Abstract
In recent years, immunotherapy has revolutionized and changed the standard of care in patients with advanced non-small cell lung cancer (NSCLC). Immune checkpoint inhibitors, fundamentally those that act by blocking the programmed cell death receptor-1 (PD-1) and its ligand the programmed cell death ligand-1 (PD-L1) have emerged as novel treatment strategies in NSCLC, demonstrating undoubted superiority over chemotherapy in terms of efficacy. Several of these immune checkpoint modulators have recently gained regulatory approval for the treatment of advanced NSCLC, such as nivolumab, atezolizumab and pembrolizumab in first-line (only the latter) and second-line settings, and more recently, durvalumab as maintenance after chemoradiotherapy in locally advanced disease. There is consensus that PD-L1 expression on tumor cells predicts responsiveness to PD-1 inhibitors in several tumor types. Hence PD-L1 expression evaluated by immunohistochemistry (IHC) is currently used as a clinical decision-making tool to support the use of checkpoint inhibitors in NSCLC patients. However, the value of PD-L1 as the ‘definitive’ biomarker is controversial as its testing is puzzled by multiple unsolved issues such as the use of different staining platforms and antibodies, the type of cells in which PD-L1 is assessed (tumor versus immune cells), thresholds used for PD-L1-positivity, or the source and timing for sample collection. Therefore, newer biomarkers such as tumor mutation burden and neoantigens as well as biomarkers reflecting host environment (microbiome) or tumor inflamed microenvironment (gene expression signatures) are being explored as more reliable and accurate alternatives to IHC for guiding treatment selection with checkpoint inhibitors in NSCLC.
Keywords: immunotherapy, immunohistochemistry (IHC), non-small cell lung cancer (NSCLC), PD-L1 expression testing, predictive biomarker
Introduction
The immune system plays an important dual role in cancer by a dynamic process called immunoediting.1,2 Most of the time, the innate and adaptive immune responses constrain tumor growth and destroy cancer cells in the so called ‘elimination’ phase or immunosurveillance. However, tumors can enter into an ‘escape’ phase through several mechanisms that confer a characteristic local immune suppression state by recruiting immunosuppressive cells, producing immunosuppressive cytokines, developing defects in tumor antigen presentation to T-cells or by expressing negative co-stimulatory molecules also called T-cell checkpoint regulators, such as the cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4), programmed cell death-1 (PD-1) and programmed cell death ligand-1 (PD-L1).1 By doing so, tumors can disrupt the normal immunity in favor of their progression. Loss of heterozygosity in human leukocyte antigen, present in almost 40% of early-stage non-small cell lung cancer (NSCLC), is a hallmark of immune escape in NSCLC evolution allowing for high subclonal neoantigen burden, apolipoprotein B mRNA editing catalytic polypeptide-like cytidine deaminase (APOBEC)-mediated mutagenesis, upregulation of cytolytic activity and expression of PD-L1.3
One of the most developed therapeutic strategies to overcome the immune evasion of tumors is the reactivation of T-cell mediated antitumor activity by modulating the immune checkpoint ligand–receptor interactions that regulate T-cell signaling. In a normal state, these immune checkpoint receptors serve to limit and extent the duration of a response on T-cell activation and prevent damage to normal tissue, providing a natural counterbalance to immune activation.4 Indeed, pharmacological design of antibodies directed against CTLA-4, PD-1 and PD-L1 pathways has already demonstrated efficacy in several trials among different tumor types5,6 and expression of PD-L1 in tumor tissue is currently harnessed to select patients for PD-(L)1 blockade therapies.7,8
Herein, we aim to review the main questions concerning immunohistochemical evaluation of PD-L1 as a biomarker in NSCLC, overview the most practical issues regarding its use as a predictive diagnostic assay in the clinic and to go over other novel predictive biomarkers of response to immune checkpoint inhibitors beyond PD-L1.
PD-L1 as predictive biomarker in NSCLC
PD-L1 is a type 1 transmembrane protein (B7-H1) that belongs to the B7 ligands family and may be expressed both on hematopoietic cells (dendritic cells, macrophages, mast cells, T-cells and B-lymphocytes) and nonhematopoietic cells, including endothelial, epithelial and tumor cells.9,10 Expression of PD-L1 on tumor cells promotes down-regulation and self tolerance of the immune system from rejecting the tumor by suppressing T-cell inflammatory activity through binding to the regulatory T-cell receptor, PD-1.11 Among ligands belonging to the B7 family, PD-L1 is the principal membrane inhibitory ligand and the most studied so far in NSCLC.12
Expression of PD-L1 by immunohistochemistry (IHC) staining has been the core strategy to select NSCLC patients for PD-(L)1 inhibitors. It is currently known that PD-L1 expression by IHC can enrich for PD-(L)1 blockade efficacy and overexpression has been associated with higher response rates and better outcomes to several checkpoint inhibitors13,14 (Table 1).
Table 1.
Outcomes of PD-1/PD-L1 axis inhibitors in NSCLC phase III clinical trials based on PD-L1 testing.
| Immune checkpoint inhibitor | PD-L1 analysisa |
Stratification by PD-L1 | PD-L1 cutoffs for outcome | Outcome |
||
|---|---|---|---|---|---|---|
| RR | PFS | OS | ||||
| First line (compared with standard first-line chemotherapy) | ||||||
|
Nivolumab
(Checkmate-026)15 |
28–8 mAb retrospective |
No | ⩾5% ⩾50% |
26% versus 33% 34% versus 39% |
4.2 m versus 5.9 m (HR 1.15) 5.4 m versus 5.8 m (HR 1.07) |
14.4 m versus 13.2 m (HR 1.02) 15.9 m versus 13.9 m (HR 0.90) |
|
Pembrolizumab
(KEYNOTE-024)16 |
22C3 mAb prospective |
No | ⩾50% | 44.8% versus 27.8% | 10.3 m versus 6 m (HR 0.50) | Not reported (HR 0.60) |
| Second line (compared with docetaxel) | ||||||
|
Nivolumab
(Checkmate-017)14 |
28–8 mAb retrospective |
No | Negative and positive | 20% versus 9% | 3.5 m versus 2.8 m (HR 0.62) | 9.2 m versus 6 m (HR 0.59) |
| <1% | 17% versus 10% | 3.1 m versus 3 m (HR 0.66) | 8.7 m versus 5.9 m (HR 0.58) | |||
| ⩾1% | 17% versus 11% | 3.3 m versus 2.8 m (HR 0.67) | 9.3 m versus 7.2 m (HR 0.69) | |||
| <5% | 15% versus 12% | 2.2 m versus 2.9 m (HR 0.75) | 8.5 m versus 6.1 m (HR 0.70) | |||
| ⩾5% | 21% versus 8% | 4.8 m versus 3.1 m (HR 0.54) | 10 m versus 6.4 m (HR 0.53) | |||
