Abstract
Pearl millet is a cereal crop known for its high tolerance to drought, heat and salinity stresses as well as for its nutritional quality. The molecular mechanism of drought tolerance in pearl millet is unknown. Here we attempted to unravel the molecular basis of drought tolerance in two pearl millet inbred lines, ICMB 843 and ICMB 863 using RNA sequencing. Under greenhouse condition, ICMB 843 was found to be more tolerant to drought than ICMB 863. We sequenced the root transcriptome from both lines under control and drought conditions using an Illumina Hi-Seq platform, generating 139.1 million reads. Mapping of sequenced reads against the foxtail millet genome, which has been relatively well-annotated, led to the identification of several differentially expressed genes under drought stress. Total of 6799 and 1253 differentially expressed genes were found in ICMB 843 and ICMB 863, respectively. Pathway and gene function analysis by KEGG online tool revealed that the drought response in pearl millet is mainly regulated by pathways related to photosynthesis, plant hormone signal transduction and mitogen-activated protein kinase signaling. The changes in expression of drought-responsive genes determined by RNA sequencing were confirmed by reverse-transcription PCR for 7 genes. These results are a first step to understanding the molecular mechanisms of drought tolerance in pearl millet and lay a foundation for its genetic improvement.
Introduction
Drought is a major abiotic stress that adversely affects agricultural productivity worldwide. Most of the cereal crops on which the world population depends for food are susceptible to drought. Water deficiency is a global concern that is expected to become worse in the next decades [1]. Therefore, understanding the mechanisms of drought tolerance in cereal crops and development of drought-tolerant varieties are key strategies to maintain yield under drought conditions. Mechanisms that overcome drought stress in plants include drought avoidance, drought tolerance, drought escape and drought recovery [2,3]. At the molecular level, plants respond to drought by inducing both regulatory and functional genes [4]. Many metabolic pathways, such as the abscisic acid (ABA) signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway and photosynthesis-associated pathways, respond to drought stress [5–7]. Recently many efforts have been made to elucidate the mechanism of drought tolerance in non-model cereal crops through molecular and genomic approaches [8–10].
Pearl millet (Pennisetum glaucum (L.) R. Br.) (2n = 2X = 14) is the world’s sixth most important cereal crop, and is primarily grown as a rain-fed crop in the low rainfall zones of Sub-Saharan Africa and the Indian subcontinent [11]. Pearl millet is a C4 cereal crop belonging to the family Poaceae and subfamily Panicoideae. It is a cross-pollinated crop with short life cycle and outbreeding nature. [12]. Compared to other cereal crops such as rice, wheat, maize and sorghum, it has high tolerance to abiotic stresses, such as drought, salinity, high temperature and soil nutrient deficiency [13]. It is mainly grown for food grain production, but in some cases, it is also grown for hay, bird feed, biofuel and forage. Pearl millet is a nutrient-rich crop; its seeds contain more protein, iron, zinc and energy than seeds of rice, wheat and maize [14].
So far only two research groups have investigated drought tolerance in pearl millet using transcriptomic approaches [15,16]. A few genes, such as PgDREB2A (Drought responsive element binding protein 2A) and PgGPx (Glutathione peroxide), have been studied for their role in drought tolerance [17,18]. However, it is expected that many more genes are involved. Recently, the pearl millet genome has been sequenced by the international pearl millet genome sequencing consortium (IPMGSC). The draft genome sequence is publicly available, and will help to understand the pearl millet drought tolerance mechanisms [19], although it needs to be further annotated.
Transcriptomic analysis using next generation sequencing (NGS) is an efficient method for exploring gene expression patterns in plants. However, RNA Sequencing (RNA-Seq) using NGS has the ability to detect differentially expressed genes (DEGs) because it has a dynamic range of expression levels [20]. Here, we used an RNA-Seq approach to understand the pathways involved in pearl millet in responses to drought stress. The data will help to design more detailed studies of drought tolerance in this species as well as develop DNA markers that could facilitate breeding more drought-tolerant varieties.
Materials and methods
Experimental material and growth condition
Seeds of pearl millet were procured from International Crop Research Institute of Semi-arid Tropics (ICRISAT), India. Two inbred lines ICMB 843 as drought tolerant and ICMB 863 as comparatively less drought tolerant having different levels of drought tolerance were selected for our study.
The experiment was conducted under controlled greenhouse conditions. Around 20 seeds of each inbred line were sown in equal volumes of soil and vermiculite in perforated terracotta pots with the size of 11 litters. Each growth experiment was done with three replications. For growth tests and RNA-Seq, drought stress was imposed on 21-day-old plants by withholding water for 5 days, while control plants were watered on alternate days.
Plants of both lines were grown as described in the “Experimental material and growth condition” subsection. SPAD values of control and drought stressed plants were measured using SPAD 502 Plus Chlorophyll Meter (Spectrum Technologies Inc., Aurora, Illinois, USA). Relative water content (RWC) was calculated as previously described [21].
RNA extraction, library preparation and sequencing
Roots of drought-stressed plants and control plants were ground in liquid nitrogen with a mortar and pestle. Total RNA was isolated with Trizol (Sigma-Aldrich Cat.no:15596026), and genomic DNA was digested with RNase-Free DNase (Qiagen). RNA degradation and potential DNA contamination were checked by gel electrophoresis. RNA integrity number (RIN) and quantitation check were performed by Agilent 2100 bioanalyzer. After the quality check procedures, mRNA from roots was enriched using oligo(dT) beads. The mRNA was fragmented randomly by adding fragmentation buffer, then the cDNA was synthesized using mRNA template and random hexamers, after which a second strand synthesis buffer (Illumina), dNTPs, RNase H and DNA polymerase I were added to initiate the second-strand synthesis. After terminal repair and sequencing adapter ligation, the double-stranded cDNA library was completed through size selection and PCR enrichment. The qualified libraries were subjected to sequencing by Illumina Hiseq through NOVOgene Co., ltd. Twelve data sets for paired-end sequencing reads in FASTQ format were submitted to SRA (Sequence Read Archive) at NCBI, and can be retrieved by the accession number SRP125789.
