB1) [196] and MDR proteins (MRP, MRP1/ABCC1) [197], could help the cells deport drugs, whereas the non-pump resistance could activate the defense against cell death. An RNA nanotechnology platform for a two-in-one RNAi delivery system was demonstrated to generate precisely controlled and biocompatible nanoparticles that silence pump and non-pump resistance genes simultaneously [198].
8. Conclusion and future perspectives
Over the last 15 years, RNAi has been widely applied as a promising therapeutic approach to a number of pathologic conditions. Progress has been made in the investigation of target genes and the development of delivery systems for RNAi. In addition, the efficiency of delivery as well as gene silencing have greatly improved over time. However, challenges remain for the successful clinical application of RNAi-based therapeutics. Safety concerns have been the principal reason that clinical trials of some RNAi therapies have been withdrawn. Innate immune activation resulting from nanocarriers and RNAi itself is another challenge. An ideally designed delivery system is crucial to achieve effective, targeted gene silencing with low or no toxicity in vivo. Future research is expected to focus on addressing these important issues. Tissue-specific ligands with greater stability and affinity are also anticipated to improve the delivery systems. On the basis of our understanding of the development of RNAi and delivery systems, we believe that these efforts will lead to a new age of molecular therapy in which patients can benefit from safe, efficient, and personalized therapy.
Acknowledgments
The University of Texas MD Anderson Cancer Center is supported in part by the National Institutes of Health through Cancer Center Support Grant P30CA016672, CA217685, P50 CA098258, UH3 TR000943, U01 CA213759, R35 CA209904), the Ovarian Cancer Research Fund, Inc., CPRIT grant RP120214, DP150091, V-Foundation, the Frank McGraw Memorial Chair in Cancer Research, and the American Cancer Society Research Professor Award.
Footnotes
Conflicts of interest
The authors declare that they have no conflicts of interest related to the topics discussed here.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials (Basel) 2017;7(4) doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa FF. Non-coding RNAs: lost in translation? Gene. 2007;386(1–2):1–10. doi: 10.1016/j.gene.2006.09.028. doi:S0378-1119(06)00614-7 [pii] 10.1016/j.gene.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 6.Mansoori B, Sandoghchian Shotorbani S, Baradaran B. RNA interference and its role in cancer therapy. Adv Pharm Bull. 2014;4(4):313–321. doi: 10.5681/apb.2014.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61(9):746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith I, Procter M, Gelber RD, Guillaume S, Feyereislova A, Dowsett M, et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369(9555):29–36. doi: 10.1016/S0140-6736(07)60028-2. doi:S0140-6736(07)60028-2 [pii] 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 12.Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. doi: 10.1056/NEJMoa062867. doi:355/23/2408 [pii] 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 13.Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3(12):739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggert US, Field CM, Mitchison TJ. Small molecules in an RNAi world. Mol Biosyst. 2006;2(2):93–96. doi: 10.1039/b515335b. [DOI] [PubMed] [Google Scholar]
- 15.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21(9):1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016;6(3):235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 18.Kozak K. Annotation and specificity of existing genome-wide small interfering RNA libraries. Nucleic Acid Ther. 2013;23(1):71–80. doi: 10.1089/nat.2012.0387. [DOI] [PubMed] [Google Scholar]
- 19.Mohr SE, Perrimon N. RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip Rev RNA. 2012;3(2):145–158. doi: 10.1002/wrna.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupaimoole R, Han HD, Lopez-Berestein G, Sood AK. MicroRNA therapeutics: principles, expectations, and challenges. Chin J Cancer. 2011;30(6):368–370. doi: 10.5732/cjc.011.10186. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie W, Zhao M, Zhou W, Guo L, Huang L, Yu W, et al. Targeting of integrin-linked kinase with small interfering RNA inhibits VEGF-induced angiogenesis in retinal endothelial cells. Ophthalmic Res. 2013;49(3):139–149. doi: 10.1159/000345070. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y, E G, Wang E, Pal K, Dutta SK, Bar-Sagi D, et al. VEGF exerts an angiogenesis-independent function in cancer cells to promote their malignant progression. Cancer Res. 2012;72(16):3912–3918. doi: 10.1158/0008-5472.CAN-11-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xi HQ, Zhang KC, Li JY, Cui JX, Gao YH, Wei B, et al. RNAi-mediated inhibition of Lgr5 leads to decreased angiogenesis in gastric cancer. Oncotarget. 2017;8(19):31581–31591. doi: 10.18632/oncotarget.15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XD, Wu Q, Yang SH. Effects of siRNA-mediated HIF-1alpha gene silencing on angiogenesis in osteosarcoma. Pak J Med Sci. 2017;33(2):341–346. doi: 10.12669/pjms.332.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MG, Jo SD, Yhee JY, Lee BS, Lee SJ, Park SG, et al. Synergistic anti-tumor effects of bevacizumab and tumor targeted polymerized VEGF siRNA nanoparticles. Biochem Biophys Res Commun. 2017 doi: 10.1016/j.bbrc.2017.05.103. [DOI] [PubMed] [Google Scholar]