| <10% | 16% versus 11% | 2.3 m versus 2.8 m (HR 0.70) | 8.2 m versus 6.1 m (HR 0.70) | |||
| ⩾10% | 19% versus 9% | 3.7 m versus 3.3 m (HR 0.58) | 11 m versus 7.1 m (HR 0.50) | |||
|
Nivolumab
(Checkmate-057)17 |
28-8 mAb retrospective |
No | Negative and positive | 19% versus 12% | 2.3 m versus 4.2 m (HR 0.92) | 12.2 m versus 9.4 m (HR 0.73) |
| <1% | 9% versus 15% | 2.1 m versus 3.6 m (HR 1.19) | 10.5 m versus 10.1 m (HR 0.87) | |||
| ⩾1% | 31% versus 12% | 4.2 m versus 4.5 m (HR 0.70) | 17.7 m versus 9 m (HR 0.58) | |||
| <5% | 10% versus 14% | 2.1 m versus 4.2 m (HR 1.31) | 9.8 m versus 10.1 m (HR 0.96) | |||
| ⩾5% | 36% versus 13% | 5 m versus 3.8 m (HR 0.54) | 19.4 m versus 8.1 m (HR 0.43) | |||
| <10% | 11% versus 14% | 2.1 m versus 4.2 m (HR 1.24) | 9.9 m versus 10.3 m (HR 0.96) | |||
| ⩾10% | 37% versus 13% | 5 m versus 3.7 m (HR 0.52) | 19.9 m versus 8 m (HR 0.40) | |||
|
Pembrolizumab
(KEYNOTE-010b)18 |
22C3 mAb prospective |
Yes | ⩾1% | 18% versus 9% | 3.9 m versus 4 m (HR 0.88) | 10.4 m versus 8.5 m (HR 0.71) |
| ⩾50% | 30% versus 8% | 5 m versus 4.1 m (HR 0.59) | 14.9 m versus 8.2 m (HR 0.54) | |||
|
Atezolizumab
(OAK)19 |
SP142 mAb prospective |
Yes | Negative and positive | 14% versus 13% | 2.8 m versus 4 m (HR 0.95) | 13.8 m versus 9.6 m (HR 0.73) |
| TC1/2/3 or IC1/2/3 | 18% versus 16% | 2.8 m versus 4.1 m (HR 0.91) | 15.7 m versus 10.3 (HR 0.74) | |||
| TC2/3 or IC2/3 | 22.5% versus 12.5% | 4.1 m versus 3.6 m (HR 0.76) | 16.3 m versus 10.8 m (HR 0.67) | |||
| TC3 or IC3 | 31% versus 11% | 4.2 m versus 3.3 m (HR 0.63) | 20.5 m versus 8.9 m (HR 0.41) | |||
| TC0 and IC0 | 8% versus 11% | 2.6 m versus 4 m (HR 1) | 12.6 m versus 8.9 m (HR 0.75) | |||
| Stage III NSCLC (compared with placebo) | ||||||
|
Durvalumab
(Pacific)20 |
SP263 mAb prospective |
No | Negative and positive | 28.4% versus 16% | 16.8 m versus 5.6 m (HR 0.52) | Not reported |
In all these studies PD-L1 analysis were made in fresh or archival FFPE biopsies.
In the KEYNOTE-010 study we only reported data of the cohort treated at the approved dose of 2 mg/kg of pembrolizumab.
HR, hazard ratio; IC, immune cell; m, months; mAb, monoclonal antibody; NSCLC, non-small cell lung cancer; OS, overall survival; PD-1, programmed cell death receptor-1; PD-L1, programmed cell death ligand-1; PFS, progression-free survival; RR, objective response rate; TC, tumor cell; TC0 or IC0, PD-L1 expression on less than 1% of tumor cells or tumor-infiltrating immune cells, respectively; TC 1/2/3 or IC 1/2/3, PD-L1 expression on 1% or more of tumor cells or tumor-infiltrating immune cells, respectively; TC 2/3 or IC 2/3, PD-L1 expression on 5% or more of tumor cells or tumor-infiltrating immune cells, respectively; IC3, PD-L1 expression on 10% or more of tumor-infiltrating immune cells; TC3, PD-L1 expression on 50% or more of tumor cells.
In advanced NSCLC, the percentage of PD-L1 positive patients [Tumor Proportion Score (TPS) ⩾1%] is found in around half of samples evaluated for PD-L1. However, differences in PD-L1 expression have been observed in the context of specific phase III clinical trials [53% (119/225) in CheckMate-017 trial and 54% (246/455) CheckMate-057 trial with nivolumab; 66% (1475/2222) in KEYNOTE-010 trial with pembrolizumab and 54% (463/850) in atezolizumab OAK trial]. Approximately a third of patients have high PD-L1 expression levels (TPS ⩾50%), but likewise, differences across studies have been detected [40% (214/541) in CheckMate-026 trial, 30.2% (500/1653) in KEYNOTE 024 trial and 16% (137/850) in OAK trial respectively]. Changes in the percentage of positive and high PD-L1 patients detected in these trials could be explained by the different antibody clones used and the variability across patient subpopulations.14–19
PD-L1 diagnostics tests
Evaluation of PD-L1 expression by IHC has overcome a cumbersome process for pathologists and oncologists, not because of the complexity of the technique itself, but for the singularity of the co-development of an assay together with a specific checkpoint inhibitor. Each of these assays uses its own PD-L1 antibody, platform, custom reagents and scoring criteria to calculate TPS on tumor cells. Some tests have been labeled as ‘companion’ diagnostic as they are a prerequisite for receiving a drug prescription, whereas others are only ‘complementary’ this is to say not required but of aid for the use of the associated drug21 (Table 2).
Table 2.
Summary of PD-L1 monoclonal antibodies and technical aspects for evaluation and agencies’ approvals in NSCLC.
| PD-L1 mAb clone | Ab host species | Automated platform | Checkpoint inhibitor (target) | PD-L1 scoring | Definition of positivity (cutoffs) | FDA status | EMA status | Indication | Cutoffs for indications |
|---|---|---|---|---|---|---|---|---|---|
| 22C3 | Mouse | Dako (Autostainer Link 48) | Pembrolizumab (PD-1) |
TC | TC ⩾1% (minimum of 100 TC) | Companion | CE mark | Second and first-line NSCLC | ⩾1% second line ⩾50% first line |
| 28-8 | Rabbit | Dako (Autostainer Link 48) | Nivolumab (PD-1) |
TC | TC ⩾1% (minimum of 100 TC) |
Complementary | CE mark | Second-line NSCLC | All comers |
| SP142 | Rabbit | Ventana (BenchMark ULTRA) | Atezolizumab (PD-L1) |
TC, IC | TC ⩾50% or IC ⩾10% (minimum of 50 TC with associated stroma) |
Complementary | CE mark | Second-line NSCLC | All comers |
| SP263 | Rabbit | Ventana (BenchMark ULTRA) | Durvalumab (PD-L1) |
TC | TC ⩾25% (minimum of 100 TC) |
FDA approval only for urothelial carcinoma | CE mark for nivolumab and pembrolizumab in NSCLC and durvalumab in urothelial carcinoma | Locally advanced NSCLC | All comers |
| 73-10 | Rabbit | Dako | Avelumab (PD-L1) |
TC | TC ⩾1% (minimum cells not defined) |
FDA approval | NA | NA | NA |
Ab, antibody; CE, European Conformity (CE)-marked; Companion, provides information that is essential for the effective use of a corresponding drug or biological product, within approved label; Complementary, provides additional information about how a drug might be used, but not required; EMA, European Medicines Agency; FDA, United States Food and Drug Administration; IC, immune cell; mAb, monoclonal antibody; NA, not available; NSCLC, non-small cell lung cancer; PD-1, programmed cell death receptor-1; PD-L1, programmed cell death ligand-1; TC, tumor cell.
Currently five clones are being used for PD-L1 IHC testing: 22C3, 28-8, SP142, SP263 and 73-10. In the majority, staining is evaluated on the cell membrane as it is presumed that the PD-(L)1 axis is only functional when it ligates a counter-receptor.22 Herein we summarize the main characteristics of the available commercial assays specific for PD-L1 testing in lung cancer.