Reads mapping and differential gene expression analysis
Raw reads obtained by sequencing were quality-checked using online tool FASTQC (Version.0.11.5). Qualified reads were mapped to the reference genomes using Genome Work bench 9.5.4 (CLC Bio. Japan). The read counts were normalized by reads per kilobase per million mapped reads (RPKM = Total exon reads/mapped reads in million X exon length in kb) for each gene and log2 transformed.
Gene expression levels were estimated by RPKM values using advanced plugins for RNA-Seq in CLC Workbench 9.5.4. Baggereley test was used to calculate P values. Genes with log2 fold changes >2 or < -2, and FDR-corrected p values <0.05 were regarded as DEGs.
Pathway enrichment analysis
The DEGs in both inbred lines of pearl millet (ICMB 843 and ICMB 863) under control and stressed conditions were mapped to the biological pathways by using online tool KEGG (Kyoto encyclopedia for genes and genomes) [22] and the pathways enriched by DEGs were studied further.
Validation of RNA-Seq data by real time RT-PCR
Seven genes were selected based on the RPKM values. Real time PCR was performed using Step-one Real-Time instrument (Applied Biosystems) and the SYBR Green I kit (Roche, Basel, Switzerland). Reactions were performed in triplicate and contained 100 ng of cDNA, 1 μL of each primer (10 μM/ μL) and 10 μL of SYBR green master mix in a final volume of 20 μL. The primer pairs listed in S1 Table were designed to yield products with the sizes of 80–200 bp based on the sequences in the foxtail millet genome mapped with the pearl millet reads. The PCR cycling conditions were as follows: an initial denaturation step of 20 s at 50 °C, 10 min at 95 °C, and 40 cycles of 15 s at 95 °C, and 1 min at 60 °C followed by melt curve analysis. The PgActin gene was used as a reference gene [23]. Relative fold differences for each sample in each experiment were calculated using the ΔΔCt method [24].
Results
Effect of drought treatment on physiology of pearl millet lines
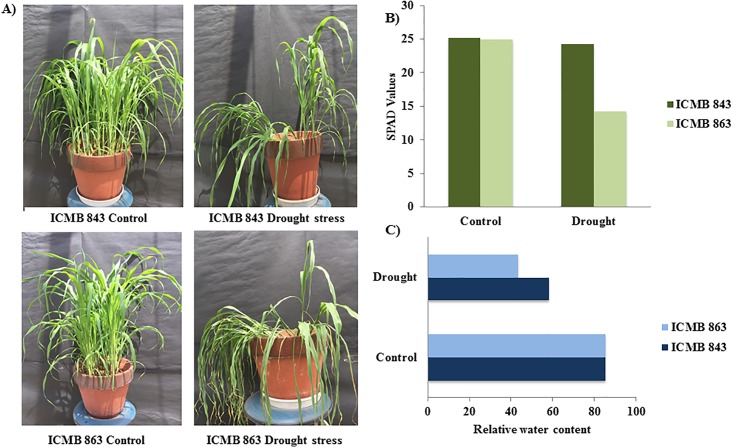
Within 5–7 days of drought stress treatment, ICMB 863 started showing drought stress symptoms such as leaf tip drying and rolling, whereas ICMB 843 did not show any of these symptoms. Both lines showed lodging during drought stress, but lodging was more severe in ICMB 863 (Fig 1A). Total chlorophyll content or greenness determined by SPAD meter (Fig 1B) showed a greater reduction of chlorophyll in ICMB 863 than in ICMB 843. These observations indicate that the drought caused a greater reduction in photosynthetic activity in ICMB 863 than in ICMB 843. Drought treatment decreased RWC significantly more in ICMB 863 than in ICMB 843 (Fig 1C). These results suggest that ICMB 843 is more tolerant to drought than ICMB 863 at seedling stage.
Fig 1. Physiological analysis of pearl millet for drought.
A) Effects of drought on the physiology of pearl millet lines (ICMB 843 and ICMB 863). B) SPAD value of pearl millet lines under and drought conditions. Values are mean ± SE (n = 20) C) Relative water content of leaf under control and drought stress. Values are mean ± SE (n = 20).
Transcriptomic profiling and mapping statistics of pearl millet
Roots are the organ that would first respond to drought stress, and drought-stressed pearl millet roots were subjected to RNA-Seq to analyze their transcriptome. Illumina sequencing resulted in generating around 139 million of raw reads. Preprocessing with the CLC genomics workbench 9.0 generated 134 million clean reads (Table 1).