- 26.Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu S, et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7(1):1692. doi: 10.1038/s41598-017-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homami A, Ghazi F. MicroRNAs as biomarkers associated with bladder cancer. Med J Islam Repub Iran. 2016;30:475. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Zheng J, Sun C, Wang L, Jin G, Xin L, et al. MicroRNA expression levels as diagnostic biomarkers for intraductal papillary mucinous neoplasm. Oncotarget. 2017 doi: 10.18632/oncotarget.17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y, et al. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med. 2015;19(4):760–769. doi: 10.1111/jcmm.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu YB, Hu JJ, Sun WJ, Duan XH, Chen X. Prognostic value of miR-141 downregulation in gastric cancer. Genet Mol Res. 2015;14(4):17305–17311. doi: 10.4238/2015.December.16.31. [DOI] [PubMed] [Google Scholar]
- 32.Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, et al. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer. 2013;132(7):1633–1645. doi: 10.1002/ijc.27840. [DOI] [PubMed] [Google Scholar]
- 33.Zhu ZM, Xu YF, Su QJ, Du JD, Tan XL, Tu YL, et al. Prognostic significance of microRNA-141 expression and its tumor suppressor function in human pancreatic ductal adenocarcinoma. Mol Cell Biochem. 2014;388(1–2):39–49. doi: 10.1007/s11010-013-1897-y. [DOI] [PubMed] [Google Scholar]
- 34.Winther M, Knudsen S, Dahlgaard J, Jensen T, Hansen A, Jensen PB, et al. Clinical Impact of a Novel MicroRNA Chemo-Sensitivity Predictor in Gastrooesophageal Cancer. PLoS One. 2016;11(2):e0148070. doi: 10.1371/journal.pone.0148070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmer F, Venderbosch S, Dijkstra JR, Vink-Borger EM, Faber C, Mekenkamp LJ, et al. MicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancer. Oncotarget. 2015;6(26):22996–23007. doi: 10.18632/oncotarget.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 37.Guan H, Zhao P, Dai Z, Liu X, Wang X. SH3GL1 inhibition reverses multidrug resistance in colorectal cancer cells by downregulation of MDR1/P-glycoprotein via EGFR/ERK/AP-1 pathway. Tumour Biol. 2016;37(9):12153–12160. doi: 10.1007/s13277-016-5092-0. [DOI] [PubMed] [Google Scholar]
- 38.Martucci NM, Migliaccio N, Ruggiero I, Albano F, Cali G, Romano S, et al. Nanoparticle-based strategy for personalized B-cell lymphoma therapy. Int J Nanomedicine. 2016;11:6089–6101. doi: 10.2147/IJN.S118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Fei T, Zheng X, Brown M, Zhang P, Liu XS, et al. An Integrative Pharmacogenomic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine. Sci Rep. 2016;6:22120. doi: 10.1038/srep22120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prussia A, Thepchatri P, Snyder JP, Plemper RK. Systematic approaches towards the development of host-directed antiviral therapeutics. Int J Mol Sci. 2011;12(6):4027–4052. doi: 10.3390/ijms12064027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6(3):206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 42.Blake SJ, Bokhari FF, McMillan NA. RNA interference for viral infections. Curr Drug Targets. 2012;13(11):1411–1420. doi: 10.2174/138945012803530161. [DOI] [PubMed] [Google Scholar]
- 43.Choi JG, Bharaj P, Abraham S, Ma H, Yi G, Ye C, et al. Multiplexing seven miRNA-Based shRNAs to suppress HIV replication. Mol Ther. 2015;23(2):310–320. doi: 10.1038/mt.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34(6):696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tchurikov NA, Fedoseeva DM, Gashnikova NM, Sosin DV, Gorbacheva MA, Alembekov IR, et al. Conserved sequences in the current strains of HIV-1 subtype A in Russia are effectively targeted by artificial RNAi in vitro. Gene. 2016;583(1):78–83. doi: 10.1016/j.gene.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Kravatsky YV, Chechetkin VR, Fedoseeva DM, Gorbacheva MA, Kretova OV, Tchurikov NA. Mutation frequencies in HIV-1 subtype-A genome in regions containing efficient RNAi targets. Mol Biol (Mosk) 2016;50(3):480–485. doi: 10.7868/S0026898416020117. [DOI] [PubMed] [Google Scholar]
- 47.Cornu TI, Mussolino C, Bloom K, Cathomen T. Editing CCR5: a novel approach to HIV gene therapy. Adv Exp Med Biol. 2015;848:117–130. doi: 10.1007/978-1-4939-2432-5_6. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Hellermann G, et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat Med. 2005;11(1):56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- 49.DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, Toudjarska I, et al. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral Res. 2008;77(3):225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Ballarin-Gonzalez B, Thomsen TB, Howard KA. Clinical translation of RNAi-based treatments for respiratory diseases. Drug Deliv Transl Res. 2013;3(1):84–99. doi: 10.1007/s13346-012-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barik S, Lu P. Therapy of respiratory viral infections with intranasal siRNAs. Methods Mol Biol. 2015;1218:251–262. doi: 10.1007/978-1-4939-1538-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malekshahi SS, Salimi V, Arefian E, Fatemi-Nasab G, Adjaminejad-Fard S, Yavarian J, et al. Inhibition of Respiratory Syncytial Virus Replication by Simultaneous Targeting of mRNA and Genomic RNA Using Dual-Targeting siRNAs. Mol Biotechnol. 2016;58(11):767–775. doi: 10.1007/s12033-016-9976-4. [DOI] [PubMed] [Google Scholar]