PD-L1 IHC 22C3 pharmDx assay
The Dako 22C3 pharmDx (Agilent Technologies/Dako, Carpinteria, CA, USA) was one of the first PD-L1 IHC kit assays to obtain regulatory approval and so far it is the only one that has gained regulatory status of ‘companion’ diagnostic by the United States Food and Drug Administration (US FDA) for the treatment with pembrolizumab both in previously treated and untreated patients with advanced NSCLC.23 A minimum number of 100 tumor cells are needed to consider a sample valid for its evaluation in formalin-fixed paraffin-embedded (FFPE) tissue using the monoclonal mouse [immunoglobulin (Ig)G1] clone 22C3 and PD-L1 positivity criteria is defined when a membrane staining (partial or complete) of tumor cells ⩾1% is observed.
PD-L1 IHC 28-8 pharmDx assay
The Dako 28-8 pharmDx (Agilent Technologies/Dako), is a qualitative PD-L1 immunohistochemical assay that gained US FDA approval as a ‘complementary’ diagnostic test as well as the European Conformity (CE)-mark certification for the use of nivolumab in the second line of advanced NSCLC.24 Clone PD-L1 28-8 (ab205921; Abcam, Cambridge, UK) is a monoclonal rabbit anti-human antibody, which binds to the extracellular domain of human PD-L1.25 As stated previously, a minimum of 100 tumor cells are needed to address PD-L1 in FFPE tissue samples and positivity is defined at TPS equal or greater than 1%.
PD-L1 IHC SP142 assay
The Ventana SP142 assay (Ventana Medical Systems Inc., Tucson, AZ, USA) is a qualitative IHC assay that uses a rabbit monoclonal anti-PD-L1 clone which binds the intracellular domain of the protein. This test is CE-marked and has been approved by the US FDA as ‘complementary’ diagnostic tool for atezolizumab treatment in patients with metastatic NSCLC whose disease progressed during or following platinum-containing chemotherapy, as well as in patients with advanced urothelial carcinoma.26 Evaluation in FFPE samples requires at least 50 viable tumor cells. Tumor-associated stroma is not required for tumor cells scoring but it is essential for scoring tumor-infiltrating immune cells (ICs). Unlike the other assays, the scoring algorithm is based on either the percentage of PD-L1 expressing tumor cells or IC of any intensity. PD-L1 expression in ⩾50% tumor cells or ⩾10% ICs may be associated with enhanced overall survival (OS) from atezolizumab.27
PD-L1 IHC SP263 assay
PD-L1 clone SP263 is a rabbit monoclonal primary antibody that binds to a transmembrane glycoprotein corresponding to amino acids 284-290 of PD-L1 protein.28 The Ventana SP263 assay (Ventana Medical Systems Inc.) is intended to assess PD-L1 expression in FFPE tissue and gained the CE-In Vitro Device certification29 and US FDA ‘complementary’ test designation30 for the identification of patients with locally advanced or metastatic urothelial carcinoma most likely to benefit from durvalumab. For durvalumab treatment, PD-L1 cell positivity is considered when plasma membrane protein staining at any intensity is observed in at least 25% of tumor cells. A minimum of 100 tumor cells are required to determine the TPS. Recently this test has gained CE-mark, not US FDA, label expansion to inform treatment decisions in NSCLC patients being considered for pembrolizumab and nivolumab based on the results of a comparison study with other currently available PD-L1 assays (22C3 and 28-8).31
PD-L1 IHC 73-10 assay
The Dako 73-10 assay, the fifth PD-L1 US FDA-approved diagnostic test, was co-developed and commercialized to support the use of avelumab therapy. Clone PD-L1 73-10 is a monoclonal rabbit antibody property of Merck (KGaA, Darmstadt, Germany) optimized to detect PD-L1 expression in FFPE samples. Although a cutoff has not been definitely determined, the pre-defined PD-L1 positivity is considered when a complete circumferential or partial linear plasma membrane staining is observed at any intensity in at least 1% of tumor cells. Since this assay is still in development, the minimum of viable tumor cells required to determine the TPS is still undefined.32
Preanalytical considerations and tissue preparation for PD-L1 testing
General recommendations for IHC, including PD-L1 antibodies, are to perform the technique on FFPE freshly cut tissue sections at a thickness of 3–5 μM and mounted on positively charged slides. For each PD-L1 staining section it is advised to process a positive control to ensure the reliability of the PD-L1 expression since variations have been observed as a function of the preanalytical process (such as time to fixation, fixation time and sample processing),32 although there are not much data currently available on the reproducibility of PD-L1 IHC assays based on these preanalytical variable factors. A sample over-fixation can be the reason for an inadequate antibody penetration impairing final reading. Therefore, time to sample fixation must be reduced to the minimum possible (optimally under 30 min) followed by a 10% neutral formalin buffering for at least 6–48 h in biopsies and 24–48 h in resection samples.33 Long-term storage of cut tissue sections and of tissue blocks should be also avoided to ensure the quality of the PD-L1 expression.34 Different studies have reported that PD-L1 expression fades with the age of the specimens used for analyses, particularly in tissue blocks older than three years35 and even older than one year.36
Comparison and standardization of different PD-L1 IHC platforms
As outlined before, several reproducible PD-L1 assays have been developed for each of the immune-inhibitors (Table 2). This is leading to the paradoxical situation where the pathologists must select between different antibodies and assay conditions according to the prescription of a drug, rather than focusing on the accuracy of the technique itself. In the effort to overcome this limitation, and reach harmonization between the assays, several initiatives are ongoing to validate their reproducibility and improve standardization for IHC scoring (Table 3).
Table 3.
Comparison studies among different PD-L1 assays.
| Study | Samples | Monoclonal Ab | Platforms | Readers | Results |
|---|---|---|---|---|---|
| IASLC BLUEPRINT phase I study
Hirsch et al.37 |
39 | 22C3, 28-8, SP142, SP263 | Dako, Ventana | 3 | 3 (22C3, 28-8 and SP263) of 4 assays similar for TC staining. SP142 lower score. IC scored poor |
| NCCN study
Rimm et al.38 |
90 | 22C3, 28-8, SP142, E1L3N | Dako, Ventana, Leica | 13 | 3 (22C3, 28-8 and E1L3N) of 4 assays appear to be interchangeable from analytical perspective. SP142 lower score |
| German harmonization study
Scheel et al.39 |
15 | 22C3, 28-8, SP142, SP263 | Dako, Ventana | 9 | Scoring on TC reproducible for all assays. Variability in staining patterns |
| Japan harmonization study
Fujimoto et al.40 |
40 | 22C3, 28-8, SP142, SP263 | Dako, Ventana | 4 | 3 (22C3, 28-8 and SP263) of 4 assays show equivalent predictive performance. SP142 lower score |
| Australian harmonization study
Hendry et al.41 |
368 | 22C3, 28-8, SP142, SP263 | Dako, Ventana | 1 | 2 (22C3 and 28-8) of 4 assays are comparable enough to use interchangeably. Agreement between IC scores were poor |
| Italian harmonization study
Marchetti et al.42 |
100 | 22C3, SP263 | Dako, Ventana | 4 centers | 22C3 pharmDx and the Ventana SP263 assays could be used interchangeably. Excellent agreement at a cutoff of ⩾50% |
| AZ500 study
Ratcliffe et al.31 |
500 | 22C3, 28-8, SP263 | Dako, Ventana | 2 | High analytical correlation among the 3 commercially available assays and multiple cutoffs |
| Swedish harmonization study
Brunnström et al.43 |
55 | 28-8 from two different vendors, 22C3, SP142, SP263 |
Dako, Ventana | 7 | Good concordance between 22C3 (from two different vendors), 28-8 and SP263. SP142 presented the highest deviation from the reference scores. Better agreement at a cutoff of ⩾50% |
| French harmonization study
Adam et al.44 |
41 | 22C3, 28-8, SP142, SP263, E1L3N | Dako, Ventana, Leica | 7 centers | 22C3, 28-8 and SP263 assays gave highly concordant TC results for ⩾50% threshold |
| 22C3 PD-L1 harmonization study
Røge et al.45 |
77 | 22C3 | Dako, Ventana, Leica | 3 | LDT protocols provide an almost identical result to the 22C3 pharmDx kit |
| 22C3 PD-L1 harmonization study
Ilie et al.46 |
120 | 22C3 | Dako, Ventana, Leica | 3 | LDT protocols on Dako and Ventana platforms showed an almost 100% concordance |
| 22C3 PD-L1 harmonization study on Ventana’s platform
Neuman et al.47 |
41 | 22C3 | Dako, Ventana | 2 | Ventana’s 22C3 protocols obtained similar results than those using the Dako 22C3 staining platform |
| IASLC BLUEPRINT phase II
Tsao et al.48 |
81 (different sample and histological types) | 22C3, 28-8, SP142, SP263, 73-10 | Dako, Ventana | 24 | 22C3, 28-8 and SP263 are comparable, SP142 detects less, while 73-10 stains more PD-L1 positive TCs Scoring of TC PD-L1 expression by pathologists on tissue samples shows strong reliability whereas scoring of PD-L1 on ICs and cytology samples may have lower reliability |
| Cytologic and histologic PD-L1 comparison study
Skov et al.49 |
86 (paired samples from histological and cytological materials) | 22C3, 28-8 | Dako | 1 | PD-L1 staining can be performed on cytological samples. A good correlation was found between 22C3 and 28-8 assays, whether applied to histological or cytological cell blocks |
Ab: antibody; IC: immune cell; LDT: laboratory developed test; NCCN: National Comprehensive Cancer Network; PD-L1, programmed cell death ligand-1; TC: tumor cell.