Table 1. Basic statistics for sequenced reads from transcriptome of pearl millet.
| Samples | Raw reads | Clean reads | Q20(%)** | Q30(%)*** | Total mapped**** |
|---|---|---|---|---|---|
| ICMB843_control1 | 11615061 | 11188437 | 93.17 | 84.48 | 14,208,882(63.48%) |
| ICMB843_control2 | 11934447 | 11340253 | 93.58 | 85.21 | 13,546,964(59.73%) |
| ICMB843_control3 | 11469576 | 11070226 | 93.44 | 84.88 | 12,913,182(58.32%) |
| ICMB843_Treated1 | 7648776 | 7433785 | 93.66 | 85.77 | 5,482,220(36.87%) |
| ICMB843_Treated2 | 13014255 | 12566731 | 94.78 | 87.97 | 6,049,930(24.07%) |
| ICMB843_Treated3 | 11678697 | 11382308 | 93.61 | 85.69 | 8,638,226(37.95%) |
| ICMB863_control1 | 10735082 | 10169174 | 93.65 | 85.86 | 110,10,118(54.17%) |
| ICMB863_control2 | 10029364 | 9297019 | 93.97 | 85.30 | 8,582,854(46.16%) |
| ICMB863_control3 | 14717884 | 14570208 | 95.05 | 85.86 | 17,240,062(59.16%) |
| ICMB863_Treated1 | 10551898 | 10249771 | 94.33 | 88.84 | 10,256,990(50.04%) |
| ICMB863_Treated2 | 16115349 | 15657629 | 94.47 | 87.65 | 19,234,298(61.42%) |
| ICMB863_Treated3 | 9614712 | 9342152 | 93.41 | 85.33 | 10,034,136(53.70%) |
| Total | 139125101 | 134267693 | 126,187.774 |
1,2,3 = Individual Biological replications of sample
**Q20(%) = Bases with the phred value >20 (in percentage)
***Q30(%) = Bases with the phred value >30 (in percentage)
****Total number of reads mapped to the genome of Setaria Italica (percentage)
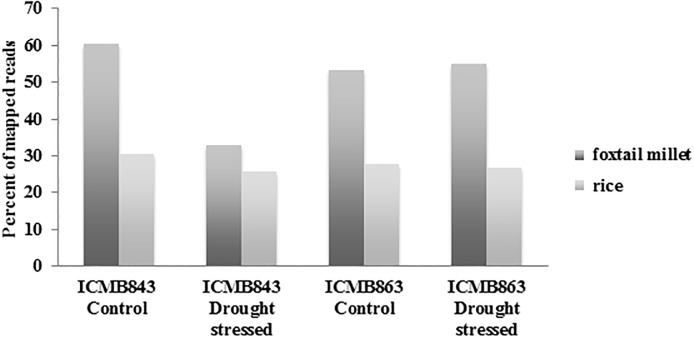
The clean reads were mapped to the genomes of two closely related members of the Poaceae family, foxtail millet (S. italica) and rice (O. sativa) [25]. These genomes were used because they have been annotated better than the pearl millet genome. About 33–61% of the reads were mapped to the foxtail millet genome, and about 25–30% of the reads were mapped to the rice genome (Fig 2). Therefore, the foxtail millet genome was used as the reference genome for further analyses. Interestingly, the percentage of mapped reads was lower in the samples from drought-stressed ICMB 843 than in the other samples (Fig 2 and Table 1). This might be because drought stress increases variation in transcripts in ICMB 843.
Fig 2. Mapping of pearl millet reads to the foxtail millet genome and the rice genome.
X axis denotes the sample names under control and drought stress and Y axis denotes the percentage of mapped genes.
Differentially expressed genes for drought stress in pearl millet
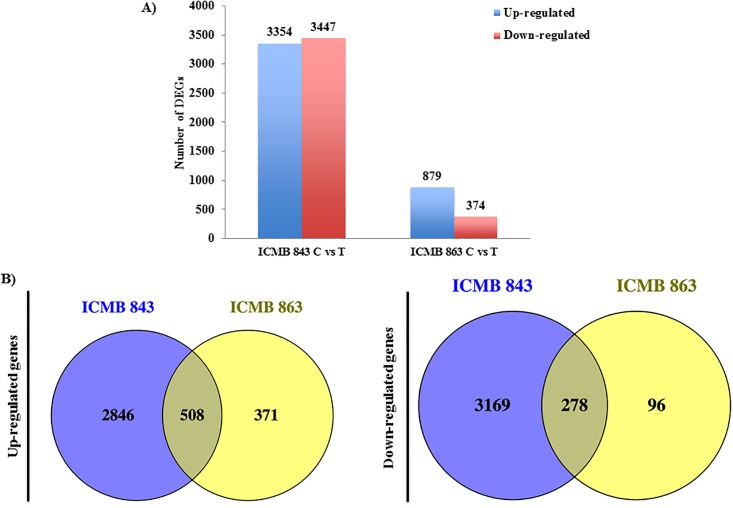
RPKM and log2 transformation were used for extracting DEGs. Drought treatment up-regulated ~4 times more genes in the drought-tolerant line ICMB 843 than in the drought-sensitive line ICMB 863, and while it down-regulated ~10 times more genes in ICMB 843 than in ICMB 863 (Fig 3A).
Fig 3. DEG analysis in comparisons study.
A) Number of DEGs in all combinations with fold change >2 or <-2 and FDR-corrected pvalue <0.05) Blue and red bars indicate up- and down- regulated respectively. C: control and T and Treated: Drought- treated B) Venn diagram showing an up-regulated (right side) and down regulated (left side) genes in lines ICMB 843 and ICMB 863 under drought treated situation.
Venn diagram highlights overlapping DEGs expressed in both ICMB 843 and ICMB 863 under drought situation (Fig 3B). In ICMB 843, 2846 genes were up-regulated and 3169 were down-regulated by drought treatment, while in ICMB 863 about 371 genes were up-regulated and 96 genes were down-regulated. Common genes that were up-regulated and down-regulated in both lines were 508 and 278 respectively.