- 53.Gane EJ. Future anti-HBV strategies. Liver Int. 2017;37(Suppl 1):40–44. doi: 10.1111/liv.13304. [DOI] [PubMed] [Google Scholar]
- 54.Ebert G, Poeck H, Lucifora J, Baschuk N, Esser K, Esposito I, et al. 5′ Triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology. 2011;141(2):696–706. 706 e691–693. doi: 10.1053/j.gastro.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto N, Sato Y, Munakata T, Kakuni M, Tateno C, Sanada T, et al. Novel pH-sensitive multifunctional envelope-type nanodevice for siRNA-based treatments for chronic HBV infection. J Hepatol. 2016;64(3):547–555. doi: 10.1016/j.jhep.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 56.Lee CH, Kim JH, Lee SW. The Role of MicroRNA in Pathogenesis and as Markers of HCV Chronic Infection. Curr Drug Targets. 2016 doi: 10.2174/1389450117666160401125213. [DOI] [PubMed] [Google Scholar]
- 57.Zekri AN, Youssef AS, El-Desouky ED, Ahmed OS, Lotfy MM, Nassar AA, et al. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol. 2016;37(9):12273–12286. doi: 10.1007/s13277-016-5097-8. [DOI] [PubMed] [Google Scholar]
- 58.Shaker OG, Senousy MA. Serum microRNAs as predictors for liver fibrosis staging in hepatitis C virus-associated chronic liver disease patients. J Viral Hepat. 2017 doi: 10.1111/jvh.12696. [DOI] [PubMed] [Google Scholar]
- 59.Jiao X, Fan Z, Chen H, He P, Li Y, Zhang Q, et al. Serum and exosomal miR-122 and miR-199a as a biomarker to predict therapeutic efficacy of hepatitis C patients. J Med Virol. 2017 doi: 10.1002/jmv.24829. [DOI] [PubMed] [Google Scholar]
- 60.Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160(6):1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 62.Hoelscher SC, Doppler SA, Dressen M, Lahm H, Lange R, Krane M. MicroRNAs: pleiotropic players in congenital heart disease and regeneration. J Thorac Dis. 2017;9(Suppl 1):S64–S81. doi: 10.21037/jtd.2017.03.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwekkeboom RF, Lei Z, Doevendans PA, Musters RJ, Sluijter JP. Targeted delivery of miRNA therapeutics for cardiovascular diseases: opportunities and challenges. Clin Sci (Lond) 2014;127(6):351–365. doi: 10.1042/CS20140005. [DOI] [PubMed] [Google Scholar]
- 64.Tadin-Strapps M, Robinson M, Le Voci L, Andrews L, Yendluri S, Williams S, et al. Development of lipoprotein(a) siRNAs for mechanism of action studies in non-human primate models of atherosclerosis. J Cardiovasc Transl Res. 2015;8(1):44–53. doi: 10.1007/s12265-014-9605-1. [DOI] [PubMed] [Google Scholar]
- 65.Zhou F, Jia X, Yang Q, Yang Y, Zhao Y, Fan Y, et al. Targeted delivery of microRNA-126 to vascular endothelial cells via REDV peptide modified PEG-trimethyl chitosan. Biomater Sci. 2016;4(5):849–856. doi: 10.1039/c5bm00629e. [DOI] [PubMed] [Google Scholar]
- 66.Zhou F, Jia X, Yang Y, Yang Q, Gao C, Hu S, et al. Nanofiber-mediated microRNA-126 delivery to vascular endothelial cells for blood vessel regeneration. Acta Biomater. 2016;43:303–313. doi: 10.1016/j.actbio.2016.07.048. [DOI] [PubMed] [Google Scholar]
- 67.Navickas R, Gal D, Laucevicius A, Taparauskaite A, Zdanyte M, Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res. 2016;111(4):322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kasner M, Gast M, Galuszka O, Stroux A, Rutschow S, Wang X, et al. Circulating exosomal microRNAs predict functional recovery after MitraClip repair of severe mitral regurgitation. Int J Cardiol. 2016;215:402–405. doi: 10.1016/j.ijcard.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 69.Eliasson L, Esguerra JL. Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol (Oxf) 2014;211(2):273–284. doi: 10.1111/apha.12285. [DOI] [PubMed] [Google Scholar]
- 70.Guglielmi V, D’Adamo M, Menghini R, Cardellini M, Gentileschi P, Federici M, et al. MicroRNA 21 is up-regulated in adipose tissue of obese diabetic subjects. Nutr Healthy Aging. 2017;4(2):141–145. doi: 10.3233/NHA-160020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaviani M, Azarpira N, Karimi MH, Al-Abdullah I. The role of microRNAs in islet beta-cell development. Cell Biol Int. 2016;40(12):1248–1255. doi: 10.1002/cbin.10691. [DOI] [PubMed] [Google Scholar]
- 72.Pishavar E, Behravan J. miR-126 as a therapeutic agent for Diabetes Mellitus. Curr Pharm Des. 2017 doi: 10.2174/1381612823666170424120121. [DOI] [PubMed] [Google Scholar]
- 73.de Gonzalo-Calvo D, van der Meer RW, Rijzewijk LJ, Smit JW, Revuelta-Lopez E, Nasarre L, et al. Serum microRNA-1 and microRNA-133a levels reflect myocardial steatosis in uncomplicated type 2 diabetes. Sci Rep. 2017;7(1):47. doi: 10.1038/s41598-017-00070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zou HL, Wang Y, Gang Q, Zhang Y, Sun Y. Plasma level of miR-93 is associated with higher risk to develop type 2 diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. 2017 doi: 10.1007/s00417-017-3638-5. [DOI] [PubMed] [Google Scholar]
- 75.Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, et al. Circulating MicroRNA-122 Is Associated With the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes. 2017;66(2):347–357. doi: 10.2337/db16-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y, Tian F, Li H, Zhou Y, Lu J, Ge Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynaecol Obstet. 2015;130(1):49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 77.Sam MR, Azadbakhsh AS, Farokhi F, Rezazadeh K, Sam S, Zomorodipour A, et al. Genetic modification of bone-marrow mesenchymal stem cells and hematopoietic cells with human coagulation factor IX-expressing plasmids. Biologicals. 2016;44(3):170–177. doi: 10.1016/j.biologicals.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 78.Wang W, Li C, Li W, Kong L, Qian A, Hu N, et al. MiR-150 enhances the motility of EPCs in vitro and promotes EPCs homing and thrombus resolving in vivo. Thromb Res. 2014;133(4):590–598. doi: 10.1016/j.thromres.2013.12.038. [DOI] [PubMed] [Google Scholar]