The first of its kind was the Blueprint PD-L1 IHC assay project, aimed to compare the analytical performance of four PD-L1 validated assays (22C3, 28-8, SP142, SP263) in 39 NSCLC samples assessed by three independent readers.37 A total of three (22C3, 28-8 and SP263) out of four assays were analytically similar for TPS but inter-observer concordance for IC staining was poor. The NCCN study by Rimm and colleagues38 compared the performance of four different antibodies (22C3, 28-8, SP142, E1L3N) in 90 resected NSCLC. The study showed that three of the four assays (22C3, 28-8 and E1L3N) evaluated by 13 independent readers were interchangeable, whereas the SP142 assay was associated with a lower score.38 Later on, three other prospective studies by Scheel and colleagues39 Fujimoto and colleagues40 and Hendry and colleagues41 tested in 15, 40 and 368 specimens respectively, the reliability of measuring PD-L1 protein expression by comparing four different clones (22C3, 28-8, SP142, SP263). In the German harmonization study by Scheel and colleagues39 although scoring of tumor cells was reproducible and no differences inter-observers were noticed for all assays; staining patterns observed were not similar in all situations. On the other hand, the scoring of ICs yielded low concordance levels. Fujimoto and colleagues40 observed only an equivalent performance between 22C3, 28-8 and SP263 assays, whereas the work by Hendry and colleagues41 concluded that apart from 22C3 and 28-8, the SP142 and SP263 assays cannot be used interchangeably in clinical practice. As an additional analysis, the IC scoring was also assessed and a poor concordance was observed.
The Italian harmonization study reported an excellent agreement concordance of 0.99 (95% confidence interval: 0.96–1) between 22C3 pharmDx and SP263 assay at a cutoff of ⩾50%.42 In a larger comparative study (n = 500 FFPE archival NSCLC specimens), Ratcliffe and colleagues compared three antibody clones (22C3, 28-8, SP263) and found high analytical concordance (percentage agreement >90%) among the three commercially available assays at multiple expression cutoff and inter-observer, expanding indications of the Ventana SP263 assay to identify patients eligible for treatment with pembrolizumab or nivolumab.31 The Swedish harmonization study by Brunnström and colleagues compared four PD-L1 antibody clones (22C3 from two different vendors, 28-8, SP142, and SP263) and investigated interrater variation among pathologists, concluding that inter-pathologist variability is higher than assay variability. A better agreement was obtained between 22C3, 28-8 and SP263 antibodies and among pathologists at a cutoff of TPS ⩾50%.43
In order to determine whether laboratory developed tests (LDTs) with different automated staining platforms could achieve an analytical performance close to the validated ones, the multicentric French harmonization study evaluated PD-L1 IHC status on 41 NSCLC samples using different PD-L1 clones (22C3, 28-8, SP142, SP263 and E1L3N) performed either with Ventana BenchMark Ultra, Bond (Leica Biosystems) or Autostainer Link 48 (Dako) and found high concordance for three of the clones (22C3, 28-8 and SP263) in tumor cells above ⩾50% threshold and IC staining.44 Røge and colleagues45 successfully developed and validated LDT protocols (Ventana, Leica Biosystems, Dako) with clone 22C3 in 77 specimens of NSCLC providing an almost identical result to that of pharmDx Assay. Ilie and colleagues,46 by using the same clone and platforms showed almost 100% concordance for LDT protocols developed on Ventana and Dako platforms and a very high inter-pathologist concordance, whereas the harmonization study on Ventanas’ platform using 22C3 by Neuman and colleagues47 found similar results than using Dako platforms (87.8% and 85.3% concordance with the Ventana Ultraview kit and Ventana Optiview kit respectively) as well as a high inter-observer and intra-observer agreement.
Recently, preliminary results of the Blueprint phase II project have been reported.48 The study aims to validate assay comparability results observed in the Blueprint phase I but in a larger cohort of 81 specimens, using a larger panel of readers and to evaluate heterogeneity of PD-L1 scores in ‘real-life’ lung cancer specimens such as core needle biopsies or fine needle aspiration cytologies prepared from the same resected tumor, comparing five PD-L1 IHC assays (22C3, 28-8, SP142, SP263 and 73-10). The results obtained so far demonstrate high reliability to PD-L1 TPS between digital pathology versus glass slides, a poor concordance of IC staining between assays and a comparable equivalence among all pathologists.
Practical aspects of PD-L1 expression testing in the clinic
Current guidelines for advanced NSCLC50,51 have recently incorporated immune checkpoint therapies in their treatment algorithms and therefore, PD-L1 biomarker testing is today a requirement in the initial molecular workup of NSCLC.