Effect of drought on biological pathways of pearl millet
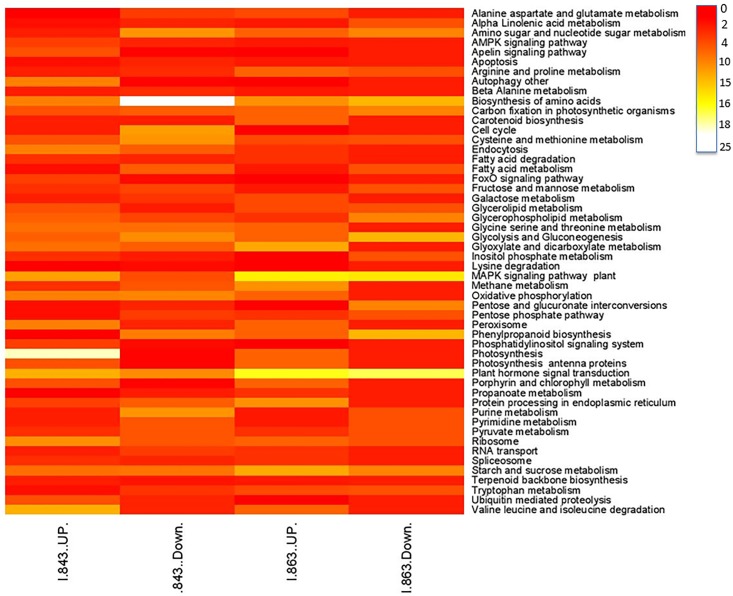
To associate the DEGs found in our RNA-Seq with known biological pathways, KEGG analysis was performed. The biological pathways enriched with up-regulated DEGs were photosynthesis, plant hormone signal transduction and MAPK signal transduction (Fig 4).
Fig 4. Heat map showing the number of DEGs enriched in different KEGG pathways.
The bar scale shows the number of DEGs associated with pathways.
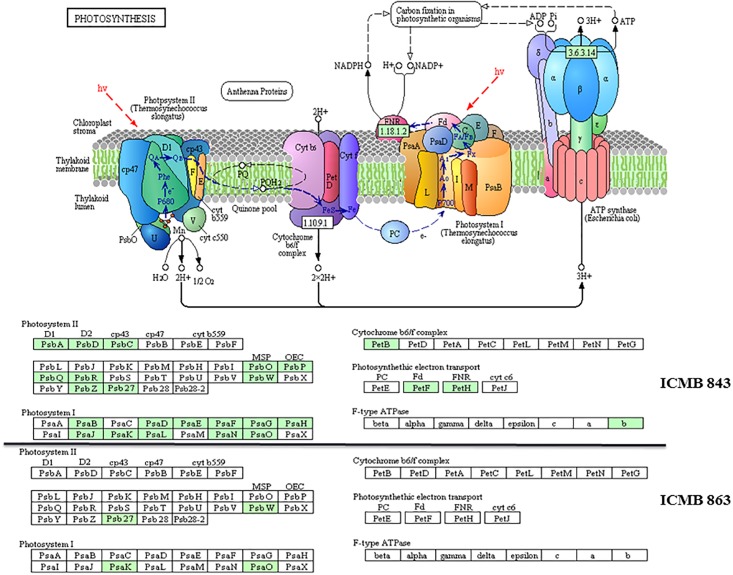
There were around 25 upregulated genes encoding components of five subunits of photosynthesis (Photosystem II, Photosystem I, cytochrome b6/f complex, photosynthetic electron transport and f-type ATPase). Most of them were up regulated in ICMB 843 but not in ICMB 863 (Fig 5).
Fig 5. DEGs involved in photosynthesis.
Genes up-regulated by drought stress are shown (25 genes in ICMB 843 and 4 in ICMB 863) in green boxes. White boxes indicate the non-drought-responsive genes. This image was generated using the online tool KEGG Mapper–Colour Pathway (http://www.genome.jp/kegg/tool/map_pathway3.html).
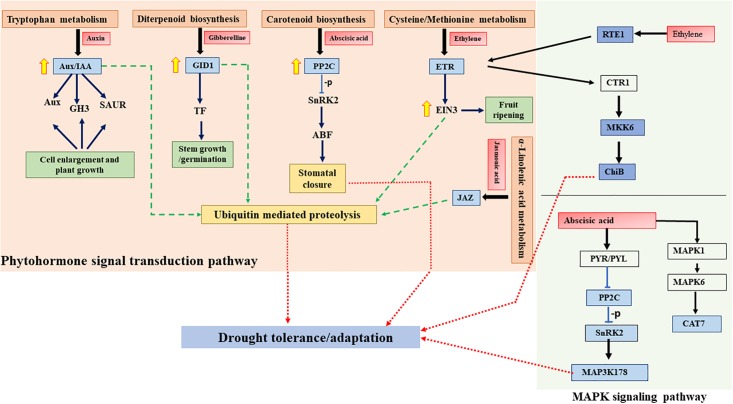
In total, 22 genes in our study shows association with these two signaling pathways. These 22 genes were mainly related to the genes with Auxin biosynthesis (Aux/IAA, GH3, SAUR), Diterpenoid biosynthesis (GID1, PIF), Carotenoid biosynthesis (PP2C, SnRK2, ABF), Ethylene biosynthesis (ETR, RTE, MKK6, ChiB) and Abscisic acids (PVR, MAP3K17/18, CAT1, MAPK1, MAPK6). Further details about the pathways and associated genes are listed in the (S2 Table). Plant hormone signaling pathway and MAPK pathways seems to be interlinked with each other as they share common byproducts that may increase tolerance to drought in pearl millet (Fig 6).
Fig 6. DEGs involved in phytohormone and MAPK signaling pathway.