- 79.Huleihel L, Sellares J, Cardenes N, Alvarez D, Faner R, Sakamoto K, et al. Modified mesenchymal stem cells using miRNA transduction alter lung injury in a bleomycin model. Am J Physiol Lung Cell Mol Physiol. 2017 doi: 10.1152/ajplung.00323.2016. ajplung 00323 02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qu Y, Zhang Q, Cai X, Li F, Ma Z, Xu M, et al. Exosomes derived from miR-181-5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med. 2017 doi: 10.1111/jcmm.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei GJ, An G, Shi ZW, Wang KF, Guan Y, Wang YS, et al. Suppression of MicroRNA-383 Enhances Therapeutic Potential of Human Bone-Marrow-Derived Mesenchymal Stem Cells in Treating Spinal Cord Injury via GDNF. Cell Physiol Biochem. 2017;41(4):1435–1444. doi: 10.1159/000468057. [DOI] [PubMed] [Google Scholar]
- 82.Pan Y, Shu X, Sun L, Yu L, Sun L, Yang Z, et al. miR196a5p modulates gastric cancer stem cell characteristics by targeting Smad4. Int J Oncol. 2017 doi: 10.3892/ijo.2017.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang L, Wang J, Li Z, Liu Y, Jiang M, Li Y, et al. Silencing stem cell factor attenuates stemness and inhibits migration of cancer stem cells derived from Lewis lung carcinoma cells. Tumour Biol. 2016;37(6):7213–7227. doi: 10.1007/s13277-015-4577-6. [DOI] [PubMed] [Google Scholar]
- 84.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hickerson RP, Vlassov AV, Wang Q, Leake D, Ilves H, Gonzalez-Gonzalez E, et al. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides. 2008;18(4):345–354. doi: 10.1089/oli.2008.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sarett SM, Kilchrist KV, Miteva M, Duvall CL. Conjugation of palmitic acid improves potency and longevity of siRNA delivered via endosomolytic polymer nanoparticles. J Biomed Mater Res A. 2015;103(9):3107–3116. doi: 10.1002/jbm.a.35413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bramsen JB, Kjems J. Chemical modification of small interfering RNA. Methods Mol Biol. 2010;721:77–103. doi: 10.1007/978-1-61779-037-9_5. [DOI] [PubMed] [Google Scholar]
- 88.Lee SJ, Son S, Yhee JY, Choi K, Kwon IC, Kim SH, et al. Structural modification of siRNA for efficient gene silencing. Biotechnol Adv. 2013;31(5):491–503. doi: 10.1016/j.biotechadv.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 89.Tokatlian T, Segura T. siRNA applications in nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2(3):305–315. doi: 10.1002/wnan.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18(4):305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 91.Bramsen JB, Kjems J. Engineering small interfering RNAs by strategic chemical modification. Methods Mol Biol. 2013;942:87–109. doi: 10.1007/978-1-62703-119-6_5. [DOI] [PubMed] [Google Scholar]
- 92.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Danquah MK, Zhang XA, Mahato RI. Extravasation of polymeric nanomedicines across tumor vasculature. Adv Drug Deliv Rev. 2011;63(8):623–639. doi: 10.1016/j.addr.2010.11.005. doi:S0169-409X(10)00287-5 [pii] 10.1016/j.addr.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 94.Raouane M, Desmaele D, Urbinati G, Massaad-Massade L, Couvreur P. Lipid conjugated oligonucleotides: a useful strategy for delivery. Bioconjug Chem. 2012;23(6):1091–1104. doi: 10.1021/bc200422w. [DOI] [PubMed] [Google Scholar]
- 95.Layek B, Lipp L, Singh J. Cell Penetrating Peptide Conjugated Chitosan for Enhanced Delivery of Nucleic Acid. Int J Mol Sci. 2015;16(12):28912–28930. doi: 10.3390/ijms161226142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Loughlin AJ, Mager I, de Jong OG, Varela MA, Schiffelers RM, El Andaloussi S, et al. Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles. Mol Ther. 2017 doi: 10.1016/j.ymthe.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Powell D, Chandra S, Dodson K, Shaheen F, Wiltz K, Ireland S, et al. Aptamer-functionalized hybrid nanoparticle for the treatment of breast cancer. Eur J Pharm Biopharm. 2017;114:108–118. doi: 10.1016/j.ejpb.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bruck J, Pascolo S, Fuchs K, Kellerer C, Glocova I, Geisel J, et al. Cholesterol Modification of p40-Specific Small Interfering RNA Enables Therapeutic Targeting of Dendritic Cells. J Immunol. 2015;195(5):2216–2223. doi: 10.4049/jimmunol.1402989. [DOI] [PubMed] [Google Scholar]