Selection of anti-PD-L1 clones and thresholds for treatment indications
Nivolumab and pembrolizumab were the first antibodies targeting PD-1 that gained US FDA and European Medicines Agency (EMA) approval for the treatment of advanced NSCLC after demonstration of statistically significant improvement in OS as compared with standard chemotherapy with docetaxel.14,17,18 Later on, in October 2016 and September 2017, atezolizumab was approved by the US FDA and EMA respectively for the same indication, being the first anti-PD-L1 therapy approved for the treatment of NSCLC.19 In a first-line setting, pembrolizumab has been approved by both agencies, the US FDA and EMA, based on a significant improvement in progression-free survival (PFS) and OS compared with platinum-based chemotherapy,16 and more recently, maintenance treatment for one year with durvalumab, an anti-PD-L1 checkpoint inhibitor, has granted breakthrough therapy designation by the US FDA to treat locally advanced unresectable NSCLC patients after chemoradiotherapy.20 Specific thresholds requisitions for PD-L1 expression have been stablished for each drug prescription in which they were co-developed (Table 2). While a minimum of 1% PD-L1 expression is required for pembrolizumab treatment in second-line treatment,18 no restriction based on any threshold is required for nivolumab or atezolizumab.14,17,19 In the first line, pembrolizumab is only indicated in those tumors with strong PD-L1 expression higher or equal to 50%16 whereas no selection based on PD-L1 expression on tumor cells is required for durvalumab treatment after chemoradiotherapy in locally advanced unresectable NSCLC.20
Incorporating PD-L1 in the molecular diagnostic workup of NSCLC
The optimal sequential approach for PD-L1 biomarker testing in advanced NSCLC is not yet well defined. In squamous NSCLC, where no other biomarkers have been identified for treatment selection, PD-L1 testing might be straight forward in terms of sample disposition. On the contrary, in nonsquamous tumors, assessment of PD-L1 might be an issue in small tumor biopsies when several other relevant biomarkers such as EGFR, ALK, ROS1 or BRAF are also required to define treatment selection. There is consistent data suggesting that oncogenic-driven tumors might have lower response to PD-(L)1inhibitors compared with the wild-type population and currently there is no approved indication, nor PD-L1 testing recommendation, for immunotherapy in this setting.17–20,52 Whether PD-L1 testing in nonsquamous tumors has to be restricted only after excluding other relevant oncogenic biomarkers, that is to say in a sequential approach, is certainly an issue that needs to be addressed individually in each center by defining customized tissue-management workflows in order to optimize the use of biological samples without impairing the time for treatment initiation.53
Optimal samples for PD-L1 testing in NSCLC
Besides the inherent variability associated to each technique and commercially available antibody, there are other important biological aspects to be considered when analyzing PD-L1 expression in NSCLC such as the tumor heterogeneity54–59 (Figure 1) or the dynamic expression evolving after therapies.60,61 These factors might explain in part the robust 10–20% of responses observed with PD-(L)1 inhibitors despite the absence or weak immunoreactivity for PD-L1 expression.13 However, recent data suggest a reasonable concordance between both metachronous (different time-points) and synchronous (different locations at the same time-point) specimens. In the KEYNOTE-010 trial, pembrolizumab provided benefit compared with docetaxel irrespective of whether archival or new tumor samples were used to assess PD-L1 expression [(hazard ratio (HR) 0.81 and 0.86 respectively].18 Likewise, in the FIR study with atezolizumab, high agreement of PD-L1 expression was observed between paired archival and fresh tumor samples at TPS ⩾50% or IC ⩾10% cutoffs.62 In one of the largest cohorts (n = 4784) of patients with advanced NSCLC screened for PD-L1 in pembrolizumab KEYNOTE-001, -010, and -024 trials, the prevalence of PD-L1 expression was similar across prior lines of therapy and different disease characteristics examined.63 On the other hand, there is reasonable concordance between PD-L1 expression among different FFPE samples from the same tumor indicating that staining of one block might be enough to capture the entire tumor heterogeneity.64,65
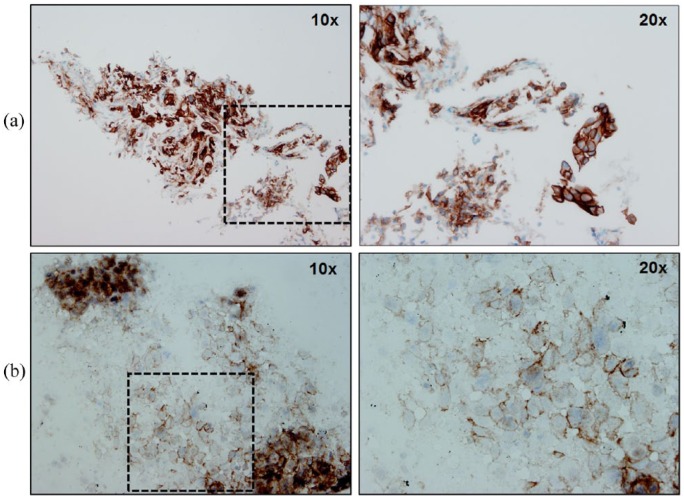
Figure 1.
Representative examples of different NSCLC cases stained with anti-PD-L1 22C3 antibody (Dako pharmDx Assay). (a) FFPE-biopsy section with strongly membranous PD-L1 staining on both tumor and stromal cells. (b) Positive membranous PD-L1 staining on a cytology specimen. Microscope images captured at ×10 and ×20 magnifications. FFPE, formalin-fixed paraffin-embedded; NSCLC, non-small cell lung cancer; PD-L1, programmed cell death ligand-1.
Analytical interpretation and reading of PD-L1 staining on tiny tumor samples can be often misleading and result in a false negative interpretation.32,55,66 Thus, defining the minimum number of viable tumor cells before testing is a crucial aspect for interpretation. Cytology specimens can be excellent material for PD-L1 testing. However, none of the assays are advocated for its use as they have not been technically validated for cytology specimens yet and have never been used in clinical trials. Nonetheless, there is growing data suggesting that PD-L1 IHC testing and quantification on non-FFPE samples such as smears, block sections or liquid based cytologies, might be as well feasible and comparable to those obtained from biopsy specimens49,67 (Figure 2). This is of particular relevance as cytology is the only source of material for diagnose in more than one third of the patients with advanced NSCLC. Another challenging issue for cytology samples is the lack of a validated scoring algorithm. There is currently not enough data available to guide how cytology specimens must be scored or whether cutoffs for positivity should be similar to those used on histologic material. Bearing in mind that a minimum of 100 tumor cells are required in the majority of PD-L1 assays, a significant number of cytology preparations would fall far below this threshold and result nonassessable. A clinical trial to assess the feasibility of PD-L1 expression testing on cytological samples as a surrogate of tissue specimens is currently ongoing [ClinicalTrials.gov identifier: NCT03092739].
Figure 2.
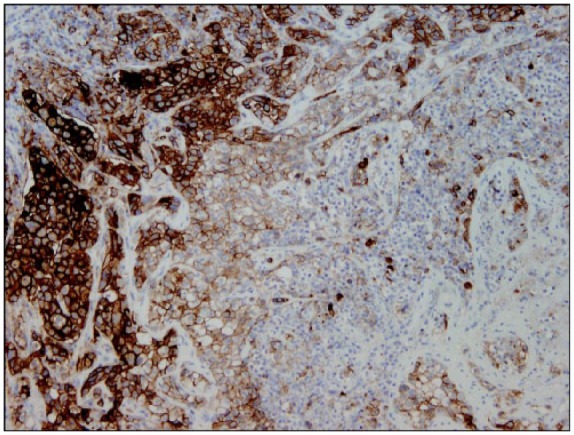
Example of a FFPE section of an advanced NSCLC depicting spatial heterogeneous pattern for PDL1 staining using 22C3 antibody (Dako pharmDx Assay). Microscope image captured at ×20 magnification. FFPE, formalin-fixed paraffin-embedded; NSCLC, non-small cell lung cancer; PD-L1, programmed cell death ligand-1.
In practical terms, the use of various types of tumor samples including fresh or archival FFPE biopsies as well as diverse FFPE samples obtained from primary or metastatic sites of the same tumor might be adequate in NSCLC to identify the best candidates for anti-PD-(L)1 therapeutic strategies. However, bearing in mind the intratumor heterogeneity and that small samples can miss to capture the entire immune contexture of the tumor, an effort should be made to select for each tumor those specimens with higher tumor representation and preferably those samples obtained after systemic treatments or radiotherapy shortly before immune checkpoint inhibitors administration to ensure a robust and reliable PD-L1 result.