The proposed pathways show phytohormone biosynthesis (light orange) and MAPK signaling pathways (light blue). This figure shows only the genes that have been associated with the pathway from transcriptome analysis. Red boxes show plant hormones, blue boxes show the genes up-regulated stress, green arrow shows genes involved in drought tolerance through ubiquitin mediated proteolysis and red arrows shows the direct involvement in the drought tolerance. Light blue lines with a bar denotes inhibition and simple arrows denotes activation. -p indicates de phosphorylation process. Aux/IAA: auxin-responsive protein IAA; GH3: Auxin-responsive GH3 family protein; SAUR: SAUR-like auxin-responsive protein family; CRE: Histidine kinase 2/3/4 (cytokinin receptor); B-ARR: Two-component response regulator ARR-B family; GID1: Alpha/beta-Hydrolases superfamily protein; TF: Phytochrome interacting factor 3 (PIF3); PP2C: Protein phosphatase 2C; SnRK2: Serine/threonine-protein kinase SRK2; ABF: ABA responsive element binding factor; ETR: Ethylene receptor; EIN3: ETHYLENE-INSENSITIVE3-like 3; JAZ: Jasmonate ZIM domain-containing protein; RTE1: Transmembrane protein 222; MKK9: Mitogen-activated protein kinase kinase 9; ChiB: Basic endochitinase B; MAP3K17/18: Mitogen-activated protein kinase kinase kinase 17/18; MAPK1: Mitogen-activated protein kinase 1; MAPK6: Mitogen-activated protein kinase 6; CAT7: Catalase.
Validation of RNA-Seq data with the quantitative real time PCR
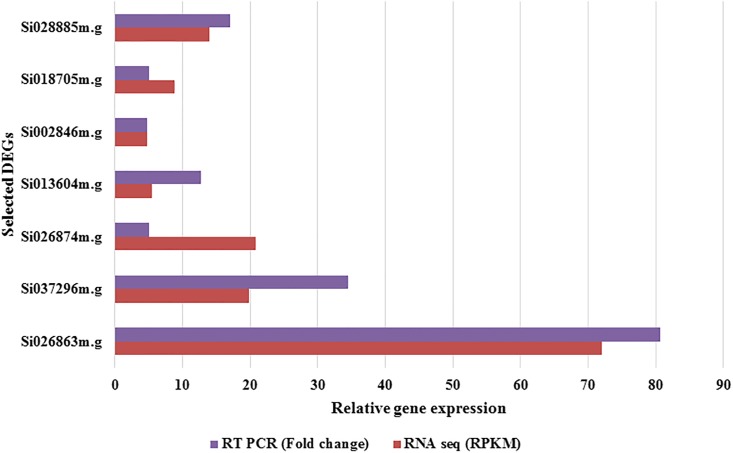
The relative gene expression patterns of the qRT-PCR results for 7 genes were consistent with RNA-Seq data (Fig 7). The annotations for the genes selected for validation are listed in (Table 2).
Fig 7. Validation of RNA-Seq result with RT PCR.
Expression of 7 randomly selected genes was examined by RT- PCR analysis. For each gene, fold changes were calculated by ΔΔCt method in the RT-PCR and with the RPKM values in the RNA-Seq, respectively.
Table 2. Annotations of the genes selected for RT-PCR.
| Name of the gene (Identifier) | Annotations | Fold (RT PCR) |
|---|---|---|
| Si028885m.g | Mixed linked glucan synthase | 1.9 |
| Si018705m.g | Chlorophyll centre-I related protein | 5.11 |
| Si002846m.g | Dehydration responsive elements binding protein 1F | 4.45 |
| Si013604m.g | Uncharacterized protein | 12.8 |
| Si026874m.g | Uncharacterized protein | 5.1 |
| Si037296m.g | Glutathione s-transferase (GSTU6) | 34.5 |
| Si026863m.g | Abiotic stress responsive factor | 80.7 |
Discussion
Many drought-responsive genes were identified by our RNA-Seq analysis. Many of these genes were associated with photosynthesis, plant hormone signaling and MAPK signaling pathway. To our knowledge, this is the first study in pearl millet for drought tolerance by RNA-Seq approach.
Drought responsive genes involved in photosynthesis
As a primary metabolic process, photosynthesis plays a crucial central role in plants life when under drought stress. One of the major effects of drought and other environmental stresses [7,26] is to damage both of the Photosystems (PS I and PS II, respectively) [27]. PS II is a complex network of protein containing several types of chlorophyll binding components. Main function of these components is to organize chlorophyll for light harvesting but PS II also acts as cofactor needed for oxidation of water [28]. In our study, we found that 25 up-regulated genes in ICMB 843 and 8 genes in ICMB 863 are involved in photosynthesis and photosynthesis-related pathways. In Arabidopsis, PS II and light harvesting complex II (LHC II) play an important role in preventing the photo-damage to PS II under drought stress [29]. Ten PS II genes were up-regulated in ICMB 843 upon drought stress. These included PsbQ, which is necessary for regulation of activity of PS II [30], PsbR, which is required for preparation of oxygen evolving complex of PS II, PsbP, PsbQ and PsbW, which stabilizes the supramolecular organization of PS II and six other genes (PsbA, PsbD, PsbC, Psb I, PsbZ, Psb27) [31]. On the other hand, in ICMB 863, only PsbW and Psb27 genes were up-regulated. As mainly these genes are related to avoid photodamage to PS II, it might be possible that on drought stress pearl millet activates the genes to maintain its structural integrity. Other up-regulated genes such as PsaB PsaD and PsaE are related to electron transport and ferredoxin binding [32]. PsaF, PsaG, PsaH, PsaK, PsaL, PsaK and PsaO have a role in binding of light harvesting complex [33] and genes encoding them were also found to be up-regulated by drought stress (Fig 5 and S2 Table). These results raise the possibility that electron transport in PS I and PS II is activated by drought stress in pearl millet.