- 99.Wang Y, Chen X, Tian B, Liu J, Yang L, Zeng L, et al. Nucleolin-targeted Extracellular Vesicles as a Versatile Platform for Biologics Delivery to Breast Cancer. Theranostics. 2017;7(5):1360–1372. doi: 10.7150/thno.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117(12):3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 102.Meng Z, Zhang X, Wu J, Pei R, Xu Y, Yang D, et al. RNAi induces innate immunity through multiple cellular signaling pathways. PLoS One. 2013;8(5):e64708. doi: 10.1371/journal.pone.0064708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meng Z, Lu M. RNA Interference-Induced Innate Immunity, Off-Target Effect, or Immune Adjuvant? Front Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 105.Schluep T, Lickliter J, Hamilton J, Lewis DL, Lai CL, Lau JY, et al. Safety, Tolerability, and Pharmacokinetics of ARC-520 Injection, an RNA Interference-Based Therapeutic for the Treatment of Chronic Hepatitis B Virus Infection, in Healthy Volunteers. Clin Pharmacol Drug Dev. 2016 doi: 10.1002/cpdd.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35(2):180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Valenzuela RA, Suter SR, Ball-Jones AA, Ibarra-Soza JM, Zheng Y, Beal PA. Base modification strategies to modulate immune stimulation by an siRNA. Chembiochem. 2015;16(2):262–267. doi: 10.1002/cbic.201402551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grimm D. Small silencing RNAs: state-of-the-art. Adv Drug Deliv Rev. 2009;61(9):672–703. doi: 10.1016/j.addr.2009.05.002. doi:S0169-409X(09)00155-0 [pii] 10.1016/j.addr.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 109.Giacca M, Zacchigna S. Virus-mediated gene delivery for human gene therapy. J Control Release. 2012;161(2):377–388. doi: 10.1016/j.jconrel.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 110.Keller M. Nanomedicinal delivery approaches for therapeutic siRNA. Int J Pharm. 2009;379(2):210–211. doi: 10.1016/j.ijpharm.2009.03.038. doi:S0378-5173(09)00180-X [pii] 10.1016/j.ijpharm.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 111.Rezaee M, Oskuee RK, Nassirli H, Malaekeh-Nikouei B. Progress in the development of lipopolyplexes as efficient non-viral gene delivery systems. J Control Release. 2016;236:1–14. doi: 10.1016/j.jconrel.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 112.Chapoy-Villanueva H, Martinez-Carlin I, Lopez-Berestein G, Chavez-Reyes A. Therapeutic silencing of HPV 16 E7 by systemic administration of siRNA-neutral DOPC nanoliposome in a murine cervical cancer model with obesity. J BUON. 2015;20(6):1471–1479. [PubMed] [Google Scholar]
- 113.Dunne PD, Dasgupta S, Blayney JK, McArt DG, Redmond KL, Weir JA, et al. EphA2 Expression Is a Key Driver of Migration and Invasion and a Poor Prognostic Marker in Colorectal Cancer. Clin Cancer Res. 2016;22(1):230–242. doi: 10.1158/1078-0432.CCR-15-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song W, Hwang Y, Youngblood VM, Cook RS, Balko JM, Chen J, et al. Targeting EphA2 impairs cell cycle progression and growth of basal-like/triple-negative breast cancers. Oncogene. 2017 doi: 10.1038/onc.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 116.Wagner MJ, Mitra R, McArthur MJ, Baze W, Barnhart K, Wu SY, et al. Preclinical Mammalian Safety Studies of EPHARNA (DOPC Nanoliposomal EphA2-Targeted siRNA) Mol Cancer Ther. 2017;16(6):1114–1123. doi: 10.1158/1535-7163.MCT-16-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan Y, Chen C, Huang Y, Zhang F, Lin G. Study of the pH-sensitive mechanism of tumor-targeting liposomes. Colloids Surf B Biointerfaces. 2017;151:19–25. doi: 10.1016/j.colsurfb.2016.11.042. [DOI] [PubMed] [Google Scholar]
- 118.Gujrati M, Vaidya AM, Mack M, Snyder D, Malamas A, Lu ZR. Targeted Dual pH-Sensitive Lipid ECO/siRNA Self-Assembly Nanoparticles Facilitate In Vivo Cytosolic sieIF4E Delivery and Overcome Paclitaxel Resistance in Breast Cancer Therapy. Adv Healthc Mater. 2016;5(22):2882–2895. doi: 10.1002/adhm.201600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iversen F, Yang C, Dagnaes-Hansen F, Schaffert DH, Kjems J, Gao S. Optimized siRNA-PEG conjugates for extended blood circulation and reduced urine excretion in mice. Theranostics. 2013;3(3):201–209. doi: 10.7150/thno.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nag OK, Awasthi V. Surface engineering of liposomes for stealth behavior. Pharmaceutics. 2013;5(4):542–569. doi: 10.3390/pharmaceutics5040542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chauhan H, Mohapatra S, Munt DJ, Chandratre S, Dash A. Physical-Chemical Characterization and Formulation Considerations for Solid Lipid Nanoparticles. AAPS PharmSciTech. 2016;17(3):640–651. doi: 10.1208/s12249-015-0394-x. [DOI] [PubMed] [Google Scholar]
- 122.Botto C, Mauro N, Amore E, Martorana E, Giammona G, Bondi ML. Surfactant effect on the physicochemical characteristics of cationic solid lipid nanoparticles. Int J Pharm. 2017;516(1–2):334–341. doi: 10.1016/j.ijpharm.2016.11.052. [DOI] [PubMed] [Google Scholar]
- 123.Lobovkina T, Jacobson GB, Gonzalez-Gonzalez E, Hickerson RP, Leake D, Kaspar RL, et al. In vivo sustained release of siRNA from solid lipid nanoparticles. ACS Nano. 2011;5(12):9977–9983. doi: 10.1021/nn203745n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu YH, Kim E, Park DE, Shim G, Lee S, Kim YB, et al. Cationic solid lipid nanoparticles for co-delivery of paclitaxel and siRNA. Eur J Pharm Biopharm. 2012;80(2):268–273. doi: 10.1016/j.ejpb.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 125.Jin J, Bae KH, Yang H, Lee SJ, Kim H, Kim Y, et al. In vivo specific delivery of c-Met siRNA to glioblastoma using cationic solid lipid nanoparticles. Bioconjug Chem. 2011;22(12):2568–2572. doi: 10.1021/bc200406n. [DOI] [PubMed] [Google Scholar]