Future prospects for immunotherapy
Understanding the molecular causes of response to immunotherapy in lung cancer and other tumors is currently one of the most important fields of research in oncology. Therefore, several other biomarkers beyond PD-L1 are being evaluated to predict better outcomes to immunotherapy.
Lung cancer has a very high rate of somatic mutations when compared with other tumors, with 8.7 mutations per megabase in adenocarcinomas (ADCs) and 9.7 in squamous cell carcinomas (SqCCs).68 However, mutation load is 10-fold higher in smokers than in never-smokers69,70 which correlates with the consistently lower tumor mutation burden (TMB) observed in NSCLC harboring known oncogenic drivers such as EGFR, ALK, ROS1, BRAF-V600E, MET exon 14 skipping mutation with the exception of BRAF non-V600E and KRAS mutant tumors.71 Recently mismatch repair deficiency, which is characterized by having high rate of somatic mutations and neoantigens has been also associated to immune checkpoint blockade response regardless of the cancers’ tissue of origin.72
High TMB is emerging as a key biomarker of sensitivity to immune checkpoint inhibitors across all lung cancers. Data regarding the potential of TMB has been retrospectively assessed in several clinical trials with PD-(L)1 inhibitors. By using whole-exome sequencing of NSCLC patients treated with pembrolizumab, higher nonsynonymous mutation load was associated with improved objective response, durable clinical benefit, and PFS.73 Efficacy also correlated with the molecular smoking signature, higher neoantigen burden, and DNA repair pathway mutations. In the phase III CheckMate-026 trial with nivolumab in chemonaïve NSCLC patients, PFS was longer in patients treated with nivolumab and high TMB regardless of PD-L1 status (HR = 0.62), albeit numbers were too small to drive conclusions.70 In an exploratory analysis of the CheckMate-032, small-cell lung cancer patients with high TMB had improved overall response rate, PFS, and OS compared with low/medium TMB for both nivolumab monotherapy and nivolumab plus ipilimumab, being the first study to evaluate the impact of TMB on outcomes with combination immunotherapy.74 In 153 patients treated with pembrolizumab in the KEYNOTE-028 and KEYNOTE-012 clinical trials, increasing mutational and neoantigen load as well as T-cell inflamed genetic signature were significantly associated with higher responses and longer PFS to pembrolizumab above cutoffs.75 More recently, TMB has been associated with improved PFS and OS with atezolizumab in first and second-line NSCLC patients enrolled in three phase II trials (POPLAR, BIRCH and FIR) suggesting that TMB may be an independent predictor of improved responsiveness to atezolizumab in advanced NSCLC.76 One of the major issues about TMB testing is the absence of a standardized cutoff, variable number of exome sequences reads, total genes included and the use of several platforms and panels for exome sequencing. Therefore, optimization of TMB cutoff and prospective validation is warranted in lung cancer.
Both, lung ADCs and SqCCs NSCLC tumors have distinct patterns of somatic genome alterations77 but share characteristic mutational signatures mainly related to smoking exposure and APOBEC, a key molecular driver inducing mutations in multiple human cancers.78,79 These specific mutational signatures have been correlated with the expression of PD-L1 and a T-cell inflamed signature in head and neck carcinomas80 suggesting that a specific mutational signature might also serve as a biomarker for immune checkpoint inhibition in cancer.
Still, not only quantity but also quality of mutations is crucial to predict the immune response. The immunogenicity of a mutant peptide depends on its affinity for binding major histocompatibility complex class I ligands so that it can be presented and recognized by cytotoxic T-lymphocytes.81 Neoantigen load does not differ between ever-smokers from lung ADCs and SqCCs tumors but is significantly lower in lung ADCs from never-smokers.77 Neoantigen intratumor heterogeneity as well as clonal neoantigens, which are associated with a higher PD-L1 expression and smoking signatures, elicit T-cell immunoreactivity and influence the response of lung cancer patients to immune checkpoint inhibitors.3
Capturing the stromal immune compartment by transcriptome analysis and expression profiles may also provide new insights into the molecular features associated with clinical response to checkpoint inhibitors. To date, no genetic immune signatures have been identified for lung cancer. In the POPLAR trial, patients with pre-existing immunity, defined as high T-effector (Teff) interferon gamma (IFNG)-associated gene expression, had an improved OS with atezolizumab (Teff/IFNG high HR 0.43 versus low Teff/IFNG low HR 1.10).7
Currently blood-based biomarkers of immune response are also being explored as a surrogate source of information alternative to tissue samples. The first data on a blood-based TMB test (bTMB) to measure TMB were recently presented in a large number of plasma samples (n = 794) from two pivotal trials with atezolizumab (phase II POPLAR and phase III OAK) and found that high bTMB was associated with a longer PFS in patients treated with atezolizumab.82 A circulating tumor DNA-TMB validation study (B-F1RST) is underway to prospectively explore the efficacy and safety of multiple targeted therapies in NSCLC patients using Foundation ACT (FACT) platform [ClinicalTrials.gov identifier: NCT03178552]. By using this blood-based testing approach, it may be possible to extend TMB testing to a broad number of patients, including those who are unable to undergo an invasive tumor biopsy. New evidence shows that primary resistance to immune checkpoint inhibitors can be due to abnormal gut microbiome composition.83 Fecal microbiota transplantation from cancer patients who responded to checkpoint inhibitors into germ-free or antibiotic-treated mice ameliorates the antitumor effects of PD-1 blockade and metabolomic whole genome shotgun sequencing demonstrates correlations between clinical responses and the relative abundance in patient’s stools of Akkermansia muciniphila, a mucin-degrading bacterium of the human intestine, offering novel avenues for manipulating the gut ecosystem.83
Perspectives and conclusions
Immunotherapy has already established a firm foothold in the landscape of NSCLC treatment. Therapeutic blockade of immune checkpoint regulators, mainly those that focus on the PD-(L)1 axis, has demonstrated improved clinical outcomes in lung cancer patients by restoring T-cell responses, hence host immunity against tumors. Even so, identifying which patients are going to derive most benefit from these agents is an issue that has still to be resolved. Despite its inherent analytic and predictive limitations, PD-L1 protein expression testing remains presently the biomarker of choice to inform clinical decision-making on treatment with immune checkpoint inhibitors. In an effort for harmonization, several cross-validation studies between platforms are ongoing and will be key to achieve standardization between IHC assays. Meanwhile, prospective studies assessing cytology specimens as an alternative to tissue samples for PD-L1 testing as well as incorporating novel and more precise immune predictive biomarkers such as TMB are warranted to validate them as reliable predictors of response to immunotherapy.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iDs: Cristina Teixidó  https://orcid.org/0000-0002-7226-6567
https://orcid.org/0000-0002-7226-6567
Noemí Reguart  https://orcid.org/0000-0001-8190-499X
https://orcid.org/0000-0001-8190-499X
Contributor Information
Cristina Teixidó, Department of Pathology, Hospital Clínic, Barcelona, SpainTranslational Genomics and Targeted Therapeutics in Solid Tumors, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Catalunya, Spain.
Noelia Vilariño, Translational Genomics and Targeted Therapeutics in Solid Tumors, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Catalunya, SpainMedical Oncology, Hospital Clínic, Barcelona, Spain.
Roxana Reyes, Medical Oncology, Hospital Clínic, Barcelona, Spain.
Noemí Reguart, Department of Medical Oncology, Hospital Clínic, Barcelona, Spain.