Drought and plant hormone signal transduction and MAPK signalling
Phytohormones, such as abscisic acid (ABA), cytokinin (CK), gibberellic acid (GA), auxin, and ethylene, and jasmonate regulate diverse processes and confers drought tolerance to plants [34]. In our analysis, we found genes associated with the biosynthesis and signalling of these phytohormones. Such genes include AUX/IAA, GH3, and SAUR. Auxin biosynthesis is known to confer drought tolerance in Rice [35], OsGH3-2 a member of the GH3 family was proved to be modulating the level of ABA and stress tolerance in rice [36], SAUR genes has not been yet functionally characterized, are altered by auxin and are hypothesized to be involved in the drought tolerance by auxin [37]. GID1 and Phytochrome interacting factor 4 are involved GA signaling and in regulating drought stress responses [38,39]. PP2C, SnRK2, and ABRF are involved in ABA signaling and in regulating drought stress [40–42]. Our study suggests that homologs of these genes are all up-regulated by drought stress in pearl millet. Biosynthesis of ethylene from 1-aminocyclopropane-1-carboxylic acid during drought is triggered to induce the senescence [43]. We found up- regulation of pearl millet homologs encoding ETR and EIN3 which were previously showed to mediate ethylene signaling and to negatively regulate drought stress [44–46]. Further studies are needed to elucidate their relevance to drought stress response of pearl millet. Biosynthesis, modification, and signaling of jasmonate during drought are comprehensively studied [47]. Up-regulation of homologs possibly encoding JAZ protein which mediates jasmonate signaling was observed in our study.
MAPK is a signal transduction pathway somewhat conserved in all eukaryotes, including yeasts, animals and plants; involvement of MAPK in drought stress responses has been already reported [48]. RNA-Seq analysis of maize found that MAPK kinase kinases (MAPKKKs) have important regulatory functions in drought tolerance in maize [49]. In our study of pearl millet, we observed the drought-responsive up-regulation of MAPK signaling pathway. Some other components of MAPK cascades are known to be involved in ABA and ethylene signaling and drought stress responses. The Upregulation of these kinases may contribute to conferring drought tolerance in case of the pearl millet (Fig 6 and S2 Table).
Conclusions
Pearl millet is a drought-tolerant cereal crop and therefore represents a novel source for investigating the molecular mechanism of drought tolerance in cereal crops. Two pearl millet inbred lines, ICMB 843 and ICMB 863, which exhibit diverse responses against drought stress were used for the study. ICMB 843 is relatively more tolerant to drought than ICMB 863 at seedling stage. RNA-Seq analysis showed that ICMB 843 activated more number of drought responsive genes than ICMB 863. Genes related to photosynthesis, plant hormone signal transduction and MAPK signalling pathways were induced by drought in pearl millet. Identified drought- responsive genes and metabolic pathways are targets for future studies in order to understand the molecular mechanism of drought tolerance in pearl millet.
Supporting information
(XLSX)
(DOCX)
Acknowledgments
We would like to thank Dr. S. K. Gupta, Senior Scientist working on pearl millet breeding at ICRISAT, India for providing seed materials and his guidance related to pearl millet physiological assay.
Data Availability
DATA in FASTQ format were submitted to SRA (Sequence read archive), NCBI (Bio project Id: PRJNA419859). The raw reads can be accessed with accession number SRR6327851 to SRR6327856 for control samples and SRR6327857 to SRR6327862 for drought-stressed samples.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Bates BC, Kundzewicz ZW, Wu S, Palutikof JP. Climate Change and Water [Internet]. Climate change and water. 2008. 210 p. http://www.citeulike.org/group/14742/article/8861411%5Cnhttp://www.ipcc.ch/publications_and_data/publications_and_data_technical_papers.shtml#.UREVW6X7Uy4
- 2.Cruz de Carvalho MH. Drought stress and reactive oxygen species. Plant Signal Behav [Internet]. 2008;3(3):156–65. Available from: http://www.tandfonline.com/doi/abs/10.4161/psb.3.3.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.By P. World ’ s largest Science, Technology & Medicine Open Access book publisher Molecular and Morphophysiological Analysis Molecular and Morphophysiological Analysis of Drought Stress in Plants of Drought Stress in Plants.