- 126.Xue HY, Tran N, Wong HL. A biodistribution study of solid lipid-polyethyleneimine hybrid nanocarrier for cancer RNAi therapy. Eur J Pharm Biopharm. 2016;108:68–75. doi: 10.1016/j.ejpb.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Naseri N, Valizadeh H, Zakeri-Milani P. Solid Lipid Nanoparticles and Nanostructured Lipid Carriers: Structure, Preparation and Application. Adv Pharm Bull. 2015;5(3):305–313. doi: 10.15171/apb.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Han Y, Li Y, Zhang P, Sun J, Li X, Sun X, et al. Nanostructured lipid carriers as novel drug delivery system for lung cancer gene therapy. Pharm Dev Technol. 2016;21(3):277–281. doi: 10.3109/10837450.2014.996900. [DOI] [PubMed] [Google Scholar]
- 129.Shao Z, Shao J, Tan B, Guan S, Liu Z, Zhao Z, et al. Targeted lung cancer therapy: preparation and optimization of transferrin-decorated nanostructured lipid carriers as novel nanomedicine for co-delivery of anticancer drugs and DNA. Int J Nanomedicine. 2015;10:1223–1233. doi: 10.2147/IJN.S77837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taratula O, Kuzmov A, Shah M, Garbuzenko OB, Minko T. Nanostructured lipid carriers as multifunctional nanomedicine platform for pulmonary co-delivery of anticancer drugs and siRNA. J Control Release. 2013;171(3):349–357. doi: 10.1016/j.jconrel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xue HY, Wong HL. Tailoring nanostructured solid-lipid carriers for time-controlled intracellular siRNA kinetics to sustain RNAi-mediated chemosensitization. Biomaterials. 2011;32(10):2662–2672. doi: 10.1016/j.biomaterials.2010.12.029. [DOI] [PubMed] [Google Scholar]
- 132.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A. 2007;104(36):14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123(Pt 8):1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 134.Gomes-da-Silva LC, Fonseca NA, Moura V, Pedroso de Lima MC, Simoes S, Moreira JN. Lipid-based nanoparticles for siRNA delivery in cancer therapy: paradigms and challenges. Acc Chem Res. 2012;45(7):1163–1171. doi: 10.1021/ar300048p. [DOI] [PubMed] [Google Scholar]
- 135.Tros de Ilarduya C, Sun Y, Duzgunes N. Gene delivery by lipoplexes and polyplexes. Eur J Pharm Sci. 2010;40(3):159–170. doi: 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 136.Yang C, Gao S, Dagnaes-Hansen F, Jakobsen M, Kjems J. Impact of PEG Chain Length on the Physical Properties and Bioactivity of PEGylated Chitosan/siRNA Nanoparticles in Vitro and in Vivo. ACS Appl Mater Interfaces. 2017;9(14):12203–12216. doi: 10.1021/acsami.6b16556. [DOI] [PubMed] [Google Scholar]
- 137.Sun P, Huang W, Kang L, Jin M, Fan B, Jin H, et al. siRNA-loaded poly(histidine-arginine)6-modified chitosan nanoparticle with enhanced cell-penetrating and endosomal escape capacities for suppressing breast tumor metastasis. Int J Nanomedicine. 2017;12:3221–3234. doi: 10.2147/IJN.S129436. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 138.Gu J, Al-Bayati K, Ho EA. Development of antibody-modified chitosan nanoparticles for the targeted delivery of siRNA across the blood-brain barrier as a strategy for inhibiting HIV replication in astrocytes. Drug Deliv Transl Res. 2017 doi: 10.1007/s13346-017-0368-5. [DOI] [PubMed] [Google Scholar]
- 139.Gao LY, Liu XY, Chen CJ, Wang JC, Feng Q, Yu MZ, et al. Core-shell type lipid/rPAA-Chol polymer hybrid nanoparticles for in vivo siRNA delivery. Biomaterials. 2014;35(6):2066–2078. doi: 10.1016/j.biomaterials.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 140.Zhao X, Li F, Li Y, Wang H, Ren H, Chen J, et al. Co-delivery of HIF1alpha siRNA and gemcitabine via biocompatible lipid-polymer hybrid nanoparticles for effective treatment of pancreatic cancer. Biomaterials. 2015;46:13–25. doi: 10.1016/j.biomaterials.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 141.Yang XZ, Dou S, Wang YC, Long HY, Xiong MH, Mao CQ, et al. Single-step assembly of cationic lipid-polymer hybrid nanoparticles for systemic delivery of siRNA. ACS Nano. 2012;6(6):4955–4965. doi: 10.1021/nn300500u. [DOI] [PubMed] [Google Scholar]
- 142.Li Y, Huang X, Lee RJ, Qi Y, Wang K, Hao F, et al. Synthesis of Polymer-Lipid Nanoparticles by Microfluidic Focusing for siRNA Delivery. Molecules. 2016;21(10) doi: 10.3390/molecules21101314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ki MH, Kim JE, Lee YN, Noh SM, An SW, Cho HJ, et al. Chitosan-based hybrid nanocomplex for siRNA delivery and its application for cancer therapy. Pharm Res. 2014;31(12):3323–3334. doi: 10.1007/s11095-014-1422-3. [DOI] [PubMed] [Google Scholar]
- 144.Zhong J, Huang HL, Li J, Qian FC, Li LQ, Niu PP, et al. Development of hybrid-type modified chitosan derivative nanoparticles for the intracellular delivery of midkine-siRNA in hepatocellular carcinoma cells. Hepatobiliary Pancreat Dis Int. 2015;14(1):82–89. doi: 10.1016/s1499-3872(15)60336-8. [DOI] [PubMed] [Google Scholar]
- 145.Ju Z, Ma J, Wang C, Yu J, Qiao Y, Hei F. Exosomes from iPSCs Delivering siRNA Attenuate Intracellular Adhesion Molecule-1 Expression and Neutrophils Adhesion in Pulmonary Microvascular Endothelial Cells. Inflammation. 2017;40(2):486–496. doi: 10.1007/s10753-016-0494-0. [DOI] [PubMed] [Google Scholar]
- 146.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 147.Lee HM, Choi EJ, Kim JH, Kim TD, Kim YK, Kang C, et al. A membranous form of ICAM-1 on exosomes efficiently blocks leukocyte adhesion to activated endothelial cells. Biochem Biophys Res Commun. 2010;397(2):251–256. doi: 10.1016/j.bbrc.2010.05.094. [DOI] [PubMed] [Google Scholar]