References
- 1. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011; 331: 1565–1570. [DOI] [PubMed] [Google Scholar]
- 2. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017; 541: 321–330. [DOI] [PubMed] [Google Scholar]
- 3. McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016; 351: 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366: 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012; 12: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. González-Cao M, Karachaliou N, Viteri S, et al. Targeting PD-1/PD-L1 in lung cancer: current perspectives. Lung Cancer 2015; 6: 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016; 387: 1837–1846. [DOI] [PubMed] [Google Scholar]
- 8. Rebelatto MC, Midha A, Mistry A, et al. Development of a programmed cell death ligand-1 immunohistochemical assay validated for analysis of non-small cell lung cancer and head and neck squamous cell carcinoma. Diagn Pathol 2016; 11: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong H, Zhu G, Tamada K, et al. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med 1999; 5: 1365–1369. [DOI] [PubMed] [Google Scholar]
- 10. Yu H, Boyle TA, Zhou C, et al. PD-L1 expression in lung cancer. J Thorac Oncol 2016; 11: 964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharpe AH, Wherry EJ, Ahmed R, et al. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol 2007; 8: 239–245. [DOI] [PubMed] [Google Scholar]
- 12. Sun Y, Wang Y, Zhao J, et al. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer 2006; 53: 143–151. [DOI] [PubMed] [Google Scholar]
- 13. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015; 372: 2018–2028. [DOI] [PubMed] [Google Scholar]
- 14. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015; 373: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017; 376: 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016; 375: 1823–1833. [DOI] [PubMed] [Google Scholar]
- 17. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015; 373: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016; 387: 1540–1550. [DOI] [PubMed] [Google Scholar]
- 19. Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017; 389: 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Antonia SJ, Villegas A, Daniel D, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med 2017; 377: 1919–1929. [DOI] [PubMed] [Google Scholar]
- 21. Milne CP, Bryan C, Garafalo S, et al. Complementary versus companion diagnostics: apples and oranges? Biomark Med 2015; 9: 25–34. [DOI] [PubMed] [Google Scholar]
- 22. Sznol M, Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res 2013; 19: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. US Food and Drug Administration. Dako PD-L1 IHC 22C3 pharmDx, http://www.accessdata.fda.gov/cdrh_docs/pdf15/P150013c.pdf (accessed 1 October 2017).
- 24. US Food and Drug Administration. Dako PD-L1 IHC 28–8 pharmDx, http://www.accessdata.fda.gov/cdrh_docs/pdf15/P150025c.pdf (accessed 1 October 2017).
- 25. Phillips T, Simmons P, Inzunza HD, et al. Development of an automated PD-L1 immunohistochemistry (IHC) assay for non-small cell lung cancer. Appl Immunohistochem Mol Morphol 2015; 23: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ventana Medical Systems. Media release. Roche announces FDA approval for VENTANA PD-L1 (SP142) Assay to support immunotherapy treatment decisions in lung cancer, http://www.ventana.com/roche-receives-fda-approval-for-pd-l1-assay-for-nsclc (accessed 1 October 2017).
- 27. Ventana Medical Systems, Inc. Ventana PD-L1 (SP142) Assay Staining of Non-Small Cell Lung Cancer Interpretation Guide 2017, http://productlibrary.ventana.com/ventana_portal/OpenOverlayServlet?launchIndex=1&objectId=741–48601015703EN (accessed 25 January 2018).
- 28. Quon C, Xia X, Smith M, et al. VENTANA anti-PD-L1 (SP263) rabbit monoclonal antibody: a high specificity and sensitivity anti-human PD-L1 antibody. J Clin Oncol 2016; 34(Suppl. 15): e14509. [Google Scholar]
- 29. Ventana Medical Systems, Inc. Ventana PD-L1 (SP263) Assay Staining of Non-Small Cell Lung Cancer Interpretation Guide 2016, http://www.roche-diagnostics.ch/content/dam/corporate/roche-dia_ch/documents/broschueren/tissue_diagnostics/Parameter/lung-pathology/PDF_VENTANAPD-L1SP263StainingofNSCLCInterpretationGuide.pdf (accessed 7 October 2017).
- 30. US Food and Drug Administration. VENTANA PD-L1 (SP263) Assay, https://www.accessdata.fda.gov/cdrh_docs/pdf16/p160046c.pdf (accessed 20 October 2017).
- 31. Ratcliffe MJ, Sharpe A, Midha A, et al. Agreement between programmed cell death ligand-1 diagnostic assays across multiple protein expression cutoffs in non-small cell lung cancer. Clin Cancer Res 2017; 23: 3585–3591. [DOI] [PubMed] [Google Scholar]
- 32. Tsao MS, Kerr KM, Dacic S, et al. IASLC atlas of PD-L1 immunohistochemistry testing in lung cancer. North Fort Myers, FL: Editorial Rx Press, 2017. [Google Scholar]
- 33. Kerr KM, Bubendorf L, Edelman MJ, et al. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann Oncol 2014; 25: 1681–1690. [DOI] [PubMed] [Google Scholar]
- 34. Cree IA, Booton R, Cane P, et al. PD-L1 testing for lung cancer in the UK: recognizing the challenges for implementation. Histopathology 2016; 69: 177–186. [DOI] [PubMed] [Google Scholar]
- 35. Calles A, Liao X, Sholl LM, et al. Expression of PD-1 and its ligands, PD-L1 and PD-L2, in smokers and never smokers with KRAS-mutant lung cancer. J Thorac Oncol 2015; 10: 1726–1735. [DOI] [PubMed] [Google Scholar]
- 36. Giunchi F, Degiovanni A, Daddi N, et al. Fading with time of PD-L1 immunoreactivity in non-small cells lung cancer tissues: a methodological study. Appl Immunohistochem Mol Morphol. Epub ahead of print 31 October 2016. DOI: 10.1097/PAI.0000000000000458. [DOI] [PubMed] [Google Scholar]
- 37. Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J Thorac Oncol 2017; 12: 208–222. [DOI] [PubMed] [Google Scholar]
- 38. Rimm DL, Han G, Taube JM, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol 2017; 3: 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scheel AH, Dietel M, Heukamp LC, et al. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol 2016; 29: 1165–1172. [DOI] [PubMed] [Google Scholar]
- 40. Fujimoto D, Sato Y, Uehara K, et al. Predictive performance of four programmed cell death ligand 1 assay systems on nivolumab response in previously treated patients with non-small cell lung cancer. J Thorac Oncol 2017; 12: 1654–1663. [DOI] [PubMed] [Google Scholar]
- 41. Hendry S, Byrne DJ, Wright GM, et al. Comparison of four PD-L1 immunohistochemical assays in lung cancer. J Thorac Oncol. Epub ahead of print 23 November 2017. DOI: 10.1016/j.jtho.2017.11.112. [DOI] [PubMed] [Google Scholar]
- 42. Marchetti A, Barberis M, Franco R, et al. Multicenter comparison of 22c3 pharmDx (Agilent) and SP263 (Ventana) assays to test PD-L1 expression for NSCLC patients to be treated with immune checkpoint inhibitors. J Thorac Oncol 2017; 12: 1654–1663. [DOI] [PubMed] [Google Scholar]
- 43. Brunnström H, Johansson A, Westbom-Fremer S, et al. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: inter-pathologist variability is higher than assay variability. Mod Pathol 2017; 30: 1411–1421. [DOI] [PubMed] [Google Scholar]
- 44. Adam J, Rouquette I, Damotte D, et al. PL04a.04: multicentric French harmonization study for PD-L1 IHC testing in NSCLC. J Thorac Oncol 2017; 12: S11–S12. [Google Scholar]