- 4.Bartels D, Sunkar R. Drought and salt tolerance in plants. CRC Crit Rev Plant Sci. 2005;24(1):23–58. [Google Scholar]
- 5.Tuteja N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal Behav [Internet]. 2007;2(3):135–8. Available from: http://www.tandfonline.com/doi/abs/10.4161/psb.2.3.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Sultan MARF, Liu XL, Zhang J, Yu F, Zhao HX. Physiological and comparative proteomic analysis reveals different drought responses in roots and leaves of drought-tolerant wild wheat (Triticum boeoticum). PLoS One. 2015;10(4):1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinheiro C, Chaves MM. Photosynthesis and drought: Can we make metabolic connections from available data? J Exp Bot. 2011;62(3):869–82. doi: 10.1093/jxb/erq340 [DOI] [PubMed] [Google Scholar]
- 8.Min H, Chen C, Wei S, Shang X, Sun M, Xia R, et al. Identification of Drought Tolerant Mechanisms in Maize Seedlings Based on Transcriptome Analysis of Recombination Inbred Lines. Front Plant Sci [Internet]. 2016;7(July). Available from: http://journal.frontiersin.org/Article/10.3389/fpls.2016.01080/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldoni E, Bagnaresi P, Locatelli F, Mattana M, Genga A. Comparative Leaf and Root Transcriptomic Analysis of two Rice Japonica Cultivars Reveals Major Differences in the Root Early Response to Osmotic Stress. Rice [Internet]. Rice; 2016;9(1):25 Available from: http://thericejournal.springeropen.com/articles/10.1186/s12284-016-0098-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song K, Kim HC, Shin S, Kim K-H, Moon J-C, Kim JY, et al. Transcriptome Analysis of Flowering Time Genes under Drought Stress in Maize Leaves. Front Plant Sci [Internet]. 2017;8(March):1–12. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2017.00267/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajaram V, Nepolean T, Senthilvel S, Varshney RK, Vadez V, Srivastava RK, et al. Pearl millet [Pennisetum glaucum (L.) R. Br.] consensus linkage map constructed using four RIL mapping populations and newly developed EST-SSRs Pearl millet [Pennisetum glaucum (L.) R. Br.] consensus linkage map constructed using four RIL mappin. BMC Genomics [Internet]. 2013;14(1):1. Available from: BMC Genomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shivhare R, Lata C. Exploration of Genetic and Genomic Resources for Abiotic and Biotic Stress Tolerance in Pearl Millet. Front Plant Sci [Internet]. 2017;7(January):1–17. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2016.02069/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vadez V, Hash T, Bidinger FR, Kholova J. II.1.5 Phenotyping pearl millet for adaptation to drought. Front Physiol. 2012;3 October(October):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Hash CT, Thirunavukkarasu N, Singh G, Rajaram V, Rathore A, et al. Mapping Quantitative Trait Loci Controlling High Iron and Zinc Content in Self and Open Pollinated Grains of Pearl Millet [Pennisetum glaucum (L.) R. Br.]. Front Plant Sci [Internet]. 2016;7(November):1–16. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2016.01636/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra RN, Reddy PS, Nair S, Markandeya G, Reddy AR, Sopory SK, et al. Isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of Pennisetum glaucum seedlings. Plant Mol Biol. 2007;64(6):713–32. doi: 10.1007/s11103-007-9193-4 [DOI] [PubMed] [Google Scholar]
- 16.Choudhary M, Jayanand, Padaria JC. Transcriptional profiling in pearl millet (Pennisetum glaucum L.R. Br.) for identification of differentially expressed drought responsive genes. Physiol Mol Biol Plants. 2015;21(2):187–96. doi: 10.1007/s12298-015-0287-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal P, Agarwal PK, Joshi AJ, Sopory SK, Reddy MK. Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Mol Biol Rep. 2010;37(2):1125–35. doi: 10.1007/s11033-009-9885-8 [DOI] [PubMed] [Google Scholar]
- 18.Islam T, Manna M, Reddy MK. Glutathione peroxidase of Pennisetum glaucum (PgGPx) is a functional Cd2+ dependent peroxiredoxin that enhances tolerance against salinity and drought stress. PLoS One. 2015;10(11):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varshney RK, Shi C, Thudi M, Mariac C, Wallace J, Qi P, et al. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol [Internet]. Nature Publishing Group; 2017;35(10):969–76. Available from: http://dx.doi.org/10.1038/nbt.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jongeneel V, Estreicher A, Baxevanis AD, Ouellette BFF, Wolfsberg TG, Landsman D, et al. EXPRESSED SEQUENCE TAGS (ESTs). Bioinforma A Pract Guid to Anal Genes Proteins. 2001;10(1):57–63. [Google Scholar]
- 21.Smart RE, Bingham GE. Smart R. E., & Bingham G. E. (1974). Rapid Estimates of Relative Water Content. Plant Physiology, 53(2), 258–260. https://doi.org/10.1104/pp.53.2.258Rapid Estimates of Relative Water Content. Plant Physiol. 1974;53(2):258–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(SUPPL.2):182–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shivhare R, Lata C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci Rep [Internet]. Nature Publishing Group; 2016;6(October 2015):1–12. Available from: http://dx.doi.org/10.1038/srep23036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 25.Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, et al. Reference genome sequence of the model plant Setaria. Nat Biotechnol [Internet]. Nature Publishing Group; 2012;30(6):555–61. Available from: http://dx.doi.org/10.1038/nbt.2196 [DOI] [PubMed] [Google Scholar]
- 26.Woo NS, Badger MR, Pogson BJ. A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods [Internet]. 2008;4(1):27 Available from: http://plantmethods.biomedcentral.com/articles/10.1186/1746-4811-4-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu CM, Zhang JH. Effects of water stress on photosystem II photochemistry and its thermostability in wheat plants. J Exp Bot [Internet]. 1999;50(336):1199–206. Available from: http://jxb.oxfordjournals.org/content/50/336/1199.full.pdf [Google Scholar]