- 148.Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32(6):2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yim N, Ryu SW, Choi K, Lee KR, Lee S, Choi H, et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat Commun. 2016;7:12277. doi: 10.1038/ncomms12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525–1541. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6(4):287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 153.Cooper JM, Wiklander PB, Nordin JZ, Al-Shawi R, Wood MJ, Vithlani M, et al. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov Disord. 2014;29(12):1476–1485. doi: 10.1002/mds.25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sterzenbach U, Putz U, Low LH, Silke J, Tan SS, Howitt J. Engineered Exosomes as Vehicles for Biologically Active Proteins. Mol Ther. 2017 doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017 doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ohno SI, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically Injected Exosomes Targeted to EGFR Deliver Antitumor MicroRNA to Breast Cancer Cells. Mol Ther. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lacko AG, Sabnis NA, Nagarajan B, McConathy WJ. HDL as a drug and nucleic acid delivery vehicle. Front Pharmacol. 2015;6:247. doi: 10.3389/fphar.2015.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mooberry LK, Sabnis NA, Panchoo M, Nagarajan B, Lacko AG. Targeting the SR-B1 Receptor as a Gateway for Cancer Therapy and Imaging. Front Pharmacol. 2016;7:466. doi: 10.3389/fphar.2016.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shahzad MM, Mangala LS, Han HD, Lu C, Bottsford-Miller J, Nishimura M, et al. Targeted delivery of small interfering RNA using reconstituted high-density lipoprotein nanoparticles. Neoplasia. 2011;13(4):309–319. doi: 10.1593/neo.101372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ding Y, Wang W, Feng M, Wang Y, Zhou J, Ding X, et al. A biomimetic nanovector-mediated targeted cholesterol-conjugated siRNA delivery for tumor gene therapy. Biomaterials. 2012;33(34):8893–8905. doi: 10.1016/j.biomaterials.2012.08.057. [DOI] [PubMed] [Google Scholar]
- 161.Pan B, Ren H, Lv X, Zhao Y, Yu B, He Y, et al. Hypochlorite-induced oxidative stress elevates the capability of HDL in promoting breast cancer metastasis. J Transl Med. 2012;10:65. doi: 10.1186/1479-5876-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tang J, Kuai R, Yuan W, Drake L, Moon JJ, Schwendeman A. Effect of size and pegylation of liposomes and peptide-based synthetic lipoproteins on tumor targeting. Nanomedicine. 2017 doi: 10.1016/j.nano.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lakhin AV, Tarantul VZ, Gening LV. Aptamers: problems, solutions and prospects. Acta Naturae. 2013;5(4):34–43. [PMC free article] [PubMed] [Google Scholar]
- 164.Li X, Zhao Q, Qiu L. Smart ligand: aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy. J Control Release. 2013;171(2):152–162. doi: 10.1016/j.jconrel.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 165.Bagalkot V, Gao X. siRNA-aptamer chimeras on nanoparticles: preserving targeting functionality for effective gene silencing. ACS Nano. 2011;5(10):8131–8139. doi: 10.1021/nn202772p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lai WY, Wang WY, Chang YC, Chang CJ, Yang PC, Peck K. Synergistic inhibition of lung cancer cell invasion, tumor growth and angiogenesis using aptamer-siRNA chimeras. Biomaterials. 2014;35(9):2905–2914. doi: 10.1016/j.biomaterials.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 167.Li L, Hou J, Liu X, Guo Y, Wu Y, Zhang L, et al. Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas. Biomaterials. 2014;35(12):3840–3850. doi: 10.1016/j.biomaterials.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 168.Binzel DW, Shu Y, Li H, Sun M, Zhang Q, Shu D, et al. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol Ther. 2016;24(7):1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee SK, Siefert A, Beloor J, Fahmy TM, Kumar P. Cell-specific siRNA delivery by peptides and antibodies. Methods Enzymol. 2012;502:91–122. doi: 10.1016/B978-0-12-416039-2.00005-7. [DOI] [PubMed] [Google Scholar]
- 170.Rengaswamy V, Zimmer D, Suss R, Rossler J. RGD liposome-protamine-siRNA (LPR) nanoparticles targeting PAX3-FOXO1 for alveolar rhabdomyosarcoma therapy. J Control Release. 2016;235:319–327. doi: 10.1016/j.jconrel.2016.05.063. [DOI] [PubMed] [Google Scholar]
- 171.Palanca-Wessels MC, Booth GC, Convertine AJ, Lundy BB, Berguig GY, Press MF, et al. Antibody targeting facilitates effective intratumoral siRNA nanoparticle delivery to HER2-overexpressing cancer cells. Oncotarget. 2016;7(8):9561–9575. doi: 10.18632/oncotarget.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Guo J, Russell EG, Darcy R, Cotter TG, McKenna SL, Cahill MR, et al. Antibody-Targeted Cyclodextrin-Based Nanoparticles for siRNA Delivery in the Treatment of Acute Myeloid Leukemia: Physicochemical Characteristics, in Vitro Mechanistic Studies, and ex Vivo Patient Derived Therapeutic Efficacy. Mol Pharm. 2017;14(3):940–952. doi: 10.1021/acs.molpharmaceut.6b01150. [DOI] [PubMed] [Google Scholar]
- 173.Jones SK, Lizzio V, Merkel OM. Folate Receptor Targeted Delivery of siRNA and Paclitaxel to Ovarian Cancer Cells via Folate Conjugated Triblock Copolymer to Overcome TLR4 Driven Chemotherapy Resistance. Biomacromolecules. 2016;17(1):76–87. doi: 10.1021/acs.biomac.5b01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 175.Wang J, Mi P, Lin G, Wang YX, Liu G, Chen X. Imaging-guided delivery of RNAi for anticancer treatment. Adv Drug Deliv Rev. 2016;104:44–60. doi: 10.1016/j.addr.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang F, Song X, Li X, Xin J, Wang S, Yang W, et al. Noninvasive visualization of microRNA-16 in the chemoresistance of gastric cancer using a dual reporter gene imaging system. PLoS One. 2013;8(4):e61792. doi: 10.1371/journal.pone.0061792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lin Q, Huang H, Chen J, Zheng G. Using Fluorescence Imaging to Track Drug Delivery and Guide Treatment Planning In Vivo. Methods Mol Biol. 2016;1444:153–166. doi: 10.1007/978-1-4939-3721-9_14. [DOI] [PubMed] [Google Scholar]