- 45. Røge R, Vyberg M, Nielsen S. Accurate PD-L1 protocols for non-small cell lung cancer can be developed for automated staining platforms with clone 22C3. Appl Immunohistochem Mol Morphol 2017; 25: 381–385. [DOI] [PubMed] [Google Scholar]
- 46. Ilie M, Khambata-Ford S, Copie-Bergman C, et al. Use of the 22C3 anti-PD-L1 antibody to determine PD-L1 expression in multiple automated immunohistochemistry platforms. PLoS One 2017; 12: e0183023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neuman T, London M, Kania-Almog J, et al. A harmonization study for the use of 22C3 PD-L1 immunohistochemical staining on ventana’s platform. J Thorac Oncol 2016; 11: 1863–1868. [DOI] [PubMed] [Google Scholar]
- 48. Tsao MS, Kerr KM, Yatabe Y, et al. PL 03.03 Blueprint 2: PD-L1 Immunohistochemistry comparability study in real-life clinical samples (BLUEPRINT 2). J Thorac Oncol 2017; 12: S1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skov BG, Skov T. Paired comparison of PD-L1 expression on cytologic and histologic specimens from malignancies in the lung assessed with PD-L1 IHC 28–8pharmDx and PD-L1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol 2017; 25: 453–459. [DOI] [PubMed] [Google Scholar]
- 50. Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016; 27: v1–v27. [DOI] [PubMed] [Google Scholar]
- 51. Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2017; 15: 504–535. [DOI] [PubMed] [Google Scholar]
- 52. Peters S, Gettinger S, Johnson ML, et al. Phase II trial of atezolizumab as first-line or subsequent therapy for patients with programmed death-ligand 1-selected advanced non-small-cell lung cancer (BIRCH). J Clin Oncol 2017; 35: 2781–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reck M, Rabe KF. Precision diagnosis and treatment for advanced non–small-cell lung cancer. N Engl J Med 2017; 377: 849–861. [DOI] [PubMed] [Google Scholar]
- 54. Kim M-Y, Koh J, Kim S, et al. Clinicopathological analysis of PD-L1 and PD-L2 expression in pulmonary squamous cell carcinoma: comparison with tumor-infiltrating T cells and the status of oncogenic drivers. Lung Cancer 2015; 88: 24–33. [DOI] [PubMed] [Google Scholar]
- 55. Ilie M, Long-Mira E, Bence C, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol 2016; 27: 147–153. [DOI] [PubMed] [Google Scholar]
- 56. McLaughlin J, Han G, Schalper KA, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol 2016; 2: 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uruga H, Bozkurtlar E, Huynh TG, et al. Programmed cell death ligand (PD-L1) expression in stage II and III lung adenocarcinomas and nodal metastases. J Thorac Oncol 2017; 12: 458–466. [DOI] [PubMed] [Google Scholar]
- 58. Paulsen EE, Kilvaer TK, Khanehkenari MR, et al. Assessing PDL-1 and PD-1 in non-small cell lung cancer: a novel immunoscore approach. Clin Lung Cancer 2017; 18: 220–233.e8. [DOI] [PubMed] [Google Scholar]
- 59. Takamori S, Toyokawa G, Okamoto I, et al. Discrepancy in programmed cell death-ligand 1 between primary and metastatic non-small cell lung cancer. Anticancer Res 2017; 37: 4223–4228. [DOI] [PubMed] [Google Scholar]
- 60. Twyman-Saint Victor C, Rech AJ, Maity A, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015; 520: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sheng J, Fang W, Yu J, et al. Erratum: expression of programmed death ligand-1 on tumor cells varies pre and post chemotherapy in non-small cell lung cancer. Sci Rep 2016; 6: 23850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chaft JE, Chao B, Akerley WL, et al. Evaluation of PD-L1 expression in metachronous tumor samples and FDG-PET as a predictive biomarker in Ph2 study (FIR) of atezolizumab (MPDL3280A). J Thorac Oncol 2015; 10(Suppl. 2): S176. [Google Scholar]
- 63. Aggarwal C, Abreu DR, Felip E, et al. Prevalence of PD-L1 expression in patients with non-small cell lung cancer screened for enrollment in KEYNOTE-001, -010, and -024. Ann Oncol 2016; 27(Suppl. 6): 1060P. [Google Scholar]
- 64. Rehman JA, Han G, Carvajal-Hausdorf DE, et al. Quantitative and pathologist-read comparison of the heterogeneity of programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer. Mod Pathol 2017; 30: 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cho JH, Sorensen SF, Choi YL, et al. Programmed death ligand 1 expression in paired non-small cell lung cancer tumor samples. Clin Lung Cancer 2015; 16: 385–390. [DOI] [PubMed] [Google Scholar]
- 66. Kitazono S, Fujiwara Y, Tsuta K, et al. Reliability of small biopsy samples compared with resected specimens for the determination of programmed death-ligand 1 expression in non–small-cell lung cancer. Clin Lung Cancer 2015; 16: 385–390. [DOI] [PubMed] [Google Scholar]
- 67. Heymann JJ, Bulman WA, Swinarski D, et al. Programmed death-ligand 1 expression in non-small cell lung carcinoma: comparison among cytology, small biopsy, and surgical resection specimens. Cancer 2017; 125: 896–907. [DOI] [PubMed] [Google Scholar]
- 68. Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012; 150: 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Peters S, Creelan B, Hellmann MD, et al. Abstract CT082: impact of tumor mutation burden on the efficacy of first-line nivolumab in stage IV or recurrent non-small cell lung cancer: an exploratory analysis of CheckMate 026. Cancer Res 2017; 77: CT082. [Google Scholar]
- 71. Spigel DR, Schrock AB, Fabrizio D, et al. Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J Clin Oncol 2016; 34: 9017. [Google Scholar]
- 72. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017; 357: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Atmaca A. OA 07.03a Impact of tumor mutation burden on the efficacy of nivolumab or nivolumab + ipilimumab in small cell lung cancer: an exploratory analysis of CheckMate 032. In: IASLC 18th world conference on lung cancer, 2017. [Google Scholar]
- 75. Cristescu R, Mogg R, Ayers M, et al. Mutational load (ML) and T-cell-inflamed microenvironment as predictors of response to pembrolizumab. ASCO-SITC clinical immuno-oncology symposium. J Clin Oncol 2017; 35: S7S. [Google Scholar]
- 76. Kowanetz M, Zou W, Shames D, et al. OA20.01 Tumor Mutation Burden (TMB) is associated with improved efficacy of atezolizumab in 1L and 2L+ NSCLC patients. J Thorac Oncol 2017; 12: S321–S322. [Google Scholar]
- 77. Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016; 48: 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alexandrov LB, Ju YS, Haase K, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016; 354: 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zehir A, Benayed R, Shah RH, et al. Erratum: mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23: 1004. [DOI] [PubMed] [Google Scholar]
- 80. Rieke DT, Messerschmidt C, Ochsenreither S, et al. Abstract e14613: association of an APOBEC mutational signature, mutational load, and BRCAness with inflammation and PD-L1 expression in HNSCC. J Clin Oncol 2017; 35: e14613. [Google Scholar]
- 81. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015; 348: 69–74. [DOI] [PubMed] [Google Scholar]
- 82. Gandara DR, Kowanetz M, Mok TSK, et al. Blood-based biomarkers for cancer immunotherapy: tumor mutational burden in blood (bTMB) is associated with improved atezolizumab (atezo) efficacy in 2L+ NSCLC (POPLAR and OAK) ESMO congress. Ann Oncol 2017; 28(Suppl. 5): v460–v496. [Google Scholar]
- 83. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 359: 91–97. [DOI] [PubMed] [Google Scholar]