- 28.Wydrzynski T, Hillier W, Conlan B. Engineering model proteins for Photosystem II function. Photosynth Res. 2007;94(2–3):225–33. doi: 10.1007/s11120-007-9271-0 [DOI] [PubMed] [Google Scholar]
- 29.Chen Y-E, Liu W-J, Su Y-Q, Cui J-M, Zhang Z-W, Yuan M, et al. Different response of photosystem II to short and long term drought stress in Arabidopsis thaliana. Physiol Plant [Internet]. 2016;(April):225–35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26918860 [DOI] [PubMed] [Google Scholar]
- 30.Thornton LE, Ohkawa H, Roose JL, Kashino Y, Keren N, Pakrasi HB. Homologs of Plant PsbP and PsbQ Proteins Are Necessary for Regulation of Photosystem II Activity in the Cyanobacterium Synechocystis 6803. Plant Cell Online [Internet]. 2004;16(8):2164–75. Available from: http://www.plantcell.org/cgi/doi/10.1105/tpc.104.023515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suorsa M, Sirpiö S, Allahverdiyeva Y, Paakkarinen V, Mamedov F, Styring S, et al. PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J Biol Chem. 2006;281(1):145–50. doi: 10.1074/jbc.M510600200 [DOI] [PubMed] [Google Scholar]
- 32.Tullberg A, Alexciev K, Pfannschmidt T, Allen JF. Photosynthetic Electron Flow Regulates Transcription of the psaB Gene in Pea (Pisum sativum L.) Chloroplasts Through the Redox State of the Plastoquinone Pool. Plant Cell Physiol [Internet]. 2000;41(9):1045–54. Available from: http://academic.oup.com/pcp/article/41/9/1045/2329569/Photosynthetic-Electron-Flow-Regulates [DOI] [PubMed] [Google Scholar]
- 33.Scheller HV, Jensen PE, Haldrup A, Lunde C, Knoetzel J. Role of subunits in eukaryotic Photosystem I. Biochim Biophys Acta—Bioenerg. 2001;1507(1–3):41–60. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson S, Kudoyarova GR, Veselov DS, Arkhipova TN, Davies WJ. Plant hormone interactions: Innovative targets for crop breeding and management. J Exp Bot. 2012;63(9):3499–509. doi: 10.1093/jxb/ers148 [DOI] [PubMed] [Google Scholar]
- 35.Jung H, Lee DK, Choi Y Do, Kim JK. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci [Internet]. Elsevier Ireland Ltd; 2015;236:304–12. Available from: http://dx.doi.org/10.1016/j.plantsci.2015.04.018 [DOI] [PubMed] [Google Scholar]
- 36.Du H, Wu N, Fu J, Wang S, Li X, Xiao J, et al. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot. 2012;63(18):6467–80. doi: 10.1093/jxb/ers300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei T, Deng K, Zhang Q, Gao Y, Liu Y, Yang M, et al. Modulating AtDREB1C Expression Improves Drought Tolerance in Salvia miltiorrhiza. Front Plant Sci [Internet]. 2017;8(January):1–17. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2017.00052/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudo M, Kidokoro S, Yoshida T, Mizoi J, Todaka D, Fernie AR, et al. Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol J. 2017;15(4):458–71. doi: 10.1111/pbi.12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Li X, He Z, Zhao X, Wang Q, Zhou B, et al. Molecular character of a phosphatase 2C (PP2C) gene relation to stress tolerance in Arabidopsis thaliana. Mol Biol Rep. 2013;40(3):2633–44. doi: 10.1007/s11033-012-2350-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang Y, Sun X, Gao S, Qin F, Dai M. Deletion of an Endoplasmic Reticulum Stress Response Element in a ZmPP2C-A Gene Facilitates Drought Tolerance of Maize Seedlings. Mol Plant [Internet]. Elsevier Ltd; 2017;10(3):456–69. Available from: http://dx.doi.org/10.1016/j.molp.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 41.Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G. SnRK2 Protein Kinases—Key Regulators of Plant Response to Abiotic Stresses. Omi A J Integr Biol [Internet]. 2011;15(12):859–72. Available from: http://www.liebertonline.com/doi/abs/10.1089/omi.2011.0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerr TCC, Abdel-Mageed H, Aleman L, Lee J, Payton P, Cryer D, et al. Ectopic expression of two AREB/ABF orthologs increase dehydration tolerance in cotton (Gossypium hirsutum). Plant Cell Environ [Internet]. 2017;1–10. Available from: http://doi.wiley.com/10.1111/pce.12906 [DOI] [PubMed] [Google Scholar]
- 43.Apelbaum A, Yang SF. Biosynthesis of stress ethylene induced by water deficit. Plant Physiol [Internet]. 1981;68(3):594–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16661963%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC425945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atwell BJ. Emission Rate in Plants. 2015;(March):2–4.
- 45.Saglam A, Terzi R, Demiralay M. Effect of polyethylene glycol induced drought stress on photosynthesis in two chickpea genotypes with different drought tolerance. Acta Biol Hung [Internet]. 2014;65(2):178–88. Available from: http://www.akademiai.com/doi/abs/10.1556/ABiol.65.2014.2.6 [DOI] [PubMed] [Google Scholar]
- 46.Sun X, Zhao T, Gan S, Ren X, Fang L, Karungo SK, et al. Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci Rep [Internet]. Nature Publishing Group; 2016;6(January):1–14. Available from: http://dx.doi.org/10.1038/srep24066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riemann M, Dhakarey R, Hazman M, Miro B, Kohli A, Nick P. Exploring Jasmonates in the Hormonal Network of Drought and Salinity Responses. Front Plant Sci [Internet]. 2015;6(December):1–16. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2015.01077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jonak C, Kiegerl S, Ligterink Wilc, BARKERt PJ, HUSKISSONt NS, by Briggs Winslow CR. Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Plant Biol. 1996;93(October):11274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Zhou M, Gao Z, Ren W, Yang F, He H, et al. RNA-seq analysis reveals MAPKKK family members related to drought tolerance in maize. PLoS One. 2015;10(11):1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(DOCX)
Data Availability Statement
DATA in FASTQ format were submitted to SRA (Sequence read archive), NCBI (Bio project Id: PRJNA419859). The raw reads can be accessed with accession number SRR6327851 to SRR6327856 for control samples and SRR6327857 to SRR6327862 for drought-stressed samples.