- 178.Sita TL, Kouri FM, Hurley LA, Merkel TJ, Chalastanis A, May JL, et al. Dual bioluminescence and near-infrared fluorescence monitoring to evaluate spherical nucleic acid nanoconjugate activity in vivo. Proc Natl Acad Sci U S A. 2017;114(16):4129–4134. doi: 10.1073/pnas.1702736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chen Y, Wang X, Liu T, Zhang DS, Wang Y, Gu H, et al. Highly effective antiangiogenesis via magnetic mesoporous silica-based siRNA vehicle targeting the VEGF gene for orthotopic ovarian cancer therapy. Int J Nanomedicine. 2015;10:2579–2594. doi: 10.2147/IJN.S78774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Veiseh O, Kievit FM, Fang C, Mu N, Jana S, Leung MC, et al. Chlorotoxin bound magnetic nanovector tailored for cancer cell targeting, imaging, and siRNA delivery. Biomaterials. 2010;31(31):8032–8042. doi: 10.1016/j.biomaterials.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang P, Moore A. In Vivo Magnetic Resonance Imaging of Small Interfering RNA Nanodelivery to Pancreatic Islets. Methods Mol Biol. 2016;1372:25–36. doi: 10.1007/978-1-4939-3148-4_2. [DOI] [PubMed] [Google Scholar]
- 182.Gottesman MM, Ling V. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 2006;580(4):998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 183.Sims JT, Ganguly SS, Bennett H, Friend JW, Tepe J, Plattner R. Imatinib reverses doxorubicin resistance by affecting activation of STAT3-dependent NF-kappaB and HSP27/p38/AKT pathways and by inhibiting ABCB1. PLoS One. 2013;8(1):e55509. doi: 10.1371/journal.pone.0055509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature. 2007;446(7137):749–757. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 185.Takara K, Sakaeda T, Okumura K. An update on overcoming MDR1-mediated multidrug resistance in cancer chemotherapy. Curr Pharm Des. 2006;12(3):273–286. doi: 10.2174/138161206775201965. [DOI] [PubMed] [Google Scholar]
- 186.Palmeira A, Vasconcelos MH, Paiva A, Fernandes MX, Pinto M, Sousa E. Dual inhibitors of P-glycoprotein and tumor cell growth: (re)discovering thioxanthones. Biochem Pharmacol. 2012;83(1):57–68. doi: 10.1016/j.bcp.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 187.Xu X, Ho W, Zhang X, Bertrand N, Farokhzad O. Cancer nanomedicine: from targeted delivery to combination therapy. Trends Mol Med. 2015;21(4):223–232. doi: 10.1016/j.molmed.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nastiuk KL, Krolewski JJ. Opportunities and challenges in combination gene cancer therapy. Adv Drug Deliv Rev. 2016;98:35–40. doi: 10.1016/j.addr.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zheng W, Yin T, Chen Q, Qin X, Huang X, Zhao S, et al. Co-delivery of Se nanoparticles and pooled SiRNAs for overcoming drug resistance mediated by P-glycoprotein and class III beta-tubulin in drug-resistant breast cancers. Acta Biomater. 2016;31:197–210. doi: 10.1016/j.actbio.2015.11.041. [DOI] [PubMed] [Google Scholar]
- 190.Mitchell MJ, Chen CS, Ponmudi V, Hughes AD, King MR. E-selectin liposomal and nanotube-targeted delivery of doxorubicin to circulating tumor cells. J Control Release. 2012;160(3):609–617. doi: 10.1016/j.jconrel.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pradhan P, Giri J, Rieken F, Koch C, Mykhaylyk O, Doblinger M, et al. Targeted temperature sensitive magnetic liposomes for thermo-chemotherapy. J Control Release. 2010;142(1):108–121. doi: 10.1016/j.jconrel.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 192.Li Y, Wang H, Wang K, Hu Q, Yao Q, Shen Y, et al. Targeted Co-delivery of PTX and TR3 siRNA by PTP Peptide Modified Dendrimer for the Treatment of Pancreatic Cancer. Small. 2017;13(2) doi: 10.1002/smll.201602697. [DOI] [PubMed] [Google Scholar]
- 193.Khanna A, Mahalingam K, Chakrabarti D, Periyasamy G. Ets-1 expression and gemcitabine chemoresistance in pancreatic cancer cells. Cell Mol Biol Lett. 2011;16(1):101–113. doi: 10.2478/s11658-010-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kars MD, Iseri OD, Gunduz U. Drug resistant breast cancer cells overexpress ETS1 gene. Biomed Pharmacother. 2010;64(7):458–462. doi: 10.1016/j.biopha.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 195.Wu M, Liu X, Jin W, Li Y, Li Y, Hu Q, et al. Targeting ETS1 with RNAi-based supramolecular nanoassemblies for multidrug-resistant breast cancer therapy. J Control Release. 2017;253:110–121. doi: 10.1016/j.jconrel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 196.Gottesman MM, Pastan IH. The Role of Multidrug Resistance Efflux Pumps in Cancer: Revisiting a JNCI Publication Exploring Expression of the MDR1 (P-glycoprotein) Gene. J Natl Cancer Inst. 2015;107(9) doi: 10.1093/jnci/djv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92(16):1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 198.Jang M, Han HD, Ahn HJ. A RNA nanotechnology platform for a simultaneous two-in-one siRNA delivery and its application in synergistic RNAi therapy. Sci Rep. 2016;6:32363. doi: 10.1038/srep32363. [DOI] [PMC free article] [PubMed] [Google Scholar]


